Significance
The central nervous system utilizes Ca2+-triggered synaptic vesicle fusion mediated by SNAREs and other synaptic proteins for neurotransmitter release into the synaptic cleft. Complexin, a small cytosolic protein, plays a dual role in regulating spontaneous minirelease and in activating Ca2+-triggered fusion, but the molecular mechanisms are still unclear. Here we found that the C-terminal domain of mammalian complexin interacts with membranes in a curvature-dependent fashion similar to other curvature-sensing proteins, such as α-synuclein. Together with a previous study of worm complexin, this finding suggests that curvature-sensing of the C-terminal domain is evolutionarily conserved. Moreover, localization to the highly curved membrane of synaptic vesicles is important for regulating spontaneous release by complexin.
Keywords: synaptic vesicle fusion, SNAREs, complexin, membrane fusion, neurotransmitter release
Abstract
In presynaptic nerve terminals, complexin regulates spontaneous “mini” neurotransmitter release and activates Ca2+-triggered synchronized neurotransmitter release. We studied the role of the C-terminal domain of mammalian complexin in these processes using single-particle optical imaging and electrophysiology. The C-terminal domain is important for regulating spontaneous release in neuronal cultures and suppressing Ca2+-independent fusion in vitro, but it is not essential for evoked release in neuronal cultures and in vitro. This domain interacts with membranes in a curvature-dependent fashion similar to a previous study with worm complexin [Snead D, Wragg RT, Dittman JS, Eliezer D (2014) Membrane curvature sensing by the C-terminal domain of complexin. Nat Commun 5:4955]. The curvature-sensing value of the C-terminal domain is comparable to that of α-synuclein. Upon replacement of the C-terminal domain with membrane-localizing elements, preferential localization to the synaptic vesicle membrane, but not to the plasma membrane, results in suppression of spontaneous release in neurons. Membrane localization had no measurable effect on evoked postsynaptic currents of AMPA-type glutamate receptors, but mislocalization to the plasma membrane increases both the variability and the mean of the synchronous decay time constant of NMDA-type glutamate receptor evoked postsynaptic currents.
Membrane fusion of synaptic vesicles is an essential process for neurotransmitter release in the nervous system (1–3). The central fusion machinery includes the SNARE (soluble NSF-attachment protein receptor) fusion proteins synaptobrevin/VAMP2 (vesicle-associated membrane protein), syntaxin-1, and SNAP-25 (4, 5), and other synaptic proteins, including synaptotagmin, complexin, Munc13, and Munc18 (6–8). Complexin is a small cytosolic protein that is evolutionarily conserved in mammals, Drosophila, and Caenorhaditis elegans, which cooperates with synaptotagmin to regulate synaptic vesicle exocytosis (9, 10).
Here we briefly summarize results that are most relevant for this work and focus on the complexin-1 isoform (referred to as Cpx). Cpx can be divided into four domains with different functions (Fig. 1A): flexible N- and C-terminal domains, an accessory domain, and a central α-helical domain (11). The N-terminal domain (residues 1–27) of Cpx plays a role in activation of fast synchronous release in murine neurons (12–14) and in isolated chromaffin cells (15). The C-terminal domain (amino acid residues 71–134) binds to phospholipids (16–18), and it is important for synaptic vesicle priming in neurons (15, 16). Cpx without the C-terminal domain does not suppress spontaneous release in neuronal cultures, but it still activates Ca2+-triggered release in neuronal cultures (16) and in a reconstituted system (19). The C-terminal domain of worm complexin preferentially interacts with curved membranes via a membrane-binding motif (20, 21). The accessory domain (amino acid residues 28–48) regulates spontaneous fusion in neurons and it suppresses Ca2+-independent fusion in reconstituted systems (12, 14, 19, 22–27). Although the accessory domain is required for regulating spontaneous release, mutations of this domain do not affect the activating function of Cpx compared with wild-type neurons in rescue experiments with Cpx knockdown (28, 29), and it can be entirely eliminated while still maintaining the activating function of Cpx in a reconstituted system (30). The α-helical central domain (amino acid residues 49–70) binds to the SNARE complex (31, 32) and is essential for all functions of Cpx in all species studied to date, including priming (16, 28, 29, 33), inhibiting spontaneous fusion (12, 19, 24, 25, 34–36), and activation of Ca2+-triggered fusion (12–14, 19, 34, 36–38).
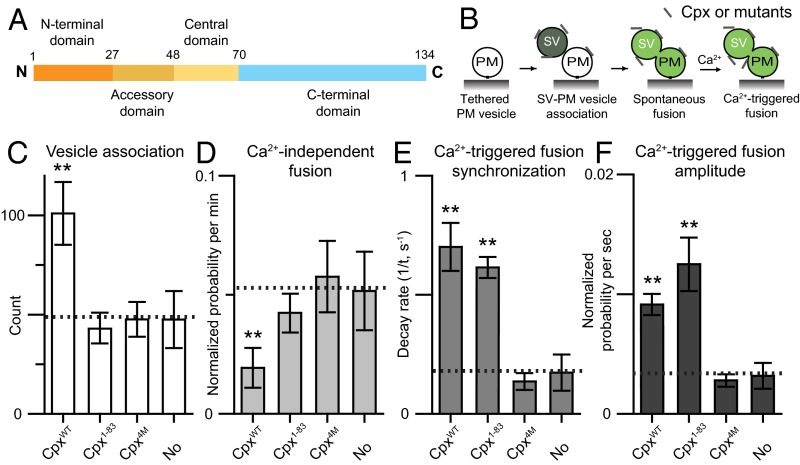
Fig. 1.
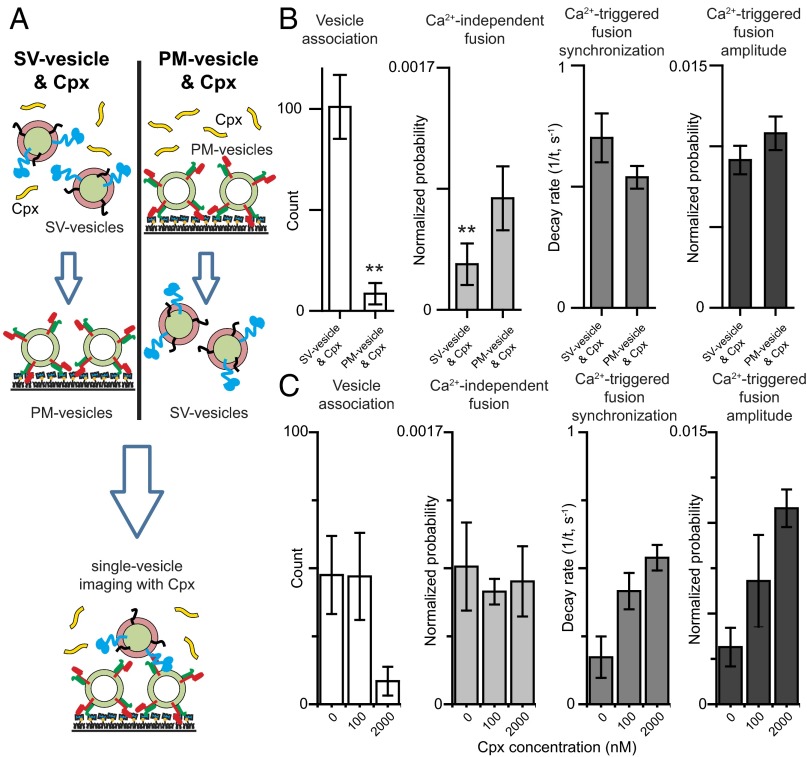
The C-terminal domain of Cpx is important for suppressing Ca2+-independent fusion, but not for Ca2+-triggered fusion. (A) Domain structure of complexin-1. (B) Schematic diagram of the single-vesicle content mixing assay (Methods). The bar graphs show the effects of 2 μM Cpx or Cpx fragments on the SV–PM vesicle association count during the first acquisition periods (Methods) (C), the average probability of Ca2+-independent fusion events per second (D), the decay rate (1/τ) of the histogram upon Ca2+ injection (E), and the amplitude of the first 1-s time bin (probability of a fusion event in that bin) upon 500 μM Ca2+ injection (F). The fusion probabilities and amplitudes were normalized with respect to the corresponding number of analyzed SV–PM vesicle pairs (Methods). Individual histograms are shown in Fig. S1. The dashed lines indicate the control without Cpx. C, D, and F show means ± SD for multiple independent repeat experiments (Table S1). E shows decay rates and error estimates computed from the covariance matrix upon fitting the corresponding histograms with a single exponential decay function using the Levenberg–Marquardt algorithm. **P < 0.01 using the Student’s t test with respect to the control experiment without Cpx (dashed line).
Taking these data together, we find that Cpx is an important regulator for synaptic vesicle fusion: it promotes Ca2+-triggered synaptic vesicle fusion in conjunction with SNAREs and synaptotagmins, while suppressing or regulating spontaneous fusion in certain species or experimental conditions. Here we reinvestigate the role of the C-terminal domain of Cpx. Using single-molecule membrane-binding assays, we find that mammalian Cpx preferentially binds to highly curved membranes in agreement with previous studies with worm complexin (20). Rescue studies of complexin-1/2 double knockdown (KD) in cultures of cortical neurons reveal that synaptic vesicle membrane localization behaves similarly to wild-type control, whereas preferential plasma membrane localization interferes with suppression of spontaneous release and increases both the variability and the mean of the fast time constant of N-methyl-d-aspartate receptor (NMDAR) -evoked postsynaptic currents (EPSCs). Taking these data together, we find that the C-terminal domain is expendable for the activation of Ca2+-triggered release, but proper membrane localization is important for regulation of spontaneous release.
Results
The C-Terminal Domain of Complexin Is Important for Suppressing Ca2+-Independent Fusion, but Not for Ca2+-Triggered Fusion in a Reconstituted System.
To study the molecular mechanisms of Cpx, several reconstituted systems have been developed (19, 39–42). As we reported previously, the C-terminal domain is important for suppression of Ca2+-independent fusion in a single-vesicle content mixing assay with reconstituted full-length neuronal SNAREs, and synaptotagmin-1 (19, 43, 44). Our fusion assay (Fig. 1B) monitors both individual Ca2+-independent and Ca2+-triggered fusion events with the same sample and it distinguishes between docking and content mixing (19). Briefly, two types of vesicles are reconstituted: vesicles with reconstituted syntaxin-1A and SNAP-25A that mimic the plasma membrane (referred to as PM vesicles) and vesicles with reconstituted synaptobrevin-2 and synaptotagmin-1 that mimic synaptic vesicles (referred to as SV vesicles). The PM vesicles are tethered on a passivated surface, SV vesicles are added and incubated in absence or presence of Cpx or Cpx fragments or Cpx mutants for a 1-min period, and then unbound SV vesicles are removed. Subsequently, Ca2+-independent fusion is monitored for a 1-min period, followed by injection of 500 μM Ca2+, and Ca2+-triggered fusion was monitored for an additional 1-min period (for more details, see Methods). Our system qualitatively reproduces the effects of synaptotagmin-1, Cpx, and their mutations, on both spontaneous release and evoked release that have been observed in cortical neuronal cultures (19, 45).
Using the constructs and protein purification methods described in this work, Cpx wild-type (CpxWT) activates Ca2+-triggered fusion and suppresses Ca2+-independent fusion in our fusion assay (Fig. 1 C–F and Fig. S1), in agreement with previous work that used different syntaxin-1A and synaptobrevin-2 expression constructs (19). We analyzed our fusion experiments by the counts of associated SV–PM vesicle pairs (Fig. 1C), the average probability of Ca2+-independent fusion per second in the absence of Ca2+ (“Ca2+-independent fusion” in Fig. 1D), the decay rate of the fusion histogram upon Ca2+ injection (“Ca2+-triggered fusion synchronization” in Fig. 1E), and the probability of Ca2+-triggered fusion during the first 1-s time bin upon Ca2+ injection (“Ca2+-triggered fusion amplitude” in Fig. 1F) (Methods). The fusion probabilities and amplitudes were normalized with respect to the corresponding number of analyzed SV–PM vesicle pairs (Table S1). Thus, the normalized fusion probabilities and amplitudes provide information about fusion of associated SV–PM vesicles, independent of the number of associated SV–PM vesicle pairs.
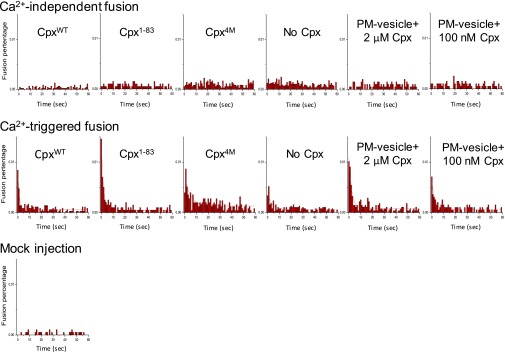
Fig. S1.
Data figure for Figs. 1 and 5. Experiments were performed in the presence of both full-length synaptotagmin-1 and neuronal SNAREs, and in the absence or presence of 2 μM Cpx, Cpx fragment, or Cpx mutant as well as preincubation of tethered PM vesicles in the presence of 2 μM and 100 nM Cpx. The Ca2+-independent fusion histograms and the 500 μM Ca2+-triggered fusion histograms (1-s time bins) were cumulated over the 1-min acquisition period for all rounds and repeat experiments and then normalized with respect to the number of analyzed SV–PM vesicle pairs (Methods). The Ca2+-triggered fusion histograms were fit to an exponential decay function and the resulting decay rates are shown in the “Ca2+-triggered fusion synchronization” bar charts in Figs. 1 and 5. The numbers of independent repeat experiments and analyzed SV–PM vesicle pairs are provided in Table S1.
Table S1.
Data summary table for the single vesicle–vesicle fusion experiments shown in Figs. 1 and 5 and Fig. S1
| Experiment | Ca2+-independent fusion events | Ca2+-triggered fusion events | No. of analyzed SV–PM vesicle pairs | Repeats n |
| CpxWT | 70 | 181 | 3,571 | 9 |
| Cpx1–83 | 79 | 142 | 1,988 | 4 |
| Cpx4M | 207 | 266 | 3,509 | 6 |
| No Cpx | 229 | 198 | 3,856 | 7 |
| PM vesicle and 2,000 nM Cpx | 127 | 210 | 2,762 | 7 |
| PM vesicle and 100 nM Cpx | 83 | 115 | 1,971 | 3 |
| Mock | 30 | 1,882 | 9 |
For definition of the repeat experiments, see Methods.
The Cpx 4M mutant (Cpx4M, R48A, R59A, K69A, and Y70A), which impairs the interaction to ternary SNARE complex (38), lost all Cpx effects on both Ca2+-independent and Ca2+-triggered fusion (Fig. 1 C–F and Fig. S1). Elimination of the C-terminal domain reduced the ability of Cpx1–83 to suppress Ca2+-independent release (Fig. 1D and Fig. S1), although there was no effect on Ca2+-triggered fusion (Fig. 1 E and F). Note that the effect of the Cpx1–83 construct on fusion is similar to that of the slightly longer Cpx1–86 construct that we previously used (19), but unlike the Cpx1–86 construct, it does not interfere with SV–PM vesicle association. Taking these data together, we find that the C-terminal domain is important for suppressing Ca2+-independent fusion, but not for activation of Ca2+-triggered fusion in our single-vesicle fusion assay.
The C-Terminal Domain of Cpx Plays a Role in Membrane Localization in Neurons.
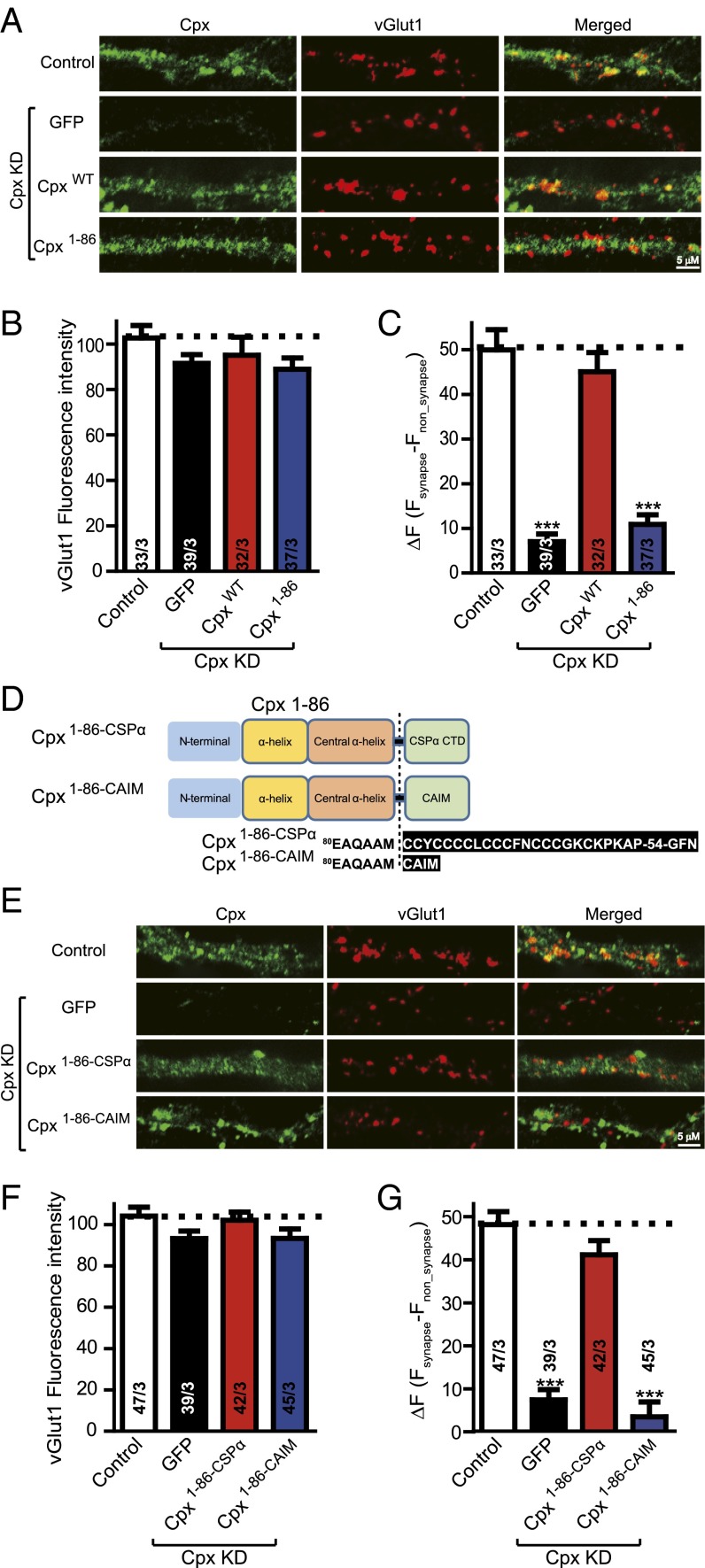
To test if the functional role of the Cpx C-terminal region is related to membrane localization in mouse dissociated cortical neuron cultures, we used lentiviral delivery of shRNA-dependent KD of Cpx and complexin-2 to generate complexin-deficient neurons and performed rescue experiments to test the function of membrane-localization chimeras (Methods). We stained neurons with antibodies to Cpx and to the vesicular glutamate transporter (vGlut1), along with fluorescently labeled secondary antibodies, to selectively visualize excitatory nerve terminals. Quantitation of immunofluorescence revealed that vGlut1-specifc fluorescence intensity was identical in all conditions (Fig. 2 A and B), indicating normal synapse formation. We measured the Cpx-specific fluorescence intensity at synaptic locations that had been labeled by vGlut1 (Fsynapse) and at locations that were randomly picked elsewhere that are not colocalized with vGlut1 locations (Fnon_synapse). We calculated ΔF = Fsynapse – Fnon_synapse to determine the subcellular location of Cpx (Fig. 2C). Although Cpx was present throughout neurons, there was a preference for synaptic localization (Fig. 2 A and C). Cpx KD (along with expression of GFP) abolished the fluorescence intensity. Wild-type (CpxWT) but not Cpx1–86 reversed the localization impairment observed in Cpx-deficient neurons (Fig. 2 A and C), suggesting that the C-terminal domain is essential for preferential subcellular localization of Cpx to synapses. However, we note that even in the absence of the C-terminal domain, Cpx1–86 still activates fast synchronous release (16) in neurons, suggesting that Cpx1–86 still interacts with trans SNARE complexes at sites of docked and primed synaptic vesicles. Moreover, such sites may not be accessible by the Cpx antibody.
Fig. 2.
The C-terminal domain of Cpx is important for subcellular location. (A) Representative images of cultured mouse cortical neurons infected with control lentivirus or with lentivirus expressing the Cpx shRNA plus either EGFP only (Cpx KD), or together with wild-type Cpx (CpxWT), or C-terminal truncated mutant Cpx (Cpx1–86). Neurons were fixed and labeled by double immunofluorescence using vGlut1 (to mark excitatory synapses) and Cpx antibodies along with fluorescently labeled secondary antibodies (Methods). The scale bar in right lower corner applies to all images. (B) Summary graphs of vGlut1-specific fluorescence intensities for all conditions as described for A. (C) Summary graphs of ΔF (Fsynapse − Fnon_synapse) of Cpx-specific fluorescence intensity for all conditions as described for A. (D) Domain structures and Cpx chimeras in which the C-terminal sequence was replaced with either the C-terminal sequence of CSPα (Cpx1–86–CSPα) or with the CAIM sequence (Cpx1–86–CAIM). (E) Representative images of cultured mouse cortical neurons infected with control lentivirus or with lentivirus expressing the Cpx shRNA plus either EGFP only (Cpx KD), or together with Cpx synaptic vesicle target mutant (Cpx1–86–CSPα), or Cpx plasma membrane target mutant (Cpx1–86–CAIM). Neurons were treated as described for A. The scale bar in right lower corner applies to all images. (F) Summary graphs of vGlut1-specific fluorescence intensities for all conditions as described for E. (G) Summary graphs of ΔF (Fsynapse − Fnon_synapse) of Cpx-specific fluorescence intensity for all conditions as described for E. Data shown in summary graphs are means ± SEM; numbers of cells/independent cultures analyzed are listed in the bars. Statistical assessments were performed by the Student’s t test comparing each condition to the indicated control experiment (***P < 0.001).
We hypothesized that the Cpx C-terminal domain plays a role in Cpx localization via preferential synaptic vesicle association. To test this hypothesis, we designed two chimeras consisting of Cpx1–86 and two different membrane localization elements (Fig. 2D): Cpx1–86–CSPα is a chimera of Cpx1–86 fused to the C-terminal palmitoylated sequence of cysteine-string protein-α (CSPα), a sequence that targets synaptic vesicles (46); and Cpx1–86–CAIM is a chimera of Cpx1–86 fused to the C-terminal isoprenylation sequence (CAIM) that targets the plasma membrane (28). Immunocytochemistry experiments, similar to above, were again performed in lentivirus-infected neurons (Fig. 2 E–G). Imaging experiments revealed that Cpx1–86–CSPα, but not Cpx1–86–CAIM, could rescue the preference of wild-type Cpx for presynaptic localization (Fig. 2 F and G). Taken together, our results suggest that the C-terminal domain plays a role in preferential membrane localization of Cpx to synaptic vesicles.
Mislocalization of Cpx Impairs Both Spontaneous AMPA Receptor-Mediated Miniature EPSCs and Affects Evoked NMDA Receptor-Mediated EPSCs.
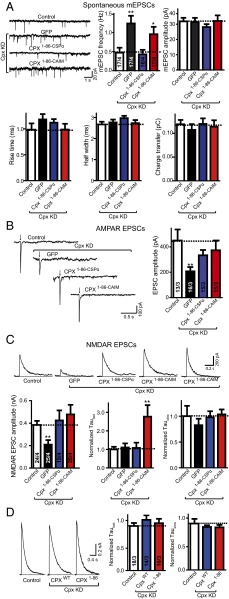
We next examined if preferential membrane association of Cpx via its C-terminal domain is functionally required. We used the same KD/rescue strategy in mouse dissociated cortical neurons as described above and measured both AMPA-receptor (AMPAR)-dependent and NMDA-receptor (NMDAR)-dependent synaptic responses. As expected, KD of Cpx alone increased spontaneous (miniature) excitatory postsynaptic currents (mEPSCs) (12). The increase in mEPSC frequency induced by Cpx KD was rescued by Cpx1–86–CSPα returning to levels similar to wild-type control but only slightly by Cpx1–86–CAIM (Fig. 3A). The mEPSC amplitude and kinetic parameters were unchanged in all conditions, suggesting that these manipulations do not affect the postsynaptic AMPAR density, and that the effects on the mEPSC frequency are because of presynaptic processes. In measurements of action-potential–evoked release mediated by AMPA-type glutamate receptors (AMPARs), both Cpx1–86–CSPα and Cpx1–86–CAIM could rescue the Cpx KD impairment similarly to Cpx1–86 (Fig. 3B). Thus, association of the C-terminal domain of Cpx with synaptic vesicle membranes is essential for suppressing spontaneous release, but it has no effect on AMPAR EPSCs.
Fig. 3.
Mislocalization of Cpx to the plasma membrane affects NMDAR EPSCs. (A) Sample trace (Upper Left) and summary graphs of the frequency and amplitude (Upper Right), and kinetics (Lower) of mEPSCs, recorded in cultured cortical neurons that were infected with a control lentivirus (control) or lentiviruses expressing complexin shRNAs (GFP) without or with coexpression of Cpx1–86–CSPα or Cpx1–86–CAIM. Neurons were infected on DIV 4–6, and analyzed on DIV 14–16. Recordings were performed in 50 μM AP-5, 100 μM picrotoxin and 1 μM tetrodotoxin. (B) Sample traces (Left) and summary graphs of the amplitude (Right) of action-potential–evoked AMPAR-mediated EPSCs monitored in control or Cpx-deficient cortical neurons as described for A, except that tetrodoxin was omitted. (C) Sample traces (Upper), summary graphs of the amplitude (Lower Left), normalized τfast decay time (Lower Center) and normalized τslow decay time (Lower Right) of action-potential–evoked NMDAR-mediated EPSCs monitored in control or Cpx-deficient cortical neurons as described for A. For Cpx1–86–CAIM two sample traces were included to illustrate the degree of variability. (D) Sample traces (Left) and summary graphs of τfast decay time (Center) and τslow decay time (Right) of action-potential evoked NMDAR-mediated EPSCs monitored in cultured cortical neurons that were infected with a control lentivirus (control) or lentiviruses expressing Cpx shRNAs with coexpression of wild-type complexin1 (CpxWT) or C-terminal truncated mutant Cpx (Cpx1–86). The decay times were normalized to control. Recordings were performed in 20 μM CNQX and 100 μM picrotoxin. Data shown in summary graphs are means ± SEM; numbers of cells/independent cultures analyzed are listed in the bars. Statistical assessments were performed by the Student’s t test comparing each condition to the indicated control experiment (*P < 0.05, **P < 0.01).
Because both Cpx1–86–CSPα and Cpx1–86–CAIM could fully support AMPAR-mediated action-potential–evoked release, we next asked if these two chimeras have the same ability in kinetically accelerating release.
In neuronal cultures, recurrent network activity randomly contaminates AMPAR EPSC measurements, so it is difficult to quantitatively analyze the asynchronous portion of AMPAR EPSCs (16, 29). Doing so may result in an overestimation of asynchronous neurotransmission. Therefore, we also measured and analyzed NMDAR-mediated EPSCs to avoid the network activity EPSCs (16, 29). Although both Cpx1–86–CSPα and Cpx1–86–CAIM rescued NMDAR-mediated EPSC amplitude similarly to wild-type control (Fig. 3C), the kinetics of NMDAR activation was slightly altered and exhibited more variability when expressing Cpx1–86–CAIM. To quantify these differences in kinetics, NMDAR-mediated EPSCs were measured and fitted by a double-exponential function. Consistent with previous results (29), no substantial kinetic difference was observed between Cpx KD and control. In contrast, Cpx1–86–CAIM but not Cpx1–86–CSPα increased both the variability and the mean of the fast time constant, τfast (Fig. 3C) (time constants were normalized to the control). The slow time constant, τslow, was unchanged in all conditions.
We next tested if the C-terminal domain of Cpx itself or its plasma membrane mislocalization produced the observed alterations in neurotransmitter release kinetics. We compared CpxWT and Cpx1–86-expressed neurons with control neurons. Deletion of the C-terminal domain (Cpx1–86) itself did not result in a difference in the kinetics of NMDAR mediated EPSCs compared with control or CpxWT expressed neurons (Fig. 3D), suggesting that plasma membrane mislocation of Cpx is responsible for the effects on the fast time constant of NMDAR-mediated EPSCs, although the C-terminal domain itself is expendable for evoked release kinetics.
The C-Terminal Domain of Mammalian Cpx Is a Membrane Curvature Sensor.
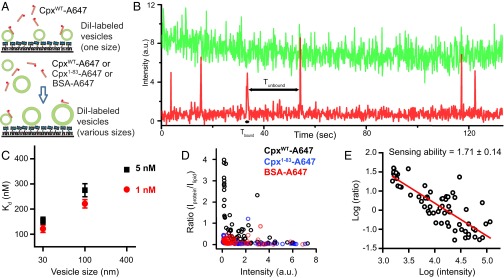
The C-terminal domain of worm complexin binds more strongly to small (diameter ∼30 nm) than to large (diameter ∼120 nm) unilamellar liposomes based on observed line broadening of NMR spectra and induced circular dichroism helicity (20). To test whether mammalian Cpx also has this curvature sensing ability, we first measured the binding constant (Kd) of labeled Cpx to different sizes of vesicles using a single-molecule binding assay. In this assay, 0.2% DiI (1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate)-labeled acidic liposomes of defined sizes were tethered on a polyethylene glycol (PEG)-coated surface through biotin-neutravidin coupling (Methods). Cpx was labeled with the organic dye Alexa 647 at residue 8 (Cpx-A647) and added in solution to the surface-tethered liposomes, and binding events were observed in real time (Fig. 4A). Liposome-bound Cpx-A647 was detected by colocalized fluorescence microscopy over a period of 120 s (Fig. 4B). The bound and unbound dwell times (respectively, Tbound and Tunbound) were plotted as histograms (Fig. S2). Using thousands of individual liposomes, Kon and Koff rates were determined by single exponential fit, and Kd was calculated (Fig. 4C). Cpx has a higher binding affinity to smaller liposomes than to larger ones, with this trend being similar at different Cpx concentrations (1 nM and 5 nM) (Fig. 4C).
Fig. 4.
The C-terminal domain of mammalian Cpx is a membrane curvature sensor. (A) Single molecule Cpx membrane binding assay. (Upper) Alexa 647-labeled Cpx was incubated with surface-tethered DiI-labeled protein-free liposomes with the same lipid composition as that of the SV vesicles with the specified diameter. Results are shown in B and C. (Lower) Alexa 647-labeled Cpx, Cpx1–83, or BSA were incubated in solution with DiI-labeled protein-free liposomes with the same lipid composition as that of SV vesicles with various diameters and liposomes were then surface-tethered. Results are shown in D and E. For details, see Methods. (B) Representative example of a fluorescence intensity time trace of single Cpx-A647 molecules (labeled with Alexa 647, red) as they bind and unbind to a single surface-tethered liposome (labeled with DiI, green). Each time a Cpx-A647 molecule binds the liposome, the red intensity jumps to a high level. When the molecule dissociates after the bound dwell time Tbound, the intensity drops back to the background level. See also Fig. S2. (C) Single-molecule membrane curvature-sensing assay using surface-tethered DiI-labeled liposomes of specified diameters and at specified Cpx concentrations. The apparent Kd was calculated by Kd = a C Koff/Kon, where a is the labeling efficiency and C is the concentration of the protein. Kon and Koff were obtained from the covariance matrix upon fitting the Tbound and Tunbound dwell time histograms with a single exponential decay function (Fig. S2 and Table S2), respectively. Experiments were performed with thousands of liposomes of defined diameter at each concentration. (D) Membrane curvature-sensing assay using incubation of liposomes and proteins in solution, followed by surface tethering. Each circle represents binding of multiple labeled Cpx (black), Cpx1-83 (blue), and BSA (red) molecule to a single DiI-labeled protein-free liposome. Mixtures of vesicles with diameters of 30, 100, and 400 nm were incubated with Cpx, Cpx1–83, and BSA, respectively. (E) Same data as in D for CpxWT plotted on a double-logarithmic scale. The slope of fitted linear function is defined as the curvature-sensing value (47).
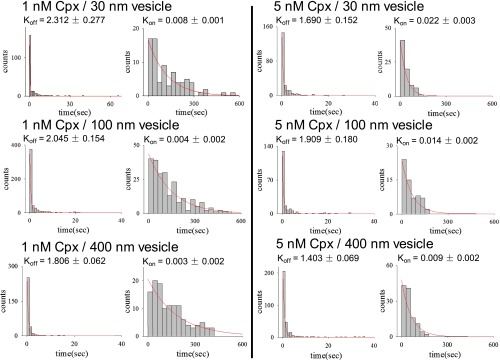
Fig. S2.
Histograms for Cpx-vesicle bound dwell times (Tbound) and unbound dwell times (Tunbound) for the specified Cpx concentrations and liposome diameters, related to Fig. 4 B and C. The on (Kon) and off (Koff) rates were obtained from by fitting the respective dwell time histograms with a single exponential decay function. The numbers of binding events for each condition are provided in Table S2.
Table S2.
Data summary table for the single-molecule binding experiments shown in Fig. 4 B and C and Fig. S2
| Experiment | No. of binding events |
| 1 nM CpxWT | |
| 30-nm vesicles | 144 |
| 100-nm vesicles | 125 |
| 400-nm vesicles | 184 |
| 5 nM CpxWT | |
| 30-nm vesicles | 161 |
| 100-nm vesicles | 251 |
| 400-nm vesicles | 185 |
To corroborate this result with a different assay where binding is measured in solution, we incubated a mixture of different sizes of liposomes ranging from 30 nm to 400 nm with labeled protein for 30 min, tethered these liposomes onto a PEGylated surface, and then removed unbound protein by buffer exchange. The ratio between labeled protein and DiI-vesicle fluorescence intensities indicates the binding probability of the labeled protein to the membrane with different curvatures, as described in ref. 47 (Fig. 4D). As expected, the Cpx density is markedly higher for liposomes with lower fluorescence intensities, suggesting that Cpx preferentially binds to smaller liposomes (Fig. 4D). As a control, the lipid binding protein BSA (48) labeled with Alexa 647 (BSA-A467) and the C-terminally truncated fragment, Cpx1–83, had no preference for liposome size (Fig. 4D). We calculated the membrane curvature “sensing” value of Cpx by plotting Cpx density versus intensity on a double-logarithmic scale and fit to a linear function (Fig. 4E), resulting in ∼1.71, a number that is similar to that of α-synuclein (∼1.4) (47). In summary, our binding assays suggest that the C-terminal domain of mammalian Cpx is a curvature sensor that preferentially localizes Cpx to highly curved membranes.
Effect of Preincubation of PM Vesicles with Cpx.
In a previous single-vesicle lipid-mixing assay, full-length wild-type Cpx reduced vesicle association (albeit in the absence of full-length synaptotagin-1) (42), in contrast to our results (Fig. 1). However, in this previous study, v-SNARE vesicles (i.e., containing reconstituted synaptobrevin-2 without synaptotagmin-1) were tethered on an imaging surface, whereas t-SNARE vesicles (containing reconstituted syntaxin-1A and SNAP-25A) were premixed in solution with Cpx and then added to the surface with tethered v-SNARE vesicles. In contrast, in our assay, SV vesicles are mixed with Cpx in solution and then added to surface-tethered PM vesicles that contain t-SNAREs. Thus, the different sequence of addition of Cpx to the different vesicles may cause the difference in vesicle association probability in the two assays. Indeed, when preincubating surface tethered PM vesicles with Cpx (Fig. 5A), SV–PM vesicle association was substantially reduced, whereas Ca2+-triggered fusion parameters were not significantly affected and there was only a small difference in Ca2+-independent fusion probability (Fig. 5B) with or without PM vesicle preincubation. As mentioned above, the amplitudes and probabilities were normalized with respect to the corresponding number of analyzed SV–PM vesicle pairs. We also tested the effect of preincubating the PM vesicles at two different Cpx concentrations and found no significant difference in the effects of Cpx on Ca2+-independent and Ca2+-triggered fusion with or without preincubation (Fig. 5C). We note that this result is in contrast to a recently published study (49), for unknown reasons.
Fig. 5.
Decrease in SV–PM vesicle association probability upon preincubating PM vesicles with Cpx. (A) Schematic diagram of the single-vesicle content mixing assay using two different preincubation methods. (Left) Standard protocol as described in Methods: SV vesicles are added simultaneously with 2 μM Cpx to the immobilized PM vesicles. (Right) Immobilized PM vesicles are preincubated with Cpx before adding SV vesicles. (B) The bar graphs show the SV–PM vesicle association count during the first acquisition periods, the average probability of Ca2+-independent fusion events per second, the decay rate (1/τ) of the histogram upon 500 μM Ca2+ injection, and the amplitude of the first 1-s time bin (probability of a fusion event in that bin) upon Ca2+ injection. The fusion probabilities and amplitudes were normalized with respect to the corresponding number of analyzed SV–PM vesicle pairs (Methods). Individual histograms are in Fig. S1. (C) The bar graphs show the effect of preincubating with PM vesicles with Cpx at specified concentrations. The vesicle association, Ca2+-independent fusion, and Ca2+-triggered amplitude bar charts show means ± SD for multiple independent repeat experiments (Table S1). The Ca2+-triggered fusion synchronization panels show decay rates and error estimates computed from the covariance matrix upon fitting the corresponding histograms with a single exponential decay function using the Levenberg–Marquardt algorithm. Statistical assessments were performed by the Student’s t test (**P < 0.01).
Discussion
Cpx and its homologs have at least two physiological functions: activation of Ca2+-triggered synchronous neurotransmitter release by cooperating with synaptotagmin, and regulation of spontaneous release in the presynaptic terminal (9, 10, 50). The activating function of complexin is conserved across all species (mammals, Drosophila, and C. elegans) and different types of Ca2+-triggered synaptic vesicle fusion studied to date (7, 12, 16, 28, 34–37, 51–53). Regulation of spontaneous release by complexin is less conserved among species and varies depending on experimental conditions: for example, in Drosophila spontaneous release increases with knockout of complexin (14, 54). Similarly, knockdown in cultured cortical neurons increases spontaneous release, although knockout of complexin in mice only affects spontaneous release depending on the particular neuronal cell type (12, 16, 27, 28). A split pair of N-terminal and central domain fragments of Cpx is sufficient to activate Ca2+-triggered release using our reconstituted single-vesicle fusion assay, suggesting that the N-terminal domain acts as an independent module within the synaptic fusion machinery (30). Moreover, the Cpx accessory domain can be entirely eliminated, and the resulting split pair of Cpx constructs still activates Ca2+-triggered fusion (i.e., a clamping process is not required for activation) (30). Cpx has two conformations when bound to the ternary SNARE complex (55, 56), one of which induces a conformational change at the membrane-proximal C-terminal end of the ternary SNARE complex that specifically depends on the N-terminal, accessory, and central domains of Cpx (56).
We studied the role of the C-terminal domain of Cpx using a combination of single-molecule membrane binding assays, single-vesicle fusion assays, and electrophysiological experiments in cortical neuronal cultures. Previous work showed that the C-terminal domain of worm complexin preferentially binds to small unilamellar liposomes (20). We extended these studies by showing that mammalian Cpx also has a strong preference for binding to highly curved membranes via the C-terminal domain (Fig. 4), suggesting that the curvature-sensing ability of complexins is an evolutionary conserved feature. Moreover, we showed that it has as curvature sensing value comparable to that of α-synuclein (47), which has been shown to localize to synaptic vesicles (57, 58).
Because there is a large difference of curvature between the plasma membrane and synaptic vesicle membrane, it is likely that the membrane curvature-sensing ability of the C-terminal domain specifically localizes Cpx to synaptic vesicles. Indeed, our studies in neuronal cultures with membrane-localizing chimeras of Cpx support this notion (Figs. 2 and 3). Taken together, our results suggest that the C-terminal domain of Cpx functions by preferentially localizing Cpx to synaptic vesicle membranes. Such localization may be important for the role of Cpx to suppress Ca2+-independent fusion in vitro (Fig. 1) and to regulate spontaneous release in neurons (Fig. 3), although Ca2+-independent release is still slightly suppressed in vitro when the C-terminal domain is eliminated (Fig. 1 and Fig. S1). Moreover, when the C-terminal domain is replaced with a plasma membrane binding element, it increases both the variability and the mean of the fast-decay time constant of NMDAR-mediated EPSCs (Fig. 3). Thus, active mislocalization of Cpx has a mild effect on NMDAR-mediated evoked release, but elimination of the entire C-terminal domain does not affect evoked release at all (Figs. 1 and 3). Finally, suppression of Ca2+-independent fusion requires a higher Cpx concentration than activation of Ca2+-triggered fusion in our vesicle fusion system (19). Taken together we speculate that the Cpx C-terminal domain functions by increasing the local concentration at sites of docked and primed synaptic vesicles, thereby effectively increasing the effect of Cpx on spontaneous release, whereas the effect on activation of Ca2+-triggered fusion already occurs at lower Cpx concentration.
Methods
Protein Purification.
Synaptobrevin-2 and syntaxin-1A.
Full-length, cysteine-free rat synaptobrevin-2 (VAMP2) and rat syntaxin-1A were expressed separately in Escherichia coli with a N-terminal, tobacco etch virus (TEV) protease-cleavable, hexa-histidine tag from plasmid pTEV5 (59). Proteins were expressed overnight at 25 °C in autoinducing media (60) in E. coli strain C43 (61). Cell pellets from 8 L of culture were suspended in 400 mL of 1× PBS (10 mM phosphate buffer, 2.7 mM potassium chloride, 137 mM sodium chloride, pH 8.0), 5 mM EDTA, and 1 mM PMSF supplemented with Complete Protease Inhibitor Mixture tablets (Roche), and lysed by three passes through an Emulsiflex C5 homogenizer at 15,000 psi. Inclusion bodies were removed by two consecutive 10-min spins at 5,000 × g in a JA-14 (Beckman Coulter) rotor, and the membrane fraction collected by centrifugation at 125,000 × g for 2 h in a Ti-45 (Beckman Coulter) rotor. Membranes were further washed by homogenization in 360 mL of buffer containing 10 mM Tris H2SO4, pH 7.5, 10 mM EDTA, 10% (wt/vol) glycerol; membranes were harvested again by centrifugation in a Ti-45 rotor at 125,000 × g for 1 h. The membrane pellet was resuspended in 20 mM Hepes, pH 7.5, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), and 10% (wt/vol) glycerol, and centrifuged for an additional hour in the same rotor. Membranes were suspended to a protein concentration of 5 mg/mL in buffer containing 20 mM Hepes, pH 7.5, 500 mM NaCl, 1 mM TCEP, 10 mM imidazole, and 10% (wt/vol) glycerol, 1 mM PMSF supplemented with 2 EDTA-free Complete Protease Inhibitor Tablets. n-Dodecylmaltoside (DDM) [2% (wt/vol)] (Anatrace) was added, and after incubation at 4 °C for 1 h with stirring, the sample was centrifuged for 35 min at 125,000 × g in a Ti-45 (Beckman Coulter) rotor, and the supernatant loaded onto a 1-mL column of Nickel-NTA agarose (Qiagen). Beads were harvested by centrifugation and poured into a column, attached to an AKTA Prime and washed with 50 mL of buffer containing 20 mM Hepes, pH 7.5, 300 mM NaCl, 1 mM TCEP, 20 mM imidazole, 110 mM n-octylglucoside (OG), and 10% (wt/vol) glycerol, and the proteins were eluted in the same buffer containing 450 mM imidazole and 1 M NaCl. The protein-containing fractions were combined and injected on a Superdex 200 HR 10/300 GL (GE Healthcare) that was equilibrated with 20 mM Hepes, pH 7.5, 300 mM NaCl, 1 mM TCEP, 110 mM OG, and 10% (wt/vol) glycerol. The peak fractions of concentration greater than 40 µM for syntaxin-1A and 80 µM for synaptobrevin-2 were then combined, and digested with 100 µg TEV protease for 45 min at ambient temperature, after which the reaction was complete and the TEV protease had precipitated. TEV was removed by centrifugation at 5,000 rpm for 10 min in an Eppendorf model 5804 R tabletop centrifuge (Eppendorf). The concentration was measured by UV absorption at 280 nm and aliquots were flash frozen in liquid nitrogen and stored at −80 °C. Note that in a previous study we used GST expression constructs for syntaxin-1A and synaptobrevin-2 (19). Comparison of Fig. 1 with our previous study shows that both sets of expression constructs produce identical results.
SNAP-25.
Cysteine-free SNAP-25A (C84S, C85S, C90S, and C92S) was expressed from plasmid pTEV5 (59) with an N-terminal TEV protease cleavable hexa-histidine tag. The proteins were expressed overnight at 25 °C in autoinducing media (60) in E. coli strain BL21 (DE3). Cells from 4 L of culture were resuspended in 200 mL of 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 20 mM imidazole supplemented with 1 mM PMSF and 4 EDTA-free protease-inhibitor mixture tablets. Cells were lysed by three passes through the Emulsiflex C5 homogenizer (Avestin) at 15,000 psi. The lysate was clarified by centrifugation in the Ti-45 rotor for 1.5 h at 125,000 × g. The supernatant was bound to a 5-mL Nickel-NTA column by stirring at 4 °C for 1 h. Beads with bound SNAP-25 were harvested by centrifugation, poured into a column, and attached to an AKTA Prime (GE Healthcare). The column was washed with 150 mL of SNAP-25 buffer containing 50 mM sodium phosphate, pH 8.0, 300 mM NaCl supplemented with 50 mM imidazole and eluted with buffer containing 50 mM sodium phosphate, pH 8.0 and 300 mM NaCl supplemented with 350 mM imidazole. The protein-containing fractions were combined, dithiothreitol (DTT) was added to 5 mM, 1 mM EDTA was added, and 150 µg of TEV protease was added to remove the hexa-histidine tag. This mixture was dialyzed against buffer containing 20 mM Hepes, pH 7.5, 100 mM NaCl, and 4 mM DTT overnight at 4 °C. The TEV-cleaved SNAP-25 was concentrated in a 15-mL Amicon ultracentrifugal concentrator with a 10,000 molecular-weight cut-off membrane (Millipore) to 5 mL and injected on the Superdex 200 (16/60) column (GE Healthcare) equilibrated with buffer containing 20 mM Hepes, pH 7.5, 100 mM NaCl, and 4 mM DTT. Protein-containing fractions were combined and the concentration of SNAP-25 was measured by absorbance at 280 nm, and aliquots were flash frozen in liquid nitrogen and stored at −80 °C.
Synaptotagmin.
Full-length rat synaptotagmin-1 was expressed from plasmid pJ414 (DNA 2.0) with a C-terminal deca-histadine tag preceded by a PreScission protease cleavage site to remove the tag, and all cysteines were changed to alanine except the cysteine residue at position 277. Synaptotagmin-1 was expressed in E. coli BL21 (DE3) by growing the cells to OD600 between 0.6 and 0.8 at 37 °C then induced at 20 °C for 12–16 h with 0.5 mM isfopropyl-β-d-thiogalactopyranoside (IPTG). The cells from 6 L of induced culture were harvested and suspended in 200 mL of buffer containing 50 mM sodium phosphate, pH 7.4, 600 mM NaCl, 2 mM DTT, and 10% (wt/vol) glycerol supplemented with PMSF to 1 mM, and two EDTA free complete protease inhibitor mixture tablets. Cells were lysed by three passes through the Emulsiflex C5 homogenizer (Avestin) at 15,000 psi. Cell debris and inclusion bodies were removed by centrifugation at 5,000 × g in a Beckman JA-20 rotor (Beckman Coulter) for 10 min. The supernatant was then centrifuged at 5,000 × g for 10 min in the same rotor. Membranes were collected by centrifugation at 125,000 × g in a Beckman Ti-45 rotor for 1 h. Membranes were washed by homogenization in 100 mL buffer containing 50 mM sodium phosphate, pH 7.4, 600 mM NaCl, 5 mM DTT, 1 mM PMSF, and 10% (wt/vol) glycerol supplemented with two EDTA-free complete protease-inhibitor mixture tablets (Roche). Membranes were harvested again by centrifugation in a Ti-45 rotor at 125,000 × g for 1 h. The pellet was suspended in 100 mL buffer containing 50 mM sodium phosphate, pH 7.4, 600 mM NaCl, 5 mM DTT, 1 mM PMSF, 10% (wt/vol) glycerol supplemented with 2 EDTA-free complete protease-inhibitor mixture tablets and frozen in two 50-mL aliquots in liquid nitrogen. One aliquot of membranes was solubilized in the presence of 1.5% (wt/vol) DDM for 1 h at 4 °C. The extract was clarified by centrifugation in a Ti-45 rotor at 125,000 × g for 35 min. The supernatant was bound to a 4-mL bed volume of Nickel-NTA beads (Qiagen) equilibrated in buffer containing 50 mM sodium phosphate, pH 7.4, 600 mM NaCl, 2 mM DTT, 10% (wt/vol) glycerol, and 1.5% DDM by stirring at 4 °C for 1 h. Beads were harvested by centrifugation and poured into a column, attached to an AKTA Prime (GE Healthcare), and washed with 50 mL of buffer containing 50 mM sodium phosphate, pH 7.4, 600 mM NaCl, 2 mM DTT, 10% (wt/vol) glycerol, and 110 mM OG, and then eluted with the same buffer containing 500 mM imidazole. Protein-containing fractions were combined and injected on a Superdex 200 (16/60) column equilibrated in 20 mM sodium phosphate, pH 7.4, 300 mM NaCl, 2 mM DTT, 110 mM OG, and 10% (wt/vol) glycerol. Peak fractions were then combined and 100 μg of PreScission protease (GE Healthcare) was added to cleave the tag overnight at 4 °C. Cleaved synaptotagmin-1 was diluted with 20 mM sodium phosphate, pH 7.4, 100 mM NaCl, 2 mM DTT, 110 mM OG, 20 µM EGTA, and 10% (wt/vol) glycerol to bring the NaCl to 100 mM then injected on a MonoS 5/50 column (GE Healthcare), washed with the same buffer, and then eluted with buffer containing 20 mM sodium phosphate, pH 7.4, 600 mM NaCl, 2 mM DTT, 110 mM OG, 20 µM EGTA, and 10% (wt/vol) glycerol. Protein-containing fractions were dialyzed against 20 mM sodium phosphate, pH 7.4, 300 mM NaCl, 2 mM DTT, 110 mM OG, and 10% (wt/vol) glycerol to lower the salt concentration. The protein concentration was measured by UV absorption at 280 nm and aliquots were flash-frozen in liquid nitrogen and stored at −80 °C.
Cpx, Cpx A8C, and the 4M mutant of Cpx.
Full-length wild-type Cpx, Cpx A8C, and the 4M mutant of complexin-1 (R48A, R59A, K69A, Y70A, C105A) were expressed separately in E. coli using BL21 (DE3) cells with a thrombin-cleavable N-terminal hexa-histidine tag from plasmid pET28a (Novagen, EMD Chemicals). Cells were grown in Luria Broth to an OD600 between 0.6 and 0.8 and protein expression was induced by the addition of IPTG to 0.5 mM for 12–16 h at 20 °C. Cells were harvested by centrifugation and resuspended in 20 mM Hepes, pH 7.5, 500 mM NaCl, 2 mM DTT (except 0.5 mM TCEP for Cpx A8C), and 10 mM imidazole supplemented with 1 mM PMSF and EDTA-free complete protease inhibitor mixture tablets. Cells were lysed by passing them through the Emulsiflex C5 homogenizer (Avestin) at 15,000 psi three times. The lysate was clarified by centrifugation in a Ti-45 rotor for 35 min at 125,000 × g. Supernatant was bound to a 4-mL bed volume of Nickel-NTA beads in batch, stirring for 1 h at 4 °C. The beads were harvested by centrifugation and poured into a column, attached to an ATKA Prime (GE Healthcare), and washed with 60 mL of buffer containing 20 mM Hepes, pH 7.5, 500 mM NaCl, 2 mM DTT (except 0.5 mM TCEP for Cpx A8C), 10 mM imidazole, and the same buffer containing 25 mM imidazole and then eluted with the same buffer containing 450 mM imidazole. Protein-containing fractions were combined and 100 units of thrombin (Haematologic Technologies) were added. The sample was then dialyzed against 500 mL of 20 mM Hepes, pH 7.5, 50 mM NaCl, and 4 mM DTT (except 0.5 mM TCEP for Cpx A8C) (Cpx buffer B) overnight at 4 °C. PMSF was added to 1 mM to quench the thrombin activity and the mixture was injected on to a MonoQ 5/50 column equilibrated with Cpx buffer B. The column was washed with 20 column volumes of Cpx buffer B and the protein eluted with a linear NaCl gradient in Cpx buffer B from 50 mM to 500 mM NaCl over 30 column volumes. The protein-containing fractions were combined and dialyzed against 20 mM Hepes, pH 7.5, 100 mM NaCl, 4 mM DTT (0.5 mM TCEP for the A8C mutant) overnight at 4 °C and then concentrated using a 3,000 molecular-weight cut-off dialysis cassette (Millipore). The protein concentration was measured by absorbance at 280 nm, and aliquots were frozen in liquid nitrogen and stored at −80 °C.
C-terminally truncated Cpx.
Cpx1–83 and Cpx1–83 E25C were expressed in E. coli as an N-terminal hexa-histidine fusion construct followed by a thrombin cleavage site (pET28a) in BL21 (DE3) cells (Novagen, EMD Chemicals). Cells were autoinduced overnight at 30 °C (60). Induced cells from 4 L of culture were harvested and resuspended in 200 mL of 20 mM Hepes pH7.5, 500 mM NaCl, 2 mM DTT, 10 mM imidazole, 6 EDTA free protease inhibitor tablets (Roche), and 1 mM PMSF. Cells were lysed by passing them three times through the Emulsiflex C5 homogenizer (Avestin) at 15,000 psi and the lysate was clarified by centrifugation in the Ti-45 rotor at 125,000 × g for 35 min. The lysate was bound to 5 mL of Ni NTA agarose (Qiagen) by stirring for 1 h at 4 °C. Beads were harvested by centrifugation and washed with 10-column volumes of 20 mM Hepes pH 7.5, 500 mM NaCl, 2 mM DTT (except 0.5 mM TCEP for Cpx1-83 E25C), 25 mM imidazole, and eluted with 20 mM Hepes pH 7.5, 500 mM NaCl, 2 mM DTT, 450 mM imidazole. Protein-containing fractions were combined, 100 units of thrombin (Haematologic Technologies) was added and the mixture was dialyzed against 500 mL of 20 mM Tris pH 8.0, 50 mM NaCl, 2 mM DTT (except 0.5 mM TCEP for Cpx1–83 E25C) overnight at 4 °C. PMSF was added to 1 mM to inactivate the thrombin and the mixture was injected on a monoQ 5/50 column (GE Healthcare) equilibrated in 20 mM Tris pH 8.0, 50 mM NaCl, 2 mM DTT; DTT was substituted by 0.5 mM TCEP for Cpx1–83 E25C. A peak in the flow through was observed and the bound protein was eluted in a linear gradient from 50 mM to 500 mM NaCl. All protein-containing fractions were analyzed by SDS/PAGE gel electrophoresis. Flow-through fractions containing pure Cpx1–83 were combined, the protein concentration was measured by absorbance at 280 nm, and aliquots were flash-frozen in liquid nitrogen and stored at −80 °C.
Fluorescent Dye Labeling for Membrane Binding Studies.
Purified Cpx A8C, Cpx1–83 E25C, and commercial BSA were labeled with Alexa 647-maleimide in the presence of 5 mM TCEP overnight, following the protocol described in ref. 62. The labeling efficiency was assessed by absorbance spectroscopy: it was ∼40% for both the Cpx A8C and Cpx1–83 E25C.
Single Vesicle Fusion Experiments.
Preparation of PM and SV vesicles.
For the single-vesicle fusion experiments (Figs. 1 and 5 and Fig. S1) we used the same membrane compositions and protein densities as previously described (19, 44). SV vesicles were reconstituted with both synaptotagmin-1 and synaptobrevin-2, whereas PM vesicles were reconstituted with syntaxin-1A and SNAP-25A. For SV vesicles the lipid composition was phosphatidylcholine (PC) (48%), phosphatidylethanolamine (PE) (20%), phosphatidylserine (PS) (12%), and cholesterol (20%). For PM vesicles the lipid composition was Brain Total Lipid Extract (Avanti Polar Lipids) supplemented 3.5 mol% PIP2, and 0.1 mol% biotinylated PE. Quantitative 31P NMR analysis by Avanti Polar Lipids of two different stocks of Brain Lipid Extract revealed the following average composition of the major components: PC (17%), PE (8%), plasmalogen PE (14%), PS (8%), and sphingomyelin (5%). Among the remaining 48% of other components of Brain Total Lipid Extract is at least 20% cholesterol (63), so at variance with the previously published protocol (44), cholesterol was not additionally supplemented. Dried lipid films were dissolved in 110 mM OG buffer containing purified proteins at protein-to-lipid ratios of 1:200 for synaptobrevin-2 and syntaxin-1A, and 1:800 for synaptotagmin-1.
A three- to fivefold excess of SNAP-25 (with respect to syntaxin-1A) and 3.5 mol% PIP2 were added to the protein–lipid mixture for PM vesicles only. Detergent-free buffer (20 mM Hepes, pH 7.4, 90 mM NaCl, and 0.1% 2-mercaptoethanol) was added to the protein–lipid mixture until the detergent concentration was at (but not lower than) the critical micelle concentration of 24.4 mM (i.e., vesicles did not yet form). For the preparation of SV vesicles, 50 mM sulforhodamine B (Invitrogen) was added to the protein–lipid mixture. The vesicles subsequently formed during size-exclusion chromatography using a Sepharose CL-4B column, packed under near constant pressure by gravity with a peristaltic pump (GE Healthcare) in a 5.5-mL column with a 5-mL bed volume, that was equilibrated with buffer V (20 mM Hepes, pH 7.4, 90 mM NaCl) supplemented with 20 µM EGTA and 0.1% 2-mercaptoethanol.. The eluent was subjected to dialysis into 2 L of detergent-free buffer V supplemented with supplemented with 20 µM EGTA, 0.1% 2-mercaptoethanol, 5 g of Bio-beads SM2, and 0.8 g/L Chelex 100 resin. After 4 h, the buffer was exchanged with 2 L of fresh buffer V containing 20 µM EGTA, 0.1% 2-mercaptoethanol and Bio-beads, and the dialysis continued for another 12 h (overnight). We note that for SV vesicles, the chromatography equilibration and elution buffers did not contain sulforhodamine, so the effective sulforhodamine concentration inside SV vesicles is considerably (up to 10-fold) lower than 50 mM.
As described previously (43), the presence and purity of reconstituted proteins was confirmed by SDS/PAGE of the vesicle preparations, and the directionality of the membrane proteins (facing outward) was assessed by chymotrypsin digestion followed by SDS/PAGE. The size distributions of the SV and PM vesicles were analyzed by cryo-EM, as described previously (39).
Single-vesicle content-mixing assay.
We used the same assay as previously described (19). SV vesicles were labeled with a soluble fluorescent content dye at a moderately self-quenching concentration; for simplicity in this work, we did not include a lipid-dye because we were exclusively interested in the exchange of content, the correlation for neurotransmitter release. The surface of the quartz slides was passivated by coating the surface with PEG molecules, which alleviated nonspecific binding of vesicles. The same protocol and quality controls (surface coverage and nonspecific binding) were used as described previously (39, 44), except that PEG-SVA (Laysan Bio) instead of mPEG-SCM (Laysan Bio) was used because it has a longer half-life. The surface was functionalized by inclusion of biotin-PEG (Laysan Bio) during pegylation. A quartz slide was assembled into a flow chamber and incubated with neutravidin (0.1 mg/mL). Biotinylated PM vesicles (diluted 100 times) were tethered by incubation at ambient temperature (25 °C) for 30 min, followed by three rounds of washing with 120 µL buffer V, to remove unbound PM vesicles; each buffer wash effectively replaces the (3 µL) flow-chamber volume more than 100 times. Upon the start of illumination and recording of the fluorescence from a particular field of view of the flow chamber, SV vesicles (diluted 100–1,000 times, depending on the acquisition round, see below) were loaded into the flow chamber and association of SV vesicles with PM vesicles was monitored for 1 min. Cpx, Cpx fragments, or Cpx mutants were added concurrently together with the SV vesicles at 2-μM concentration. While continuing the recording, the flow chamber was washed three times (120 µL of buffer V and including the Cpx, Cpx fragments, or Cpx mutant at 2 μM concentration) to remove unbound SV vesicles. Subsequently, we continued recording for 1 min to monitor Ca2+-independent fusion events. Thus, in contrast to a previous version of this assay, where SV vesicles where incubated with tethered PM vesicles for at least 30 min (43), monitoring of fusion events was started right after the 1-min SV–PM vesicle association period and buffer washes. Subsequently, Ca2+-solution was injected into the flow chamber consisting of 500 µM Ca2+, 500 nM Cy5 dye molecules (used as an indicator for the arrival of Ca2+ in the evanescent field), and Cpx, Cpx fragments, or Cpx mutant at 2-μM concentration in buffer V. Ca2+-triggered fusion events were monitored within the same field-of-view upon injection for a 1-min period. The injection was performed at a speed of 66 µL/s by a motorized syringe pump (Harvard Apparatus) using a withdrawal method similar to the one described previously (19, 39). All experiments were carried out at ambient temperature (25 °C).
Multiple acquisition rounds and repeats for the single-vesicle fusion experiments.
To increase the throughput of the assay and make better use of the vesicle samples, after intensive washing (3 × 120 µL) with buffer V (which includes 20 μM EGTA to remove Ca2+ from the sample chamber), we repeated the entire acquisition sequence (SV vesicle loading, counting the number of associated SV–PM vesicles, monitoring of Ca2+-independent fusion, Ca2+ injection, and monitoring of Ca2+-triggered fusion) in a different imaging area within the same flow chamber. Five such acquisition rounds were performed with the same sample chamber. SV vesicles were diluted 1,000× for the first and second acquisition rounds, 200× for the third and fourth acquisition rounds, and 100× for the fifth acquisition round to offset the slightly increasing saturation of the surface with SV vesicles. The entire experiment (each with five acquisition rounds) was then repeated n times (Table S1), using different flow chambers (referred to as the “repeat experiment”). Among the specified number of repeats, there are at least three different protein preparatons and vesicle reconstitutions, so the variations observed in the bar charts reflect sample variations as well as variations among different flow chambers.
Instrument set-up for the single-vesicle fusion experiments.
All experiments were performed on a prism-type total internal reflection fluorescence (TIRF) microscope using 532 nm (green) laser (CrystaLaser) excitation. Two observation channels were created by a 640-nm single-edge dichroic beamsplitter (FF640-FDi01-25X36, Shemrock): one channel was used for the fluorescence-emission intensity of the content dyes and the other one for that of the Cy5 dyes that are part of the injected Ca2+-solution. The two channels were recorded on two adjacent rectangular areas (45 × 90 μm2) of a charge-coupled device (CCD) camera (iXon+ DV 897E, Andor Technology). The imaging data were recorded with the smCamera software developed by Taekjip Ha, The Johns Hopkins University, Baltimore, MD. Our procedure resulted in a time series of images over a total of 3 min, consisting of the subsequent 1-min periods of vesicle association, Ca2+-independent, and Ca2+-triggered fusion, plus 5-s intervals for buffer exchanges. The arrival time of Ca2+ was determined by monitoring of the Cy5 channel.
Data analysis of the single-vesicle fusion experiments.
We detected content dye fluorescent spots in a particular imaging area with the smCamera program obtained from Taekjip Ha, The Johns Hopkins University, Baltimore, MD. The appearance of a content dye fluorescence spot corresponds to an association of a SV vesicle to a surface-tethered PM vesicle. However, only those spots were subsequently analyzed and counted that showed exactly one stepwise increase of the fluorescence intensity during the SV–PM vesicle association period of 1 min that precedes the subsequent fusion periods. Thus, this procedure excludes SV vesicles that underwent Ca2+-independent fusion during the vesicle-association period because fusion leads to a second stepwise fluorescence intensity increase. Moreover, for data-acquisition rounds within the same sample chamber, this procedure excludes SV vesicles that were already associated during a previous acquisition round. Stepwise increases in the fluorescence-intensity time traces were automatically detected by the computer program described in ref. 64, and manually checked to ensure correct performance of the automated procedure. Collectively, we refer this selected set of content dye fluorescent spots as the “analyzed SV–PM vesicle pairs.”
In the subsequent Ca2+-independent and Ca2+-triggered fusion periods of 1 min each, a second stepwise increase in content dye fluorescence intensity was counted as a Ca2+-independent and Ca2+-triggered fusion event, respectively. Histograms for Ca2+-independent and Ca2+-triggered fusion event occurrence were then generated with a time bin of 1 s, Ca2+-triggered fusion histograms were synchronized by the appearance of fluorescence intensity of the Cy5 dye molecules that are part of the injected Ca2+-solution. Histograms were cumulated over all acquisition rounds and repeat experiments. The cumulated histograms were subsequently normalized by dividing the histograms by the total number of analyzed SV–PM vesicle pairs for a particular condition (i.e., the sum of the number of analyzed SV–PM vesicle pairs for all acquisition rounds and repeat experiments for a particular condition) (Fig. S1 and Table S1).
In the bar charts in Figs. 1 and 5, the vesicle association count refers to the number of associated SV–PM vesicle pairs only during the first acquisition round, averaged over for all repeat experiments for a particular condition. We only used the first acquisition round for the vesicle association count because in subsequent rounds, the imaging surface becomes progressively saturated so a small number of vesicle-association events may be missed in these subsequent rounds. The average probability of Ca2+-independent fusion events per second is calculated by dividing the overall Ca2+-independent fusion probability of the cumulated and normalized histogram during the 1-min observation period by 60. The amplitude is the probability of Ca2+-triggered fusion in the cumulated and normalized histogram during the first time bin (1 s) upon Ca2+ injection. The error bars for the probabilities and amplitudes were calculated as SDs for the n repeat experiments (Table S1) by using individually cumulated (but globally normalized) histograms for all rounds of a particular repeat experiment. The histograms of the Ca2+-triggered fusion probability (Fig. S1) were fit to a single exponential decay function using the Levenberg–Marquardt algorithm. The degree of synchronization was described by the decay rate and error bars for the decay rate were obtained from the covariance matrix of the fit.
Single-Molecule Protein–Liposome Binding Assays.
For the single-molecule protein-vesicle binding assays (Fig. 4 and Fig. S2) a mixture of POPC (1-palmitoyl-2-dioleoyl-sn-glycero-3-phosphatidylcholine), DOPS (1,2-dioleoyl-sn-glycero-3-phosphatidylserine), cholesterol, POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), DiI, and Biotin-PE (at molar ratio of 48.9:10:20:20:1:0.1) in chloroform was first dried in a vacuum and was then resuspended in buffer V (20 mM Hepes, pH 7.4, 90 mM KCl) to result in a total lipid concentration of 10 mM. Protein-free large unilamellar liposomes at different sizes (30-, 100-, and 400-nm diameters) were prepared by extrusion through polycarbonate filters (Avanti Polar Lipids).
For the protein–liposome binding experiments shown in Fig. 4 B and C, unilamellar liposomes with the specified diameter were tethered on a surface through biotin-streptavidin coupling (65). For these experiments, Cpx was labeled at the eighth amino acid residue with Alexa 647 dye. Binding events between the vesicles and Cpx-A647 were monitored in real time at ambient temperature for 3∼10 min right after 1∼20 nM Cpx-A647 was loaded with buffer V containing glucose, trolox, and glucose oxidase.
For the protein–liposome binding experiments shown in Fig. 4 D and E, 5 µM Cpx-A647, Cpx1–83-A647, or BSA-A647 were preincubated with 5 µM liposomes in solution at ambient temperature for 30 min. The unilamellar liposomes were prepared as described above, but an equimolar mixture of different sizes (30-, 100-, and 400-nm diameters) was used. Images were acquired after protein-bound liposomes were tethered on surface through biotin-streptavidin coupling. Unbound liposomes were then removed through extensive washing with vesicle-free buffer.
For all protein–liposome binding experiments, labeled proteins were excited with 633-nm laser light and emission intensity was detected in the range 650–790 nm. Labeled liposomes were excited with 562-nm laser light and emission intensity was detected in the range 590–620 nm. Images of both channels were acquired simultaneously using a dichroic beam splitter on an EMCCD camera (Andor 897). A TIRF microscope was used for both single-vesicle and single-molecule experiments with the configuration shown in figure 2 and figure S1 in ref. 65.
Neuronal Culture Experiments.
Neuronal cultures were obtained from mouse cortex as described previously (29). Briefly, mouse cortices were dissected at postnatal day 0, dissociated by 0.25% trypsin digestion for 12 min at 37 °C, plated on poly-L-lysine–coated circular glass coverslips (12-mm diameter), and cultured in MEM (GIBCO) supplemented with 2% (vol/vol) B27 (GIBCO), 0.5% (wt/vol) glucose, 100 mg/L transferrin, 5% (vol/vol) FBS, and 2 mM Ara-C (Sigma). Neurons were infected with lentiviruses at days in vitro (DIV) 4–6 and analyzed at DIV 14–16. All animal procedures were performed in accordance with South-Central University for Nationalities animal use rules and the requisite approvals of animal use committees.
Production of lentiviruses.
Lentiviral expression vector and three helper plasmids (pRSV-REV, pMDLg/pRRE, and vesicular stomatitis virus G-protein expression vector) were cotransfected into HEK293T cells (ATCC), at 6, 2, 2, and 2 μg of DNA per 25 cm2 culture area, respectively, using Polyethylenimine (PEI) (66), and cell-culture supernatants containing the viruses were collected 48 h after transfection and used for infection of neurons. All steps were performed under level II biosafety conditions.
Immunocytochemistry and imaging experiments.
Mouse cortical neurons infected with various lentiviruses were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100, incubated with anticomplexin (polyclonal; L669) and anti-vGlut1 [monoclonal; N28/9 (Neuromab)] primary antibodies in PBS with 5% BSA, washed, and stained with polyclonal anticomplexin and monocolonal anti-vGlut1 and visualized using Alexa Fluor-488 goat anti-rabbit and Alexa Fluor-546 goat anti-mouse secondary antibodies (Molecular Probes). Images were acquired by using a Nikon C2 confocal microscope equipped with a 60× oil-immersion objective. Identical settings were applied to all samples in each experiment. We measured the average pixel intensities by manually tracing each dendrite, with a >twofold background signal. We measured the Cpx-specific fluorescence intensity at synaptic locations that had been labeled by vGlut1 (Fsynapse) and at locations that were randomly picked elsewhere that are not colocalized with vGlut1 locations (Fnon_synapse). We calculated ΔF = Fsynapse – Fnon_synapse to determine the subcellular location of Cpx.
Electrophysiological recordings.
Electrophysiological recordings were performed in whole-cell patch-clamp mode using concentric extracellular stimulation electrodes. Evoked synaptic responses were triggered by a bipolar electrode placed 100–150 mm from the soma of neurons recorded. Patch pipettes were pulled from borosilicate glass capillary tubes (Warner Instruments) using a P97 pipette puller (Sutter). The resistance of pipettes filled with intracellular solution varied between 3 and 5 MOhm. After formation of the whole-cell configuration and equilibration of the intracellular pipette solution, the series resistance was adjusted to 8–10 MOhm. Synaptic currents were monitored with an EPC10 amplifier (HEKA). Single extracellular stimulus pulses (90 μA, 1 ms) were controlled with a Model 2100 Isolated Pulse Stimulator (A-M Systems) for all evoked EPSCs measurements. The whole-cell pipette solution contained 120 mM CsCl, 5 mM NaCl, 1 mM MgCl2, 10 mM Hepes, 10 mM EGTA, 0.3 mM Na-GTP, 3 mM Mg-ATP, and 5 mM QX-314 (pH 7.2, adjusted with CsOH). The bath solution contained 140 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, and 10 mM glucose (pH 7.4, adjusted with NaOH). AMPAR-mediated EPSCs were pharmacologically isolated by adding the NMDA receptor blockers AP-V (50 μM) and the GABAA-receptor blockers picrotoxin (100 μM) to the extracellular solution. NMDA-receptor mediated EPSCs were recorded at a holding potential of +40 mV with 20 μM CNQX, 100 μM picrotoxin, and 15 μM glycine and by raising the MgCl2-concentration to 4 mM. Spontaneous mEPSCs were monitored in the presence of tetrodotoxin (TTX; 1 μM) to block action potentials. Miniature events were analyzed in Clampfit 10 (Molecular Devices) using the template matching search and a minimal threshold of 5 pA and each event was visually inspected for inclusion or rejection by an experimenter blind to the recording condition. For the measurement of the synaptic or nonsynaptic localized Cpx fluorescence intensity, we manually chose fluorescent spots signal along the dendrite.
Acknowledgments
We thank Dr. Yeon-Kyun Shin for critical review of the manuscript, and Dr. Taekjip Ha for software of single particle/molecule microscopy experiments. This work was supported by the 973-Program Funding (Grant 2015CB856304); the National Science Foundation of China (Grant 31300892); National Science Foundation of Hubei Grants 2014CFA027 and 2014CFB455 (to X.Y.); and National Institutes of Health Grant R37-MH63105 (to A.T.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609917113/-/DCSupplemental.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15(7):658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490(7419):201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395(6700):347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 5.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92(6):759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 6.Geppert M, et al. Synaptotagmin I: A major Ca2+ sensor for transmitter release at a central synapse. Cell. 1994;79(4):717–727. doi: 10.1016/0092-8674(94)90556-8. [DOI] [PubMed] [Google Scholar]
- 7.McMahon HT, Missler M, Li C, Südhof TC. Complexins: Cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83(1):111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 8.Hata Y, Slaughter CA, Südhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366(6453):347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 9.Mohrmann R, Dhara M, Bruns D. Complexins: Small but capable. Cell Mol Life Sci. 2015;72(22):4221–4235. doi: 10.1007/s00018-015-1998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trimbuch T, Rosenmund C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat Rev Neurosci. 2016;17(2):118–125. doi: 10.1038/nrn.2015.16. [DOI] [PubMed] [Google Scholar]
- 11.Pabst S, et al. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J Biol Chem. 2000;275(26):19808–19818. doi: 10.1074/jbc.M002571200. [DOI] [PubMed] [Google Scholar]
- 12.Maximov A, Tang J, Yang X, Pang ZPP, Südhof TC. Complexin controls the force transfer from SNARE complexes to membranes in fusion. Science. 2009;323(5913):516–521. doi: 10.1126/science.1166505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue M, et al. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat Struct Mol Biol. 2010;17(5):568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue M, et al. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol. 2007;14(10):949–958. doi: 10.1038/nsmb1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhara M, et al. Complexin synchronizes primed vesicle exocytosis and regulates fusion pore dynamics. J Cell Biol. 2014;204(7):1123–1140. doi: 10.1083/jcb.201311085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaeser-Woo YJ, Yang X, Südhof TC. C-terminal complexin sequence is selectively required for clamping and priming but not for Ca2+ triggering of synaptic exocytosis. J Neurosci. 2012;32(8):2877–2885. doi: 10.1523/JNEUROSCI.3360-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiler F, Malsam J, Krause JM, Söllner TH. A role of complexin-lipid interactions in membrane fusion. FEBS Lett. 2009;583(14):2343–2348. doi: 10.1016/j.febslet.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diao J, et al. Complexin-1 enhances the on-rate of vesicle docking via simultaneous SNARE and membrane interactions. J Am Chem Soc. 2013;135(41):15274–15277. doi: 10.1021/ja407392n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai Y, et al. Complexin inhibits spontaneous release and synchronizes Ca2+-triggered synaptic vesicle fusion by distinct mechanisms. eLife. 2014;3:e03756. doi: 10.7554/eLife.03756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snead D, Wragg RT, Dittman JS, Eliezer D. Membrane curvature sensing by the C-terminal domain of complexin. Nat Commun. 2014;5:4955. doi: 10.1038/ncomms5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wragg RT, et al. Synaptic vesicles position complexin to block spontaneous fusion. Neuron. 2013;77(2):323–334. doi: 10.1016/j.neuron.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishnakumar SS, et al. Re-visiting the trans insertion model for complexin clamping. eLife. 2015;4:e04463. doi: 10.7554/eLife.04463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kümmel D, et al. Complexin cross-links prefusion SNAREs into a zigzag array. Nat Struct Mol Biol. 2011;18(8):927–933. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313(5787):676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 25.Giraudo CG, et al. Alternative zippering as an on-off switch for SNARE-mediated fusion. Science. 2009;323(5913):512–516. doi: 10.1126/science.1166500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radoff DT, et al. The accessory helix of complexin functions by stabilizing central helix secondary structure. eLife. 2014;3:e04553. doi: 10.7554/eLife.04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trimbuch T, et al. Re-examining how complexin inhibits neurotransmitter release: SNARE complex insertion or electrostatic hindrance? eLife. 2014;3:e02391. doi: 10.7554/eLife.02391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Cao P, Südhof TC. Deconstructing complexin function in activating and clamping Ca2+-triggered exocytosis by comparing knockout and knockdown phenotypes. Proc Natl Acad Sci USA. 2013;110(51):20777–20782. doi: 10.1073/pnas.1321367110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Kaeser-Woo YJ, Pang ZPP, Xu W, Südhof TC. Complexin clamps asynchronous release by blocking a secondary Ca(2+) sensor via its accessory α helix. Neuron. 2010;68(5):907–920. doi: 10.1016/j.neuron.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai Y, et al. N-terminal domain of complexin independently activates calcium-triggered fusion. Proc Natl Acad Sci USA. 2016;113(32):E4698–E4707. doi: 10.1073/pnas.1604348113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bracher A, Kadlec J, Betz H, Weissenhorn W. X-ray structure of a neuronal complexin-SNARE complex from squid. J Biol Chem. 2002;277(29):26517–26523. doi: 10.1074/jbc.M203460200. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33(3):397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 33.Cai H, et al. Complexin II plays a positive role in Ca2+-triggered exocytosis by facilitating vesicle priming. Proc Natl Acad Sci USA. 2008;105(49):19538–19543. doi: 10.1073/pnas.0810232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin JA, Hu Z, Fenz KM, Fernandez J, Dittman JS. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr Biol. 2011;21(2):97–105. doi: 10.1016/j.cub.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hobson RJ, Liu Q, Watanabe S, Jorgensen EM. Complexin maintains vesicles in the primed state in C. elegans. Curr Biol. 2011;21(2):106–113. doi: 10.1016/j.cub.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huntwork S, Littleton JT. A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci. 2007;10(10):1235–1237. doi: 10.1038/nn1980. [DOI] [PubMed] [Google Scholar]
- 37.Reim K, et al. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell. 2001;104(1):71–81. doi: 10.1016/s0092-8674(01)00192-1. [DOI] [PubMed] [Google Scholar]
- 38.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126(6):1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 39.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. eLife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malsam J, et al. Complexin arrests a pool of docked vesicles for fast Ca2+-dependent release. EMBO J. 2012;31(15):3270–3281. doi: 10.1038/emboj.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malsam J, et al. The carboxy-terminal domain of complexin I stimulates liposome fusion. Proc Natl Acad Sci USA. 2009;106(6):2001–2006. doi: 10.1073/pnas.0812813106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon TY, et al. Complexin and Ca2+ stimulate SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15(7):707–713. doi: 10.1038/nsmb.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyoung M, et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc Natl Acad Sci USA. 2011;108(29):E304–E313. doi: 10.1073/pnas.1107900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyoung M, Zhang Y, Diao J, Chu S, Brunger AT. Studying calcium-triggered vesicle fusion in a single vesicle-vesicle content and lipid-mixing system. Nat Protoc. 2013;8(1):1–16. doi: 10.1038/nprot.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525(7567):62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou P, Bacaj T, Yang X, Pang ZPP, Südhof TC. Lipid-anchored SNAREs lacking transmembrane regions fully support membrane fusion during neurotransmitter release. Neuron. 2013;80(2):470–483. doi: 10.1016/j.neuron.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen MB, et al. Membrane curvature sensing by amphipathic helices: A single liposome study using α-synuclein and annexin B12. J Biol Chem. 2011;286(49):42603–42614. doi: 10.1074/jbc.M111.271130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charbonneau DM, Tajmir-Riahi HA. Study on the interaction of cationic lipids with bovine serum albumin. J Phys Chem B. 2010;114(2):1148–1155. doi: 10.1021/jp910077h. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, Zhu Y, Shin YK. Pre-incubation of t-SNAREs with complexin I increases content-mixing efficiency. Biochemistry. 2016;55(26):3667–3673. doi: 10.1021/acs.biochem.6b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neher E. Complexin: Does it deserve its name? Neuron. 2010;68(5):803–806. doi: 10.1016/j.neuron.2010.11.038. [DOI] [PubMed] [Google Scholar]
- 51.Cao P, Yang X, Südhof TC. Complexin activates exocytosis of distinct secretory vesicles controlled by different synaptotagmins. J Neurosci. 2013;33(4):1714–1727. doi: 10.1523/JNEUROSCI.4087-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue M, et al. Complexins facilitate neurotransmitter release at excitatory and inhibitory synapses in mammalian central nervous system. Proc Natl Acad Sci USA. 2008;105(22):7875–7880. doi: 10.1073/pnas.0803012105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X, et al. Evolutionary conservation of complexins: From choanoflagellates to mice. EMBO Rep. 2015;16(10):1308–1317. doi: 10.15252/embr.201540305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jorquera RA, Huntwork-Rodriguez S, Akbergenova Y, Cho RW, Littleton JT. Complexin controls spontaneous and evoked neurotransmitter release by regulating the timing and properties of synaptotagmin activity. J Neurosci. 2012;32(50):18234–18245. doi: 10.1523/JNEUROSCI.3212-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin L, Kim J, Shin YK. Complexin splits the membrane-proximal region of a single SNAREpin. Biochem J. 2016;473(14):2219–2224. doi: 10.1042/BCJ20160339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi UB, Zhao M, Zhang Y, Lai Y, Brunger AT. Complexin induces a conformational change at the membrane-proximal C-terminal end of the SNARE complex. eLife. 2016;5:e16886. doi: 10.7554/eLife.16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burré J, et al. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329(5999):1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diao J, et al. Native α-synuclein induces clustering of synaptic-vesicle mimics via binding to phospholipids and synaptobrevin-2/VAMP2. eLife. 2013;2:e00592. doi: 10.7554/eLife.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59(3):231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 61.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260(3):289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 62.Weninger K, Bowen ME, Chu S, Brunger AT. Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci USA. 2003;100(25):14800–14805. doi: 10.1073/pnas.2036428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katyal SL, Barilaro L, Hanin I. Lipid composition of different areas of murine brain: Effects of lipid extraction procedures. Lipids. 1985;20(3):201–203. doi: 10.1007/BF02534255. [DOI] [PubMed] [Google Scholar]
- 64.Diao J, Zhao M, Zhang Y, Kyoung M, Brunger AT. Studying protein-reconstituted proteoliposome fusion with content indicators in vitro. BioEssays. 2013;35(7):658–665. doi: 10.1002/bies.201300010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diao J, et al. A single vesicle-vesicle fusion assay for in vitro studies of SNAREs and accessory proteins. Nat Protoc. 2012;7(5):921–934. doi: 10.1038/nprot.2012.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang W, et al. An optimized method for high-titer lentivirus preparations without ultracentrifugation. Sci Rep. 2015;5:13875. doi: 10.1038/srep13875. [DOI] [PMC free article] [PubMed] [Google Scholar]