Significance
Vesicular monoamine transporters (VMATs) are the targets of numerous psychoactive drugs, and play a critical role in the overall process of synaptic transmission by replenishing depleted monoamine stores in synaptic vesicles. VMATs transport monoamines in a process that involves exchange of two H+ per substrate. Here we show that two potent inhibitors of VMAT2, tetrabenazine and reserpine, bind to different conformations of the protein. The transition that generates a reserpine-binding site requires a proton gradient across the membrane. Here we emulate the effect of the proton gradient by tinkering with residues that form the cytoplasmic gate. These findings provide vital information about the conformational dynamics of a mammalian H+-coupled antiporter. Such conformational transitions constitute essential steps in all transport processes.
Keywords: neurotransmitter transporter, ion coupling, reserpine, tetrabenazine
Abstract
Neurotransporters located in synaptic vesicles are essential for communication between nerve cells in a process mediated by neurotransmitters. Vesicular monoamine transporter (VMAT), a member of the largest superfamily of transporters, mediates transport of monoamines to synaptic vesicles and storage organelles in a process that involves exchange of two H+ per substrate. VMAT transport is inhibited by the competitive inhibitor reserpine, a second-line agent to treat hypertension, and by the noncompetitive inhibitor tetrabenazine, presently in use for symptomatic treatment of hyperkinetic disorders. During the transport cycle, VMAT is expected to occupy at least three different conformations: cytoplasm-facing, occluded, and lumen-facing. The lumen- to cytoplasm-facing transition, facilitated by protonation of at least one of the essential membrane-embedded carboxyls, generates a binding site for reserpine. Here we have identified residues in the cytoplasmic gate and show that mutations that disrupt the interactions in this gate also shift the equilibrium toward the cytoplasm-facing conformation, emulating the effect of protonation. These experiments provide significant insight into the role of proton translocation in the conformational dynamics of a mammalian H+-coupled antiporter, and also identify key aspects of the mode of action and binding of two potent inhibitors of VMAT2: reserpine binds the cytoplasm-facing conformation, and tetrabenazine binds the lumen-facing conformation.
Neuronal communication is dependent on the transmission of nerve impulses through chemical synapses, junctions at which electrical signals are relayed from one neuron to another via classical neurotransmitters. Transport and storage of neurotransmitters in secretory vesicles, mediated by vesicular neurotransmitter transporters, allow the regulated exocytosis of neurotransmitters to the synaptic cleft (1–4). Vesicular transporters of cationic neurotransmitters include the vesicular monoamine transporters (VMATs) and acetylcholine transporter (5, 6). Two monoamine transporters, VMAT1 and VMAT2, are responsible for the uptake of dopamine, serotonin, adrenaline, noradrenaline, and histamine in a process that involves the exchange of two protons for one substrate molecule (1–3, 7).
The VMATs belong to the major facilitator superfamily (MFS) of secondary transporters. Analysis of the available crystal structures of MFS transporters reveals multiple conformations during the transport cycle (8), consistent with the alternating access model, which best describes the general transport mechanism (9). A remarkable body of biochemical and biophysical data on the H+-coupled β-galactoside symporter LacY, one of the best-studied MFS transporters, provides strong support for this model (10, 11). Despite these impressive advances, the precise molecular mechanism of coupling between the movements of proton and substrate remains elusive. Protonation and deprotonation of key residues are suggested to function as a “molecular switch” that induces conformational changes (8, 10). In H+-coupled symporters, binding of substrate and generation of the ternary complex allow for the conformational switch between lumen-facing (Clum) and cytoplasm-facing (Ccyt) states. In contrast, in H+-coupled antiporters, protonation and substrate binding are thought to be mutually exclusive and thus the conformational switch can presumably occur after binding of either one (12).
In addition to the native substrates, VMATs interact with many clinically relevant drugs, including the psychostimulant 3,4-methylene-dioxymethamphetamine and the parkinsonian toxin 1-methyl-4-phenylpyridinium (MPP+) (7, 13, 14). For example, heterologous expression of VMATs protects mammalian and yeast cells against MPP+ toxicity by sequestering the toxin in vesicles and away from its primary site of action in mitochondria (7, 15).
The best-characterized inhibitors of VMATs are reserpine and tetrabenazine (TBZ) (1). Reserpine is an indole alkaloid, an antipsychotic and antihypertensive drug rarely used today (16, 17). TBZ is a clinically relevant drug that is used for treatment of hyperkinetic disorders associated with Huntington’s disease and Tourette’s syndrome (18).
Reserpine is a high-affinity competitive inhibitor of VMATs (19, 20). Its binding has been investigated in detail in chromaffin granules from bovine adrenal medulla (21, 22) and in proteoliposomes reconstituted with purified transporter (23). Binding of [3H]reserpine is accelerated upon imposition of a proton electrochemical gradient across the membrane, suggesting that high-affinity binding sites are recruited upon imposition of the gradient (22–24).
In contrast to reserpine, TBZ depletes predominantly central rather than peripheral amine stores (25) because of a significantly greater specificity for VMAT2 over VMAT1 (26). TBZ is a noncompetitive inhibitor of transport that prevents binding of reserpine and substrates. Consequently, reserpine and substrates inhibit [3H]TBZ binding only at concentrations 100 times higher than their respective KD and KM values (22, 27). A general mechanism of VMAT2 inhibition by TBZ has been proposed that involves two major steps, namely initiation of the TBZ-bound conformation, followed by immobilization of the transporter by generation of a dead-end complex of TBZ with the transporter (28). However, the molecular nature of these processes, like the mechanism of proton coupling, remains to be elucidated.
Here we identify mutants that alter the nature of the conformational equilibrium and mimic the protonated state of VMAT2. We show that these mutants eliminate the requirement for a proton gradient normally necessary for reserpine binding and for reserpine inhibition of TBZ binding, and that they have a higher propensity to adopt a Ccyt state, according to accessibility studies. These data suggest that the residues at these positions are essential for keeping the cytoplasmic gate closed. Moreover, we speculate that TBZ binding prevents reserpine and substrate binding by inducing a closed state in wild-type VMAT2 that cannot undergo proton-induced conformational changes.
Results
Accessibility of Reserpine to Its Binding Site Is Dramatically Enhanced by Modifications Close to the Cytoplasmic End of Transmembrane 11.
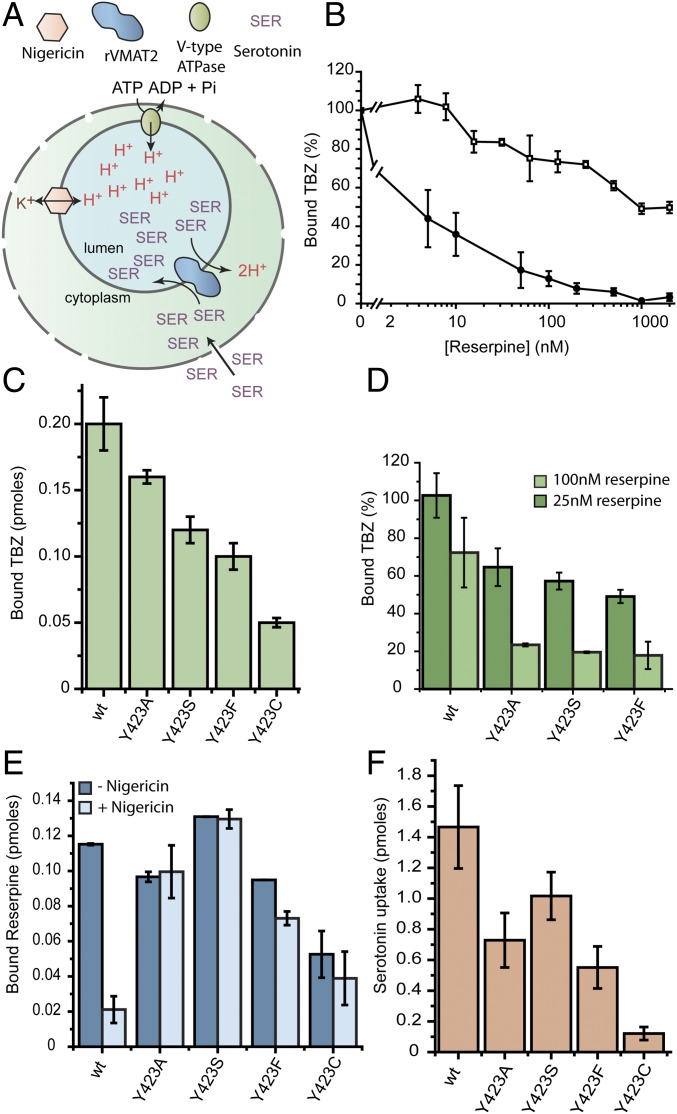
We previously identified two clusters of residues that appear to function as molecular hinge points about which the two six-transmembrane–helix bundles flex and straighten to open and close the pathways on either side of the membrane as required for transport (29). Specifically, the data suggested that polar residues in transmembrane (TM)2 and TM11 create a hydrophilic cluster in one of the anchor points, whereas the other anchor point involves hydrophobic interactions between TM5 and TM8. To further understand their role and to map the pathways leading from the hinge points, we initiated a study of the accessibility of Cys replacements along TM11. To perform such studies, rat VMAT2 (rVMAT2) was expressed in HEK293 cells where the wild-type protein binds [3H]dihydrotetrabenazine, an analog of the noncompetitive inhibitor tetrabenazine, and accumulates [3H]serotonin into vesicles in a process that is dependent on the proton gradient generated by a V-type ATPase (Fig. 1A). To assay for both activities, the plasma membrane was permeabilized with digitonin, and the cells were then challenged with the radiolabeled compounds. To generate the vesicular pH gradient necessary for the transport reaction, ATP was supplied along with serotonin. The ionophore nigericin (Nig) was used to collapse the pH gradient (Fig. 1A).
Fig. 1.
Site-directed mutagenesis of Y423 favors the reserpine-binding conformation. (A) Cartoon illustrating the experimental system. (B) Digitonin-permeabilized cells transfected with either wild-type (□) or Y423C (●) rVMAT2 were assayed for [3H]TBZ binding ability in the presence of increasing concentrations of reserpine. (C) [3H]TBZ binding to digitonin-permeabilized whole cells. (D) [3H]TBZ binding to whole cells expressing various replacements at position 423, following incubation with 25 nM (dark green) or 100 nM (light green) reserpine. (E) Digitonin-permeabilized cells supplemented with ATP were assayed for [3H]reserpine binding with (light blue) or without (dark blue) the addition of the ionophore nigericin. (F) [3H]Serotonin transport into digitonin-permeabilized whole cells expressing various replacements at position 423. Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
Characterization of the Cys replacements in TM11, and specifically of Y423C, revealed unforeseen effects on their binding of TBZ and reserpine. TBZ is a mixed-type inhibitor that prevents binding of reserpine or of transportable substrates to the wild-type protein, as suggested by the finding that they both inhibit TBZ binding only at concentrations that are orders of magnitude higher than their respective KD and KM, (22, 27) (see also Fig. S1 for a more detailed study). In particular, reserpine, which binds to wild-type rVMAT2 with a KD of ∼5 nM (30), inhibited TBZ binding to wild-type rVMAT2 by ∼50% only when present at concentrations as high as ∼1 to 2 μM (Fig. 1B, □). Surprisingly, however, TBZ binding to Y423C rVMAT2 was 50% inhibited at <10 nM reserpine (Fig. 1B, ●), suggesting that the mutation generates a conformation in which the accessibility of reserpine to its binding site is dramatically enhanced. Other replacements at the same position (Y423A/S/F) displayed robust TBZ binding (Fig. 1C) and led also to a pronounced increase in TBZ sensitivity (Fig. 1D).
Fig. S1.
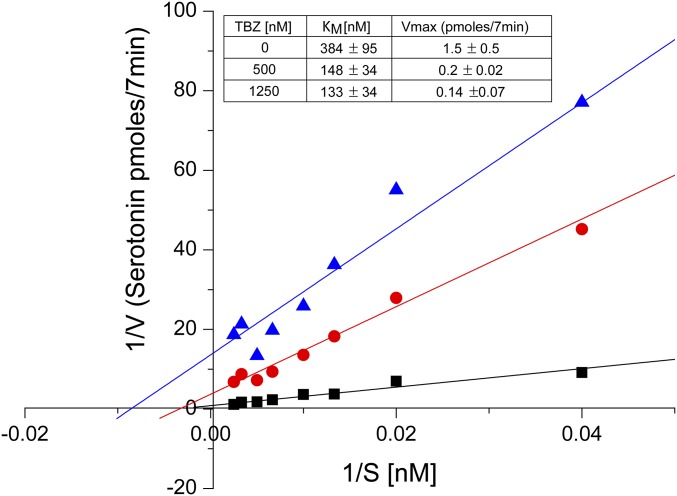
TBZ is a mixed-type inhibitor. Noncompetitive inhibitors are a special rare case of mixed inhibitors in which only the Vmax is changed. In the case of TBZ, the KM values change about twofold, whereas the Vmax changes 10-fold (Inset). In this manuscript, as well as in previous literature, the term “noncompetitive inhibitor” is freely used for the mode of action of TBZ. In the experiment shown in this figure, cells were incubated with increasing concentrations of [3H]serotonin without TBZ (solid black square) or with 500 nM TBZ (solid red circle) or 1,250 nM TBZ (solid blue triangle). KM and Vmax values shown in the table were calculated using Origin 9.2 software (OriginLab) and are averages of three experiments.
The above findings suggest that in the mutants of position 423, TBZ can no longer prevent binding of reserpine. We therefore hypothesized that the mutations induced a conformation that favors binding of reserpine. To test this hypothesis directly, we assayed [3H]reserpine binding in the presence and absence of a proton electrochemical gradient. This gradient was generated by adding ATP to stimulate the V-ATPase, and was collapsed by the addition of nigericin, a K+/H+ ionophore. As expected from previous results (24), robust binding was observed to wild-type rVMAT2 in the presence of a proton gradient (>0.11 pmol) but binding was reduced in its absence (0.02 ± 0.008 pmol) (Fig. 1E). In contrast, the various replacements at position 423 allowed [3H]reserpine to bind to practically the same extent in both the presence and absence of a proton gradient (Fig. 1E). We hypothesize that mutations at position 423 mimic the effects of the proton gradient and modify the conformational equilibrium in such a way that allows reserpine to bind more readily. Notably, the replacements can still support TBZ binding, and three of them still display robust serotonin transport (Fig. 1 C and F, respectively).
Homology Modeling Predicts That Y423 Contributes to the Cytoplasmic Gate.
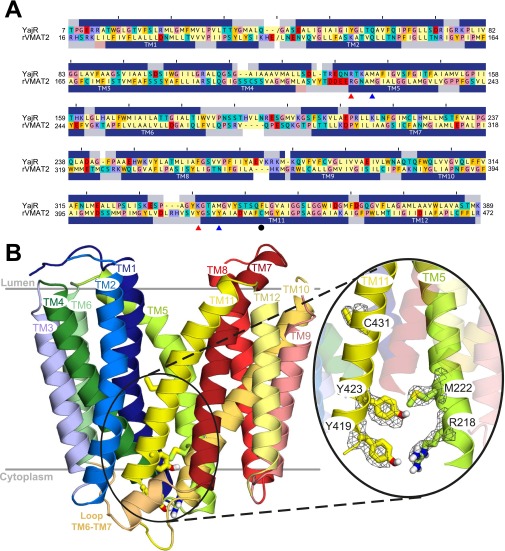
To understand the molecular basis of the unique behavior observed in the Y423 mutants, we constructed a homology model of rVMAT2 in a lumen-facing conformation, using a recently reported structure of a putative H+-dependent transporter, YajR, as a template (31) (Materials and Methods and Fig. S2). According to both the previous model based on LacY (29) and the new model based on YajR, Y423 is located in the cytoplasmic end of TM11 (Fig. 2A). Analysis of the model revealed that Y423 is located very close to M222, which is in TM5. Interestingly, we were able to identify a second set of interactions involving Y419 and R218, just one helical turn away from Y423 and M222, respectively (Fig. 2A and Fig. S2). A recent study that analyzed all available crystal structures in the MFS concluded that the cytoplasmic gate is formed by residues in TMs 4, 5, 10, and 11 (32). This led us to test the hypothesis that the above network of interactions connecting the cytoplasmic ends of TM5 and TM11 contributes to the cytoplasmic gate in VMAT2.
Fig. S2.
Lumen-facing model of rVMAT2 generated using YajR as a template. (A) Sequence alignment between YajR and rVMAT2 excluding the N and C termini and the long loop between TM1 and TM2 (indicated by “/”). The positions of C431 (solid black circle), the Y–R pair (solid red triangle), and the Y–M pair (solid blue triangle) are indicated. The secondary structure assignment from DSSP (62) for the template is shown as blue bars above the alignment, whereas the secondary structure prediction for rVMAT2 generated using PSIPRED is shown below. Sequences are colored according to the chemical nature of the residues: Aliphatic (Asp, Ile, Leu, Met, Val) are in pale yellow; polar uncharged (Asn, Gln, Ser, Thr) and cysteine are in cyan; aromatic (Phe, Trp, Tyr) are in yellow-orange; acidic (Asp, Glu) are in red; basic (Lys, Arg, His) are in purple; and glycine and proline are in pink. (B) Overview of the homology model showing all of the transmembrane helices and highlighting the residues studied here as sticks. The position of a native cysteine, C431, is also shown. The mesh indicates the density of side-chain atoms in 300 MODELLER models, as obtained using VolMap plugin v1.1 in VMD (63) after aligning their backbone atoms. The figure was made using PyMOL v1.6 (Schrödinger).
Fig. 2.
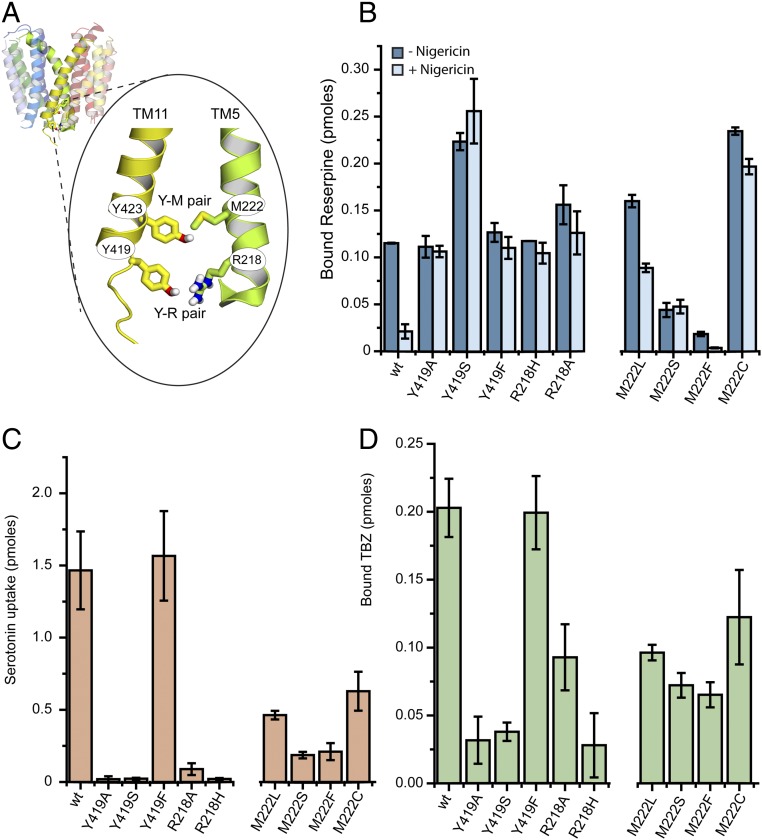
R218, M222, Y419, and Y423 contribute to the cytoplasmic gate. (A) Homology model of rVMAT2 based on the structure of YajR (PDB ID code 3WDO) in a lumen-facing conformation. TM5 and TM11 are shown as opaque. R218 and M222 from TM5, and Y419 and Y423 from TM11, are represented as sticks. The close-up view indicates the predicted interactions between these residues as part of the cytoplasmic gate. (B) [3H]Reserpine binding by whole cells expressing different replacements at positions 218, 222, and 419, as described in Fig. 1. (C and D) [3H]Serotonin transport (C) and [3H]TBZ binding (D) to digitonin-permeabilized cells. Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
Mutagenesis Studies Validate the Proposed Roles.
To test the above hypothesis, we generated various replacements at positions 419, 218, and 222 and tested the effect of each replacement on the ability to bind [3H]reserpine in the absence of a proton gradient. Mutating these residues should shift the equilibrium of the transporter to more cytoplasm-facing, reserpine-binding states even in the absence of a proton gradient, similar to the effect of mutations at Y423 (Fig. 1E). The results in Fig. 2B show that indeed, unlike wild-type rVMAT2, the Ala, Ser, and Phe replacements at position 419 allowed efficient [3H]reserpine binding that was independent of the presence (−Nig) and absence (+Nig) of a proton gradient. Moreover, as shown in Fig. 2B, binding of reserpine to R218A/H was also independent of the generation of a proton gradient, supporting a central role of Tyr419 and Arg218 (Y–R) in the intracellular gate. Unlike the mutations at position 423, replacements in the Y–R positions had a profound effect on serotonin transport and TBZ binding (Fig. 2 C and D). Both R218 replacements resulted in only very low (R218A) or no (R218H) transport. Both mutations, especially R218H, also had an inhibitory effect on TBZ binding. In the case of position 419, the Ala and Ser replacements displayed practically no transport activity (Fig. 2C) and, correspondingly, very low TBZ binding (Fig. 2D), whereas Y419F retained wild-type–like activity. These results point to the significance of an aromatic residue at position 419, and raise the possibility that Y419 and R218 interact via cation–π interactions that the His replacement (R218H) is too short to achieve.
Similar studies with the potential counterpart of Y423 revealed that mutating M222 (TM5) to Ser generated a protein that binds [3H]reserpine equally efficiently in the presence and absence of a proton gradient (Fig. 2B). Replacement with Leu, a hydrophobic residue, retained some of the dependence of [3H]reserpine binding on the proton gradient, as nigericin inhibited by ∼50% (Fig. 2B). Replacement with Phe, a bulky aromatic residue, strongly impaired binding (Fig. 2B). Similar to the mutations at position Y423, M222 mutants were still able to transport serotonin, albeit to lower levels (Fig. 2C), and had no substantial effect on TBZ binding ability (Fig. 2D).
The similarity of the phenotypes prompted us to speculate that the above residues act as two “locks,” the “inner lock” being Y423–M222 and the “outer lock” composed of Y419–R218 (Fig. 2A). Notably, although replacements in all of the above positions allow binding of reserpine independent of the proton gradient, only replacements at the Y419–R218 positions have a strong effect on TBZ binding and serotonin transport. This raises the question of why the hypothetical inner-lock mutations do not affect the latter processes despite affecting the former. We suggest that replacements at positions 423–222 moderately affect the conformational equilibrium, so that a high-affinity binder such as reserpine can still shift the equilibrium. This suggestion is supported by the fact that a binder with lower affinity, such as serotonin, inhibits TBZ binding in mutants of Y419 and R218 but not in mutants of Y423 or M222 (Fig. S3). Moreover, inhibition of TBZ binding (and therefore a high ratio of reserpine to TBZ binding) by modification of the Y–R positions seems to indicate here a lack of ability to complete the transport cycle (Table S1).
Fig. S3.
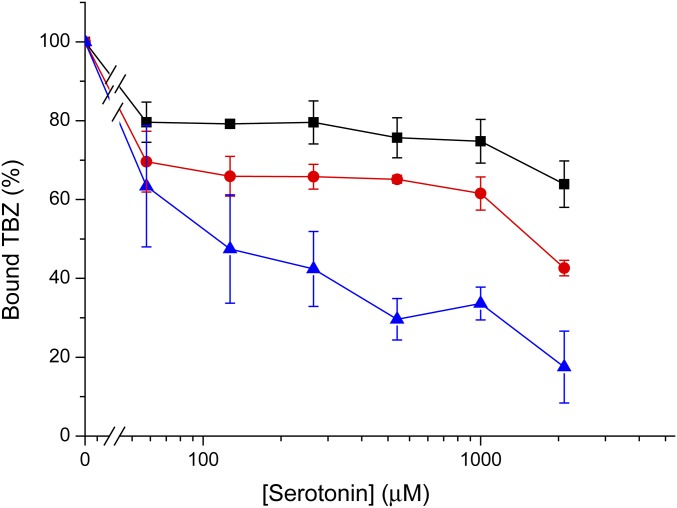
TBZ binding inhibition by serotonin. Cells transfected with the wild-type (solid black square) or mutants Y423A (solid red circle) and R218A (solid blue triangle) were assayed for [3H]TBZ binding ability following incubation with the indicated concentrations of serotonin. Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
Table S1.
Correlation between ligand binding, serotonin transport activity, and accessibility to MTSEA
| Strain | Binding ratio, reserpine/TBZ | Serotonin uptake, pmol per 20 min | IC50 for MTSEA, μM |
| wild-type | 0.57 ± 0.02 | 1.47 ± 0.27 | 205 ± 78 |
| Y419F | 0.64 ± 0.37 | 1.57 ± 0.31 | 80 ± 8 |
| R218A | 1.26 ± 0.01 | 0.09 ± 0.04 | 57 ± 7 |
| Y419A | 3.50 ± 0.67 | 0.02 ± 0.02 | 9 ± 0.3 |
| R218H | 5.57 ± 0.88 | 0.02 ± 0.01 | 24 ± 0.3 |
| Y419S | 5.88 ± 1.36 | 0.02 ± 0.01 | 1.9 × 10−4 ± 0.05 |
Accessibility of the Gating Residues.
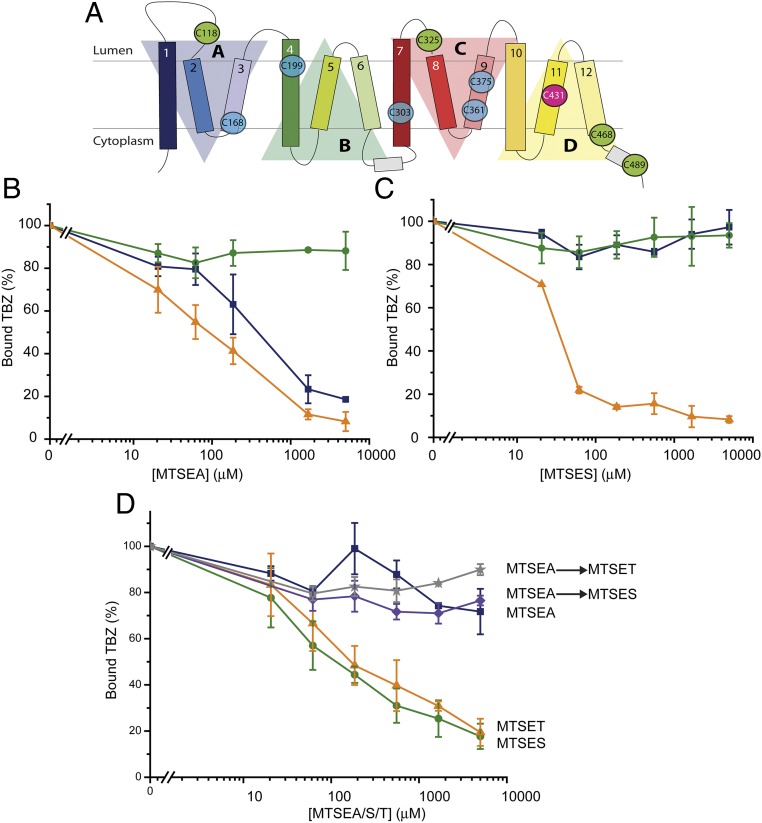
The role of the Cys residues in human VMAT2 has been studied in detail (33). Sulfhydryl reagents such as N-ethylmaleimide (NEM) or the methanethiosulfonate (MTS)-based compounds inhibit TBZ binding and serotonin transport in digitonin-permeabilized cells (33), but only 1 of the 10 Cys residues (the position equivalent to C431 in TM11 in the rat homolog) (Fig. 3A) is responsible for the majority (∼80%) of this effect in human VMAT2. The secondary target (responsible for ∼20% inhibition) is one or more of the four cysteines not within the TM domains. All of the Cys residues are conserved in rVMAT2 at the corresponding positions and, as will be shown below, C431 in TM11 is the primary target also in rVMAT2. Because the fully Cys-less human VMAT2 is known to be inactive (33), we constructed a partially Cys-less mutant in rVMAT2 that can serve as a background for Cys accessibility studies. In this mutant, referred to as Δ5, we replaced all of the non-TM cysteines with Ser (C118, C325, C468, and C489) (Fig. 3A, green circles) whereas C431 was replaced with Ala (Fig. 3A, pink circle). As expected from the results with human VMAT2, TBZ binding to wild-type rVMAT2 (Fig. 3A, solid blue square), but not to Δ5 (Fig. 3A, solid green circle), was inhibited by the membrane-permeant sulfhydryl reagent 2-aminoethyl methanethiosulfonate (MTSEA) (Fig. 3B). Specifically, binding to the wild-type was inhibited by ∼50% at around 0.5 mM MTSEA, and residual activity of about 20% persisted even at 5 mM reagent concentration. Addition of up to 5 mM membrane-impermeant, negatively charged reagent (2-sulfonatoethyl) methanethiosulfonate (MTSES) (Fig. 3C) also did not inhibit TBZ binding to either the wild-type (Fig. 3C, solid blue square) or Δ5 constructs (Fig. 3C, solid green circle). In these digitonin-permeabilized cells, the vesicles are right-side-out (Fig. 1A). Thus, membrane-impermeant reagent/MTSES is only accessible to the cytoplasmic side, whereas the permeant reagent/MTSEA can access the lumen. Because only MTSEA reacts with the wild-type protein, the difference in wild-type reactivity to MTSEA and MTSES suggests that the native Cys431 is accessible from the lumen and/or the membrane, and not from the cytoplasm. Importantly, these data show that Δ5 rVMAT2 provides a good template for studying accessibility and reactivity of Cys replacements.
Fig. 3.
Gating residues are exposed to the cytoplasmic side. (A) Location of the 10 native Cys residues. Five cysteines are irreplaceable (blue circles). C431 (pink circle) is the primary target for inhibition by sulfhydryl reagents. (B and C) Cells were assayed for [3H]TBZ binding ability following incubation with different sulfhydryl reagents: MTSEA (B) and MTSES (C) effect on wild-type rVMAT2 (solid blue square), partial Cys-less (∆5) (solid green circle), and Δ5-Y423C (solid orange triangle). (D) [3H]TBZ binding to Δ5-M222C is inhibited by MTSES (solid green circle) and MTSET (solid orange trianlge), but not by MTSEA (solid blue square). Incubation with MTSEA before the addition of MTSES (purple diamond) and MTSET (gray star) protects against inhibition. Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
We first studied the accessibility of two of the gate positions using Cys replacements in this Δ5 background. A Cys introduced at position 423 (Y423C) was highly accessible to both MTSEA and MTSES (Fig. 3 B and C, solid orange triangle). This finding is most striking for MTSES, which, as mentioned, did not inhibit TBZ binding to wild-type rVMAT2 even at 5 mM (Fig. 3C, solid blue square), whereas 50 μM MTSES inhibited 80% of the TBZ binding activity of Y423C. A similar picture was observed with a Cys replacement at position 222 (M222C). Specifically, TBZ binding activity of this mutant was inhibited by ∼50% using around 100 μM [2-(trimethylammonium)ethyl] methanethiosulfonate (MTSET) or MTSES (Fig. 3D, solid green circle and solid orange triangle). Interestingly, MTSEA does not inhibit TBZ binding to M222C (Fig. 3D, solid blue square) even at 5 mM. However, this effect is not due to a lack of accessibility but rather to a limitation of our assay, which is based on a functional consequence of the reaction, namely on TBZ binding. Indeed, pretreatment with MTSEA (0.5 mM) prevents inhibition by either MTSES or MTSET (Fig. 3D, purple diamond and gray star). Thus, position 222 reacts also with MTSEA but does not prevent TBZ binding, suggesting that the smaller MTSEA adduct (similar to lysine) is compatible with the same interactions formed by the native Met side chain whereas the larger adducts of MTSET and MTSES are not compatible. Taken together, the reactivity to the membrane-impermeant reagents (MTSES and MTSET) indicates that, unlike the native cysteines, Y423C and M222C are accessible to the cytoplasm.
Mutation of the Cytoplasmic Gating Residues Increases the Sensitivity of Native Cysteines to Permeant and Impermeant MTS Reagents.
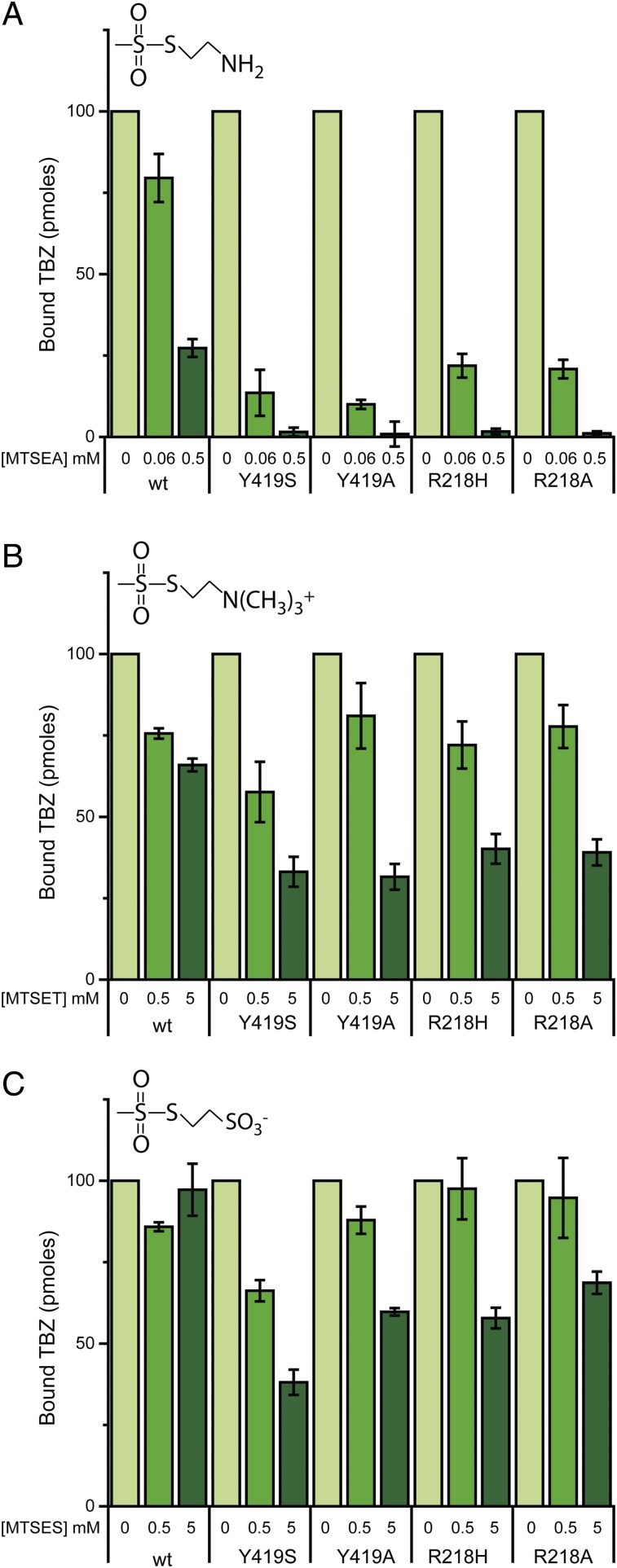
The reserpine binding results described above suggest that mutations in the proposed cytoplasmic gate shift the conformational equilibrium such that the reserpine-binding site becomes accessible. Indeed, when the sensitivity to MTSEA was tested in the background of Ala, Ser, or Phe substitutions in the Y–R and Y–M positions, we detected a dramatic increase in the apparent accessibility/reactivity to MTSEA for the native cysteines. Specifically, whereas TBZ binding to the wild-type was only partially inhibited even at 5 mM MTSEA, most of the mutants displayed a much higher sensitivity (Fig. 4A and Fig. S4). Notably, the increase in accessibility correlates well with the ability of the mutants to transport serotonin. That is, whereas mutants that lacked any transport ability (Y419A/S, R218A/H) (Fig. 2C) displayed drastic increases in apparent reactivity (Fig. 4A, Fig. S4 B and D, and Table S1), mutants that retained transport activity (Y423A/S/F, Fig. 1D; M222L/S/F/C, Fig. 2C) exhibited only small increases in sensitivity or none at all (Y419F) (Fig. S4B). Remarkably, mutants of Y419 and R218 displayed an increased sensitivity also to the impermeant MTS reagents (MTSET and MTSES). Specifically, whereas 5 mM MTSET inhibited only ∼30% activity of the wild-type, binding in mutants Y419A/S and R218A/H was inhibited ∼60 to 70% (Fig. 4B). Similarly, 5 mM MTSES had no effect on TBZ binding to the wild-type but had a significant (40 to 60%) effect on the activity of the Y419A/S and R218A/H mutants (Fig. 4C).
Fig. 4.
Mutating residues in the cytoplasmic gate increases the sensitivity to permeant and impermeant sulfhydryl reagents. Cells were assayed for [3H]TBZ binding ability following incubation with the indicated concentrations of (A) MTSEA, (B) MTSET, and (C) MTSES. Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
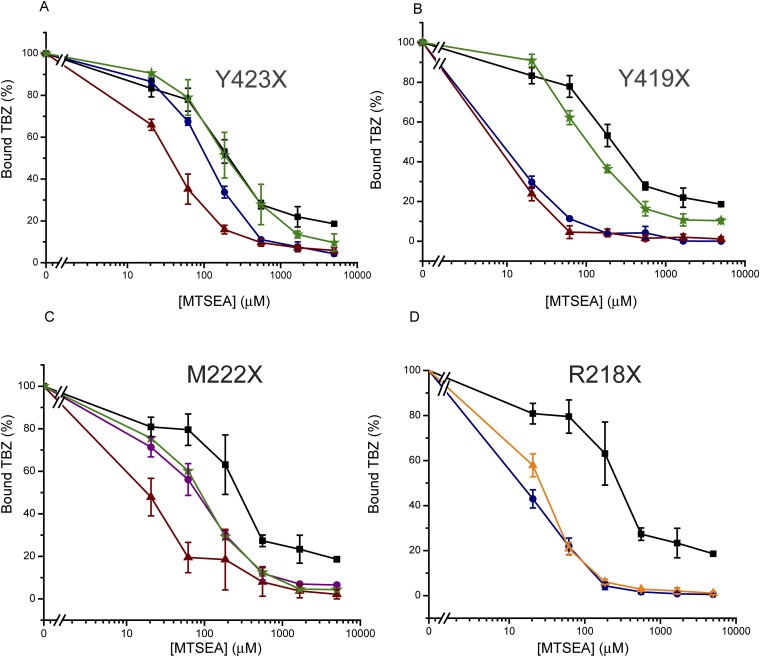
Fig. S4.
Mutating the gating residues changes the sensitivity to MTSEA. Cells were assayed for [3H]TBZ binding ability following incubation with increasing concentrations of MTSEA. (A) Mutations at position 423: wild-type (solid black square), Y423A (solid blue circle), Y423S (solid red triangle), and Y423F (green star). (B) Mutations at position 419: wild-type (solid black square), Y419A (solid blue circle), Y419S (solid red triangle), and Y419F (green star). (C) Mutations at position 222: wild-type (solid black square), M222S (solid red triangle), M222L (solid purple circle), and M222F (green star). (D) Mutations at position 218: wild-type (solid black square), R218A (solid blue circle ), and R218H (solid orange triangle ). Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
These results support the notion that the mutations that inhibit TBZ binding and serotonin transport (Y419A/S, R218A/H) do so by shifting the conformational equilibrium of the transporter to the Ccyt conformation, whereas the destabilization of the lumen-facing state by mutations at Y423 and M222 has a more moderate effect.
Protonation of D33 Shifts the Conformational Equilibrium.
The results presented above reveal that mutating the cytoplasmic gating residues uncouples the dependency of reserpine binding from the proton gradient. This finding supports our previous hypothesis that, in parallel with its energetic role, the proton gradient favors a reserpine-accessible conformation (24). Thus, we tested the possibility that protonation of one of the essential membrane-embedded carboxyl residues in rVMAT2 allows the conformational change necessary for reserpine binding. D33 (TM1) and E313 (TM7) are both highly conserved within the DHA12 subfamily (34), and have both been found to be essential for transport activity (29, 35). Moreover, we recently demonstrated the importance of the equivalent residues in a bacterial homolog of rVMAT2 (34), and highlighted them both as putative proton-binding sites.
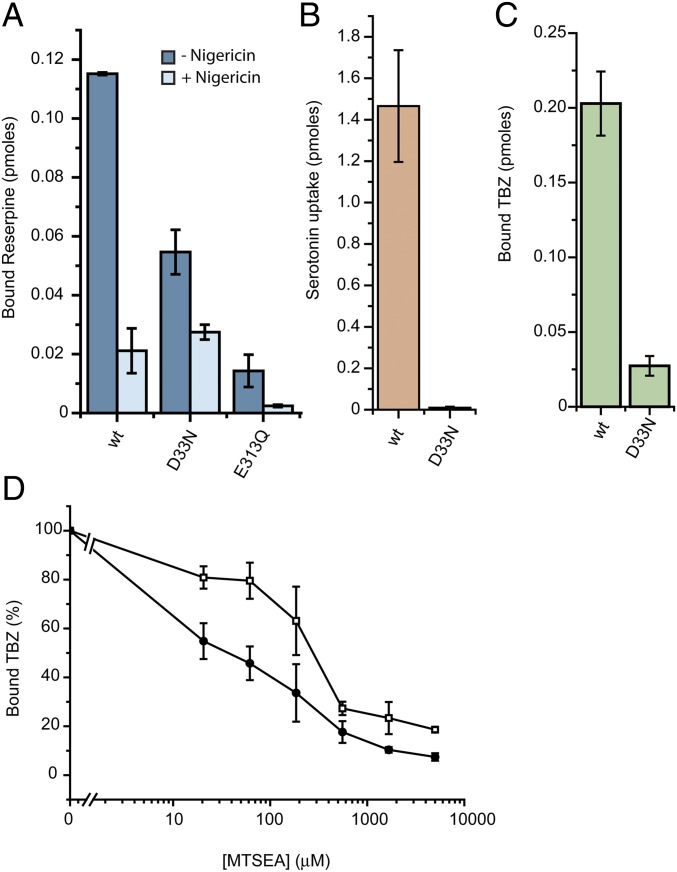
Hence, we mutated E313 and D33 to neutral residues (Q and N, respectively) and tested the mutants for reserpine binding. E313Q showed marginal activity that was insufficient for further characterization (Fig. 5A). On the other hand, D33N, although it bound only marginal levels of TBZ and showed no detectable levels of serotonin transport (Fig. 5 B and C), displayed robust levels of reserpine binding to levels about 50% of the wild-type (Fig. 5A). Reserpine binding to D33N was not fully independent of the proton gradient, as shown by the finding that nigericin inhibits ∼50%, but it was less sensitive than binding to the wild-type, which was inhibited ∼90% (Fig. 5A). Furthermore, the D33N mutation led to a 10-fold higher sensitivity to inhibition by MTSEA (IC50 ∼30 μM compared with ∼300 μM) (Fig. 5D), suggesting a conformational shift in D33N compared with wild-type rVMAT2. Taken as a whole, these results suggest that D33 contributes to the proton transport mediating the conformational change that generates a high-affinity binding site for reserpine.
Fig. 5.
Mutating D33 affects the conformation of rVMAT2. (A–C) Cells expressing rVMAT2-D33N or -E313Q were assayed for [3H]reserpine binding (A), [3H]serotonin transport (B), and [3H]TBZ binding (C) as described in Fig. 1. (D) [3H]TBZ binding to cells expressing the wild-type (□) or rVMAT2-D33N (●) following incubation with increasing concentrations of MTSEA. Results presented are averages of at least two repeats, each point in duplicate; error bars indicate SE.
Discussion
According to the alternating access mechanism, a transporter must be able to occupy at least three distinctive conformations: inward-facing, outward-facing, and occluded (9). The transition between the different conformations is accompanied by the opening and closing of molecular gates, resulting in a transporter that does not open simultaneously to both sides (36). In this work, our goal was to study the conformational states during the transport cycle of rVMAT2. We used two powerful pharmacological tools: TBZ, a noncompetitive inhibitor that prevents binding of substrates, and reserpine, a competitive inhibitor. TBZ binds to the transporter under a wide variety of conditions, including to the detergent-solubilized protein. On the other hand, to induce a reserpine-accessible state, a conformational change is needed that in turn requires translocation of at least one proton (24).
Using these tools, and guided by a reliable molecular model of rVMAT2, we identified four residues from TM5 and TM11 as belonging to the cytoplasmic gate, and therefore key in defining the conformational equilibrium of rVMAT2. Specifically, our structural model predicts a Y–M pair (Y423 and M222) and a Y–R pair (Y419 and R218), of which the latter is closest to the cytoplasm (Fig. 2A and Fig. S2B). The involvement of residues from TM5 and TM11 in the cytoplasmic gate of other MFS transporters (32, 37–40) is consistent with this proposed role.
Mutation of residues in these four positions (Y–M and Y–R) enhanced the ability of reserpine to inhibit binding of TBZ. In the case of Y423 substitutions, this effect was up to 100-fold (Fig. 1 B and D). Moreover, mutation at any of the four positions facilitated reserpine binding in the absence of the proton electrochemical gradient (Figs. 1E and 2B), from which we deduced that the mutants adopt a conformation similar to the one generated by the proton gradient. Together, these results suggest a link between the protonation event and the major conformational change of the transporter. By mutating one of the residues comprising the cytoplasmic gate, we apparently emulate this conformational change and artificially create the reserpine-accessible conformation.
Despite the consistency of the effects on reserpine binding at all four positions in the proposed gate, we noted some interesting differences depending on the particular substitution involved. Some of the mutations (Y419A/S, R218H) shift the transporter to the reserpine-binding conformation but render it incapable of completing the transport cycle or switching to a TBZ-binding conformation (Fig. 2 B–D). A significant increase in the sensitivity to impermeant MTS reagents (Fig. 4 B and C), suggesting a conformational shift, supports the notion that the mutants favor the cytoplasm-facing state. The Ala substitution R218A is not compatible with serotonin transport but still allows TBZ binding (Fig. 2 B–D). Y419F behaves essentially as the wild-type, consistent with a requirement for aromatic interactions at this position. Notably, Y419S consistently binds twice the amount of reserpine as the wild-type (in the presence of a proton gradient), suggesting that even in the presence of a proton gradient the equilibrium of the wild-type is not completely shifted to the cytoplasm-facing configuration.
In the predicted “inner” Y–M positions, replacements at position 423 (Fig. 1) and, to a lesser extent, at 222 (Fig. 2) clearly affect reserpine binding in the absence of a gradient but have a smaller effect on the ability to bind TBZ or to transport serotonin.
We suggest that the mutations in all four positions, by modifying the interactions in the cytoplasmic gate, effectively destabilize the lumen-facing state and shift the equilibrium so as to increase the proportion of cytoplasm-facing transporters.
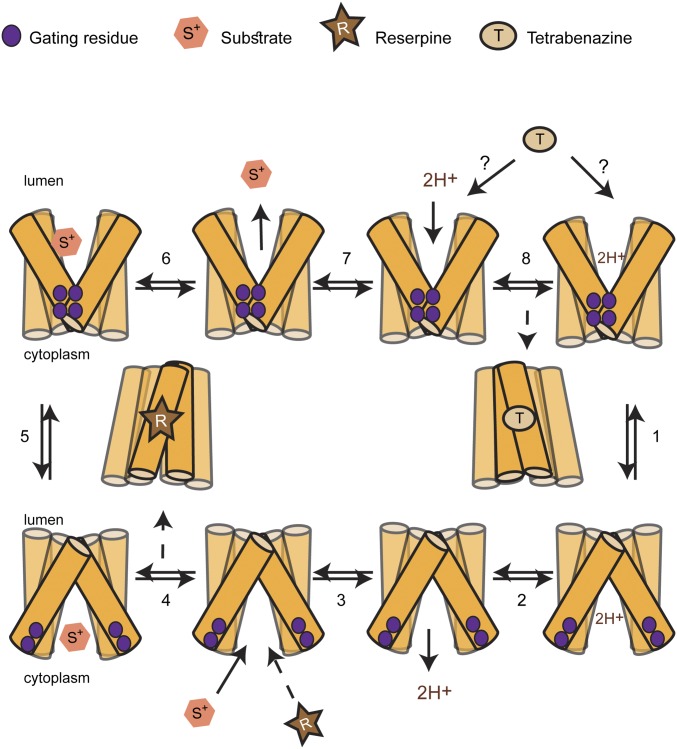
It is notable that the mutants that appear to favor the cytoplasm-facing conformation (mainly Y419S and R218H) are still capable of binding reserpine but not TBZ (Fig. 2). We therefore hypothesize that whereas reserpine binds to the cytoplasm-facing conformation, TBZ binds to a cytoplasm-closed conformation (Fig. 6). Such conformational selectivity is extremely useful, as it allows specific states in the transport cycle to be isolated biochemically. Such a selective inhibition is also a very rare property, and has been described, to our knowledge, for only three other transporters. One such example is the plasma membrane serotonin transporter SERT, where the noncompetitive inhibitor ibogaine stabilizes a cytoplasm-facing conformation, whereas other competitive inhibitors, such as cocaine, bind at the substrate-binding site in the cytoplasm-closed state (41, 42). The other two examples are the ADP:ATP translocator protein (43) and the human erythrocyte glucose transporter (44). Based on our data, therefore, we suggest using reserpine and TBZ binding as “markers” of the rVMAT2 conformational states.
Fig. 6.
Schematic of the proposed transport cycle. For simplicity, only six TMs are shown. In the absence of a proton gradient, the dominant population is of the lumen-facing conformation, as indicated by the transporter’s ability to bind TBZ but not reserpine. Binding of protons enables the conformational switch to the cytoplasm-facing conformation (step 1), whereas binding of substrate enables the change to the lumen-facing conformation (step 5). Binding of TBZ locks the transporter in a conformation that appears incompatible with substrate binding, and is therefore presumably not cytoplasm-facing. Binding of reserpine also locks the transporter in a dead-end conformation, but reserpine binding competes with substrate binding, and therefore the reserpine-bound conformation is presumably cytoplasm-facing. The dashed line for R indicates competition with substrate.
To ensure coupling between H+ and substrate transport, proton binding and release must facilitate unique conformational changes that in turn allow substrate binding or release (Fig. 6). Protonation has been shown to induce conformational changes in other proton-coupled MFS transporters. For example, in FucP, protonation of a key carboxyl was suggested to facilitate the transition from extracellular- to cytoplasm-facing (45, 46); in EmrD, decreasing the pH led to opening of the cytoplasmic cleft (47); in LacY, deprotonation enabled a conformational change of the empty transporter (10, 48, 49); and a recent study suggested a protonation event as a molecular switch between conformations in LmrP (50). Given that mutating D33 to Asn resulted in a very similar behavior to that of the gating mutants (Fig. 5), namely no serotonin transport and TBZ binding yet normal reserpine binding even in the absence of ΔµH+, we speculate that protonation of D33 may contribute to the opening of the cytoplasmic gate during the transition between lumen-facing and cytoplasm-facing states.
Together, our results provide important mechanistic insights into the transport cycle of rVMAT2 (Fig. 6). Specifically, we propose that access of reserpine to the binding site is from the cytoplasmic side, because the mutations that perturb the interaction of the cytoplasmic gate appear to increase accessibility from the cytoplasm (Fig. 6). In contrast, because the noncompetitive inhibitor TBZ does not require a proton gradient for binding, we therefore propose that TBZ binds to a site accessible from the lumen side (Fig. 6). This suggestion is supported also by the finding that mutants that bind reserpine in the absence of a proton gradient also do not bind TBZ (Fig. 2), suggesting a complementary behavior. Once bound, TBZ presumably then prevents binding of substrates by inhibiting the opening of the cytoplasmic gate. Whether gate opening is prevented by inhibiting H+ translocation or by TBZ interacting directly with conformationally important residues remains to be determined. Our finding that two potent inhibitors of rVMAT2 can be used to assess the conformation of VMAT2 should provide an important tool to aid in such studies in the future.
Materials and Methods
Plasmids.
The rat VMAT2 gene (SLC18A2), with a hemagglutinin (HA) tag in the second loop between positions 96 and 105 as well as 10 histidine residues at the C terminus (51), was cloned into the pcDNA3.1 plasmid (Invitrogen) as described previously (29). Site-directed mutagenesis was done with the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). The sequences of all constructs were verified by DNA sequencing.
Cell Culture and Transfections.
HEK293 cells were grown at 37 °C under a 5% CO2 atmosphere in DMEM with 10% (vol/vol) FBS (Biological Industries) and 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 0.25 µg/mL amphotericin B.
For rVMAT2 expression, cells grown in 10-cm plates to 40–50% confluence were transfected with 7 µg of plasmid DNA coding for wild-type or mutant rVMAT2 using PEI transfection reagent (Sigma). After 40–48 h, cells were treated with trypsin and collected, centrifuged for 3 min at 800 × g at 4 °C, and washed twice with PBS. The cell pellet was frozen in liquid air and stored at −70 °C.
Reserpine Binding Assays.
Cells from one 10-cm plate were thawed at 37 °C and resuspended with 5 mL wash buffer (140 mM KCl, 5 mM glucose, 5 mM MgCl2, 20 mM Hepes) supplemented with 5 mM ATP and permeabilized by the addition of 10 mM digitonin for 5 min. The binding reaction was started by the addition of 5 nM [3H]reserpine (ViTrax Radiochemicals; 18 Ci/mmol), and cells were then incubated for 15 min at 37 °C. Reactions were stopped by quickly diluting the cells with ice-cold binding buffer and collected by quick centrifugation (12,700 × g, 1 min). The cells were then solubilized in wash buffer supplemented with 15 μg/mL DNaseI (Sigma), protease inhibitor mixture (Sigma), and 1 mM PMSF using 2% (wt/vol) dodecyl-maltoside (DDM; Glycon). After 1 h of shaking at 4 °C, cells were centrifuged for 10 min at 21,000 × g and incubated with Ni-NTA beads (Qiagen) for 1 h at 4 °C. Beads were then spun down (1,300 × g, 1 min) and washed twice with wash buffer containing 0.08% DDM. After 20 min in 250 μL elution buffer with 450 mM imidazole at room temperature, beads were spun down and 200-μL aliquots were sampled. Radioactivity was measured using liquid scintillation.
TBZ Binding and Serotonin Transport.
For [3H]TBZ binding and [3H]serotonin transport assays, we used MultiScreen FB plates (Millipore). Cells from one 10-cm plate were thawed at 37 °C and resuspended with 5-mL wash buffer, and 150 μL was added to each well. The cells were then incubated with 200-μL wash buffer supplemented with 10 mM digitonin (and 5 mM ATP in transport experiments) for 10 min at 37 °C. The digitonin-containing buffer was then removed, and the reactions were started by the addition of wash buffer with 5 nM [3H]TBZ (ViTrax Radiochemicals; 77 Ci/mmol) or 100 nM [3H]serotonin (PerkinElmer Life Sciences; 27.8 Ci/mmol) and 5 mM ATP for 20 min at 37 °C. Reactions were terminated by washing the cells twice with 200 μL of ice-cold wash buffer. The filters were incubated for at least 2 h with Opti-Fluor scintillation fluid (PerkinElmer Life Sciences), and radioactivity was measured using liquid scintillation.
For accessibility assays, after permeabilization with digitonin, cells were incubated with wash buffer and the indicated concentrations of MTS reagents for 10 min at 37 °C. The buffer was then replaced with TBZ binding buffer. For TBZ inhibition, cells were incubated with the indicated concentrations of reserpine for 10 min at 37 °C before the addition of [3H]TBZ.
Homology Modeling of rVMAT2 in a Lumen-Facing State.
To build a molecular model of rVMAT2, the crystal structure of a putative H+-driven Escherichia coli transporter, YajR (PDB ID code 3WDO) (31), in an outward-facing conformation, was used as a template. In preparation for modeling, a preliminary pairwise sequence alignment of YajR and rVMAT2 was constructed using AlignMe server v1.1 (52) in PST mode (53). To prevent misalignments, the long TM1–TM2 loop of rVMAT2 (residues 55–125) was removed from the input for the initial alignment and therefore not modeled. Adjustments to the AlignMe alignment were then made in the following residue ranges of the rVMAT2 sequence: 52–54, 126–128, 198–199, 210–212, 277–285, 325–353, 357–380, and 400–415. These adjustments were selected to optimize the agreement with PSIPRED v3.2 (54) secondary structure and TOPCONS (55) transmembrane predictions, and to reduce the number of residues in generously allowed and disallowed regions of the Ramachandran plot in the corresponding models, as evaluated by PROCHECK v3.5.4 (56). In addition, conservation patterns were obtained for the template and target sequences using the ConSurf server (57); the conservation profiles were mapped onto each structure to aid the alignment of conserved and variable residues. After each adjustment to the alignment described above, the effect on the per-residue and global ProQM (58) score was tracked on the corresponding model. In the final alignment, the coverage was excellent (Fig. S2), ∼18% of the residues were identical, and the similarity reached ∼45%, suggesting an expected accuracy of 1–2 Å in the backbone (59, 60).
MODELLER v9.13 (61) was used to build the homology models of rVMAT2 with α-helical constraints for residues 195–205 and 343–346 and distance restraints between the atoms (residue1:atom and residue2:atom) D33:O∂1 and S197:Oγ, K139:N and Q143:Oε1, Q143:Nε2 and D427:O∂1, N147:N∂2 and D427:O∂2, and Y342:OH and D400:O∂2, to preserve polar interactions as described previously (29). All distance restraints were defined as Gaussians with a minimum at 3.5 Å and an SD of 0.1 Å. Three hundred iterations of MODELLER were carried out, and the representative rVMAT2 model was selected as that with the lowest Molpdf energy value of MODELLER (i.e., that which best fits all of the input constraints) as well as the highest PROCHECK and global ProQM scores. The final representative model was of excellent quality according to PROCHECK, with 99.4% of residues in the favored and additionally allowed regions of the Ramachandran plot. The global ProQM score was 0.717, which is a significant improvement over the score of 0.616 obtained for an earlier model built using LacY, the closest available template at the time (29). For reference, the ProQM scores of the templates, YajR and LacY, are 0.741 and 0.729, respectively. The final rVMAT2 model is available from the Protein Model DataBase (https://bioinformatics.cineca.it/PMDB/) with accession number PM0080553. The side-chain orientations of R218, M222, Y419, and Y423 in the selected model were confirmed to be characteristic of all 300 models based on analysis of the average atomic densities (Fig. S2).
Acknowledgments
We thank Dina Listov, Yifat Levine, and Shai Fainsod for help during the course of this work. S.S. is Mathilda Marks-Kennedy Professor of Biochemistry at the Hebrew University of Jerusalem. This research was supported by National Institutes of Health Grant NS16708 (to S.S.); Israel-USA Binational Science Foundation Grant 2013198 (to S.S.); and the Division of Intramural Research of the National Institute of Neurological Disorders and Stroke, NIH (L.R.F. and A.V.-J.). A.V.-J. is a recipient of the L’Oreal Chile-UNESCO Women in Science Fellowship and the L’Oreal-UNESCO Rising Talent Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The rVMAT2 model reported in this paper has been deposited in the Protein Model DataBase, https://bioinformatics.cineca.it/PMDB/ (accession no. PM0080553).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605162113/-/DCSupplemental.
References
- 1.Schuldiner S, Shirvan A, Linial M. Vesicular neurotransmitter transporters: From bacteria to humans. Physiol Rev. 1995;75(2):369–392. doi: 10.1152/physrev.1995.75.2.369. [DOI] [PubMed] [Google Scholar]
- 2.Eiden LE. The vesicular neurotransmitter transporters: Current perspectives and future prospects. FASEB J. 2000;14(15):2396–2400. doi: 10.1096/fj.00-0817rev. [DOI] [PubMed] [Google Scholar]
- 3.Blakely RD, Edwards RH. Vesicular and plasma membrane transporters for neurotransmitters. Cold Spring Harb Perspect Biol. 2012;4(2):a005595. doi: 10.1101/cshperspect.a005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons SM. Transport mechanisms in acetylcholine and monoamine storage. FASEB J. 2000;14(15):2423–2434. doi: 10.1096/fj.00-0203rev. [DOI] [PubMed] [Google Scholar]
- 5.Erickson JD, Varoqui H. Molecular analysis of vesicular amine transporter function and targeting to secretory organelles. FASEB J. 2000;14(15):2450–2458. doi: 10.1096/fj.00-0206rev. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhry FA, Edwards RH, Fonnum F. Vesicular neurotransmitter transporters as targets for endogenous and exogenous toxic substances. Annu Rev Pharmacol Toxicol. 2008;48:277–301. doi: 10.1146/annurev.pharmtox.46.120604.141146. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, et al. A cDNA that suppresses MPP+ toxicity encodes a vesicular amine transporter. Cell. 1992;70(4):539–551. doi: 10.1016/0092-8674(92)90425-c. [DOI] [PubMed] [Google Scholar]
- 8.Yan N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem Sci. 2013;38(3):151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 10.Kaback HR. A chemiosmotic mechanism of symport. Proc Natl Acad Sci USA. 2015;112(5):1259–1264. doi: 10.1073/pnas.1419325112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50(45):9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuldiner S. Competition as a way of life for H(+)-coupled antiporters. J Mol Biol. 2014;426(14):2539–2546. doi: 10.1016/j.jmb.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yelin R, Schuldiner S. The pharmacological profile of the vesicular monoamine transporter resembles that of multidrug transporters. FEBS Lett. 1995;377(2):201–207. doi: 10.1016/0014-5793(95)01346-6. [DOI] [PubMed] [Google Scholar]
- 14.Schuldiner S, Steiner-Mordoch S, Yelin R, Wall SC, Rudnick G. Amphetamine derivatives interact with both plasma membrane and secretory vesicle biogenic amine transporters. Mol Pharmacol. 1993;44(6):1227–1231. [PubMed] [Google Scholar]
- 15.Gros Y, Schuldiner S. Directed evolution reveals hidden properties of VMAT, a neurotransmitter transporter. J Biol Chem. 2010;285(7):5076–5084. doi: 10.1074/jbc.M109.081216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser HS. Reserpine: A tragic victim of myths, marketing, and fashionable prescribing. Clin Pharmacol Ther. 1996;60(4):368–373. doi: 10.1016/S0009-9236(96)90193-9. [DOI] [PubMed] [Google Scholar]
- 17.Stitzel RE. The biological fate of reserpine. Pharmacol Rev. 1976;28(3):179–208. [PubMed] [Google Scholar]
- 18.Kenney C, Jankovic J. Tetrabenazine in the treatment of hyperkinetic movement disorders. Expert Rev Neurother. 2006;6(1):7–17. doi: 10.1586/14737175.6.1.7. [DOI] [PubMed] [Google Scholar]
- 19.Kanner BI, Fishkes H, Maron R, Sharon I, Schuldiner S. Reserpine as a competitive and reversible inhibitor of the catecholamine transporter of bovine chromaffin granules. FEBS Lett. 1979;100(1):175–178. doi: 10.1016/0014-5793(79)81158-8. [DOI] [PubMed] [Google Scholar]
- 20.Jonasson J, Rosengren E, Waldeck B. Effects of some pharmacologically active amines on the uptake of arylalkylamines by adrenal medullary granules. Acta Physiol Scand. 1964;60(1–2):136–140. doi: 10.1111/j.1748-1716.1964.tb02876.x. [DOI] [PubMed] [Google Scholar]
- 21.Deupree JD, Weaver JA. Identification and characterization of the catecholamine transporter in bovine chromaffin granules using [3H]reserpine. J Biol Chem. 1984;259(17):10907–10912. [PubMed] [Google Scholar]
- 22.Scherman D, Henry JP. Reserpine binding to bovine chromaffin granule membranes. Characterization and comparison with dihydrotetrabenazine binding. Mol Pharmacol. 1984;25(1):113–122. [PubMed] [Google Scholar]
- 23.Stern-Bach Y, Greenberg-Ofrath N, Flechner I, Schuldiner S. Identification and purification of a functional amine transporter from bovine chromaffin granules. J Biol Chem. 1990;265(7):3961–3966. [PubMed] [Google Scholar]
- 24.Rudnick G, Steiner-Mordoch SS, Fishkes H, Stern-Bach Y, Schuldiner S. Energetics of reserpine binding and occlusion by the chromaffin granule biogenic amine transporter. Biochemistry. 1990;29(3):603–608. doi: 10.1021/bi00455a002. [DOI] [PubMed] [Google Scholar]
- 25.Carlsson A. Drugs which block the storage of 5-hydroxytryptamine and related amines. In: Erspamer V, editor. Handbook of Experimental Pharmacology. Vol 19. Springer; Berlin: 1965. pp. 529–592. [Google Scholar]
- 26.Peter D, Jimenez J, Liu Y, Kim J, Edwards RH. The chromaffin granule and synaptic vesicle amine transporters differ in substrate recognition and sensitivity to inhibitors. J Biol Chem. 1994;269(10):7231–7237. [PubMed] [Google Scholar]
- 27.Scherman D, Jaudon P, Henry JP. Characterization of the monoamine carrier of chromaffin granule membrane by binding of [2-3H]dihydrotetrabenazine. Proc Natl Acad Sci USA. 1983;80(2):584–588. doi: 10.1073/pnas.80.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugolev Y, Segal T, Yaffe D, Gros Y, Schuldiner S. Identification of conformationally sensitive residues essential for inhibition of vesicular monoamine transport by the noncompetitive inhibitor tetrabenazine. J Biol Chem. 2013;288(45):32160–32171. doi: 10.1074/jbc.M113.502971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaffe D, Radestock S, Shuster Y, Forrest LR, Schuldiner S. Identification of molecular hinge points mediating alternating access in the vesicular monoamine transporter VMAT2. Proc Natl Acad Sci USA. 2013;110(15):E1332–E1341. doi: 10.1073/pnas.1220497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuldiner S, Liu Y, Edwards RH. Reserpine binding to a vesicular amine transporter expressed in Chinese hamster ovary fibroblasts. J Biol Chem. 1993;268(1):29–34. [PubMed] [Google Scholar]
- 31.Jiang D, et al. Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc Natl Acad Sci USA. 2013;110(36):14664–14669. doi: 10.1073/pnas.1308127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fowler PW, et al. Gating topology of the proton-coupled oligopeptide symporters. Structure. 2015;23(2):290–301. doi: 10.1016/j.str.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiriot DS, Ruoho AE. Mutagenesis and derivatization of human vesicle monoamine transporter 2 (VMAT2) cysteines identifies transporter domains involved in tetrabenazine binding and substrate transport. J Biol Chem. 2001;276(29):27304–27315. doi: 10.1074/jbc.M103947200. [DOI] [PubMed] [Google Scholar]
- 34.Yaffe D, et al. Functionally important carboxyls in a bacterial homologue of the vesicular monoamine transporter (VMAT) J Biol Chem. 2014;289(49):34229–34240. doi: 10.1074/jbc.M114.607366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merickel A, Rosandich P, Peter D, Edwards RH. Identification of residues involved in substrate recognition by a vesicular monoamine transporter. J Biol Chem. 1995;270(43):25798–25804. doi: 10.1074/jbc.270.43.25798. [DOI] [PubMed] [Google Scholar]
- 36.Forrest LR, Krämer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807(2):167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Solcan N, et al. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 2012;31(16):3411–3421. doi: 10.1038/emboj.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyalomhe O, Herrick DZ, Cafiso DS, Maloney PC. Closure of the cytoplasmic gate formed by TM5 and TM11 during transport in the oxalate/formate exchanger from Oxalobacter formigenes. Biochemistry. 2014;53(49):7735–7744. doi: 10.1021/bi5012173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moradi M, Enkavi G, Tajkhorshid E. Atomic-level characterization of transport cycle thermodynamics in the glycerol-3-phosphate:phosphate antiporter. Nat Commun. 2015;6:8393. doi: 10.1038/ncomms9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smirnova I, et al. Transient conformers of LacY are trapped by nanobodies. Proc Natl Acad Sci USA. 2015;112(45):13839–13844. doi: 10.1073/pnas.1519485112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulling S, et al. The mechanistic basis for noncompetitive ibogaine inhibition of serotonin and dopamine transporters. J Biol Chem. 2012;287(22):18524–18534. doi: 10.1074/jbc.M112.343681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobs MT, Zhang YW, Campbell SD, Rudnick G. Ibogaine, a noncompetitive inhibitor of serotonin transport, acts by stabilizing the cytoplasm-facing state of the transporter. J Biol Chem. 2007;282(40):29441–29447. doi: 10.1074/jbc.M704456200. [DOI] [PubMed] [Google Scholar]
- 43.Buchanan BB, Eiermann W, Riccio P, Aquila H, Klingenberg M. Antibody evidence for different conformational states of ADP, ATP translocator protein isolated from mitochondria. Proc Natl Acad Sci USA. 1976;73(7):2280–2284. doi: 10.1073/pnas.73.7.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorga FR, Lienhard GE. Equilibria and kinetics of ligand binding to the human erythrocyte glucose transporter. Evidence for an alternating conformation model for transport. Biochemistry. 1981;20(18):5108–5113. doi: 10.1021/bi00521a003. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Ke M, Gong H. Protonation of Glu(135) facilitates the outward-to-inward structural transition of fucose transporter. Biophys J. 2015;109(3):542–551. doi: 10.1016/j.bpj.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467(7316):734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 47.Steed PR, Zou P, Trone KE, Mchaourab HS. Structure and pH-induced structural rearrangements of the putative multidrug efflux pump EmrD in liposomes probed by site-directed spin labeling. Biochemistry. 2013;52(45):7964–7974. doi: 10.1021/bi4012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holyoake J, Sansom MS. Conformational change in an MFS protein: MD simulations of LacY. Structure. 2007;15(7):873–884. doi: 10.1016/j.str.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Andersson M, et al. Proton-coupled dynamics in lactose permease. Structure. 2012;20(11):1893–1904. doi: 10.1016/j.str.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masureel M, et al. Protonation drives the conformational switch in the multidrug transporter LmrP. Nat Chem Biol. 2014;10(2):149–155. doi: 10.1038/nchembio.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elbaz Y, Danieli T, Kanner BI, Schuldiner S. Expression of neurotransmitter transporters for structural and biochemical studies. Protein Expr Purif. 2010;73(2):152–160. doi: 10.1016/j.pep.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamm M, Staritzbichler R, Khafizov K, Forrest LR. AlignMe—A membrane protein sequence alignment web server. Nucleic Acids Res. 2014;42(Web Server issue):W246–W251. doi: 10.1093/nar/gku291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamm M, Staritzbichler R, Khafizov K, Forrest LR. Alignment of helical membrane protein sequences using AlignMe. PLoS One. 2013;8(3):e57731. doi: 10.1371/journal.pone.0057731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 55.Bernsel A, Viklund H, Hennerdal A, Elofsson A. TOPCONS: Consensus prediction of membrane protein topology. Nucleic Acids Res. 2009;37(Web Server issue):W465–W468. doi: 10.1093/nar/gkp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 57.Glaser F, et al. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 58.Ray A, Lindahl E, Wallner B. Model quality assessment for membrane proteins. Bioinformatics. 2010;26(24):3067–3074. doi: 10.1093/bioinformatics/btq581. [DOI] [PubMed] [Google Scholar]
- 59.Olivella M, Gonzalez A, Pardo L, Deupi X. Relation between sequence and structure in membrane proteins. Bioinformatics. 2013;29(13):1589–1592. doi: 10.1093/bioinformatics/btt249. [DOI] [PubMed] [Google Scholar]
- 60.Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91(2):508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 62.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]