Significance
Individuals with more education tend to live longer. Genetic variants have been discovered that predict educational attainment. We tested whether a “polygenic score” based on these genetic variants could make predictions about people’s lifespan. We used data from three cohort studies (including >130,000 participants) to examine the link between offspring polygenic score for education and parental longevity. Across the studies, we found that participants with more education-linked genetic variants had longer-living parents; compared with those with the lowest genetic education scores, those with the highest scores had parents who lived on average 6 months longer. This finding suggests the hypothesis that part of the ultimate explanation for the extended longevity of better-educated people is an underlying, quantifiable, genetic propensity.
Keywords: genetics, education, longevity, prediction, polygenic score
Abstract
Educational attainment is associated with many health outcomes, including longevity. It is also known to be substantially heritable. Here, we used data from three large genetic epidemiology cohort studies (Generation Scotland, n = ∼17,000; UK Biobank, n = ∼115,000; and the Estonian Biobank, n = ∼6,000) to test whether education-linked genetic variants can predict lifespan length. We did so by using cohort members’ polygenic profile score for education to predict their parents’ longevity. Across the three cohorts, meta-analysis showed that a 1 SD higher polygenic education score was associated with ∼2.7% lower mortality risk for both mothers (total ndeaths = 79,702) and ∼2.4% lower risk for fathers (total ndeaths = 97,630). On average, the parents of offspring in the upper third of the polygenic score distribution lived 0.55 y longer compared with those of offspring in the lower third. Overall, these results indicate that the genetic contributions to educational attainment are useful in the prediction of human longevity.
Educational attainment is a mainstay in debates concerning social inequality and life outcomes (1, 2). Individual differences in educational attainment have been linked to variation in life chances and longevity: those with more education tend to be healthier (3), richer in adulthood (4), more upwardly socially mobile (2), and longer-lived (5, 6). Because education influences—and is influenced by—various personal characteristics and social factors (7, 8), it has been difficult to disentangle the precise reasons for its prediction of key life outcomes (9). Despite it being widely used in studies as a social-environmental variable, differences in education are under substantial genetic influence, with heritability frequently estimated at 60% and above in family studies (10–12), and 20–30% in molecular genetic studies (13, 14). Some specific education-associated genetic variants have also been uncovered in genome-wide association studies (GWAS) (15–17). The present study uses previously-discovered genetic correlates of education to predict variation in arguably the most important life outcome of all: longevity.
The association of educational outcomes—measured either by attained qualifications or by duration of full-time education—with longevity is well established in the scientific literature (e.g., refs. 18 and 19). The high value placed upon educational qualifications in society and in the labor market forms one possible explanation for this link: the higher-level occupations and socioeconomic positions afforded by better education allow greater access to health-improving resources and surroundings. However, education also acts as a signal for personal characteristics with which it is phenotypically correlated, such as general cognitive ability (20, 21), motivation (22), and health (23), in addition to aspects of a person’s socio-economic background (24). Thus, according to two nonmutually exclusive views, educational attainment might cause improvements in longevity via social mechanisms, or might itself be caused by preexisting—partly heritable—factors that also increase longevity.
Some evidence for the latter view—that some of the variance in educational attainment and longevity is caused by preexisting factors—comes from the pervasive genetic correlations of education with many other longevity-linked traits, indicating that these traits are substantially associated with the same genetic variants. For example, one study used linkage-disequilibrium (LD) regression analysis to show that educational attainment was significantly genetically correlated with lifespan-limiting conditions like cardiovascular disease and stroke (25). In addition, educational attainment is strongly genetically correlated with general cognitive ability (13, 14, 26), itself a well-replicated phenotypic (27) and genetic (28) correlate of longevity.
The Current Study
In this study, we tested whether the genetic variants associated with educational attainment are associated with longevity. We thus assessed the extent to which the genetic contributions to educational outcomes, which are preexisting and nonsocial, are related to a key health outcome. To do so, we used the established technique of testing for associations between genotyped subjects and their phenotyped relatives (in this case, the lifespan of parents) (29).
Offspring genetic variants, such as the Alzheimer’s-linked APOE e4 allele, have also been linked to parental longevity in candidate gene studies (30) and more recently in a GWAS (31; see ref. 32 for a similar analysis of epigenetic markers). Moreover, higher genetic risk for conditions, such as cardiovascular disease, diabetes, and Alzheimer’s disease, has been related to earlier parental mortality (33). Because the expected allelic effect of one allele in parents is 0.5 alleles in offspring (31), precise predictions can be made of the effect of alleles and polygenic scores on traits in the offspring themselves.
Here, we used summary data from an independent GWAS of educational attainment (15) to create polygenic profile scores (34). These scores quantify the extent to which each participant carried the genetic variants known to be associated with higher educational attainment (in the GWAS, education was measured as the number of years of education). We then linked these polygenic profile scores to data on the participants’ parents’ age at death. Our hypothesis was that offspring with polygenic profiles for higher educational attainment would have longer-living parents. We did not make a specific prediction about whether any effect would be stronger in fathers or mothers. We performed the analysis in three large, independent cohorts to test the replicability of the result, and meta-analytically combined the three estimates. The cohorts were Generation Scotland (35, 36) (n = 17,542), UK Biobank (37) (n = 116,425), and the Estonian Biobank (38) (n = 7,950).
As a sensitivity analysis, we tested whether our results still held when taking into account parental fertility: that is, when including as a numerical covariate the number of siblings that each participant reported. This was because of a possible biasing effect whereby parents with higher numbers of offspring, and thus linearly proportionate greater likelihood of the parental phenotype being included in the study, might have different genetic propensities for educational attainment. Finally, we compared the predictive value of the educational polygenic profile score for parental mortality with the predictive values for a number of other polygenic profile scores indexing phenotypes that are known to relate to mortality risk.
Results
A summary of the parental data, including number of deaths, for each of the three cohorts is presented in Table 1, and the cohorts are described more fully in Materials and Methods.
Table 1.
Descriptive statistics for parents across the three samples
| Parent | Status | Generation Scotland | UK Biobank | Estonian Biobank | |||
| n | Mean age, y (SD) | n | Mean age, y (SD) | n | Mean age, y (SD) | ||
| Mother | Alive | 10,340 | 64.4 (12.5) | 45,333 | 78.4 (8.1) | 3,247 | 63.0 (13.2) |
| Dead | 6,330 | 73.3 (12.5) | 69,990 | 74.9 (12.1) | 2,682 | 73.5 (12.7) | |
| Father | Alive | 7,923 | 62.9 (11.5) | 25,915 | 77.9 (7.3) | 2,327 | 60.9 (11.9) |
| Dead | 8,467 | 69.7 (11.9) | 85,419 | 71.3 (11.7) | 3,744 | 68.1 (12.9) | |
Phenotypic and Genetic Correlations.
Before testing our main hypotheses, we assessed the phenotypic and genetic correlations between the two principal variables under investigation: offspring education and parental longevity. We ran these analyses in UK Biobank, by far the largest of the three cohorts involved in the present study, alone. Phenotypically, offspring education (for which the median was 15 y, the mean was 15.11 y, and the range was 7–20 y) was significantly and substantially associated with both mother’s longevity [hazard ratio (HR) per additional year of offspring education = 0.897, 95% confidence interval (CI) = (0.889, 0.904), P = 3.08 × 10−126] and father’s longevity [HR per additional year of offspring education = 0.893, 95% CI = (0.886, 0.899), P = 1.74 × 10−161]. We then assessed the genetic correlation (rg) between educational attainment and parental longevity, using LD score regression (Materials and Methods) on GWAS summary scores from two previous studies of these phenotypes (15, 31). The genetic correlation was estimated at rg = 0.447 (SE = 0.080, P = 2.23 × 10−08) for mother’s longevity, and rg = 0.392 (SE = 0.056, P = 2.82 × 10−12) for father’s longevity. Thus, there were substantial relations, both phenotypic and genetic, between the two variables of interest. These were in the same effect size range as genetic correlations found in previous studies between educational attainment and a range of other health phenotypes (25).
Polygenic Profile Score Analysis.
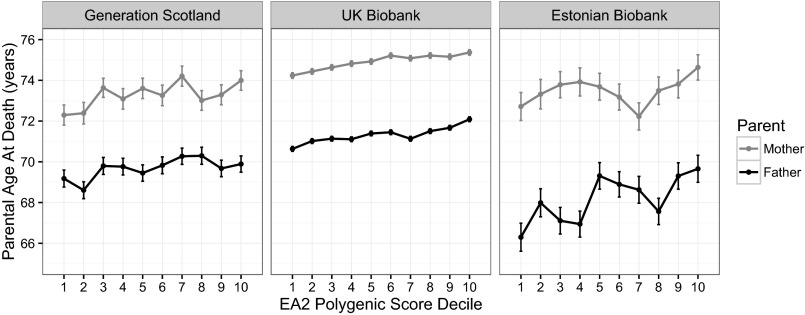
The polygenic scores for educational attainment were built using the previous GWAS results (15) and applied to the participants in Generation Scotland, UK Biobank, and the Estonian Biobank. Fig. 1 provides descriptive data for each sample, showing the age at mortality of each parent depending on each decile of the education polygenic risk score; in general, higher polygenic scores were associated with older age at death. However, this illustration only includes parents who had died. To take into account all of the data, we calculated the associations between offspring polygenic scores and parental longevity using Cox proportional hazard models (Table 2). For mothers, the HRs were not significantly different from zero in the smaller samples, but were highly significant in UK Biobank. For fathers, the results were significant in all three samples. In all cases, the point estimate was in the hypothesized direction: higher polygenic profile score was associated with lower parental mortality risk.
Fig. 1.
Parental age at death by education polygenic score decile in each cohort. Error bars in all three plots represent ±1 standard error of the mean. Note that this plot does not include data from participants whose mother and/or father was living at the time of assessment.
Table 2.
Results from Cox proportional hazard models predicting parental mortality risk from offspring education polygenic profile score
| Parent | Cohort | n Offspring | n Parental deaths | HR (95% CI) | SE | P value |
| Mother | Generation Scotland | 16,670 | 6,330 | 0.954 (0.907, 1.001) | 0.024 | 0.056 |
| UK Biobank | 115,323 | 69,990 | 0.976 (0.968, 0.984) | 0.004 | 1.52 × 10−10 | |
| Estonian Biobank | 5,929 | 2,682 | 0.979 (0.940, 1.018) | 0.020 | 0.280 | |
| Meta-analysis | 137,922 | 79,702 | 0.976 (0.968, 0.983) | 0.004 | 8.21 × 10−10 | |
| Father | Generation Scotland | 16,390 | 8,467 | 0.932 (0.891, 0.973) | 0.021 | 0.0007 |
| UK Biobank | 111,334 | 85,419 | 0.975 (0.969, 0.981) | 0.003 | 2.05 × 10−13 | |
| Estonian Biobank | 6,097 | 3,744 | 0.942 (0.909, 0.975) | 0.017 | 0.0003 | |
| Meta-analysis | 133,821 | 97,630 | 0.973 (0.967, 0.979) | 0.004 | 1.73 × 10−18 |
Hazard ratios are expressed per SD of polygenic profile score. Meta-analytic rows are in bold. All models adjusted for offspring sex, genotyping array, and SNP principal components as described in Materials and Methods.
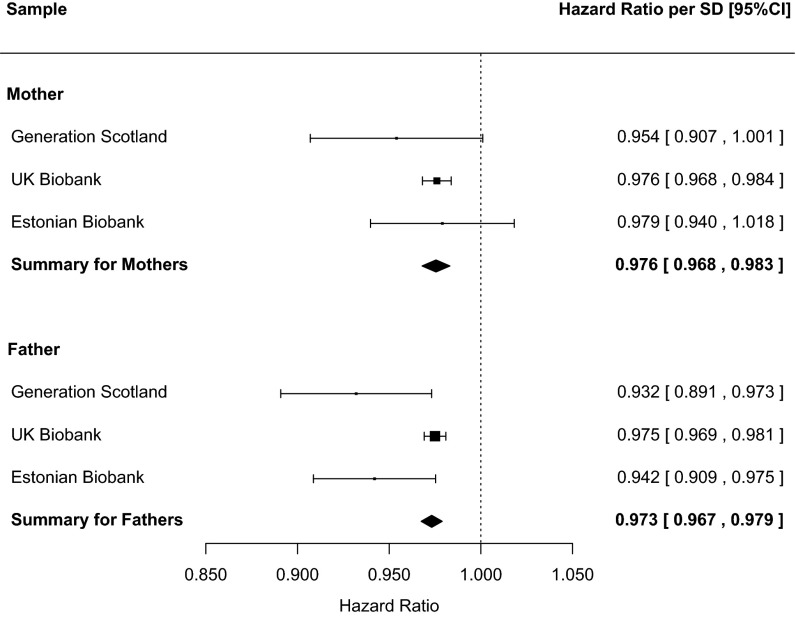
We combined the scores across the three cohorts, separately for mothers and for fathers, using a fixed-effects meta-analysis. The meta-analytic results showed that a 1 SD higher polygenic profile score for education was associated with an ∼2.5% lower mortality risk in mothers [HR = 0.976, 95% CI = (0.968, 0.983), P = 8.21 × 10−10] and fathers [HR = 0.973, 95% CI = (0.967, 0.979), P < 1.73 × 10−18]. A forest plot is shown in Fig. 2. Splitting the education polygenic profile score into tertiles, we calculated that, in the UK Biobank sample, mothers of children in the highest tertile lived on average 40.8 y beyond age 40, compared with 40.2 y in the lowest tertile (a difference of 0.6 y). The corresponding values for fathers were 35.1 y beyond age 40 in the highest tertile and 34.6 y in the lowest (a difference of 0.5 y).
Fig. 2.
Forest plot for the fixed-effects meta-analysis of the association between educational polygenic profile score and mortality risk for mothers and fathers across the three samples. HRs are for a 1 SD higher polygenic profile score.
We next ran the sensitivity analysis including number of siblings as a covariate in the model predicting parental mortality from offspring genotype. Again, this analysis was run in UK Biobank alone. The inclusion of this covariate made little difference to the UK Biobank results reported above: the association between educational polygenic profile score and mother’s longevity was HR per 1 SD higher polygenic score = 0.978, 95% CI = (0.970, 0.985), P = 3.29 × 10−09 ΔHR from original model = 0.002; the association with father’s longevity was HR = 0.977, 95% CI = (0.970, 0.984), P = 9.86 × 10−12, ΔHR = 0.002.
Other Polygenic Scores.
To provide context for the effect size of the education polygenic profile score’s association with parental mortality, we tested associations of parental longevity with a series of other offspring polygenic profile scores. We ran these analyses only in UK Biobank, using the same methods as those for the educational polygenic profile score analyses. The polygenic profile scores were for five known phenotypic predictors of mortality risk: height, body mass index (BMI), cardiovascular disease, major depressive disorder (MDD), and smoking (39–43).
The hazard ratios for predicting mortality for either parent from offspring polygenic profiles for height and for MDD were both near to 1 and nonsignificant (P > 0.28). For mothers, there was no significant relation with offspring smoking genetic risk (P = 0.07). However, scores for BMI, cardiovascular disease and, for fathers, smoking, made significant predictions of mortality risk (P < 5.85 × 10−06). The effect sizes for each of these genetic predictors were similar to that for education (approximately a 2% difference in mortality risk per 1 SD difference in the score) (Table S1). Thus, the score composed of genetic variants weighted toward their relation to educational attainment made similar-sized predictions of longevity risk to genetic scores weighted toward alleles linked to other well-established risk factors for mortality. Note that a number of other polygenic associations with mortality were addressed in the UK Biobank sample in a previous study, using somewhat different methods (33).
Table S1.
Hazard ratios (HRs) for maternal and paternal longevity predicted by noneducation polygenic profile scores in UK Biobank
| Parent | Offspring polygenic profile score | GWAS source | HR | SE | P value |
| Mother | Height | Wood et al. (39) | 1.004 | 0.004 | 0.287 |
| BMI | Locke et al. (40) | 1.020 | 0.004 | 9.23 × 10−11 | |
| Cardiovascular disease | Schunkert et al. (41) | 1.017 | 0.004 | 5.13 × 10−06 | |
| MDD | Ripke et al.; MDD Working Group of the Psychiatric Genomics Consortium (42) | 1.001 | 0.004 | 0.717 | |
| Smoking | Tobacco and Genetics Consortium (43) | 1.010 | 0.004 | 0.072 | |
| Father | Height | Wood et al. (39) | 0.999 | 0.004 | 0.726 |
| BMI | Locke et al. (40) | 1.020 | 0.003 | 5.84 × 10−06 | |
| Cardiovascular Disease | Schunkert et al. (41) | 1.024 | 0.003 | 1.52 × 10−12 | |
| MDD | Ripke et al.; MDD Working Group of the Psychiatric Genomics Consortium (42) | 0.998 | 0.003 | 0.547 | |
| Smoking | Tobacco and Genetics Consortium (43) | 1.020 | 0.004 | 1.08 × 10−08 |
HRs are per 1 SD increase in the relevant polygenic profile score. Rows with statistically significant values are highlighted in bold. Models included the same covariates as in the main analysis of educational attainment polygenic profile score.
Discussion
This study found that offspring polygenic profiles for education were robustly associated with parental longevity: those with more genetic variants related to better educational qualifications had longer-living parents. We tested the study’s principal hypothesis across three large cohorts, totaling over 130,000 participants. The associations were of broadly similar effect size in all three cohorts. Meta-analytically, there was a substantial and strongly significant overall prediction, which was similar for males and for females: individuals with 1 SD higher polygenic profile score for a college degree had parents who were at ∼2.5% lower risk of mortality. Put another way, parents with offspring in the upper third of the polygenic score distribution lived an average of 0.55 y longer than those in the lower third. The results—which were comparable to the effect sizes from other known predictors of mortality, such as cardiovascular disease and smoking, and which were bolstered by the finding of a moderate-sized genetic correlation between the two variables—suggest the hypothesis that the ultimate reason education predicts mortality is, in part, because of an underlying, quantifiable, genetic propensity.
Why do genetic variants related to educational attainment predict parental mortality? There are a number of possible mechanisms—both genetically and environmentally mediated—that might explain the result. First, these genetic variants might improve cognitive or personality phenotypes, such as intelligence, motivation, and conscientiousness, thus improving educational attainment; the higher quality of life and environment afforded by a better education might then improve health and reduce mortality risk. The effects of the genetic score might manifest directly on the parents’ behavior (to the extent that they are shared between parents and offspring), or have indirect effects via greater offspring resources and ability to care for aging parents (44). Our analysis could not test between these direct and indirect possibilities.
Second, the genetic variants related to education might also affect other variables that themselves lower mortality risk. This could occur in the absence of a causal pathway involving education itself. For example, individuals with long-term illnesses are at greater risk of educational failure (23), and also tend to perform more poorly on tests of cognitive ability (45); such health complaints, which might partly be genetic in origin, could also increase mortality risk. Third, and unlike the above mechanisms that all posit a mediated mechanism (that is, genetic variants related to educational attainment have effects on some factor that leads to additional longevity), biological pleiotropy (46) might play a role: in this view, the genetic variants reflect a general “system integrity” (47), whereby genotypes related to better physical health (and thus lower mortality risk) are also related to better neural health (and thus better educational performance).
Testing the relative contributions of each of the three possible mechanisms described above—which are not mutually exclusive—will require a finer-grained analysis of the complex pathways that link the education-associated genetic variants to longevity, via mediating traits, conditions, and behaviors. In this study, we compared the size of the association of longevity with the educational polygenic score with that for polygenic scores for height, BMI, cardiovascular disease, major depressive disorder, and smoking. Future analyses could get closer to the mechanism by assessing the degree of overlap and generality among genetic predictors of longevity. Because GWAS studies provide a deeper knowledge of the specific, causal genetic variants that are linked to education and to longevity (15–17, 31), we will be able to address their biological and social mechanisms in greater detail, improving our understanding of precisely why scores for, for example, height make no prediction of longevity but those for education do. In any case, regardless of the underlying mechanisms, the polygenic profile score for education showed predictive value. As GWAS sample sizes for education and related variables increase, and more genetic variants are uncovered (48), we would expect steadily to obtain improved genetic predictions of longevity.
The longevity prediction made by the polygenic profile for educational attainment was substantially smaller than that for the phenotype of educational attainment. This is to be expected for two reasons. First, polygenic profiles only explain small amounts of variance in their respective phenotypes because of the power of the original GWAS studies to detect SNPs with significant associations (as noted, we expect this to improve with larger, future GWAS studies), and do not include nonadditive genetic variants that may also be important in explaining heritability. Second, educational attainment is far from completely heritable (12), being influenced by social and environmental factors that may also be predictive of parental mortality. However, the finding that the longevity-predicting power of a genetic profile for education (a variable often thought of as “social”) compares favorably in effect size to polygenic profiles for cardiovascular disease and BMI (variables that are medical in nature) supports the importance of educational attainment as a general indicator of health and social status.
Our method, using parental longevity as an outcome variable predicted by offspring genotype, allowed considerably higher power compared with studies of genotype and mortality in the same individuals; such data are more difficult to collect because they require follow-up of genotyped individuals until their own death. This method, combined with the large sample sizes, our replication and meta-analysis, and the inclusion of parents who were still alive as censored data points in the proportional hazard models, substantially improved our results’ robustness. Our effect size estimates for the main analysis were similar in samples from the United Kingdom and from Estonia, indicating that the education-related genetic variants make predictions across different cultures (although further replication in other groups will be necessary). Finally, our results appeared robust to parental fertility: they were only slightly altered after adjustment for number of siblings.
Although the effects found here were broadly consistent across cultures, the samples were not fully representative of the populations from which they were drawn. All samples were restricted to individuals of White European ancestry. Whereas this reduces bias due to population stratification within each sample, it does make the results less generalizable, and samples of participants with different genetic backgrounds may show different results. In addition, self-selection effects (49) mean that those with more education, higher intelligence, higher socioeconomic status, higher conscientiousness, and closer proximity to testing centers were probably more likely to participate. The concomitant restriction of range potentially led to downward bias in our effect sizes. A more subtle consequence of self-selection is that many of the above characteristics might make these self-selected individuals more likely to benefit, in terms of health or other life outcomes such as longevity, from higher educational attainment (that is, their genetic propensities for education may interact with other traits). However, in such a conceptualization, the educational variants are still the ultimate explanation for some of the variance in longevity.
No measures of parental educational attainment were available in our samples, precluding an analysis testing whether there were any incremental associations of the polygenic score beyond phenotypic education, or whether any effects of the genetic score were entirely mediated by educational attainment. The advantages of our parental-proxy method are noted above, but we may have underestimated the effects: we would expect exactly double the effect size for polygenic prediction of an individual’s own longevity from their genetic profile (31). Finally, although we adjusted for each study participant’s number of siblings to control for fertility differences, by definition all of the individuals whose ages at mortality were analyzed in the present study (i.e., the parents) had children. It remains possible that the associations studied here would be different for individuals with no children, who may have also had systematically different polygenic profiles for education.
Conclusion
This study used molecular genetic methods in three large samples, finding that a polygenic profile score for education in offspring made a statistically significant prediction of parental longevity. In the general population, there are genetic variants that relate not only to important predictors of health outcomes (in this case, education), but also with the health outcomes themselves (in this case, longevity). Combining the polygenic profile for educational attainment with profiles for other important cognitive, personality, and health-related traits will enable future studies to make better predictions of longevity, with applications in epidemiological research and beyond.
Materials and Methods
Educational Attainment “Discovery” GWAS.
The educational attainment polygenic scores were built using summary data from the largest GWAS meta-analysis of educational attainment to date (15). To reduce the possibility of sample overlap or cryptic relatedness affecting the polygenic scores, the GWAS data were reanalyzed after excluding all United Kingdom-based cohorts for predictions into the independent United Kingdom cohorts. Similarly, the Estonian Biobank data were excluded from a second reanalysis of the GWAS data. Data from 23andMe, used in the original meta-analysis, were not available for the calculation of polygenic scores.
Independent Sample 1: Generation Scotland: The Scottish Family Health Study.
Participants.
Generation Scotland: the Scottish Family Health Study (35, 36) is a cohort study of participants recruited in the Glasgow, Tayside, Ayrshire, Arran, and northeast areas of Scotland. Initially, 7,953 probands aged 35–65 y were recruited either through their general medical practitioner (95% of probands) or via direct publicity and word-of-mouth. Their family members were also invited to take part, resulting in a final sample of 24,084 participants with an age range of 18–100 y.
All components of Generation Scotland received ethical approval from the National Health Service Tayside Committee on Medical Research Ethics (Research Ethics Committee Reference no. 05/S1401/89). Generation Scotland has been granted Research Tissue Bank status by the Tayside Committee on Medical Research Ethics (Research Ethics Committee Reference no. 10/S1402/20), providing generic ethical approval for a wide range of uses within medical research.
Genotyping.
Generation Scotland participants were genotyped with either the HumanOmniExpressExome8v1-2_A or HumanOmniExpressExome-8v1_A. Quality control was carried out in PLINK v1.9b2c (50, 51). SNPs were removed if they had a missingness rate >2% or a Hardy–Weinberg Equilibrium test at P < 10−06, leaving a total of 561,125 autosomal SNPs for analysis. Duplicate samples were removed. Individuals were removed based on gender mismatch and missingness (>2% of genotypes missing). The subsequent dataset was combined with the 1,092 individuals of the 1000 Genomes population (52) before principal components being calculated in GCTA (53). Outliers, defined by being more than 6 SDs away from the mean of the first two principal components, were removed (54). This left a sample of 20,032 participants. Individuals who appeared in both the UK Biobank and Generation Scotland studies (n = 174) were excluded from the latter study. After merging with the available covariate data, 17,542 participants had age at death or censoring information in at least one parent.
Independent Sample 2: UK Biobank.
Participants.
Data stem from the baseline wave of the UK Biobank Study (37) (www.ukbiobank.ac.uk). Analyses were performed under data applications 8304 and 10279. The UK Biobank sample was substantially larger than our other two studies: it contains around 500,000 community dwelling men and women in the United Kingdom, who were recruited between 2006 and 2010. Here, we used data from 116,425 participants (aged 40–73 y) who had genetic data and at least one parent’s longevity data available for analyses. Ethical approval for UK Biobank was granted by the Research Ethics Committee (11/NW/0382).
Genotyping.
Details on the UK Biobank genotyping procedure and quality-control steps that were included for the current analyses have been reported previously (31). Briefly, of the 152,729 participants with genetic data available as of August 2015, 116,425 were retained after exclusions based on SNP missingness, relatedness, gender mismatch, non-British ancestry, and previously reported quality control failure for the UK BiLEVE study.
Independent Sample 3: Estonian Biobank.
Participants.
The Estonian Biobank (38) is the population-based biobank of the Estonian Genome Center at the University of Tartu (EGCUT). For this study, 51,380 volunteer participants (aged 18–103 y) were recruited between 2002 and 2011. The cohort included ∼5% of the adult population from all counties of Estonia. At recruitment, the participants completed an extensive questionnaire on health, lifestyle, and genealogy and provided a blood sample. Approval for the Estonian Biobank was given by the Research Ethics Committee of the University of Tartu. All participants signed a broad informed consent form at recruitment.
Genotyping.
In total, DNA samples from >16,000 participants have been genotyped with various genome-wide arrays. In 2011, the subset of individuals selected to be genotyped with the Illumina OmniExpress chip, intentionally included 1,200 individuals who had died by that time, as well as 500 women and 250 men who were 80 y old or older at that time. The rest of this genotyped sample (in total 7,950 subjects after removing close relatives) consists of random population controls.
Parental Longevity Phenotype.
In all three independent cohorts, parental longevity was assessed for both mothers and fathers. To account for premature deaths as a result of external causes, such as accidents, and in particular, the Second World War (55), we excluded individuals who died prematurely (<40 y). Parents who were alive at the baseline wave of the respective studies were treated as censored observations. Age at censoring was calculated as the cohort’s baseline year of assessment minus the parent’s year of birth. Parents whose age at censor was <40 y were excluded. In Generation Scotland, a small number of outliers (n = 26) with an age-at-death/censor >100 y were removed.
Statistical Analyses.
LD score regression (genetic correlation).
To assess the genetic correlation between the two primary phenotypes of interest, we used LD score regression (56), which allows genetic correlations to be calculated using GWAS summary data alone (without raw genotype or phenotype data). It does not matter for LD score regression whether there is sample overlap between the studies. We calculated the genetic correlation (rg) using the summary data for educational attainment from the GWAS that also served as the basis for our polygenic profile scores (see below) (15) and using the summary data for parental longevity (specifically, the Martingale residuals from Cox proportional hazards models of parental lifespan) from a recent GWAS in the UK Biobank sample (31). LD score regression was used with all its default settings.
Polygenic scoring.
The results from the educational attainment GWAS analyses were carried forward into polygenic score models in the three independent cohorts using the PRSice software (57). We built polygenic profile scores based on the genotyped SNP data in Generation Scotland (nSNPs = 561,125), UK Biobank (nSNPs = 672,491), and EGCUT (nSNPs = 628,325). The optimal threshold determined from the previous analysis (15), specifically P = 1.00 (that is, inclusion of all SNPs) was used for all analyses in all samples.
We tested whether the polygenic profile score was related to the participant’s (that is, the offspring’s) own educational duration. To do this, we included the score in a linear regression analysis predicting the offspring’s years of education alongside covariates of age, sex, and 15 SNP-based principal components to account for population stratification. The polygenic profile score was significantly related to years of education in Generation Scotland (n = 17,814; standardized β = 0.132, SE = 0.008, P < 2.20 × 10−16), UK Biobank (n = 119,167; standardized β = 0.106, SE = 0.003, P < 2.20 × 10−16), and EGCUT (n = 7,959; standardized β = 0.118, SE = 0.011, P = 1.77 × 10−26). Note that, for Generation Scotland, a kinship matrix was fitted to account for the structure of relatedness (as in the main analysis; see below) and for UK Biobank, years of education were calculated in accordance with the protocol used in the previous GWAS meta-analysis (15). Overall, the polygenic profile score had a small, positive relation to offspring’s own educational duration.
The respective effect sizes for the SNP risk alleles were multiplied by the number of risk alleles (0, 1, or 2) carried by the participants in Generation Scotland, UK Biobank, and EGCUT. The sum score across all SNPs within each threshold yielded a polygenic score for each participant in the three cohorts. These scores are proxy measures (∼50% accurate) of the parental genetic risk scores.
Polygenic prediction of mortality.
The educational attainment polygenic scores were modeled against the age at death for the parents of the independent cohorts. Cox proportional hazards models were run, using a pedigree-derived kinship matrix to account for relatedness in Generation Scotland. Covariates included sex, and the first 15 SNP-based principal components to account for population stratification. Analyses were conducted in R using the “survival” (58), “kinship2” (59), and “coxme” (60) libraries.
Meta-analysis.
Fixed-effects meta-analysis was performed across the three cohorts, separately for mothers and fathers, using the metafor package for R (61).
Acknowledgments
We thank the cohort participants and team members who contributed to these studies. Generation Scotland received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR030060). R.E.M., S.J.R., S.P.H., W.D.H., G.D., D.C.L., D.J.P., C.R.G., and I.J.D. are members of the University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, which is supported by funding from the Biotechnology and Biological Sciences Research Council, the Medical Research Council, and the University of Edinburgh as part of the cross-council Lifelong Health and Wellbeing initiative (MR/K026992/1) and Age UK as part of the Disconnected Mind Project. The research was conducted using the UK Biobank Resource. The Estonian Genome Center at the University of Tartu work was supported through the Estonian Genome Center of University of Tartu by the Targeted Financing from the Estonian Ministry of Science and Education (SF0180142s08), the Development Fund of the University of Tartu (Grant SP1GVARENG), the European Regional Development Fund to the Centre of Excellence in Genomics (EXCEGEN; Grant 3.2.0304.11-0312), and through FP7 Grant 313010. This research was facilitated by the Social Science Genetic Association Consortium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
3A complete list of the Social Science Genetic Association Consortium members can be found in the Supporting Information.
See Commentary on page 13269.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605334113/-/DCSupplemental.
Contributor Information
Collaborators: Aysu Okbay, Jonathan P. Beauchamp, Mark A. Fontana, James J. Lee, Tune H. Pers, Cornelius A. Rietveld, Patrick Turley, Guo-Bo Chen181, Valur Emilsson, S. Fleur W. Meddens, Sven Oskarsson, Joseph K. Pickrell, Kevin Thom, Pascal Timshel, Ronald de Vlaming, Abdel Abdellaoui, Tarunveer S. Ahluwalia, Jonas Bacelis, Clemens Baumbach, Gyda Bjornsdottir, Johannes H. Brandsma, Maria Pina Concas, Jaime Derringer, Nicholas A. Furlotte, Tessel E. Galesloot, Giorgia Girotto, Richa Gupta, Leanne M. Hall, Sarah E. Harris, Edith Hofer, Momoko Horikoshi, Jennifer E. Huffman, Kadri Kaasik, Ioanna P. Kalafati, Robert Karlsson, Augustine Kong, Jari Lahti, Sven J. van der Lee, Christiaan de Leeuw, Penelope A. Lind, Karl-Oskar Lindgren, Tian Liu, Massimo Mangino, Jonathan Marten, Evelin Mihailov, Michael B. Miller, Peter J. van der Most, Christopher Oldmeadow, Antony Payton, Natalia Pervjakova, Wouter J. Peyrot, Yong Qian, Olli Raitakari, Rico Rueedi, Erika Salvi, Börge Schmidt, Katharina E. Schraut, Jianxin Shi, Albert V. Smith, Raymond A. Poot, Beate St Pourcain, Alexander Teumer, Gudmar Thorleifsson, Niek Verweij, Dragana Vuckovic, Juergen Wellmann, Harm-Jan Westra, Jingyun Yang, Wei Zhao, Zhihong Zhu, Behrooz Z. Alizadeh, Najaf Amin, Andrew Bakshi, Sebastian E. Baumeister, Ginevra Biino, Klaus Bønnelykke, Patricia A. Boyle, Harry Campbell, Francesco P. Cappuccio, Gail Davies, Jan-Emmanuel De Neve, Panos Deloukas, Ilja Demuth, Jun Ding, Peter Eibich, Lewin Eisele, Niina Eklund, David M. Evans, Jessica D. Faul, Mary F. Feitosa, Andreas J. Forstner, Ilaria Gandin, Bjarni Gunnarsson, Bjarni V. Halldórsson, Tamara B. Harris, Andrew C. Heath, Lynne J. Hocking, Elizabeth G. Holliday, Georg Homuth, Michael A. Horan, Jouke-Jan Hottenga, Philip L. de Jager, Peter K. Joshi, Astanand Jugessur, Marika A. Kaakinen, Mika Kähönen, Stavroula Kanoni, Liisa Keltigangas-Järvinen, Lambertus A.L.M. Kiemeney, Ivana Kolcic, Seppo Koskinen, Aldi T. Kraja, Martin Kroh, Zoltan Kutalik, Antti Latvala, Lenore J. Launer, Maël P. Lebreton, Douglas F. Levinson, Paul Lichtenstein, Peter Lichtner, David C.M. Liewald, LifeLines Cohort Study, Anu Loukola, Pamela A. Madden, Reedik Mägi, Tomi Mäki-Opas, Riccardo E. Marioni, Pedro Marques-Vidal, Gerardus A. Meddens, George McMahon, Christa Meisinger, Thomas Meitinger, Yusplitri Milaneschi, Lili Milani, Grant W. Montgomery, Ronny Myhre, Christopher P. Nelson, Dale R. Nyholt, William E.R. Ollier, Aarno Palotie, Lavinia Paternoster, Nancy L. Pedersen, Katja E. Petrovic, David J. Porteous, Katri Räikkönen, Susan M. Ring, Antonietta Robino, Olga Rostapshova, Igor Rudan, Aldo Rustichini, Veikko Salomaa, Alan R. Sanders, Antti-Pekka Sarin, Helena Schmidt, Rodney J. Scott, Blair H. Smith, Jennifer A. Smith, Jan A. Staessen, Elisabeth Steinhagen-Thiessen, Konstantin Strauch, Antonio Terracciano, Martin D. Tobin, Sheila Ulivi, Simona Vaccargiu, Lydia Quaye, Frank J.A. van Rooij, Cristina Venturini, Anna A.E. Vinkhuyzen, Uwe Völker, Henry Völzke, Judith M. Vonk, Diego Vozzi, Johannes Waage, Erin B. Ware, Gonneke Willemsen, John R. Attia, David A. Bennett, Klaus Berger, Lars Bertram, Hans Bisgaard, Dorret I. Boomsma, Ingrid B. Borecki, Ute Bultmann, Christopher F. Chabris, Francesco Cucca, Daniele Cusi, Ian J. Deary, George V. Dedoussis, Cornelia M. van Duijn, Johan G. Eriksson, Barbara Franke, Lude Franke, Paolo Gasparini, Pablo V. Gejman, Christian Gieger, Hans-Jörgen Grabe, Jacob Gratten, Patrick J.F. Groenen, Vilmundur Gudnason, Pim van der Harst, Caroline Hayward, David A. Hinds, Wolfgang Hoffmann, Elina Hypponen, William G. Iacono, Bo Jacobsson, Marjo- Riitta Järvelin, Karl-Heinz Jöckel, Jaakko Kaprio, Sharon L.R. Kardia, Terho Lehtimäki, Steven F. Lehrer, Patrik K.E. Magnusson, Nicholas G. Martin, Matt McGue, Andres Metspalu, Neil Pendleton, Brenda W.J.H. Penninx, Markus Perola, Nicola Pirastu, Mario Pirastu, Ozren Polasek, Danielle Posthuma, Christine Power, Michael A. Province, Nilesh J. Samani, David Schlessinger, Reinhold Schmidt, Thorkild I.A. Sørensen, Tim D. Spector, Kari Stefansson, Unnur Thorsteinsdottir, A. Roy Thurik, Nicholas J. Timpson, Henning Tiemeier, Joyce Y. Tung, André G. Uitterlinden, Veronique Vitart, Peter Vollenweider, David R. Weir, James F. Wilson, Alan F. Wright, Dalton C. Conley, Robert F. Krueger, George Davey Smith, Albert Hofman, David I. Laibson, Sarah E. Medland, Michelle N. Meyer, Jian Yang, Magnus Johannesson, Peter M. Visscher, Tõnu Esko, Philipp D. Koellinger, David Cesarini, and Daniel J. Benjamin
References
- 1.Breen R, Goldthorpe JH. Class, mobility and merit: The experience of two British birth cohorts. Eur Sociol Rev. 2001;17(2):81–101. [Google Scholar]
- 2.Breen R, Karlson KB. Education and social mobility: New analytical approaches. Eur Sociol Rev. 2014;30(1):107–118. [Google Scholar]
- 3.Zajacova A, Hummer RA, Rogers RG. Education and health among U.S. working-age adults: A detailed portrait across the full educational attainment spectrum. Biodemogr Soc Biol. 2012;58(1):40–61. doi: 10.1080/19485565.2012.666122. [DOI] [PubMed] [Google Scholar]
- 4.Harmon C, Oosterbeek H, Walker I. The returns to education: Microeconomics. J Econ Surv. 2003;17(2):115–156. [Google Scholar]
- 5.Brown DC, et al. The significance of education for mortality compression in the United States. Demography. 2012;49(3):819–840. doi: 10.1007/s13524-012-0104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montez JK, Hayward MD. Cumulative childhood adversity, educational attainment, and active life expectancy among U.S. adults. Demography. 2014;51(2):413–435. doi: 10.1007/s13524-013-0261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinch CN, Galloway TA. Schooling in adolescence raises IQ scores. Proc Natl Acad Sci USA. 2012;109(2):425–430. doi: 10.1073/pnas.1106077109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie SJ, Bates TC. Enduring links from childhood mathematics and reading achievement to adult socioeconomic status. Psychol Sci. 2013;24(7):1301–1308. doi: 10.1177/0956797612466268. [DOI] [PubMed] [Google Scholar]
- 9.Deary IJ, Johnson W. Intelligence and education: Causal perceptions drive analytic processes and therefore conclusions. Int J Epidemiol. 2010;39(5):1362–1369. doi: 10.1093/ije/dyq072. [DOI] [PubMed] [Google Scholar]
- 10.Bartels M, Rietveld MJ, Van Baal GCM, Boomsma DI. Heritability of educational achievement in 12-year-olds and the overlap with cognitive ability. Twin Res. 2002;5(6):544–553. doi: 10.1375/136905202762342017. [DOI] [PubMed] [Google Scholar]
- 11.Rowe DC, Vesterdal WJ, Rodgers JL. Herrnstein’s syllogism: Genetic and shared environmental influences on IQ, education, and income. Intelligence. 1998;26(4):405–423. [Google Scholar]
- 12.Shakeshaft NG, et al. Strong genetic influence on a UK nationwide test of educational achievement at the end of compulsory education at age 16. PLoS One. 2013;8(12):e80341. doi: 10.1371/journal.pone.0080341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krapohl E, Plomin R. Genetic link between family socioeconomic status and children’s educational achievement estimated from genome-wide SNPs. Mol Psychiatr. 2015;21(3):437–443. doi: 10.1038/mp.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marioni RE, et al. Molecular genetic contributions to socioeconomic status and intelligence. Intelligence. 2014;44(100):26–32. doi: 10.1016/j.intell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okbay A, et al. LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rietveld CA, et al. LifeLines Cohort Study GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rietveld CA, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci USA. 2014;111(38):13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lleras-Muney A. The relationship between education and adult mortality in the United States. Rev Econ Stud. 2005;72:189–221. [Google Scholar]
- 19.Nordahl H, et al. Education and cause-specific mortality: The mediating role of differential exposure and vulnerability to behavioral risk factors. Epidemiology. 2014;25(3):389–396. doi: 10.1097/EDE.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 20.Deary IJ, Strand S, Smith P, Fernandes C. Intelligence and educational achievement. Intelligence. 2007;35(1):13–21. [Google Scholar]
- 21.Strenze T. Intelligence and socioeconomic success: A meta-analytic review of longitudinal research. Intelligence. 2007;35(5):401–426. [Google Scholar]
- 22.Schoon I. A transgenerational model of status attainment: The potential mediating role of school motivation and education. Natl Inst Econ Rev. 2008;205(1):72–82. [Google Scholar]
- 23.Case A, Fertig A, Paxson C. The lasting impact of childhood health and circumstance. J Health Econ. 2005;24(2):365–389. doi: 10.1016/j.jhealeco.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Marks GN. Education, Social Background and Cognitive Ability: The Decline of the Social. Routledge; New York: 2014. [Google Scholar]
- 25.Hagenaars SP, et al. METASTROKE Consortium International Constorium for Blood Pressure GWAS SpiroMeta Consortium CHARGE Consortium Pulmonary Group CHARGE Consortium Aging and Longevity Group Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N = 112,151) Mol Psychiatr. 2016;21(6):758–767. [Google Scholar]
- 26.Calvin CM, et al. Multivariate genetic analyses of cognition and academic achievement from two population samples of 174,000 and 166,000 school children. Behav Genet. 2012;42(5):699–710. doi: 10.1007/s10519-012-9549-7. [DOI] [PubMed] [Google Scholar]
- 27.Calvin CM, et al. Intelligence in youth and all-cause-mortality: Systematic review with meta-analysis. Int J Epidemiol. 2011;40(3):626–644. doi: 10.1093/ije/dyq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arden R, et al. The association between intelligence and lifespan is mostly genetic. Int J Epidemiol. 2016;45(1):178–185. doi: 10.1093/ije/dyv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wacholder S, et al. The kin-cohort study for estimating penetrance. Am J Epidemiol. 1998;148(7):623–630. doi: 10.1093/aje/148.7.623. [DOI] [PubMed] [Google Scholar]
- 30.Reed T, Carmelli D, Robinson TS, Rinehart SA, Williams CJ. More favorable midlife cardiovascular risk factor levels in male twins and mortality after 25 years of follow-up is related to longevity of their parents. J Gerontol A Biol Sci Med Sci. 2003;58(4):367–371. doi: 10.1093/gerona/58.4.m367. [DOI] [PubMed] [Google Scholar]
- 31.Joshi PK, et al. Variants near CHRNA3/5 and APOE have age- and sex-related effects on human lifespan. Nat Commun. 2016;7:11174. doi: 10.1038/ncomms11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell JT, et al. MuTHER Consortium Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8(4):e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilling LC, et al. Human longevity is influenced by many genetic variants: Evidence from 75,000 UK Biobank participants. Aging (Albany, NY) 2016;8(3):547–560. doi: 10.18632/aging.100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell SM, et al. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith BH, et al. Generation Scotland: The Scottish Family Health Study; a new resource for researching genes and heritability. BMC Med Genet. 2006;7(1):74. doi: 10.1186/1471-2350-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith BH, et al. Cohort Profile: Generation Scotland: Scottish Family Health Study (GS:SFHS). The study, its participants and their potential for genetic research on health and illness. Int J Epidemiol. 2013;42(3):689–700. doi: 10.1093/ije/dys084. [DOI] [PubMed] [Google Scholar]
- 37.Collins R. What makes UK Biobank special? Lancet. 2012;379(9822):1173–1174. doi: 10.1016/S0140-6736(12)60404-8. [DOI] [PubMed] [Google Scholar]
- 38.Leitsalu L, et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. 2015;44(4):1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 39.Wood AR, et al. Electronic Medical Records and Genomics (eMERGE) Consortium MIGen Consortium PAGE Consortium LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locke AE, et al. LifeLines Cohort Study ADIPOGen Consortium AGEN-BMI Working Group CARDIoGRAMplusC4D Consortium CKDGen Consortium GLGC ICBP MAGIC Investigators MuTHER Consortium MIGen Consortium PAGE Consortium ReproGen Consortium GENIE Consortium International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schunkert H, et al. Cardiogenics CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ripke S, et al. Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torssander J. From child to parent? The significance of children’s education for their parents’ longevity. Demography. 2013;50(2):637–659. doi: 10.1007/s13524-012-0155-3. [DOI] [PubMed] [Google Scholar]
- 45.Biessels GJ, Deary IJ, Ryan CM. Cognition and diabetes: A lifespan perspective. Lancet Neurol. 2008;7(2):184–190. doi: 10.1016/S1474-4422(08)70021-8. [DOI] [PubMed] [Google Scholar]
- 46.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: Challenges and strategies. Nat Rev Genet. 2013;14(7):483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deary IJ. Looking for ‘system integrity’ in cognitive epidemiology. Gerontology. 2012b;58(6):545–553. doi: 10.1159/000341157. [DOI] [PubMed] [Google Scholar]
- 48.Flint J, Munafò M. Schizophrenia: Genesis of a complex disease. Nature. 2014;511(7510):412–413. doi: 10.1038/nature13645. [DOI] [PubMed] [Google Scholar]
- 49.Johnson W, Brett CE, Calvin C, Deary IJ. Childhood characteristics and participation in Scottish Mental Survey 1947 6-Day Sample Follow-ups: Implications for participation in aging studies. Intelligence. 2016;54:70–79. [Google Scholar]
- 50.Purcell S, Chang CC. 2015 PLINK 1.9 package. Available at https://www.cog-genomics.org/plink2. Accessed October 7, 2016.
- 51.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amador C, et al. Generation Scotland Recent genomic heritage in Scotland. BMC Genomics. 2015;16:437. doi: 10.1186/s12864-015-1605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corley JE, Crang JA, Deary IJ. Childhood IQ and in-service mortality in Scottish army personnel during World War II. Intelligence. 2009;37:238–242. [Google Scholar]
- 56.Bulik-Sullivan B, et al. ReproGen Consortium Psychiatric Genomics Consortium Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Therneau T. 2015a A package for survival analysis in R. R package, v2.38. Available at cran.r-project.org/package=survival. Accessed October 7, 2016.
- 59.Therneau T, Atkinson E, Sinnwel J, Schaid D, McDonnell A. 2014 kinship2: Pedigree functions. R package, v1.6.0. Available at cran.r-project.org/package=kinship2. Accessed October 7, 2016.
- 60.Therneau T. 2015b coxme: Mixed effects Cox models. R package, v2.2-5. Available at cran.r-project.org/package=coxme. Accessed October 7, 2016.
- 61.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36(3):1–48. [Google Scholar]