Significance
Defects in myosin function are linked to a number of widespread and debilitating diseases, including asthma, chronic obstructive pulmonary disease, and hypertrophic cardiomyopathy. We report here the discovery of an allosteric site that modulates myosin motor function with high specificity that opens the path toward new therapeutic solutions. Identification of specific antimyosin drugs that significantly alter a motor’s function is an imperative first step toward the development of targeted and effective treatments for such diseases. Highly specific drugs against different members of the superfamily would also provide exquisite tools to investigate in cells their functional role. Additionally, detailed, high-resolution studies of the interaction of drugs with their myosin targets provide insights into the molecular mechanism of motor function.
Keywords: myosin, actin, drug design, molecular motor, specific allosteric drugs
Abstract
Direct inhibition of smooth muscle myosin (SMM) is a potential means to treat hypercontractile smooth muscle diseases. The selective inhibitor CK-2018571 prevents strong binding to actin and promotes muscle relaxation in vitro and in vivo. The crystal structure of the SMM/drug complex reveals that CK-2018571 binds to a novel allosteric pocket that opens up during the “recovery stroke” transition necessary to reprime the motor. Trapped in an intermediate of this fast transition, SMM is inhibited with high selectivity compared with skeletal muscle myosin (IC50 = 9 nM and 11,300 nM, respectively), although all of the binding site residues are identical in these motors. This structure provides a starting point from which to design highly specific myosin modulators to treat several human diseases. It further illustrates the potential of targeting transition intermediates of molecular machines to develop exquisitely selective pharmacological agents.
Myosins comprise a family of ATP-dependent motor proteins capable of producing directed force via interaction with their track, the F-actin filament. Force production by these motors powers numerous cellular processes such as muscle contraction, intracellular transport, and cell migration and division (1). Several myosins have also been linked to genetic disorders where either gain or loss of motor function can lead to disease. These motor proteins represent promising targets for the development of drugs modulating force production in cells, tissues, and muscle (2–4). Here we report a selective, small-molecule inhibitor of smooth muscle myosin (SMM) able to induce muscle relaxation. This mechanism of action has potential relevance for many diseases where smooth muscle contractility is central to the pathophysiology, such as asthma (5, 6) and chronic obstructive pulmonary disease (7).
Smooth muscle contractility can be activated through different pathways. Existing airway smooth muscle relaxants, such as β-adrenergic agonists and muscarinic antagonists, ultimately inhibit the activity of SMM. However, they do so via specific upstream signaling pathways. Direct inhibition of SMM contractility has the advantage of relaxing contracted smooth muscle regardless of the molecular stimulus driving it. Moreover, application of SMM inhibitors to the airway provides a means of selectively modulating contractility of these tissues by delivering a high local concentration of drug. We thus set about identifying selective inhibitors of SMM that can effectively relax muscle in vivo, leading to the discovery of a highly selective, small-molecule inhibitor, CK-2018571 (CK-571).
The detailed inhibitory mechanism of CK-571 was elucidated by a combination of in vitro characterization of the step in which the drug traps the motor and determination of the high-resolution structure of SMM cocrystallized with CK-571. The drug targets an intermediate state that occurs during the recovery stroke, the large conformational rearrangement that enables repriming of the motor. Blocking this critical transition thus results in efficient inhibition of force production. The SMM/CK-571 structure not only reveals how the drug stops the motor but also provides important insights about drug specificity. Our study establishes that the drug works via an inhibitory mechanism that has not been previously described for a molecular motor, revealing a potentially powerful therapeutic approach for certain human diseases.
Results
Characterization of a Highly Selective SMM Inhibitor, CK-571.
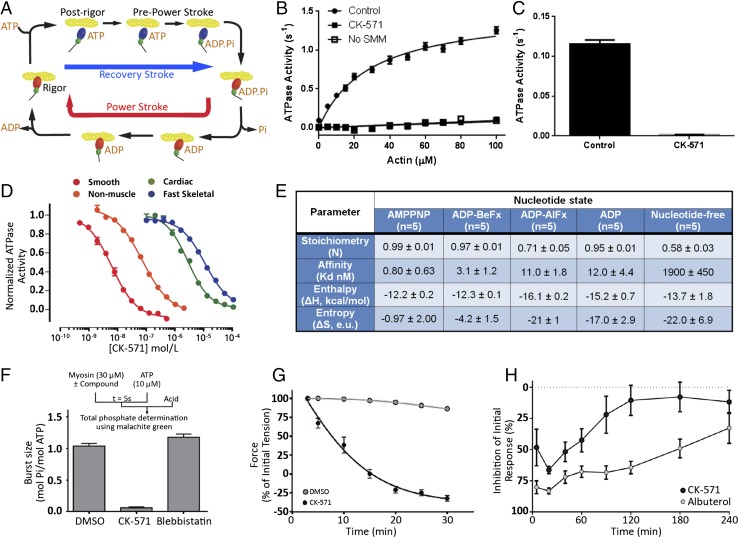
Muscle contraction is powered by the myosin molecular motor that produces force during its powerstroke (Fig. 1A). To promote tissue relaxation, inhibitors must stabilize myosin conformations that interact weakly with actin (Fig. 1A). High-throughput screens against SMM and extensive medicinal chemical optimization led to compound CK-571. As shown in Fig. 1B, the actin-activated ATPase of SMM is reduced to background levels in the presence of saturating concentrations of CK-571. The basal ATPase activity of SMM is also reduced more than 30 times upon CK-571 binding (Fig. 1C). Interestingly, the compound displays an IC50 of ∼9 nM and a high degree of selectivity for SMM compared with cardiac and striated muscle myosins, for which the IC50 is 280 and 1,255-fold lower, respectively (Fig. 1D). Notably, CK-571 also has a significantly lower affinity for nonmuscle myosin II (NMM, ∼8.5-fold less), which is remarkable given their high sequence homology to SMM (Fig. S1).
Fig. 1.
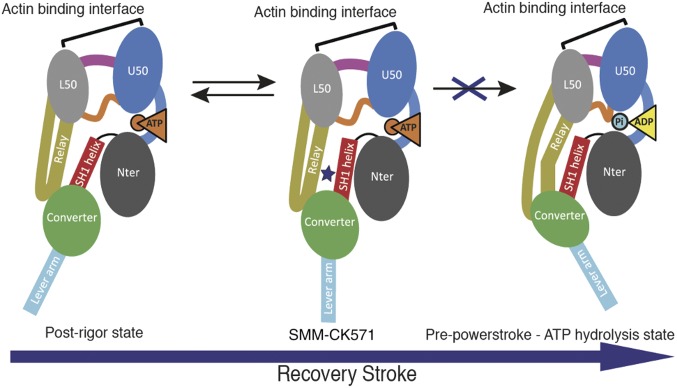
Identification of a specific SMM inhibitor. (A) Chemo-mechanical cycle of myosin. Myosin motors generate force upon releasing hydrolysis products when attached to F-actin (Powerstroke, red arrow). A swing of the distal region of the motor, the lever arm, is associated with force generation. At the end of the stroke, nucleotide-free myosin is strongly attached to F-actin (Rigor). Myosin detaches from the filament upon ATP binding [Postrigor (PR)] and then undergoes transitions that reprime the lever arm during the recovery stroke (blue arrow). Hydrolysis stabilizes the Prepowerstroke (PPS) state. Rebinding to F-actin in the PPS state triggers a series of conformational changes associated with force production that also trigger Pi and ADP release (Powerstroke, red arrow). (B) Inhibition of the actin-activated ATPase of chicken gizzard SMM S1 (0.35 µM) by 5 µM CK-571 (n = 6). Control reaction (2% DMSO) fit to a KM of 29 µM and VMax of 1.5 s−1 (data are mean ± SEM for n = 6). (C) The basal ATPase of chicken gizzard SMM S1 (1.8 µM) is reduced from 0.12 ± 0.0036 to 0.0013 ± 0.00032 (mean ± SD, n = 6) by 25 µM CK-571. (D) CK-571 specifically inhibits the steady-state actin-activated MgATPase activity of SMM over other Myosin II isoforms. (IC50 ± SD from triplicate samples: smooth, 9.0 ± 4.8 nM; nonmuscle IIB, 76 ± 4.9 nM; cardiac, 2,600 ± 240 nM; fast skeletal, 11,300 ± 840 nM). (E) CK-571 binding to SMM is nucleotide-dependent and is stronger in the presence of AMPPNP in the binding pocket. (F) CK-571 inhibits ATP hydrolysis as indicated by the reduced phosphate burst size in single turnover chemical quench experiments (mol Pi/mol myosin ± SD from 5 to 10 samples: DMSO, 1.04 ± 0.043; CK-571, 0.06 ± 0.016; blebbistatin, 1.18 ± 0.05). (G) CK-571 relaxes smooth muscle independent of calcium activation, as shown by the ability to relax skinned tail artery rings preactivated by thiophosphorylation (n = 8–12, plotted is mean ± SEM). (H) CK-571 relaxes airway smooth muscle in vivo, inhibiting MCh-induced bronchoconstriction in naïve dogs following dry powder insufflation (100 µg/kg). For comparison, the clinically relevant β2-adrenergic agonist albuterol was dosed by nebulization (10 µg/kg) (data are mean ± SEM, n = 4).
Fig. S1.
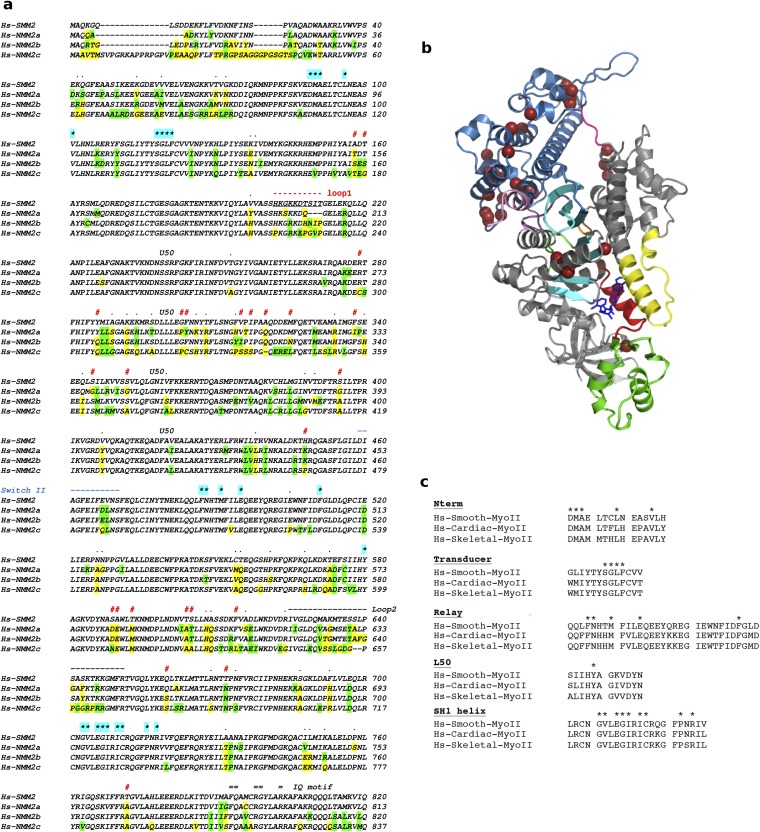
(A) Sequence alignment of the heavy chain of Hs-SMM2 and Hs-NMM2a, -b, and -c. Residues that are not conserved are shown in yellow; similar residues that are not conserved are green. Residues involved in the binding of CK-571 are labeled with a star highlighted cyan: none of these residues are different in sequence between SMMs and NMMs. Human SMM2 and NMM2a, NMM2b, and NMM2c share, respectively, 83%, 84%, and 75% sequence identity and 91%, 91%, and 88% sequence similarity in their MD. Note the main sequence differences noted with the symbol # and the sequence for loop1 and switch II indicated, respectively, in red and blue. In between these two elements, the sequence corresponds to residues of the U50 subdomain. The kinetics of the allosteric rearrangements occurring in the recovery stroke depends on the full sequence of the myosin motor and determines the specific kinetic rates of ATP hydrolysis. In transient kinetic studies, the rate that measures both the recovery stroke isomerization and hydrolysis (k+3 + k-3) is faster for SMM2 (43) (k+3 + k-3 = 50 s−1) compared with NMM2A (44) (k+3 + k-3 = 14 s−1) and NMM2B (45) (k+3 + k-3 = 16.7 s−1). Differences in the U50 subdomain or the loop1 sequence are the most likely candidates to influence the overall dynamics in this transition, in particular by controlling the rearrangements necessary in the transducer for this transition (Fig. S4C). Thus, specificity in the inhibition depends on the dynamics of this reversible transition that is modulated by residues that are far from the drug binding site. (B) Residues that are not conserved between Hs-SMM2 and NMM2 and are likely candidates to modulate the recovery stroke are shown with brown spheres on the SMM/CK-571 structure (see also Movie S4). (C) Sequence alignment of Human Skeletal, Cardiac, and Smooth Myosin II. Residues involved in CK-571 binding are labeled with a star. Note that they are all identical in these different myosins.
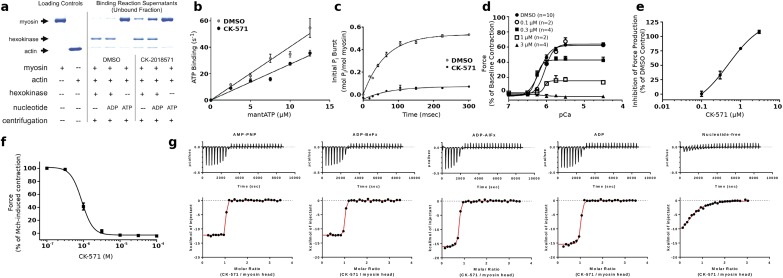
A major conformational change, the powerstroke, occurs when myosin is strongly bound to actin and drives the swing of the myosin lever arm during force production (Fig. 1A). The myosin motor cycle also has states (blue, Fig. 1A) of low affinity for actin when ATP is bound. While populating these states, the lever arm undergoes a recovery stroke to prepare the motor for producing force upon actin rebinding. A series of in vitro assays established that CK-571 traps myosin in one of the ATP-bound intermediates of the recovery stoke in which the motor is dissociated from actin. Indeed, we show that the drug binds to a state of low affinity for actin (Fig. S2A) with minimal effect on ATP binding to nucleotide-free actin-bound SMM (Fig. S2B). CK-571 binds to myosin with the highest affinity in the presence of the nonhydrolyzable ATP analogs AMPPNP (0.8 ± 0.63 nM) or ADP.BeFx (3.1 ± 1.2 nM), with somewhat weaker binding observed in the presence of ADP (12 ± 4.4 nM) or the ADP.Pi analog ADP.AlFx (11 ± 1.8 nM) (Fig. 1E and Fig. S2B). The binding of nucleotide is critical for formation of a high-affinity drug–myosin complex, as the affinity is decreased >2,000-fold in the absence of nucleotide (1,900 ± 450 nM) (Fig. 1E and Fig. S2G). AlFx, a Pi analog that promotes the formation of prepowerstroke (PPS) states (with ADP.Pi trapped), probably hinders CK-571 binding, as the observed stoichiometry is only 0.71 and the apparent Kd is similar to that measured for ADP alone (i.e., 11 nM) (Fig. 1E). Importantly, the drug also prevents ATP cleavage, as shown by quench flow experiments and reduced level of Pi burst (Fig. 1F and Fig. S2C). This inhibitor thus traps the motor in an ATP state that cannot cleave ATP. CK-571 prevents completion of the recovery stroke and precludes the motor from forming states that would rebind to F-actin and produce force. By preventing the population of force-generating states, CK-571 binding relaxes muscle in vitro and in vivo (Fig. 1 G and H and Fig. S2D). In summary, CK-571 binding traps myosin in a state that cannot strongly bind F-actin and which traps ATP rather than ADP.Pi in the active site (Fig. 1F). With CK-571, the absence of ATP cleavage and thus the absence of Pi possibly being released prevent productive actin association necessary for force production. This mechanism of myosin inhibition, which stops the motor before ATP cleavage, differs drastically from those previously described for modulators of myosin activity, such as blebbistatin (Fig. 1F) or omecamtiv mecarbil, both of which are compatible with the recovery stroke. This mechanism is particularly efficient at preventing any productive interaction of the motor with the actin filament, precluding any force generation.
Fig. S2.
Characterization of the mechanism of CK-571 in vitro. (A) CK-571 (50 µM) inhibits SMM in a weak actin-binding state, as indicated by the presence of equal or greater quantities of SMM in the supernatants from actin-binding reactions containing CK-571 compared with DMSO. This is most obvious for the motor bound with ADP, which allows strong binding to F-actin in the absence of drug but not in its presence. In the presence of ATP and saturating CK-571, essentially all SMMs are dissociated. The drug binding site is not as easily reachable when no nucleotide is bound and to a lesser extent when ADP is bound. This limits the degree to which drug binding prevents actin binding in these states. (B) CK-571 has minimal effects on the rate of ATP binding to nucleotide-free actin-bound SMM (CK-571 = 2.8 µM−1·s−1; DMSO = 4.0 µM−1·s−1) as measured using mantATP (plotted is mean ± SEM for five replicates). (C) CK-571 slows the rate of ATP hydrolysis (CK-571 = 4 s−1; DMSO = 16 s−1) and reduces the size of the phosphate burst (CK-571 = 0.1; DMSO = 0.53) as measured by chemical quench flow using a 10-fold excess of ATP (plotted is mean ± SD for four replicates). (D) CK-571 inhibits Ca2+-dependent force production in skinned rat caudal artery rings (plotted is mean ± SEM). (E) Dose–response of CK-571 inhibition in skinned rat caudal artery rings at pCa 4.5 (plotted is mean ± SEM; EC50 = 0.49 µM). (F) CK-571 dose-dependently relaxes intact rat tracheal rings precontracted with 3 µM MCh (plotted is mean ± SEM; n = 12; EC50 = 0.90 µM; Hill slope = 2.9). (G) ITC experiments in the presence of AMPPNP, ADP-BeFx, ADP-AlFx, and ADP and in the absence of nucleotide.
Relaxation of Smooth Muscle by CK-571.
Therapeutically relevant SMM inhibitors must be able to promote tissue relaxation. Importantly, the inhibition of SMM-catalyzed ATP hydrolysis by CK-571 effectively relaxes smooth muscle tissue in vitro and in vivo. CK-571 inhibits force production in detergent-permeabilized skinned artery rings stimulated to contract by increasing concentrations of calcium in a dose-responsive fashion (Fig. S2 D and E). In addition, CK-571 promotes relaxation in thiophosphorylated skinned artery rings, demonstrating that the compound can relax precontracted tissue and supporting a direct effect on myosin rather than upstream signaling pathways acting through MLCK (Fig. 1G). CK-571 relaxes rat tracheal rings that have been precontracted with the muscarinic receptor agonist methacholine (MCh) with similar potency, highlighting the ability of CK-571 to penetrate and relax intact smooth muscle tissue (Fig. S2F). Relaxation of skinned and intact tissues requires higher concentrations (approximately 1 µM), consistent with a need to inhibit >95% of the SMM to achieve relaxation. The therapeutic potential of this mechanism is further supported by the ability of CK-571 to inhibit MCh-induced bronchoconstriction in naïve dogs following dry powder delivery of CK-571 (achieved dose, 100 μg/kg). The magnitude of the inhibition achieved is comparable to the clinically relevant short-acting β2-adrenergic agonist albuterol (Fig. 1H). Given that CK-571 requires higher doses and has a shorter duration of action, potency and metabolic stability are key areas for further optimization.
Mechanism for This Efficient and Highly Selective Myosin Inhibitor.
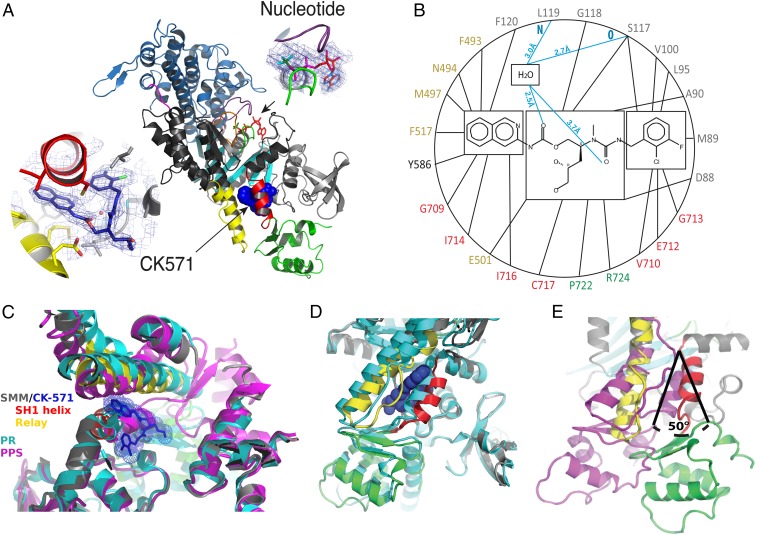
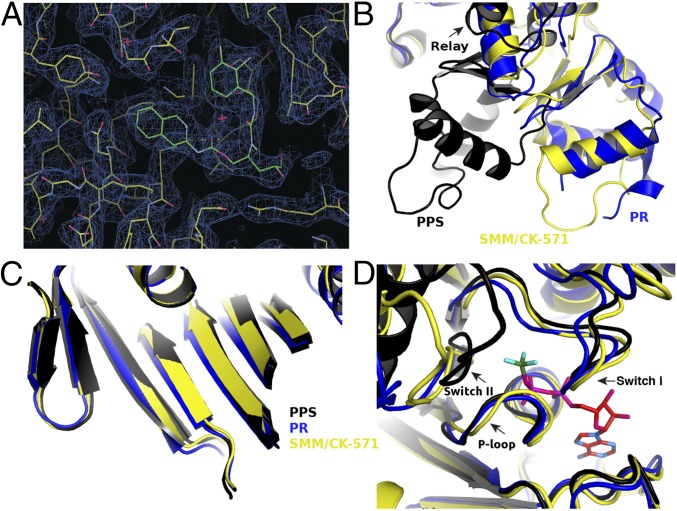
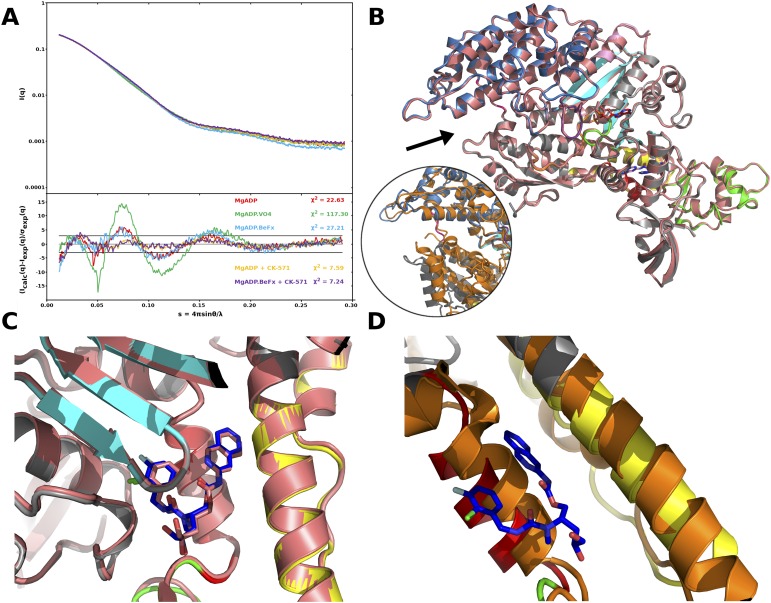
The structural basis of CK-571 action and specificity was revealed by the crystal structure of the SMM motor domain (MD) bound to CK-571 [SMM/CK-571; Protein Data Bank (PDB) ID code 5T45] at 2.8 Å resolution (Table S1). The electron density shows unambiguously that the inhibitor binds in an unexpected pocket of the MD (Figs. 2A and 3A). CK-571 is held in between two structural elements that control the lever arm position, the Relay and the SH1 helix (Fig. 2A and Movie S1), burying 1,067 Å2 of surface area in an internal pocket. It adopts a compact conformation in which the isoquinoline carbamate and the chloro-fluoro-phenyl moieties come close together via a ∼60° bend in the central aliphatic chain of CK-571 (Fig. 2A, Inset). The inhibitor has a predominant hydrophobic character and interacts via a complex network of mostly apolar contacts (Fig. 2B). There are no direct H-bonds established with protein residues. Importantly, the drug binding site of CK-571 is not accessible in either the postrigor or the PPS states (Fig. 2 C and D), as the Relay and the SH1 helix make strong interactions in these two structural states that correspond to the beginning and the end of the recovery stroke. These structural insights and the functional studies presented previously (that show that ATP binding is not affected by CK-571 whereas ATP cleavage cannot occur) show that CK-571 binds a myosin-ATP intermediate that is populated during the recovery stroke. Thus, a pocket between the Relay and the SH1 helix must open during this transition. Once bound, CK-571 traps the motor in an intermediate state, which prevents the motor from exploring states at the end of the recovery stroke that are necessary for ATP hydrolysis. Small-angle X-ray scattering (SAXS) studies were performed to demonstrate that the X-ray SMM/CK-571 structure corresponds to the state populated by the drug in solution. The SMM motor with CK-571 and either ADP or ADP.BeFx bound adopts a similar conformation in solution that also fits the scattering curve calculated from the SMM/CK-571 structure (Fig. S3A). Note that we also solved the structure of SMM/CK-571 with ADP bound. The structure is the same as that described earlier with ADP.BeFx bound. The structure supports the findings that CK-571 stabilizes myosin in an actin-unbound state, as the actin-binding pocket is open and thus the actin interface is not adequate for strong binding to F-actin (Fig. S3B). The finding that both ATP analogs and ADP can allow drug binding in this intermediate state is consistent with the early stages of the recovery stroke being explored in the absence of F-actin when ADP is bound in the active site. In contrast, the scattering curve is quite different when SMM is in the PPS state (with ADP.VO4 bound), confirming that the drug prevents access to PPS. Details of the structure compared with previously characterized ATP states prior or after the recovery stroke are shown in Figs. 2 C–E and 3. The intermediate state populated by CK-571 has similarities with the postrigor state (PR) populated at the beginning of the recovery stroke because of the position of its lever arm (in the down position). However, other MD elements are more similar to the conformation myosin motors adopt at the end of the recovery stroke. In particular, the intermediate state trapped by CK-571 differs substantially from the PR, as the central β-sheet of the MD in this intermediate adopts a conformation close to that found in states at the end of the recovery stroke (PPS; Fig. 3C and Movies S2 and S3). This β-sheet is a major part of the transducer (1) that coordinates several allosteric sites within the motor, in particular the actin-binding interface and the nucleotide-binding site. Moreover, the switch II element near the γ-phosphate of ATP has moved toward the conformation it adopts in the PPS state that allows direct interaction with the nucleotide necessary for hydrolysis. However, it remains far from this final conformation, as it adopts a position intermediate between the two end structures of the recovery stroke (PR and PPS; Figs. 1A and 3D). This result indicates that in contrast to what had been proposed from some Molecular Dynamics simulations (8), sensing the nucleotide via switch II does not correspond to the first event that would guide the repriming of the lever arm. The recovery stroke of the lever arm is not driven by keeping strong interactions between the Relay and SH1 helix, as previously proposed (8). Indeed, this mechanism (8) cannot explain how a pocket would open during the recovery stroke to allow CK-571 binding. Overall, the SMM/CK-571 structure reveals that CK-571 traps the motor in an intermediate state in which switch II stays too far from the nucleotide to stabilize the water near the γ-phosphate that allows cleavage of ATP.
Table S1.
Data collection and refinements statistics
| Crystal | SMM/CK-571 MgADP.BeFx | SMM/CK-571 MgADP | |
| Data collection | |||
| Space group | P21212 | P21212 | |
| Cell dimensions | |||
| a, b, c, Å | 72.94, 202.17, 67.02 | 73.19, 202.97, 67.56 | |
| α, β, γ, ° | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | |
| Resolution, Å | 50.00–2.65 (2.74–2.65) | 49.68–2.67 (2.77–2.67) | |
| No. reflections | |||
| Total | 202,529 | 202,527 | |
| Unique | 29,002 | 29,000 | |
| Completeness, % | 99.0 (93.8) | 99.0 (91.0) | |
| Redundancy | 6.98 (6.95) | 7.0 (6.6) | |
| Rsym | 10.2 (76.2) | 10.2 (80.72) | |
| I /σ | 14.23 (2.17) | 14.23 (1.97) | |
| Refinement | |||
| Resolution, Å | 44.35–2.80 (2.91–2.80) | 49.68–2.67 (2.77–2.67) | |
| Rwork/Rfree, % | 21.89/27.11 (29.56/37.05) | 18.47/24.40 (25.86/33.85) | |
| rmsd | |||
| Bond lengths, Å | 0.010 | 0.009 | |
| Bond angles, ° | 1.18 | 1.243 | |
| Number of atoms | |||
| Nonhydrogen, protein | 5,800 | 5,781 | |
| Drug | 35 | 35 | |
| Water | 84 | 54 | |
| Nucleotide | 31 | 27 | |
| PDB ID code | 5T45 | 5M05 |
Values in parentheses are for highest resolution shell. The structures were solved by molecular replacement with data from a single crystal.
Fig. 2.
An allosteric pocket opens to bind CK-571 during the recovery stroke. (A) Overall view of the myosin MD of the SMM/CK-571 structure in cartoon representation. CK-571 is shown in blue spheres and is located 22 Å away from the nucleotide (Right Inset: red sticks, ADP; green sticks, BeFx). CK-571 (Left Inset: 2Fo-Fc electron density) interacts with residues of the SH1 helix (red), Relay (yellow), and the Nter subdomain (light gray) as well as with Y586 from the L50 subdomain (dark gray). The other two subdomains of the motor are the U50 (blue) and converter (green). (B) Residues of the CK-571 binding site (black lines, apolar contacts; blue lines, polar contacts). Residues are indicated with the same color code as in A. (C) Comparison of the CK-571 binding pocket with the myosin PR (PDB ID code 3I5F, cyan) and PPS (PDB ID code 1BR1, magenta) states. Only the SH1 helix and the relay conformation in the SMM/CK-571 structure are compatible with the drug binding. (D) Comparison of the SMM/CK-571 structure with the myosin PR state (PDB ID code 3I5F, cyan). In the SMM/CK-571 structure, rearrangements of the Relay (yellow) and a 10° rotation of the SH1 helix (red) allow the formation of a cleft in the molecule that harbors the drug (blue spheres). (E) In the SMM/CK-571 structure, the converter is in a down position and is rotated by 50° with respect to the position it occupies in the PPS state (PDB ID code 1BR1, magenta). To compare the three structures, the proteins were aligned in C, D, and E using the N-ter domain as a reference.
Fig. 3.
Structural features of SMM2 MD in complex with CK-571 that traps an intermediate of the recovery stroke. (A) 2Fo–Fc electron density omit map, contoured at 0.7 σ, depicting the CK-571 binding site. (B) The SMM/CK-571 structure is an intermediate of the recovery stroke between the PR and PPS structures. Note in particular the difference in position of the converter in these structures. It is in a down position in the SMM/CK-571 structure (yellow), close to the position found in the PR state structures (blue), and differs greatly from the up position found in PPS structures (black). The position of the subdomains when CK-571 is bound differs from those found in previously described PR structures. Interestingly, the relay (shown in this figure) and L50 subdomain (see Movie S2) corresponds to an intermediary position between the PR and the PPS structures. (C) The central beta sheet conformation (which is part of the transducer) controls rearrangements between subdomains of the motor (1). Although the lever arm position in SMM/CK-571 is closer to the PR than the PPS position, the central beta-sheet conformation of SMM/CK-571 (yellow) differs from that of the PR state (blue), whereas its position is quite similar to that found in the PPS state (black) (Movie S2). (D) Nucleotide binding site in the SMM/CK-571, PR, and PPS structures. Note that the nucleotide binding elements switch I and P-loop are in position to bind MgATP tightly. However, in the SMM/CK-571 structure, the switch II position differs from the special PPS position necessary to promote ATP hydrolysis (black). This switch II position is far away from the nucleotide, in an intermediate position between the PR and PPS states (PPS, PDB ID code 1BR1, black; PR, PDB ID code 3I5F, blue; SMM/CK-571 is in yellow with the nucleotide in red sticks).
Fig. S3.
CK-571 stabilizes myosin in an actin-unbound state. (A) SAXS analysis of the SMM/CK-571 structure. The SMM S1 fragment was analyzed by SAXS in several nucleotide-bound conditions (MgADP, MgADP.VO4, MgADP.BeFx, MgADP + CK-571, and MgADP.BeFx + CK-571) that allow population of different states of the motor cycle. The scattering pattern of the SMM S1 fragment that corresponds to the SMM/CK-571 crystal structure we have obtained was calculated and fit against the experimental scattering curves. (Upper) Experimental scattering intensities. (Lower) Reduced residuals of the least-squares fits on a linear scale. Two horizontal lines define the ±3 σ range. The χ2 value reflects the discrepancy between theoretical and experimental curves. Note how similar the MgADP + CK-571 and MgADP.BeFx + CK-571 curves (purple, yellow) are and how they differ from that of MgADP.VO4 (green). (B) Comparison between the SMM/CK571 structures bound to MgADP.BeFx (colored) and MgADP (salmon). The two structures can be superimposed with an overall root-mean-square deviation (rmsd) of 0.32 Å for 674 Cα atoms. Both structures have an open actin-binding cleft (black arrow). This actin-binding cleft is compared with that found in the rigor-like state in which the cleft is closed (Inset, orange, PDB ID code 3I5G), after superimposition of the U50 subdomains. Note the difference in position of the L50 subdomain (gray/orange). (C) Detail of the CK-571 binding pocket for both SMM/CK-571 structures with either ADP or ADP.BeFx bound. (D) The CK-571 binding pocket in the rigor state (orange) is closed, lowering the affinity of the drug for this state.
Intriguingly, sequence analysis reveals that all 24 residues forming the inhibitor’s binding site are strictly conserved among SMM, NMM, fast skeletal, and cardiac muscle myosin II, thus indicating that the inhibitor’s selectivity is not based on specific sequence recognition at the drug site (Fig. 1D and Fig. S1). Furthermore, no sequence differences exist among the residues located along the putative pathways by which the compound would reach this site (Movie S4). In summary, comparison of the SMM/CK-571 structure with other myosin II structures (Table S2), SAXS data, and functional characterization of the motor inhibited by CK-571 demonstrate that the drug traps myosin in an intermediate ATP state of the recovery stroke incapable of ATP hydrolysis and thus blocks the motor from cycling, killing force production (Fig. 4).
Table S2.
Myosin II structures used for analysis and comparison
| PDB code | Myosin structure |
| PR, before the recovery stroke | |
| 3I5F | Squid skeletal muscle myosin II in postrigor |
| 1SR6 | Argopecten skeletal muscle myosin II in postrigor |
| 2OTG | Placopecten skeletal muscle myosin II in postrigor |
| 2MYS | Chicken skeletal muscle myosin II in postrigor |
| 4DB1 | Human beta cardiac muscle myosin II in postrigor |
| 4P7H | Human beta cardiac muscle myosin II in postrigor |
| PPS, after the recovery stroke | |
| 1QVI | Argopecten skeletal muscle myosin II in PPS |
| 1BR1 | Chicken smooth muscle myosin II in PPS |
| 2YCU | Human non muscle myosin IIC in PPS |
Fig. 4.
MD rearrangements upon the recovery stroke. The myosin MD is functionally made up of four subdomains linked by connectors that rapidly change their conformation upon transitions in the motor cycle. In particular, switch II (orange) is found close to the nucleotide. The Relay (yellow) and the SH1 helix (red) connect the MD to the converter (green) and control its swing. The converter and the adjacent neck region constitute the lever arm that amplifies MD rearrangements linked to nucleotide or actin binding. Binding of CK-571 (blue star) blocks the motor isomerization in an intermediate of the recovery stroke. Thus, unlike prevailing models, lever arm repriming corresponds to an uncoupling of these connectors and leads to transducer rearrangements that are not initiated by a switch II closure. Lever arm swing in this step is not controlled by mechanical control of subdomain orientation by coupled connectors.
CK-571 Reveals the Existence of an Allosteric Pocket Adjacent to the Binding Site of the Myosin Activator, Omecamtiv Mecarbil.
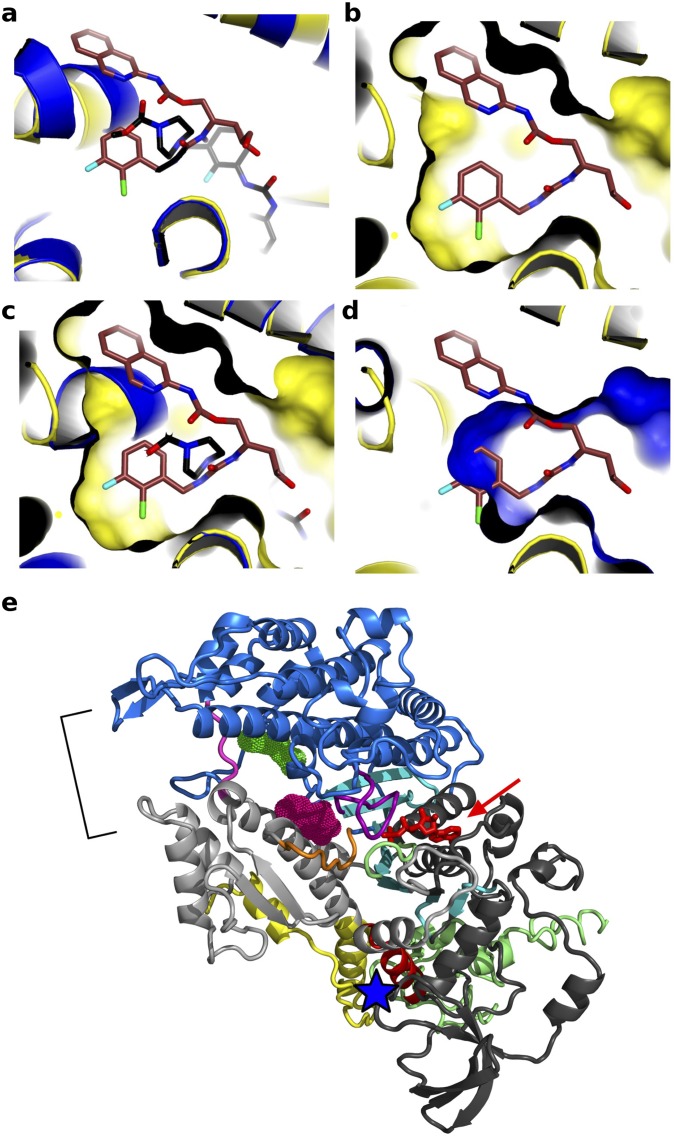
A structure of cardiac myosin II bound to the activator omecamtiv mecarbil (2) was recently solved (9) with the motor in the PR state (Fig. 1A). This activator of cardiac myosin increases force production by the heart by increasing the transition of cardiac myosin from a weakly to strongly actin-bound state, as evidenced by a faster rate of Pi release (2, 10), and by slowing down the following steps of the powerstroke. In the crystal structure, omecamtiv mecarbil binds to cardiac myosin (9) in a surface pocket accessible in the PR state (Fig. 1A). The omecamtiv mecarbil pocket only slightly overlaps with the CK-571 pocket. Indeed, most of the CK-571 site is not accessible in the PR state and opens up only during the recovery stroke (Fig. 2C and Fig. S4). The most deeply buried omecamtiv mecarbil groups (carboxymethyl-piperazine and fluoro-benzene rings) are located close to the SH1 helix where the CK-571 binding site opens during the transition (Fig. S4A). The rest of the omecamtiv mecarbil groups (amino-carbamoyl and methyl-pyridinyl) are located closer to the surface of the molecule near the converter domain. In contrast, the binding site shown for CK-571 is much deeper and quite different from that of omecamtiv mecarbil (Fig. S4). It is unlikely that in fact omecamtiv mecarbil would occupy the CK-571 pocket during the recovery stroke. Unlike CK-571, omecamtiv mecarbil does not block the motor in an intermediate of the recovery stroke but increases the population of the end states of the recovery stroke (PPS), in which the lever arm is up. It must thus occupy a pocket that is accessible in the PPS state. This is not the case for the CK-571 pocket, which is nonexistent in the PPS state. In contrast to CK-571, omecamtiv mecarbil is compatible with the recovery stroke, accelerates Pi release, and then slows the following steps of the powerstroke (10). Thus, cardiac muscle contracts more strongly when the drug is present (2) and omecamtiv mecarbil acts as an activator of force production. Together, the omecamtiv mecarbil and CK-571 binding sites on myosin thus highlight the potential of targeting the internal pockets found near the SH1 helix and the Relay to design highly specific activators and inhibitors of myosin motors for the treatment of numerous diseases in which myosin motors are a target.
Fig. S4.
Modulators of myosin activity differently influence the recovery stroke and its ability to produce force. Shown is a representation of the CK-571 (brown) and omecamtiv mecarbil (PDB ID code 4PA0, black) binding pockets. The binding site for CK-571 (orange) is distinct from that of myosin activator omecamtiv mecarbil (blue). Note that there is poor superimposition between the structural elements that surround the ligand binding pockets (A) and none of the rings of CK-571 fit in the pocket in which omecamtiv mecarbil binds (B–D). Note in particular how the binding surface of CK-571 (B) differs from that of omecamtiv mecarbil (D). (E) Representation of the Dictyostelium discoideum myosin II structure (PDB ID codes 2JHR and 1YV3) in a PPS state showing that blebbistatin (pink) and pentabromopseudilin (green) occupies surface pockets of the PPS state that are close to the ATP (red arrow) or the actin binding site (bracket), respectively. These sites are distant from the CK-571 site (blue star). Note that no pocket is available for CK-571 binding in the PPS state.
Discussion
The atomic structure of SMM/CK-571 reveals that the drug binds in an allosteric pocket of the myosin motor that opens up transiently during the recovery stroke of the myosin motor.
The drug introduces itself in a pocket that has not been found in the previously described structures of the motor. Furthermore, this pocket becomes available transiently when the ATP-bound motor explores states of the recovery stroke. The intermediate state of the recovery stroke is then stabilized and traps the motor in a structural state from which myosin cannot bind actin nor progress toward ATP hydrolysis. Drug binding blocks the recovery stroke transition via stabilization of an intermediate through interactions with two main connectors (Relay and SH1 helix) that link the motor and the lever arm. It thus prevents the motor from reaching the states at the end of the recovery stroke (PPS) that are required to allow ATP cleavage. This efficiently blocks the motor in a state that is inappropriate for force production upon interaction with actin, as ATP is not cleaved, affinity for F-actin is low, and the lever arm is not primed. Previously described myosin inhibitors, such as blebbistatin or pentabromopseudilin (3, 11, 12), trap the motor in the PPS state at the beginning of the force production event (Fig. S4E). These drugs do not prevent ATP cleavage and bind surface pockets available in the PPS state of cytoplasmic and muscle myosin II with lower affinity and specificity (13) compared with CK-571. In contrast, stabilization of an intermediate state during the recovery stroke by CK-571 blocks not only the motor rearrangements required for lever arm repriming but also those needed for the closure of the nucleotide binding site that are required for ATP hydrolysis. By keeping the myosin heads away from any possible interaction with F-actin, precluding ATP hydrolysis and repriming of the lever arm, CK-571 prevents these heads from participating in force production. CK-571 exemplifies an innovative and efficient mechanism to achieve complete relaxation of smooth muscle.
Interestingly, all of the residues of the CK-571 binding pocket are conserved among the different human myosin IIs (cytoplasmic, smooth, skeletal, and cardiac). Thus, the selectivity of CK-571 must result from residues outside the binding pocket that modulate the kinetics and energy landscape of the recovery stroke (Fig. S1). What differs among myosins is the ability of the drug to exploit a transient pocket formed during the recovery stroke. These differences may arise from small sequence changes between these molecular motors that influence the lifetimes and the nature of the intermediate states. CK-571 inhibits SMM2 with ∼eightfold greater potency than its close homolog, NMM, despite the lack of any sequence differences within the drug-binding pocket or any of the surrounding amino acids. CK-571 provides a remarkable demonstration of the principle of allosteric modulation of enzyme function by a small molecule with high specificity and high affinity. The SMM/CK-571 structure reveals a way to achieving specificity by targeting transient pockets likely to have different lifetimes in different myosins with sequence differences even remote from the drug-binding site. Even highly homologous myosins can be specifically inhibited as long as they have kinetics that differ significantly from each other. The SMM/CK-571 structure provides important insights into the allosteric mechanism of the recovery stroke transition, the critical step in which the ATP-bound motor, detached from actin, primes its lever arm to prepare myosin for its powerstroke. During this isomerization, the lever arm undergoes a drastic reorientation while the SH1 helix and Relay connectors explore different conformations. The trapped intermediate in the SMM/CK-571 structure sheds light on the recovery stroke mechanism, refuting models supporting tight coupling between the nucleotide binding site and the lever arm. Weak interactions between the Relay and SH1 helix are necessary for pocket opening and CK-571 binding. Models in which switch II interactions with the nucleotide direct the swing of the converter through a strong coupling mechanism between the Relay and SH1 helix cannot explain how CK-571 may bind. Rearrangements of the Relay and the SH1 helix during lever arm priming can allow the formation of a large pocket in between them, which traps the drug in place (Movies S1–S4). In the rigor, PR, and PPS states (Fig. 1A), these two connectors interact directly, reducing the ability for this allosteric pocket to be formed in these states and thus resulting in poor affinity of these structural states for CK-571.
Molecular simulations have provided drastically different models for the recovery stroke. A coupling mechanism discussed earlier in which the active site (switch II closure) would trigger reorientation of the converter is not compatible with the SMM/CK-571 structure (8, 14). In contrast, the structure is consistent with a Targeted Molecular Dynamics approach, in which the converter rotates in the first stage of the transition before switch II closure (15, 16). Transient time-resolved FRET data are also compatible with this scenario (17), as are previous studies that indicate the potential melting of the SH1 helix in this transition (18, 19). Furthermore, the SMM/CK-571 structure shows that the lever arm repriming does not occur via switch II sensing of the γ-phosphate. This structure reveals that the recovery stroke (a large 9-nm movement) necessary to reprime the motor is likely to capture thermal energy to explore and populate uncoupled states that allow a lever arm swing. This differs from a mechanism in which such large movement would occur via the use of chemical energy linked to formation of bonds along a pathway from the active site to the lever arm that would trigger precise allosteric rearrangements to control lever arm movement. The emerging model of the recovery stroke thus differs greatly from current ones that suggest allosteric communication between the nucleotide binding site and a distal part of the motor to reprime the lever arm.
The recovery stroke is an essential step of the cycle of all myosins. Highly specific drugs that trap intermediates of this transition can thus be designed for other myosins on the basis of the SMM/CK-571 structure. Structural studies with several myosin isoforms, and in particular the reverse motor myosin VI, have shown that the conformation of the motor in the states prior and after the recovery stroke are similar, even when the direction of the lever arm swing differs (20). Selectivity among different myosin classes may be achieved by exploiting sequence differences in the CK-571 binding pocket (Fig. S5). This allosteric binding site has thus the potential to guide the design of novel class-specific myosin inhibitors. In particular, myosin VI and myosin X are interesting pharmaceutical targets due to their recently established role in cell migration and metastasis (21–24).
Fig. S5.
Sequence alignment of Hs-Smooth Myosin II (Hs-SMM) and different human myosins. Out of 24 residues that directly interact with CK-571, 10 are not conserved in HsMyoVI, 10 are not conserved in MyoXVIIIa, 7 are not conserved in MyoVIIa, 5 are not conserved in MyoVa, and 7 are not conserved in MyoX. Residues involved in CK-571 binding are labeled with a star. Residues that are similar but not identical are in blue; residues that are not conserved are in yellow.
Methods
Crystal Structure Determination.
Chemicals.
CK-571 was synthesized by the Medicinal Chemistry group of Cytokinetics. Other chemicals were from commercial sources and generally American Chemical Society (ACS) grade or higher.
High-throughput screens were conducted using chemically diverse libraries of commercially available small molecules, resulting in the identification of an initial hit with modest biochemical potency (IC50 = 17 µM) against SMM. Through extensive medicinal chemical optimization (>3,000 analogs in series), the intrinsic biochemical potency of the chemical series was greatly improved, resulting in an IC50 of ∼9 nM for compound CK-571. Inhibitors with higher selectivity for SMM versus NMMs were sought during optimization.
Purification of myosins.
Native chicken SMM was purified from cryoground gizzards (Pel-Freez Biologicals) using previously described methods (25). Soluble chicken gizzard subfragment 1 (S1) was obtained by limited papain digestion of filamentous myosin terminated with 1 mM iodoacetic acid. Recombinant chicken SMM MD (26) (residues Met1-790) and MD + essential light chain (MDE heavy chain residues Met1-Leu819, GenBank NP_990605.2; Essential Light Chain M15645.1) were expressed with a C-terminal FLAG tag following a short linker (sequence, GSDYKDDDDK). The chicken MDE protein was prepared by coexpression of the untagged chicken essential light chain. Recombinant human SMM MDE (heavy chain residues Met1-Leu820, isoform SM-A, GenBank NP_002465) was expressed with a C-terminal 6xHis tag. Human NMM IIB and essential light chain (MDE, heavy chain residues Met1-Pro843, isoform 2, GenBank NP_005955.3) were expressed with a C-terminal FLAG tag (sequence, DYKDDDDK). The human MDE proteins were prepared by coexpression of the untagged human SMM essential light chain. The isoforms of the recombinant human SMM and NMMs both lacked the seven-amino acid insert in loop 1. FLAG-tagged recombinant proteins were purified from baculovirus-infected SF9 cells by ammonium sulfate precipitation and affinity chromatography as described (26). Rabbit fast skeletal and bovine cardiac myosins were purified from native tissues (Pel-Freez Biologicals) and digested in their filamentous forms with chymotrypsin to produce S1 fragments (27). For measuring specificity and potency, soluble myosin fragments were covalently cross-linked to excess bovine cardiac actin using carbodiimides 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/N-hydroxysuccinimide esters (NHS) (28, 29) and flash-frozen at –80 °C before use in ATPase assays.
Crystallization and structure determination.
Purified chicken SMM MD (residues 1–790) at 110 µM was incubated with 2 mM MgADP and 2 mM BeFx for 30 min on ice. The protein was mixed with CK-571 at a final concentration of 547 µM in the presence of 10% (vol/vol) DMSO, and then it was incubated for 1 h on ice. Samples were centrifuged at 11,000 × g for 15 min before crystallization. Hanging drops of SMM/CK-571 were set up by mixing equimolar ratio of the complex and of the reservoir containing 5% (wt/vol) Peg8K, 50 mM Bicine (pH 8.2), and 10% (vol/vol) DMSO. First, crystals appeared after 10 d at 4 °C and were subsequently optimized using microseeding technique. The optimized crystals were cryo-cooled in a final solution containing 8% (wt/vol) Peg8K, 50 mM Bicine (pH 8.2), 10% (vol/vol) DMSO, 25% (vol/vol) glycerol, and 547 µM CK-571. Similarly, crystals were obtained with protein containing only MgADP bound in the active site.
X-ray datasets were collected at the Proxima 1 beamline at the Synchrotron SOLEIL at 100K and at a wavelength of 0.97857 Å. The diffraction datasets were indexed and scaled with XDS (30). Molecular replacement solution was obtained with Phaser, CCP4 suite (31, 32), using PDB ID code 1BR1 (26) as the search model. The region of the converter, the Relay, and the SH1 helix were excluded from the search model. These regions were subsequently built in electron density using the ARP/wARP program (33, 34). The coordinate and geometry constraint files for the ligand were created with Sketcher of the CCP4 suite and Elbow of the Phenix suite (35, 36). Model building and refinement were carried out with the Coot (37) and Buster programs (38). In the final Ramachandran plot of the MgADP.BeFx structure, 96.29%, 3.28%, and 0.43% of residues were in the favored, allowed, and outliers regions, respectively. Figures and movies were made using PyMol (Version 1.5.0.4; www.pymol.org).
Note that crystals with CK-571 bound can be obtained in the presence of MgADP as well as other ATP analogs, and the structure solved with ADP bound is essentially the same as that we report here with MgADP.BeFx bound (see Fig. S3 and statistics on Table S1).
SAXS experiments.
SAXS data were collected on the SWING beamline (synchrotron SOLEIL). Purified SMM S1 fragment was incubated with 2 mM MgADP for 30 min on ice. The protein was then incubated with either 5 mM CK-571 or 10% DMSO for 1 h on ice, followed by the incubation with 2 mM vanadate or 2 mM BeFx for 30 min on ice when necessary. All samples were centrifuged at 20,000 × g for 10 min at 4 °C before the analysis. We injected 40 µL of the protein at 3 mg/mL between two air bubbles using the autosampler robot. We averaged 35 frames of 1.5-s exposure, and buffer scattering was subtracted from the sample data. The SMM/CK-571 S1 model was built using the SMM/CK-571 MD and the helix and ELC from previous smooth myosin structures (PDB ID code 1BR1) using the converter position as a reference. The theoretical SAXS curve of this model was calculated with CRYSOL (39) and compared based on the quality of their fits against the different experimental curves.
Biochemical Assays.
ATPase assays.
Steady-state ATPase assays were performed at 22 °C in PM12 buffer (12 mM K-Pipes, 2 mM MgCl2, pH 6.8) containing 250 µM ATP. Basal and actin-stimulated ATPase activities were measured using a spectrophotometric ATPase assay that couples ADP production to the oxidation of NADH using pyruvate kinase and lactate dehydrogenase (2). SMM S1 concentration was estimated by OD280 in 6 M guanidine HCl using a molar absorption coefficient of 95,793 M−1·cm−1 calculated based on the sequence of chicken MYH11 (1–843) and light chain MYL6. Specificity and intrinsic potency of CK-571 was determined using EDC/NHS-cross-linked Acto-S1 proteins (see Crystal Structure Determination, Purification of myosins) as well as a highly sensitive spectrofluorometric ATPase assay that couples ADP production to the conversion of Amplex Red into resorufin using pyruvate kinase, pyruvate oxidase, and horseradish peroxidase while regenerating ATP. Hydrolysis rates were normalized using reactions containing an equivalent concentration of DMSO (activity, 100%) and fit with a four-parameter logistic curve (GraphPad Prism) without additional background subtraction or data manipulation.
Actin cosedimentation assay.
Myosin binding to actin was measured by depletion of soluble myosin from binding reactions using 3 µM recombinant chicken SMM MDE (1–819) and 6 µM bovine cardiac actin in PM12 buffer. ATP and ADP were present at 1 mM where indicated, and hexokinase (10 U/mL) and glucose (2 mM) were added to nucleotide-free and ADP reactions to deplete residual ATP. Phalloidin (6 µM) was included to stabilize actin filaments. Reactions were allowed to equilibrate at 22 °C for 10 min before centrifugation (540,000 × g, 30 min), with ATP added just before centrifugation to minimize hydrolysis. Supernatants were analyzed by SDS/PAGE followed by staining with Coomassie brilliant blue.
Transient kinetics.
Chemical hydrolysis of ATP was measured in PM12 buffer at 25 °C by rapid mixing quench-flow (Biologic SFM/400, Bio-logic Inc.) under multiple turnover conditions and by manual quench in single turnover conditions (as shown in Fig. 1F). Native chicken gizzard SMM S1 (15 µM) was preincubated with DMSO or CK-571 in DMSO (25 µM) before rapid mixing with ATP (150 µM), aging, and quenching with 0.6 M perchloric acid. ATP hydrolysis was monitored using malachite green to quantify total acid-labile phosphate (40). The effect of CK-571 on ATP hydrolysis was further verified by measuring the burst size under single turnover conditions. Myosin (30 µM) was preincubated with DMSO (2%) or CK-571 in DMSO (50 µM), manually mixed 1:1 with ATP (10 µM), aged for 5 s, and then quenched by addition of perchloric acid (0.3 M final) before phosphate detection using malachite green.
Mant-ATP binding was measured in PM12 buffer at 25 °C by rapid mixing stopped flow (SF61DX2, TdK Scientific) (Fig. S2C). Mant-ATP fluorescence was monitored by excitation at 360 nm while monitoring emission through a 400-nm long pass filter. We mixed 2 μM SMM S1 and 6 μM F-actin to form the rigor state, and 6 μM CK571 or DMSO was also mixed before addition of Mant-ATP at different concentrations.
Isothermal titration calorimetry.
Isothermal titration calorimetry (ITC) experiments were carried out using a Micro-Cal Auto ITC HT microcalorimeter (Microcal Inc., now Malvern, Inc.) at 10 °C. A solution of 125 μM CK-571 in 12 mM Pipes (pH 6.8), 2 mM MgCl2, 5 mM β-mercaptoethanol, and 3% DMSO (pH 6.8) was titrated into the sample cell, which contained 8 μM chicken gizzard SMM S1 in the same buffer. SMM S1 concentration was estimated by OD280 in 6 M guanidine HCl using a molar absorption coefficient of 95,793 M−1·cm−1 calculated based on the sequence of chicken MYH11 (1–843) and light chain MYL6. Injections (10 µL) were made every 300 s. To correct for the heats of dilution and slight buffer mismatches between the titrant and sample, the average heat signal from the last three injections at the end of the experiment (when binding was saturated) was subtracted from all values. Data collection and analysis was performed using the modified Origin software included with the instrument, using a single binding-site model. Nucleotides and nucleotide analogs were present at 2 mM. To measure affinity in the absence of nucleotide, apyrase (Sigma A-6535) was included in the myosin sample at 17 µg/mL.
Tissue and in Vivo Assays.
Animal care and welfare.
Animals used in this study were maintained in accordance with the Guide for the Care and Use of Laboratory Animals of the Institute (National Research Council) (42) and under the supervision of the Institutional Animal Care and Use Committee of Cytokinetics for rodent studies or Lovelace Respiratory Research Institute for canine studies.
Skinned ring contractility.
Endothelium-denuded rat tail artery segments were cut into 3-mm helical rings, mounted on an isometric force transducer with a resting tension of 0.5 g, and incubated for 30 min at room temperature in normal Hepes–Tyrode buffer [135 mM NaCl, 6 mM KCl, 1.2 mM MgCl2, 12 mM Hepes, 2.5 mM CaCl2, 0.21% glucose (wt/vol), pH 7.4]. Tissues were skinned by incubation with skinning solution [30 mM TES, 50 mM KCl, 5 mM K2EGTA, 5.1% sucrose (wt/vol), 0.5 mM DTE, 1% Triton X-100, pH 6.9] for 1 h at room temperature. Tissues were preincubated in assay buffer (30 mM TES, 5.6 mM MgCl2, 75 mM K-propionate, 3.8 mM ATP, 16 mM creatine phosphate, 15 U/mL creatine phosphokinase, 0.5 mM DTT, 4 mM K-EGTA, and sufficient calcium to yield the desired free calcium concentrations, pH 6.9) containing DMSO or CK-571 in DMSO for 15 min before the addition of calcium. The force generated at the plateau of each calcium condition was recorded, and data were presented as a percent change from the baseline values (41). Dose–response data were fit with a three-parameter equation (assumed Hill slope, 1).
Thiophosphorylation assay.
Triton-permeabilized endothelium-denuded rat tail artery preparations (see Skinned ring contractility) were prepared and mounted as above and then incubated in rigor solution containing ATP-γ-S (1 mM) for 10 min. Tissues were preincubated with CK-571 for 15 min before the addition of ATP. ATP-induced contraction was measured for 60 min, and the relaxation was expressed as percentage of the maximum force.
Intact tracheal ring contractility.
Rat trachea were dissected and placed into cold Kreb’s–Henseleit buffer (117.5 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.18 mM MgSO4, 2.5 mM CaCl2, 25 mM NaHCO3, 11 mM glucose) equilibrated with 95% O2/5% CO2. Trachea were cut into 2-mm rings and mounted in a tissue bath (Radnoti LLC) containing Kreb’s–Henseleit buffer equilibrated with 95% O2/5% CO2 at 37 °C. Rings were stretched to a baseline isometric tension of 2 g and then induced to contract with a submaximal dose of MCh (3 µM). Rings were relaxed by treatment with ascending doses of CK-571 in DMSO. Force values were normalized to the initial MCh-induced value for each ring. Dose–response data were fit with a four-parameter equation.
Bronchoconstriction measurements in naïve dogs.
Naïve beagles (10.5–14 kg, 2.6–3 y of age) were used for this study. The response of naïve beagles (n = 8, 10.5–14 kg, 2.6–3 y of age) to increasing 1/2 log doses of inhaled MCh (0.3, 1, 3, 5, 10, 30, 50, 100, etc. mg/mL), was characterized to determine the concentration that induced a 200–250% increase in pulmonary resistance. A subset of four animals (two male, two female) were selected for further study. An abbreviated MCh dose–response (up to 3 doses) was performed to confirm the dose of MCh that induces a 200–250% increase in pulmonary resistance. Fifteen minutes later, a single dose of MCh was again given to reconfirm the response. The response to this challenge was used to compare all subsequent MCh challenges following test article treatment. Treatment with inhaled CK-571 (100 µg/kg via insufflator) or albuterol (10 µg/kg via nebulizer) was given 15 min following the single MCh challenge. MCh challenge was repeated at 5, 20, 40, 60, 90, 120, 180, and 240 min posttest article treatment. The repeated MCh challenges were aimed at evaluating the duration of action of the test article. The dogs were rested at least ∼7–14 d in between each dose to allow time for washout of the previous dose.
Supplementary Material
Acknowledgments
We thank Pierre Legrand and Andrew Thompson, as well as beamline scientists of Proxima 1 (Source Optimisée de Lumière d'Énergie Intermédiaire du Lure synchrotron) for excellent support during data collection. A.M.H. was supported by grants from Fondation de la Recherche Médicale, Agence Nationale de la Recherche, Association Française Contre les Myopathies, and Ligue Contre le Cancer. The A.M.H. team is part of Laboratoire d'Excellence CelTisPhyBio:11-LBX-0038, which is part of the Initiative d'Excellence Paris Science Lettres (ANR-10-IDEX-0001-02 PSL).
Footnotes
Conflict of interest statement: S.C. and X.Q. are former employees and potential shareholders of Cytokinetics, Inc. J.J.H., G.C., P.-P.L., B.P.M., and F.I.M. are current employees and shareholders of Cytokinetics, Inc. E.B., K.R., and C.R. received research support from Cytokinetics, Inc.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5T45 and 5M05).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609342113/-/DCSupplemental.
References
- 1.Sweeney HL, Houdusse A. Structural and functional insights into the Myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- 2.Malik FI, et al. Cardiac myosin activation: A potential therapeutic approach for systolic heart failure. Science. 2011;331(6023):1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond LM, Tumbarello DA, Kendrick-Jones J, Buss F. Small-molecule inhibitors of myosin proteins. Future Med Chem. 2013;5(1):41–52. doi: 10.4155/fmc.12.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green EM, et al. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351(6273):617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berair R, Hollins F, Brightling C. Airway smooth muscle hypercontractility in asthma. J Allergy (Cairo) 2013;2013:185971. doi: 10.1155/2013/185971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowell ML, Lavoie TL, Solway J, Krishnan R. Airway smooth muscle: A potential target for asthma therapy. Curr Opin Pulm Med. 2014;20(1):66–72. doi: 10.1097/MCP.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 7.Wedzicha JA, Decramer M, Seemungal TA. The role of bronchodilator treatment in the prevention of exacerbations of COPD. Eur Respir J. 2012;40(6):1545–1554. doi: 10.1183/09031936.00048912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koppole S, Smith JC, Fischer S. The structural coupling between ATPase activation and recovery stroke in the myosin II motor. Structure. 2007;15(7):825–837. doi: 10.1016/j.str.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Winkelmann DA, Forgacs E, Miller MT, Stock AM. Structural basis for drug-induced allosteric changes to human β-cardiac myosin motor activity. Nat Commun. 2015;6:7974. doi: 10.1038/ncomms8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, White HD, Belknap B, Winkelmann DA, Forgacs E. Omecamtiv Mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry. 2015;54(10):1963–1975. doi: 10.1021/bi5015166. [DOI] [PubMed] [Google Scholar]
- 11.Allingham JS, Smith R, Rayment I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat Struct Mol Biol. 2005;12(4):378–379. doi: 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- 12.Fedorov R, et al. The mechanism of pentabromopseudilin inhibition of myosin motor activity. Nat Struct Mol Biol. 2009;16(1):80–88. doi: 10.1038/nsmb.1542. [DOI] [PubMed] [Google Scholar]
- 13.Limouze J, Straight AF, Mitchison T, Sellers JR. Specificity of blebbistatin, an inhibitor of myosin II. J Muscle Res Cell Motil. 2004;25(4-5):337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- 14.Elber R, West A. Atomically detailed simulation of the recovery stroke in myosin by Milestoning. Proc Natl Acad Sci USA. 2010;107(11):5001–5005. doi: 10.1073/pnas.0909636107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H, Ma L, Yang Y, Cui Q. Mechanochemical coupling in the myosin motor domain. I. Insights from equilibrium active-site simulations. PLOS Comput Biol. 2007;3(2):e21. doi: 10.1371/journal.pcbi.0030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daily MD, Yu H, Phillips GN, Jr, Cui Q. Allosteric activation transitions in enzymes and biomolecular motors: Insights from atomistic and coarse-grained simulations. Top Curr Chem. 2013;337:139–164. doi: 10.1007/128_2012_409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nesmelov YE, et al. Structural kinetics of myosin by transient time-resolved FRET. Proc Natl Acad Sci USA. 2011;108(5):1891–1896. doi: 10.1073/pnas.1012320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houdusse A, Kalabokis VN, Himmel D, Szent-Györgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: A novel conformation of the myosin head. Cell. 1999;97(4):459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 19.Himmel DM, et al. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: Further insights into the mechanics of the motor. Proc Natl Acad Sci USA. 2002;99(20):12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweeney HL, Houdusse A. Myosin VI rewrites the rules for myosin motors. Cell. 2010;141(4):573–582. doi: 10.1016/j.cell.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 21.Dunn TA, et al. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006;169(5):1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida H, et al. Lessons from border cell migration in the Drosophila ovary: A role for myosin VI in dissemination of human ovarian cancer. Proc Natl Acad Sci USA. 2004;101(21):8144–8149. doi: 10.1073/pnas.0400400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knudsen B. Migrating with myosin VI. Am J Pathol. 2006;169(5):1523–1526. doi: 10.2353/ajpath.2006.060712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adhes Migr. 2011;5(5):421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellers JR, Pato MD, Adelstein RS. Reversible phosphorylation of smooth muscle myosin, heavy meromyosin, and platelet myosin. J Biol Chem. 1981;256(24):13137–13142. [PubMed] [Google Scholar]
- 26.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94(5):559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 27.Margossian SS, Lowey S. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 1982;85(Pt B):55–71. doi: 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- 28.Mornet D, Bertrand R, Pantel P, Audemard E, Kassab R. Structure of the actin-myosin interface. Nature. 1981;292(5821):301–306. doi: 10.1038/292301a0. [DOI] [PubMed] [Google Scholar]
- 29.Bonafé N, Chaussepied P. A single myosin head can be cross-linked to the N termini of two adjacent actin monomers. Biophys J. 1995;68(4) Suppl:35S–43S. [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collaborative Computational project, Number 4 The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 10):1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bricogne G, et al. BUSTER version 2.10.1 Cambridge. Global Phasing Ltd.; United Kingdom: 2011. [Google Scholar]
- 39.Svergun DI, Barberato C, Koch MHJ. CRYSOL—A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 40.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem. 1988;171(2):266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 41.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 42.Wilson DP, Sutherland C, Walsh MP. Ca2+ activation of smooth muscle contraction: Evidence for the involvement of calmodulin that is bound to the triton insoluble fraction even in the absence of Ca2+ J Biol Chem. 2002;277(3):2186–2192. doi: 10.1074/jbc.M110056200. [DOI] [PubMed] [Google Scholar]
- 43.Cremo CR, Geeves MA. Interaction of actin and ADP with the head domain of smooth muscle myosin: implications for strain-dependent ADP release in smooth muscle. Biochemistry. 1988;37:1969–1978. doi: 10.1021/bi9722406. [DOI] [PubMed] [Google Scholar]
- 44.Kovács M, Wang F, Hu A, Zhang Y, Sellers JR. Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J Biol Chem. 2003;278:38132–38140. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- 45.Rosenfeld SS, Xing J, Chen L, Sweeney HL. Myosin IIb is unconventionally conventional. J Biol Chem. 2003;278:27449–27455. doi: 10.1074/jbc.M302555200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.