Significance
Oncogenic mutations in the BRAF protein kinase occur in a large number of patients with lung cancer, the number one cause of cancer mortality worldwide. Despite the relatively high frequency (2–7%) of various oncogenic BRAF alleles in human lung adenocarcinoma (LA), the biological and clinical relevance of these mutations and the optimal therapeutic strategy to improve outcomes remains poorly defined. We report the preclinical efficacy of a next-generation BRAF kinase inhibitor across a spectrum of clinically relevant BRAF-mutant nonsmall cell lung cancer (NSCLC) models. Our results provide biological insight into the regulation of BRAF oncogene dependence and identify strategies to improve outcomes in patients with BRAF-mutant lung cancer.
Keywords: BRAF, lung, cancer, targeted therapy
Abstract
Oncogenic activation of protein kinase BRAF drives tumor growth by promoting mitogen-activated protein kinase (MAPK) pathway signaling. Because oncogenic mutations in BRAF occur in ∼2–7% of lung adenocarcinoma (LA), BRAF-mutant LA is the most frequent cause of BRAF-mutant cancer mortality worldwide. Whereas most tumor types harbor predominantly the BRAFV600E-mutant allele, the spectrum of BRAF mutations in LA includes BRAFV600E (∼60% of cases) and non-V600E mutant alleles (∼40% of cases) such as BRAFG469A and BRAFG466V. The presence of BRAFV600E in LA has prompted clinical trials testing selective BRAF inhibitors such as vemurafenib in BRAFV600E-mutant patients. Despite promising clinical efficacy, both innate and acquired resistance often result from reactivation of MAPK pathway signaling, thus limiting durable responses to the current BRAF inhibitors. Further, the optimal therapeutic strategy to block non-V600E BRAF-mutant LA remains unclear. Here, we report the efficacy of the Raf proto-oncogene serine/threonine protein kinase (RAF) inhibitor, PLX8394, that evades MAPK pathway reactivation in BRAF-mutant LA models. We show that PLX8394 treatment is effective in both BRAFV600E and certain non-V600 LA models, in vitro and in vivo. PLX8394 was effective against treatment-naive BRAF-mutant LAs and those with acquired vemurafenib resistance caused by an alternatively spliced, truncated BRAFV600E that promotes vemurafenib-insensitive MAPK pathway signaling. We further show that acquired PLX8394 resistance occurs via EGFR-mediated RAS-mTOR signaling and is prevented by upfront combination therapy with PLX8394 and either an EGFR or mTOR inhibitor. Our study provides a biological rationale and potential polytherapy strategy to aid the deployment of PLX8394 in lung cancer patients.
Oncogenic mutations in the BRAF serine/threonine protein kinase occur in a wide spectrum of solid tumor malignancies, most notably melanoma, colorectal cancer, and lung adenocarcinoma (1). Mutant BRAF promotes tumor growth by hyperactivating the RAF-MEK-ERK signaling cascade (2–5). BRAFV600E is the most common oncogenic form of BRAF in most tumor types, and selective RAF inhibitors such as vemurafenib and dabrafenib have demonstrated clinical efficacy in melanoma and LA harboring BRAFV600E (6–9). Despite these findings, ∼15% of BRAF mutant cancers do not respond to BRAF inhibitors, and the majority of patients who achieve a response will inevitably acquire resistance to these targeted agents, predominantly through reactivation of MAPK pathway signaling (4, 10–16).
The clinical efficacy of RAF inhibitors depends on the degree of suppression of MAPK pathway output (8). Vemurafenib and dabrafenib paradoxically activate the MAPK pathway in cells with oncogenic RAS or increased upstream receptor signaling, thereby enhancing cellular proliferation that can promote cutaneous squamous cell carcinomas and keratoacanthomas that often harbor RAS mutations, and potentially other RAS-driven malignancies (17–25). Combination therapy with a MEK inhibitor is one strategy to limit this reactivation and has shown substantial clinical efficacy in BRAFV600E-mutant melanoma and LA (26, 27). These results are encouraging and improve clinical outcomes and the standard of care. However, increased adverse events and therapeutic resistance limit the clinical utility of RAF plus MEK inhibitor polytherapy (26, 27).
Recently, next-generation RAF inhibitors that evade MAPK pathway reactivation were developed as an alternative strategy to potentiate the suppression of MAPK pathway output in BRAFV600E-driven cancers (28, 29). These RAF inhibitors, such as PLX8394, were shown to be effective in BRAFV600E and RAS-mutant melanoma and colorectal cancer cells (28). Unlike melanoma, however, non-V600 BRAF mutant alleles comprise a significant proportion (∼40%) of mutations in LA (30–32). Specifically, mutations in the P loop of BRAF are present at relatively high frequencies in LA: ∼13% of the BRAF-mutant cases harbor BRAFG469A and ∼22% exhibit BRAFG466V (30–32). Cancer cells that harbor non-V600 BRAF mutations appear to be less sensitive to vemurafenib, likely due to these aberrant BRAF oncoproteins signaling as vemurafenib-insensitive dimers (33). Thus, alternative strategies to more effectively block MAPK pathway activity are required to fully suppress the growth of non-V600 mutant BRAF LA. Using in vitro and in vivo models, we tested the preclinical efficacy of PLX8394 in LA cells expressing either endogenous BRAFV600E or non-V600 mutant forms of oncogenic BRAF and sought to identify mechanisms of resistance to PLX8394 that can potentially be overcome with a rational polytherapy strategy.
Results
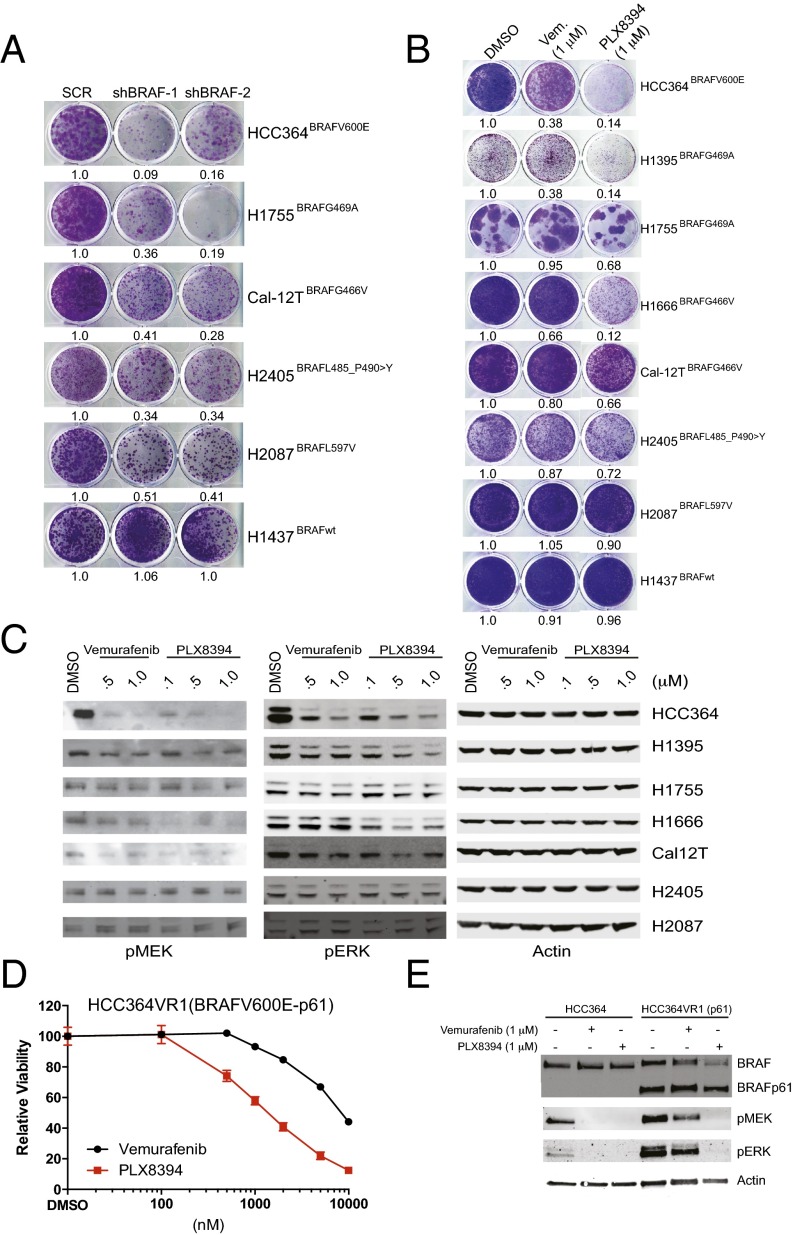
We first tested the hypothesis that LA cell growth depended on the expression of either mutant BRAFV600E or a non-V600 mutant BRAF allele. We found that shRNA-mediated silencing of BRAF expression impaired growth in a panel of BRAF-mutant LA cell lines (n = 5), whereas no substantial impact on growth was observed in LA cells harboring wild-type BRAF (Fig. 1A). These data reveal the proliferative requirement for certain non-V600 forms of mutant BRAF in LA cells. Consistent with these findings, silencing BRAF expression in non-V600 mutant BRAF LA cells suppressed MAPK pathway signaling as measured by the levels of phosphorylated ERK (p-ERK) in cellular lysates (Fig. S1).
Fig. 1.
The effects of PLX8394 RAF inhibitor treatment in BRAFV600E and non-V600 BRAF-mutant LA cells in vitro. (A) BRAF-mutant LA cell lines are addicted to each oncoprotein. Shown is the cell growth upon shRNA-mediated knockdown, conducted and analyzed as reported (4). (B and C) The effects of RAF inhibitor treatment with each indicated agent on growth (B) and signaling (C) in each BRAF-mutant LA cell line. The experiment was conducted and analyzed as reported (4), using the indicated drug doses. Vem, vemurafenib. (D and E) The effects of RAF inhibitor treatment with each indicated agent on growth (D) and signaling (E) in BRAFV600E LA cells with acquired vemurafenib resistance that express a vemurafenib-insensitive, truncated form of BRAFV600E. The experiment was conducted and analyzed as reported (4), using the indicated drug doses. All data shown represent three independent experiments, mean ± SEM, P < 0.01.
Fig. S1.
Knockdown of BRAF expression in BRAF-mutant lung cancer cell lines. Shown is the degree of shRNA-mediated knockdown of BRAF by immunoblot analysis in select LA cell lines studied in Fig. 1, conducted and analyzed as reported (4).
We next investigated the effects of BRAF inhibition with either the clinically approved BRAF inhibitor vemurafenib or the investigational BRAF inhibitor PLX8394 on cell growth and MAPK pathway signaling (using p-MEK and p-ERK levels as immunoblot analysis readouts) in seven LA cell lines expressing either endogenous V600E or non-V600 mutant forms of BRAF. We found that PLX8394 more significantly impaired tumor cell growth (Fig. 1B) and suppressed MAPK signaling (Fig. 1C) compared with vemurafenib treatment. We noted that neither vemurafenib nor PLX8394 substantially suppressed cell growth or MEK-ERK signaling in H2087 or H2405 cells that harbor more rare BRAF variants, similar to our findings in H1437 LA cells with wild-type BRAF. Collectively, these data suggest enhanced therapeutic utility of PLX8394 in lung cancer cells harboring several of the common forms of oncogenic BRAF present in LA.
We recently showed that BRAFV600E LA cells (HCC364) can acquire vemurafenib resistance by expressing an aberrant (truncated) splice variant of BRAFV600E that is vemurafenib-insensitive and constitutively dimerized (4). We tested whether the enhanced efficacy of PLX8394 versus vemurafenib extended to these vemurafenib-insensitive cells (HCC364VR1). We found that PLX8394 effectively suppressed cell growth and MAPK pathway signaling in these cells (Fig. 1 D and E). These data demonstrate the ability of PLX8394 to block MAPK pathway output and vemurafenib-insensitive LA cell growth, even in the context of constitutively dimerized forms of mutant BRAF. Thus, PLX8394 warrants further study as a promising strategy to overcome acquired vemurafenib resistance in LA patients with truncated BRAFV600E.
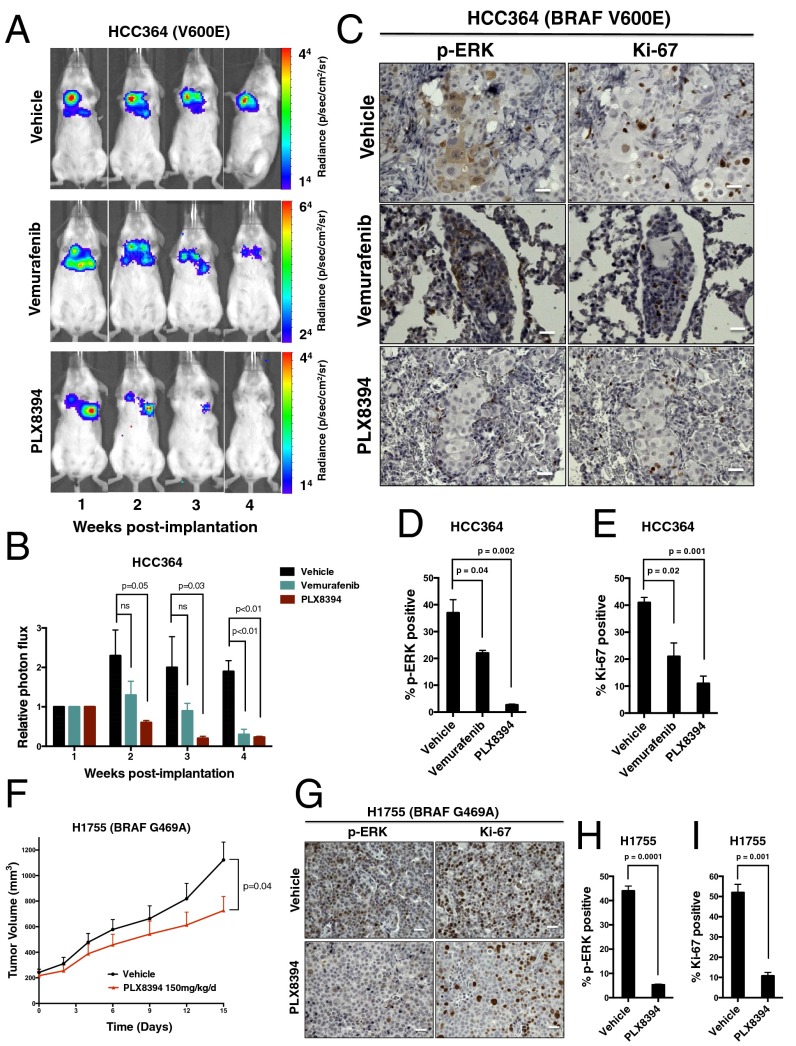
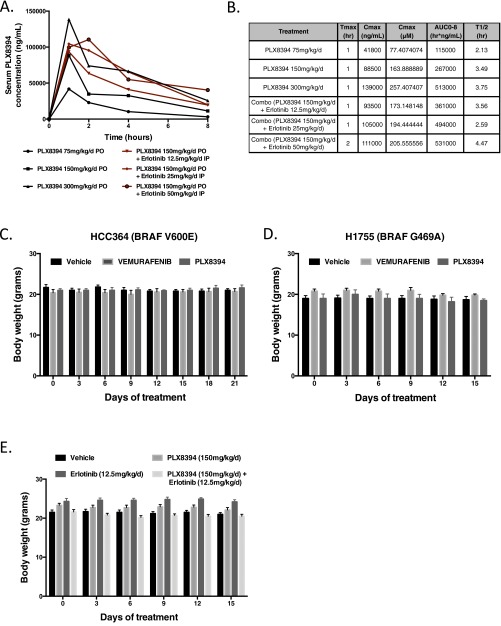
We next tested the effects of PLX8394 on LA tumor growth in vivo. HCC364 cells (the only patient-derived BRAFV600E LA cell line available) failed to establish s.c. tumors when implanted into the flanks of immunocompromised mice. Thus, we used an orthotopic lung cancer model system to assess the preclinical efficacy of BRAF inhibition in BRAFV600E LA in vivo. Specifically, we surgically implanted HCC364 cells stably expressing firefly Luciferase into the left lung parenchyma of immunodeficient mice and monitored tumor growth and response to RAF inhibitor treatment by using a bioluminescent-based imaging (BLI) system (Fig. 2 A and B). We found that PLX8394, at a dose of 150 mg⋅kg−1⋅d−1 substantially suppressed tumor growth, MAPK pathway signaling as measured by immunohistochemical (IHC) analysis of p-ERK levels, and tumor cell proliferation as measured by IHC analysis of Ki-67 levels in tumor explants (Fig. 2 C–E). Vemurafenib treatment was also effective in our orthotopic in vivo model, as expected. However, more rapid and substantial initial antitumor responses were observed with PLX8394 treatment, an effect that was associated with an enhanced and durable suppression of ERK phosphorylation and tumor cell proliferation (Fig. 2 B–E). Pharmacokinetic (PK) studies showed that PLX8394 (at 150 mg⋅kg−1⋅d−1) yielded plasma concentrations of >150 μM, with no overt toxicity in animals (Fig. S2).
Fig. 2.
The effects of PLX8394 RAF inhibitor treatment in BRAFV600E and non-V600 BRAF mutant LA cells in vivo. Representative BLI images (A) and relative photon flux (B) of HCC364 BRAFV600E-bearing mice treated with either vehicle (n = 10 mice), vemurafenib 50 mg⋅kg−1⋅d−1 (n = 10 mice), or PLX8394 150 mg⋅kg−1⋅d−1 (n = 10 mice). P value calculated with one-way ANOVA. (C) Representative IHC analysis of p-ERK and Ki-67 in tumors harvested from orthotopically implanted HCC364 mice. Data shown in the % positive p-ERK (D) and % positive Ki-67 (E) cells in explanted HCC364 tumors from mice treated with vehicle, vemurafenib, or PLX8394 for 3 wk. Data represent mean ± SEM of four tumors from each treatment cohort. P values calculated with one-way ANOVA. (F) H1755 tumor growth curve comparing vehicle (n = 10 mice) and PLX8394 (n = 10 mice) treated mice. P values calculated with Student’s t test. (G) Representative IHC analysis of p-ERK and Ki-67 in s.c.-implanted H1755 mice. Shown is the % positive p-ERK (H) and % positive Ki-67 (I) cells in explanted H1755 tumors from mice treated with either vehicle or PLX8394 for 15 d. Data represent mean ± SEM of four tumors from each treatment cohort. P values were calculated with Student’s t test. (Scale bars: 100 μm.)
Fig. S2.
Pharmacokinetics and toxicity of PLX8394 in vivo. (A) PLX8394 was given orally at 75 mg⋅kg−1⋅d−1, 150 mg⋅kg−1⋅d−1, or 300 mg⋅kg−1⋅d−1, and serum concentration (ng/mL) was measured at 0, 1, 2, 4, and 8 h after treatment (black lines). PLX8394 150 mg⋅kg−1⋅d−1 was given orally with erlotinib 12.5 mg⋅kg−1⋅d−1, 25 mg⋅kg−1⋅d−1, or 50 mg⋅kg−1⋅d−1, and serum concentration (ng/mL) was measured at 0, 1, 2, 4, and 8 h after treatment (red lines). (B) In vivo pharmacokinetics of PLX8394 monotherapy or in combination with erlonitib. AUC0-8, area under the curve 0–8 h; Cmax (μM), maximum serum concentration in μM; Cmax (ng/mL), maximum serum concentration in ng/mL; T1/2, half-life; Tmax, time to maximum concentration. (C and D) Weights derived from mice bearing either orthotopically (HCC364 cells) or s.c. (H1755 cells) implanted tumor cells and treated with either vehicle, vemurafenib, or PLX8394 for 21 (orthotopic) or 15 (s.c.) days, respectively. (E) Weights derived from mice treated with vehicle, erlotinib 12.5 mg⋅kg−1⋅d−1, PLX8394 150 mg⋅kg−1⋅d−1, or combination PLX8394 150 mg⋅kg−1⋅d−1 and erlotinib 12.5 mg⋅kg−1⋅d−1 for 14 d.
We further tested the preclinical efficacy of PLX8394 in vivo against tumors with non-V600 mutant BRAF. We first selected cell lines that represented the two most common non-V600 BRAF mutant subtypes (H1755 BRAFG469A and H1666 BRAFG466V). Of these cell lines, only H1755 cells grew as s.c. xenografts. Although H1755 cells were relatively insensitive to vemurafenib treatment in vitro, they showed sensitivity to PLX8394 treatment (Fig. 1). The BRAFG469A allele present in H1755 cells represents a significant fraction of the non-V600 BRAF mutations present in LA patients (31, 32). We found that PLX8394 treatment suppressed tumor growth, RAF-MEK-ERK signaling, and tumor cell proliferation in these H1755 xenograft tumors (Fig. 2 F–I). We did not observe overt toxicity in mice treated with PLX8394 at a dose of 150 mg⋅kg−1⋅d−1 (Fig. S2 C and D). Altogether, our in vitro and in vivo data provide a rationale for the clinical testing of PLX8394 in patients whose tumors harbor BRAFV600E (full length or truncated) and certain non-V600 mutant forms of BRAF that are recurrently present in LA.
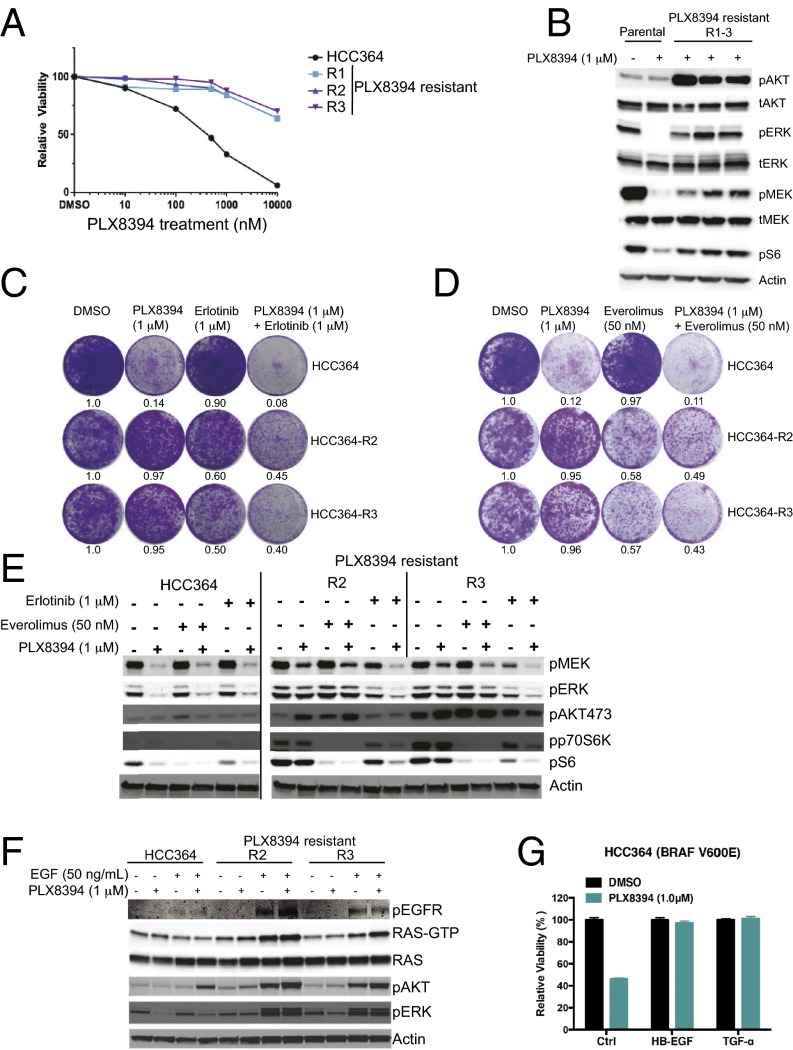
We next sought to identify potential mechanisms that may cause acquired resistance to PLX8394 treatment in BRAF-mutant LA cells. Using previous methods (4), we derived three independent PLX8394-resistant sublines from the only commercially available (at the time of our study) BRAFV600E mutant LA cell line, HCC364 (Fig. 3A; cell lines R1–R3). Immunoblot analysis revealed that each resistant subline, which is continuously grown in the presence of PLX8394 (1 μM), harbored increased levels of p-AKT and persistent phosphorylation of MEK, ERK, and S6, compared with the parental HCC364 cells treated with PLX8394 (Fig. 3B). These findings suggested that AKT and MEK-ERK signaling, potentially converging on mTORC1 and S6 activation through established mechanisms (34), could underlie PLX8394 resistance in these cells. We did not test combined MEK plus PI-3 kinase inhibitor treatment in these cells, because this combination therapy approach has proven largely infeasible clinically because of the lack of an adequate therapeutic index and substantial toxicities (35). Our prior work demonstrated that acquired vemurafenib resistance in HCC364 cells is caused by persistent EGFR signaling during BRAF inhibitor treatment (4); this persistent EGFR signaling did not result from an overall increase in EGFR expression but instead resulted from sustained secretion of EGFR ligands that stimulated EGFR activation and downstream signaling to promote resistance (4). Thus, we hypothesized that a similar EGFR-mediated mechanism may promote acquired PLX8394 resistance in these nonsmall cell lung cancer (NSCLC) cells, such that EGFR activates MEK-ERK and PI-3 kinase signaling to converge on mTORC1-S6 activation to promote resistance.
Fig. 3.
mTORC1 activation downstream of receptor kinase signaling promotes acquired PLX8394 RAF inhibitor resistance in LA cells. (A and B) BRAFV600E LA sublines with acquired PLX8394 resistance, where A shows cell growth and B shows signaling pathway activation by immunoblot analysis, both conducted and analyzed as reported (4). (C–E) The effects of inhibitor treatment with each indicated agent on growth (C and D) and signaling (E) in each indicated BRAF-mutant LA cell line. The experiment was conducted and analyzed as reported (4), using the indicated drug doses. (F) The effects of EGF stimulation on RAS-GTP levels and p-ERK and p-AKT levels in each indicated cell line, using the indicated concentration of EGF and each drug. The experiment was conducted and analyzed as reported (4). All data shown represent three independent experiments, mean ± SEM, P < 0.01. (G) HCC364 cells were treated with DMSO or PLX8394 following pretreatment with an EGF ligand (HB-EGF or TGF-α). Representative of three independent experiments.
To investigate this hypothesis, we tested the effects of both the EGFR inhibitor erlotinib and the mTORC1 inhibitor everolimus on growth and signaling in the HCC364 PLX8394-resistant sublines (using the R2 and R3 sublines as representative models), making comparisons to the parental cells. If the hypothesis is true, we reasoned that EGFR or mTORC1 inhibition should suppress cell growth in each of the resistant sublines. We found that treatment with erlotinib (at a concentration we showed suppresses p-EGFR levels in HCC364 cells; ref. 4) or everolimus alone suppressed cell growth and MAPK, PI-3 kinase, and mTORC1-S6 signaling in the PLX8394-resistant cells, with little impact on cell growth in the parental cells (Fig. 3 C–E). We noted that everolimus treatment increased the levels of p-AKTS473, indicating everolimus-induced mTORC2 activation as observed (34). Further, we found that combined treatment with either erlotinib or everolimus plus PLX8394 modestly cooperated to suppress cell growth and S6 phosphorylation (Fig. 3 C–E), suggesting that a more complete suppression of MAPK pathway signaling achieved by PLX8394 treatment is required for a maximal therapeutic effect. Consistent with these findings, we found that following serum starvation, the PLX8394-resistant sublines (R2, R3) were hyperresponsive to epidermal growth factor (EGF) stimulation, which increased the levels of active (GTP-bound) RAS and of p-ERK and p-AKT, compared with the parental cells under identical conditions (Fig. 3F). The relatively small increase in GTP-bound RAS levels and signaling that we observed upon EGF stimulation in the serum-starved HCC364 parental cells is consistent with prior findings, indicating profound feedback inhibition of receptor tyrosine kinase signaling in BRAF-mutant cells (36). This feedback inhibition of receptor kinase signaling therefore appears to be relieved in the cells with acquired PLX8394 resistance in this system. To explore this mechanism further, we stimulated serum-starved HCC364 (PLX8394-sensitive) cells with EGFR ligands, HB-EGF and TGFα, which were previously shown to increase p-ERK and p-AKT in this system (4). We found that HCC364 cells that were pretreated with either HB-EGF or TGFα had decreased sensitivity to PLX8394 compared with cells not treated with ligand (Fig. 3G). Together, our findings indicate an important role for EGFR activation (likely via EGFR ligand stimulation) in driving downstream signaling through RAS and the MEK-ERK and PI-3 kinase pathways to converge on mTORC1-S6 signaling and promote acquired PLX8394 resistance in these BRAFV600E LA cells. Our findings suggest that a clinical inhibitor of either EGFR or mTORC1 can overcome this resistance in BRAFV600E LA cells, offering two potential clinical strategies to combat PLX8394 resistance in BRAF-mutant LA patients.
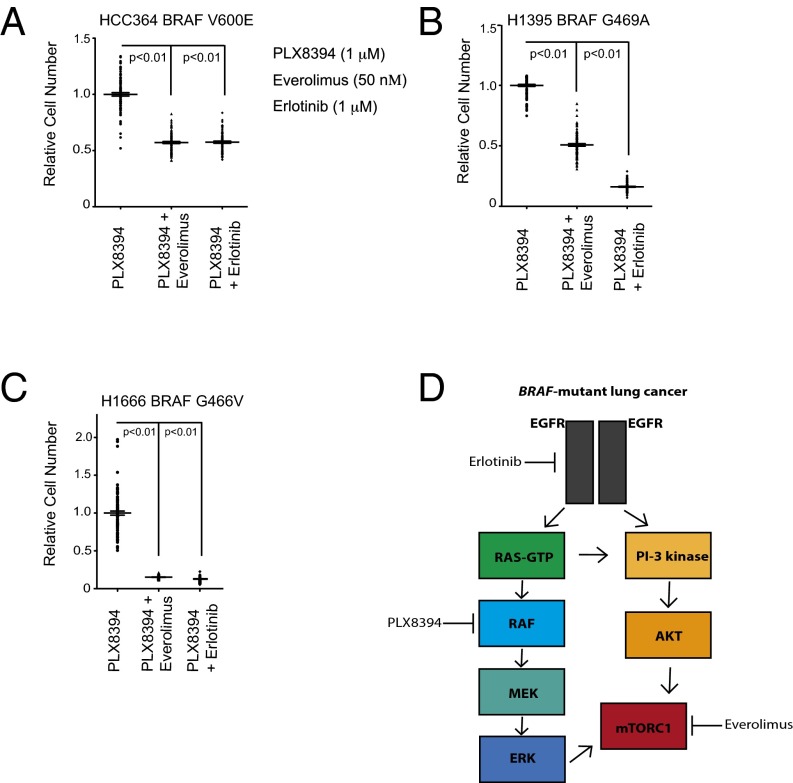
Based on these findings, we reasoned that inhibition of either EGFR with erlotinib or mTORC1 with everolimus would forestall acquired PLX8394 resistance in the parental (treatment-naïve) BRAF-mutant LA cells. To address this hypothesis, we used an established acquired resistance assay (4, 37, 38) in representative LA cell lines from our panel that express either V600E or non-V600 mutant forms of BRAF. Using HCC364 (BRAFV600E) cells, we found that cotreatment with either erlotinib or everolimus (at concentrations that suppress signaling; Fig. 3) plus PLX8394 substantially impaired the development of acquired resistance, whereas erlotinib or everolimus had little impact on cell growth when used as monotherapy (Fig. 4A). These findings were extended across both H1395 (BRAFG469A) and H1666 (BRAFG466V) LA cell lines (Fig. 4 B and C); in both of these two models, erlotinib in combination with PLX8394 appeared to be particularly effective at preventing acquired resistance. In vivo pharmacokinetic (PK) and toxicity studies showed that PLX8394 combined with erlotinib (at standard doses for mouse in vivo studies; refs. 37 and 39) yielded plasma erlotinib concentrations of >1 μM and PLX8394 concentrations of >150 μM, with no overt toxicity in animals (Fig. S2), suggesting the feasibility of this polytherapy. These data provide a potential rational polytherapy strategy to forestall acquired PLX8394 resistance in BRAF-mutant (V600 and non-V600) LA.
Fig. 4.
Upfront polytherapy with PLX8394 plus either erlotinib or everolimus impairs the onset of acquired resistance in both V600E and non-V600 BRAF-mutant LA cell lines. (A–C) The effects of combined inhibition of EGFR with erlotinib or of mTORC1 with everolimus on acquired resistance in each indicated BRAF-mutant LA cell line, using the drug concentrations indicated in A. Acquired resistance was measured by using an assay we validated and published (4). The experiment was conducted and analyzed as reported (4). All data shown represent three independent experiments, mean ± SEM, P < 0.01. (D) Schematic model summarizing our findings and the therapeutic strategies informed by our study. The testing of rational polytherapy with an FDA-approved mTORC1 or EGFR inhibitor may forestall the onset of resistance to concurrent treatment with third-generation RAF inhibitors such as PLX8394, as clinical trials (for instance, NCT02428712) of these agents in lung cancer patients are initiated.
Discussion
Altogether, our findings provide several insights. First, we show that LA cells with noncanonical BRAF mutations depend on these BRAF oncoproteins for growth. Second, RAF inhibitors that evade MAPK pathway reactivation, such as PLX8394, may more effectively block the growth of certain BRAF-mutant LAs, including those with BRAFV600E, BRAFG469A, and BRAFG466V and those with vemurafenib-insensitive truncated forms of BRAFV600E that limit response to the current Food and Drug Administration (FDA)-approved BRAF inhibitors (vemurafenib, dabrafenib). Third, we reveal EGFR-RAS-mTORC1 signaling as a mechanism of acquired resistance to PLX8394 in LA cells and provide a rational strategy to overcome this resistance (Fig. 4D). Collectively, our work is a prelude to the future understanding of how mTORC1 signaling is engaged to drive resistance to PLX8394 in human cancer and also for the mechanism-based clinical deployment of these agents in early phase clinical trials (for instance, NCT02428712).
Methods
Cell Lines and Culture Reagents.
The HCC364 cell line was kindly provided by David Solit, Memorial Sloan Kettering Cancer Center (MSKCC), New York. Cal-12T, H1395, H1755, H1666, H2405, H2087 and H1437 cells were purchased from ATCC. Cells were maintained at 37 °C in a humidified atmosphere at 5% CO2 grown in RPMI medium 1640 supplemented with 10% (vol/vol) FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin.
Generation of Drug-Resistant Cell Lines.
To generate PLX8394-resistant lines, HCC364 cells were exposed to an increasing concentration of PLX8394 from 500 nM to 10 μM. Individual populations of cells were confirmed to be drug resistant by standard cell viability assays. The genetic identity of each subclone with that of the parental cells was verified by short-tandem repeat analysis conducted at Johns Hopkins University according to established protocols.
Cell Viability Assay.
Three thousand to 5,000 cells were plated per well in 96-well plates 24 h before drug treatment. Viable cells were determined 72 h after drug treatment by using CellTiter-Glo luminescent cell viability reagent according to the manufacturer’s protocols (Promega). Each assay consisted of four replicate wells and was repeated at least three times. Data were expressed as percentage of the cell viability of control cells. The data were graphically displayed by using GraphPad Prism version 5.0 for Mac (GraphPad Software Inc.). For EGFR ligand experiments, 2.5 × 105 HCC364 cells were plated in a six-well dish, serum starved for 6 h, and then stimulated with HB-EGF (Prospec) or TGF-α (R&D) with PLX8394. Each assay was performed in triplicate. Quantification was performed with ImageJ software.
Clonal Outgrowth Assay.
Clonal outgrowth studies were conducted in 96-well format by using 100 wells for each condition and conducted in triplicate. Resistant wells were assayed after 2 mo of treatment and are plotted relative to PLX8394-treated cells.
Western Blot Analysis.
Two hundred thousand cells were seeded per well in six-well plates 24 h before drug treatment, and whole-cell lysates were prepared by using RIPA buffer [10 mM Tris⋅Cl (pH 8.0), 1 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl] supplemented with protease inhibitor and phosphatase inhibitor (Roche) and clarified by sonication and centrifugation. Equal amounts of protein were separated by 4–15% of SDS/PAGE and were transferred onto nitrocellulose membranes (Bio-Rad) for protein blot analysis. Membranes were incubated with primary antibody overnight and were washed and incubated with secondary antibody. Membranes were exposed by using either a fluorescence system (Li-cor) or a chemiluminescent reagent and images captured and bands quantified by using an ImageQuant LAS 4000 instrument (GE Healthcare).
Antibodies.
Anti–phospho-MEK, anti–phospho-ERK, anti–phospho-AKT, anti-phospho-S6, and anti–phospho-P70S6K antibodies were purchased from Cell Signaling Technology. Anti-BRAF antibody for Western blotting was purchased from Santa Cruz Biotechnology. The anti–β-Actin (A2228) antibody was purchased from Sigma-Aldrich.
Orthotopic Lung Xenografts in Immunodeficient Mice.
Six- to eight-week-old female SCID CB.17 mice were purchased from Taconic. Specific pathogen-free conditions and facilities were approved by the American Association for Accreditation of Laboratory Animal Care. Surgical procedures were reviewed and approved by the UCSF Institutional Animal Care and Use Committee (IACUC), protocol no. AN107889-01A. To prepare cell suspensions for thoracic injection, adherent tumor cells were briefly trypsinized, quenched with 10% FBS RPMI media, and resuspended in PBS. Cells were pelleted again and mixed with Matrigel matrix (BD Bioscience 356237) on ice for a final concentration of 1.0 × 105 cells per μL. The Matrigel-cell suspension was transferred into a 1-mL syringe and remained on ice until the time of implantation. For orthotopic injection, mice were placed in the right lateral decubitus position and anesthetized with 2.5% inhaled isoflurane. A 1-cm surgical incision was made along the posterior medial line of the left thorax, fascia and adipose tissue layers were dissected and retracted to expose the lateral ribs, intercostal space, and the left lung parenchyma. Upon recognition of left lung respiratory variation, a 30-guage hypodermic needle was used to advance through the intercostal space ∼3 mm into the lung tissue. For human cancer cell line HCC364, care was taken to inject 10 μL (1.0 × 106 cells) of cell suspension directly into the left lung. The needle was rapidly withdrawn, and mice were observed for pneumothorax. Visorb 4/0 polyglycolic acid sutures were used for primary wound closure of the fascia and skin layer. Mice were observed after the procedure for 1–2 h, and body weights and wound healing were monitoring weekly.
For drug treatments, orthotopically implanted tumors were allowed to grow for 1 wk before treatment. Mice were treated with either vehicle, vemurafenib, or PLX8394 at the start of week 2 (postimplantation day 8) and continued on therapy until the end of week 4 (postimplantation day 28).
In Vivo Bioluminescence Imaging.
Mice were imaged at the UCSF Preclinical Therapeutics Core starting on postinjection day 7 with a Xenogen IVIS 100 bioluminescent imaging system. Before imaging, mice were anesthetized with isoflurane and i.p. injection of 200 μL of d-Luciferin at a dose of 150 mg/kg body weight was administered. Weekly monitoring of bioluminescence of the engrafted lung tumors was performed until week 5. Radiance was calculated automatically by using Living Image Software following demarcation of the thoracic cavity (ROI) in the supine position. The radiance unit of photons per s−1/cm2⋅sr−1 is the number of photons per second that leave a square centimeter of tissue and radiate into a solid angle of one steradian (sr).
s.c. Xenograft Studies.
All animal experiments were conducted under UCSF IACUC-approved animal protocol no. AN107889-01A. H1755 tumor xenografts were generated by injection of 5 × 106 cells in a 50/50 mixture for matrigel and PBS into 6- to 8-wk-old female NOD/SCID mice. Mice were randomized to treatment groups once tumors reached an average size of 150 mm3.
For drug treatments, H1755 cells were s.c. implanted and allowed to grow to ∼200 mm3 (4 wk after implantation). Mice were then treated with vehicle, vemurafenib, or PLX8394 for 15 d.
In Vivo Compound Formulation.
PLX8394 and vemurafenib was obtained from Plexxikon. The vehicle for daily oral gavage was PEG 400 [20% (vol/vol)], tocopheryl polyethylene glycol succinate (TPGS) [5% (vol/vol)], water [75% (vol/vol)]. Vemurafenib was dissolved in the PEG [20% (vol/vol)], TPGS [5% (vol/vol)], and water [75% (vol/vol)] and dosed at 50 mg⋅kg−1⋅d−1. Vemurafenib was given daily by oral gavage. PLX8394 was dissolved in PEG 400 [20% (vol/vol)], TPGS [5% (vol/vol)], and water [75% (vol/vol)] and vortexed continuously throughout the dosing period. PLX8394 was given daily by oral gavage at a dose of 150 mg⋅kg−1⋅d−1.
In Vivo Pharmacokinetic Studies.
Immunodeficient CB.17 SCID mice were treated with PLX8394 monotherapy at three doses, 75, 150, or 300 mg⋅kg−1⋅d−1, and ∼50 μL of blood was collected from the tail vein following anesthetization with inhaled isoflurane. The final blood collection was via cardiac puncture. Whole blood was serially collected at 0, 1, 2, 4, and 8 h after PLX8394 administration and transferred to a plasma separator tube with lithium heparin (BD catalog no. 365985), and centrifuged for 10 min at 1,000 × g. Sample collection and pharmacokinetic analysis of combination treatment with PLX8394 150 mg⋅kg−1⋅d−1 and erlotinib 12.5, 25, or 50 mg⋅kg−1⋅d−1 was performed in an identical fashion to PLX8394 monotherapy.
Immunostaining (IHC): Orthotopic Lung Tissue and s.c. Xenografts.
For orthotopic lung studies, mice were killed at the primary endpoint (4 wk). Lungs were harvested en bloc and dissected along the mediastinum to separate the right and left lung lobes. Lungs were fixed in 10% neutral buffered formalin for 72 h, embedded in paraffin, and 5- to 10-μm sections were prepared. Sections were subsequently deparaffinized and incubated with antibodies directed against phospho-ERK antibody (Cell Signaling Technology), Ki-67 antibody (Leica Biosystems), and cleaved caspase-3 antibody (Cell Signaling Technology) overnight.
Statistical Methods.
For in vivo studies, one-way ANOVA was used to calculate P values for vehicle, vemurafenib, and PLX8394 orthotopic HCC364 studies including relative photon flux, percent (%) p-ERK positive, and % Ki-67 positive in tumor tissue. Student’s t test was used to compare H1755 s.c. xenograft growth, % p-ERK positive, and % Ki-67 positive tumor tissue treated with vehicle and PLX8394.
Acknowledgments
Funding was provided by NIH, Pew-Stewart Charitable Trust, Kinship-Searle Foundation, and the Van Auken Foundation (T.G.B.).
Footnotes
Conflict of interest statement: G.B. and A.R. are employees of Plexxikon, Inc., which produced vemurafenib and PLX8394. T.G.B. is a consultant to Driver Group, Novartis, Astellas, Natera, Array Biopharma, Ariad, Teva, Astrazeneca, and a recipient of research grants from Servier and Ignyta.
This article is a PNAS Direct Submission. P.A.B. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610456113/-/DCSupplemental.
References
- 1.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pratilas CA, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68(22):9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci USA. 2014;111(7):E748–E757. doi: 10.1073/pnas.1320956111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji H, et al. Mutations in BRAF and KRAS converge on activation of the mitogen-activated protein kinase pathway in lung cancer mouse models. Cancer Res. 2007;67(10):4933–4939. doi: 10.1158/0008-5472.CAN-06-4592. [DOI] [PubMed] [Google Scholar]
- 6.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci USA. 2010;107(33):14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Planchard D, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: A single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(5):642–650. doi: 10.1016/S1470-2045(16)00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long GV, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. 2015;47(3):250–256. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudin CM, Hong K, Streit M. Molecular characterization of acquired resistance to the BRAF inhibitor dabrafenib in a patient with BRAF-mutant non-small-cell lung cancer. J Thorac Oncol. 2013;8(5):e41–42. doi: 10.1097/JTO.0b013e31828bb1b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson DB, et al. Acquired BRAF inhibitor resistance: A multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur J Cancer. 2015;51(18):2792–2799. doi: 10.1016/j.ejca.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell. 2011;19(1):11–15. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Goetz EM, Garraway LA. Mechanisms of resistance to mitogen-activated protein kinase pathway inhibition in BRAF-mutant melanoma. Am Soc Clin Oncol Educ Book. 2012:680–684. doi: 10.14694/EdBook_AM.2012.32.189. [DOI] [PubMed] [Google Scholar]
- 16.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 20.Anforth RM, et al. Cutaneous manifestations of dabrafenib (GSK2118436): A selective inhibitor of mutant BRAF in patients with metastatic melanoma. Br J Dermatol. 2012;167(5):1153–1160. doi: 10.1111/j.1365-2133.2012.11155.x. [DOI] [PubMed] [Google Scholar]
- 21.Callahan MK, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367(24):2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberholzer PA, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol. 2012;30(3):316–321. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366(3):207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmer L, et al. Atypical melanocytic proliferations and new primary melanomas in patients with advanced melanoma undergoing selective BRAF inhibition. J Clin Oncol. 2012;30(19):2375–2383. doi: 10.1200/JCO.2011.41.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews MC, et al. BRAF inhibitor-driven tumor proliferation in a KRAS-mutated colon carcinoma is not overcome by MEK1/2 inhibition. J Clin Oncol. 2013;31(35):e448–e451. doi: 10.1200/JCO.2013.50.4118. [DOI] [PubMed] [Google Scholar]
- 26.Long GV, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 27.Planchard D, et al. Dabrafenib plus trametinib in patients with previously treated BRAF-mutant metastatic non-small cell lung cancer: An open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17(7):984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015;526(7574):583–586. doi: 10.1038/nature14982. [DOI] [PubMed] [Google Scholar]
- 29.Girotti MR, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell. 2015;27(1):85–96. doi: 10.1016/j.ccell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchetti A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29(26):3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 32.Paik PK, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29(15):2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao Z, et al. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015;28(3):370–383. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu T, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18(8):2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 36.Lito P, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22(5):668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blakely CM, et al. NF-κB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Reports. 2015;11(1):98–110. doi: 10.1016/j.celrep.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hrustanovic G, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21(9):1038–1047. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bivona TG, et al. FAS and NF-κB signalling modulate dependence of lung cancers on mutant EGFR. Nature. 2011;471(7339):523–526. doi: 10.1038/nature09870. [DOI] [PMC free article] [PubMed] [Google Scholar]