Significance
Directed enzyme evolution is a powerful approach for discovering new catalysts with applications in green chemistry and elsewhere. However, “hits” in sequence space are rare: If too few members of a library are examined, the chances of success of a campaign are limited. Ultrahigh-throughput screening in emulsion droplets has dramatically increased the odds but requires a fluorescent reaction product that triggers selection of hits. We now introduce an absorbance-based microfluidic droplet sorter that broadens the scope of assays to those producing UV/Vis-active chromophores and demonstrate its usefulness by evolving a dehydrogenase based on the screening of half a million library members. Making ultrahigh-throughput screening possible for previously inaccessible reactions enables much wider use of microfluidic droplet sorters for laboratory evolution.
Keywords: protein engineering, directed evolution, microfluidics, ultrahigh-throughput, emulsion droplets
Abstract
Ultrahigh-throughput screening, in which members of enzyme libraries compartmentalized in water-in-oil emulsion droplets are assayed, has emerged as a powerful format for directed evolution and functional metagenomics but is currently limited to fluorescence readouts. Here we describe a highly efficient microfluidic absorbance-activated droplet sorter (AADS) that extends the range of assays amenable to this approach. Using this module, microdroplets can be sorted based on absorbance readout at rates of up to 300 droplets per second (i.e., >1 million droplets per hour). To validate this device, we implemented a miniaturized coupled assay for NAD+-dependent amino acid dehydrogenases. The detection limit (10 μM in a coupled assay producing a formazan dye) enables accurate kinetic readouts sensitive enough to detect a minimum of 1,300 turnovers per enzyme molecule, expressed in a single cell, and released by lysis within a droplet. Sorting experiments showed that the AADS successfully enriched active variants up to 2,800-fold from an overwhelming majority of inactive ones at ∼100 Hz. To demonstrate the utility of this module for protein engineering, two rounds of directed evolution were performed to improve the activity of phenylalanine dehydrogenase toward its native substrate. Fourteen hits showed increased activity (improved >4.5-fold in lysate; kcat increased >2.7-fold), soluble protein expression levels (up 60%), and thermostability (Tm, 12 °C higher). The AADS module makes the most widely used optical detection format amenable to screens of unprecedented size, paving the way for the implementation of chromogenic assays in droplet microfluidics workflows.
Directed evolution has arguably become the most popular method to generate enzymes with improved and altered activities (1–5), but the success of this approach is dependent on exploring a maximum of combinatorial diversity (6). Therefore, screening of large libraries of variants is essential and technologies to trawl sequence space efficiently are required. Massive scale-down of assay volumes by compartmentalization of library members in water-in-oil emulsion droplets has recently led to the development of ultrahigh-throughput screening platforms that use small volumes (typically picoliters) and allow sorting of more than 106 variants per hour (7–9) (cf., schematic in Fig. 1A). The key technical module to make this possible is a microfluidic droplet sorter that has so far relied exclusively on fluorescent readouts (fluorescence-activated droplet sorting, FADS) (10, 11). By alternatively transforming microfluidic emulsions to double emulsions (12) or hydrogel beads equipped with polyelectrolyte shells (13), sorting in standard flow cytometers also becomes possible. However, to date, all ultrahigh-throughput screening campaigns implemented in microcompartmentalized formats (including microcapillary arrays) have so far relied on detection of a fluorescent product (13–20). When assays that lack this type of readout are to be used (e.g., involving instead the widely used absorbance detection), droplet screening is currently impossible: Colony screening assays (relying on precipitation of insoluble product and thus encumbered by poor dynamic range and assay quality) or microwell-plate screening with sophisticated robots (higher quality but expensive in terms of capital expenditure and running costs) remain the only options. In the present work, we remedy this unsatisfactory situation and report a microfluidic absorbance detection module suitable for ultrahigh-throughput microdroplet sorting that was validated by successful enzyme evolution. The target chemistry chosen is the NAD+-dependent deamination of amino acids catalyzed by phenylalanine dehydrogenase (PheDH), a representative of the large class of oxidoreductases (21, 22). Variants of PheDH are selected by absorbance-activated droplet sorting (AADS), based on the ability of our device to deliver high levels of enrichment (∼2,800-fold).
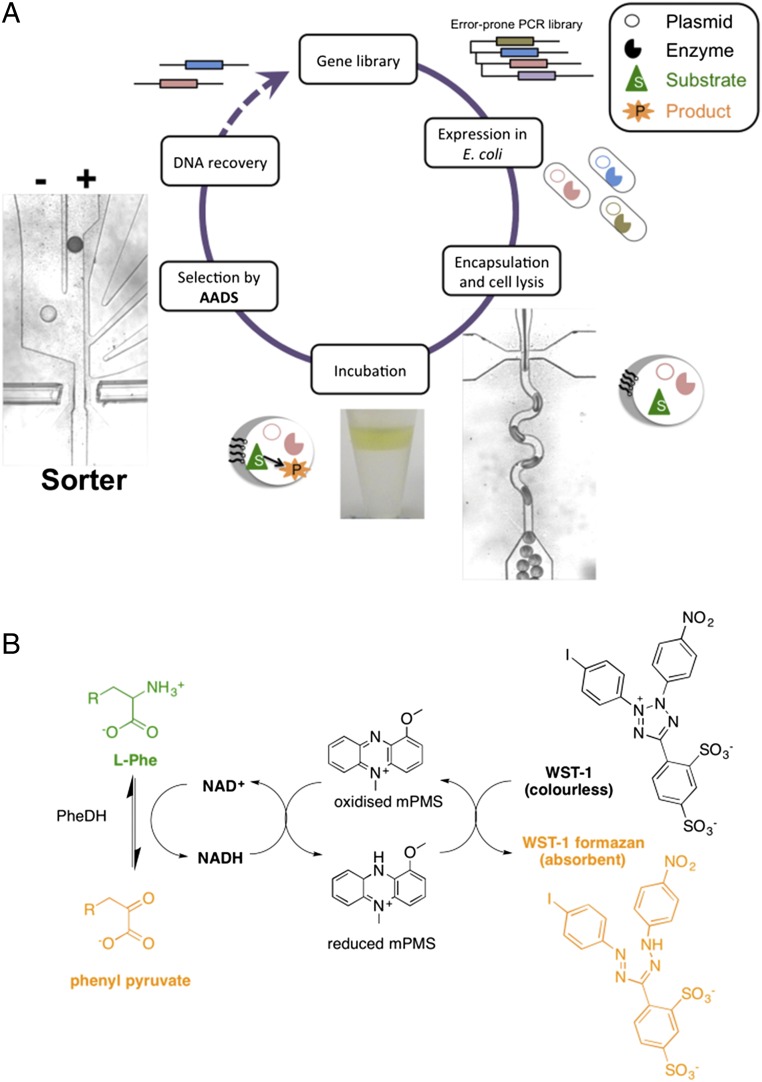
Fig. 1.
Directed evolution cycle based on sorting of microdroplets by their absorbance. (A) Droplets are generated to encapsulate single cells together with a lysis agent and substrates, incubated in tubing, and reinjected into the sorting module. This AADS module relies on head-on fiber optics embedded in the microdroplet sorting device that are used to measure on-the-fly transmittance as droplets pass in-between the fibers. (B) Scheme for the reaction of the NAD+-dependent PheDH from Rhodococcus sp. M4 (PheDH). The reduction of the cofactor NAD+ to NADH in the deamination direction is detected by a coupled assay involving the electron coupling reagent 1-methoxy-5-methylphenazinium methyl sulfate (mPMS) and the reduction of the water-soluble tetrazolium salt 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium (WST-1) to give the absorbing dye WST-1 formazan.
Results
Design of a Microfluidic Sorter Based on Absorbance Detection.
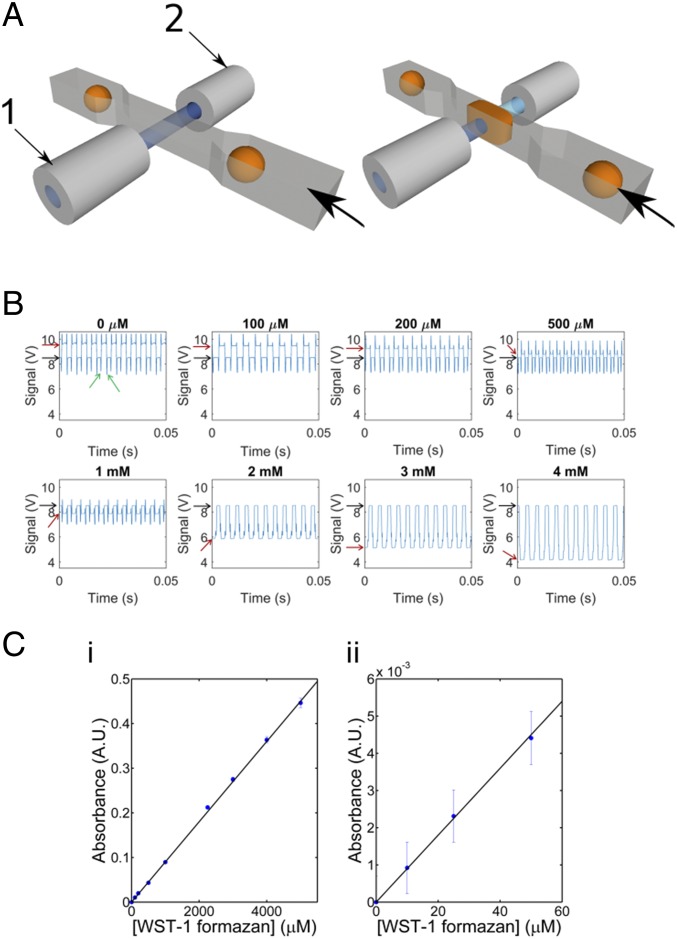
The conversion of l-phenylalanine (l-Phe) to phenyl pyruvate by PheDH was detected in a coupled reaction, in which the oxidation of NADH leads to the coupled reduction of a water-soluble tetrazolium salt, WST-1, to give a formazan dye (in the presence of the electron coupling reagent mPMS; Fig. 1B). To detect the formazan product, an absorbance setup was designed, in which two optical fibers—aligned face-to-face across the droplet channel (Fig. 2A)—were embedded into a planar microfluidic chip (SI Appendix, Fig. S1 A and B). Whenever a droplet passes through the interrogation volume, a decrease in transmittance relative to its dye concentration is recorded and used as the basis of a sorting decision. To detect small transmittance changes, the power of the LED light source was maximized so that the photodiode detector worked close to its saturation level and the estimated detection limit for WST-1 formazan was ∼7.5 μM (SI Appendix, A.1). We experimentally verified the sensitivity of the setup by measuring dilutions of the dye. Readouts from the photodetector for droplets with increasing concentrations of WST-1 formazan from 0 to 4 mM are shown in Fig. 2B (larger plots are available in SI Appendix, Fig. S2). The transmittance was calculated from the average voltage corresponding to the center of the droplets, away from the signal spikes originating from refraction at the oil–water interface of droplets crossing the detection chamber. Although the transmittance values for droplets decrease with increasing WST-1 formazan, the signal for the oil remains constant. In practice, the droplet transmittance is greater than that of the oil at low WST-1 formazan concentration and less than that of the oil at high concentrations (SI Appendix, Fig. S3). From these data, an absorbance calibration plot was constructed (Fig. 2C), showing excellent linearity over a wide range of concentrations (based on monitoring >200 droplets, with low coefficients of variation <5%). The sensitivity depends on good fiber alignment minimizing stray light, which, in our case, was measured to be <0.5% (determined with a strongly absorbing food dye). The experimental limit of detection (Fig. 2) was thus found to be around 10 μM WST-1 formazan, nearly coinciding with the previous estimate and suggesting that the device is optimally implemented.
Fig. 2.
AADS for droplet microfluidics. (A) Schematic of the head-on fiber optics on both sides of a microchannel. One fiber (1) is coupled to a light source, whereas the one facing it across the droplet channel (2) is coupled to a photodetector. When a droplet passes through the interrogation volume, the intensity of light detected decreases concomitantly with its concentration of a chromogenic dye. (B) Dilution series for the detection of WST-1 formazan. Series of droplets read in sequence for several concentrations (here covering a range from 0 to 4 mM WST-1). The scatter signal at the edges of droplets (marked by green arrows) defines the boundaries of an absorbance measurement in one droplet. The transmittance value for each droplet is read as the average value in-between these edges. The relative positions of the transmittance values for the droplet content and the oil phase (indicated as red vs. black arrows on the left) cross over around 650 µM WST-1 formazan (see SI Appendix, Fig. S2 for larger versions of the plots shown here), because the droplet signal is greater than the invariant baseline signal for the oil below this concentration and less than that above. For the sorting of libraries, 1 mM WST-1 formazan was added to minimize the signal spikes caused by scattering at droplet boundaries at low dye concentrations: For [WST-1 formazan] >2 mM, the spikes all but disappear. (C) Calibration plot showing the linearity of absorbance across a range of WST-1 formazan concentrations using the readout shown in A, in two different concentration regimes: (i) between 0 and 5 mM and (ii) from 0 to 50 μM (glycine–KOH buffer, 100 mM, pH 10, 20 °C).
Operation and Calibration of the Absorbance Sorter.
During AADS screening campaigns, improved enzyme variants producing more absorbing product than the wild-type catalyst have to be detected and sorted by activation of on-chip electrodes that pull droplets toward the sorting channel dielectrophoretically (11, 23). To validate the microfluidic operation of the sorter, mixtures of droplets containing (i) PBS buffer or, alternatively, (ii) dilutions of a strongly absorbing black food dye were generated, stored in tubing, and reinjected into the sorting chip. An open-source Arduino board (Arduino Due) was used to monitor the voltage signal of the detector in real time and to compare this voltage to a threshold value below which a trigger was released (SI Appendix, A.5). This trigger signal, in turn, activated the high-voltage AC field applied to the electrodes (SI Appendix, Fig. S4 A and B). Snapshots of a droplet in the process of being sorted (recognizable by the encapsulated black dye) are displayed in Fig. 3A (see Movie S1).
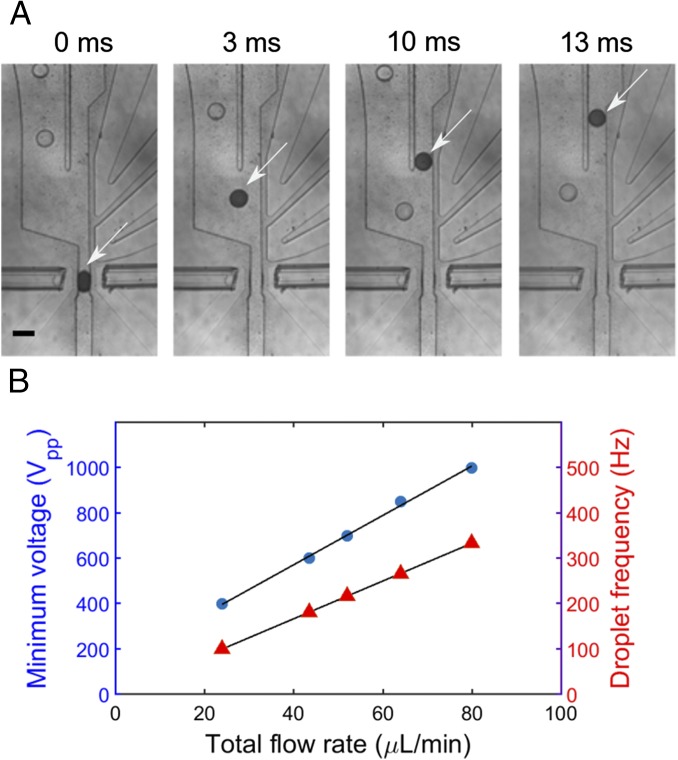
Fig. 3.
Operation of AADS. (A) Snapshots of microdroplets selectively directed toward the sorting channel by triggering embedded electrodes on their absorbance signal. The white arrows on each snapshot track a droplet as it is being pushed toward the sorting channel. Optical fibers were inserted into guide microchannels and adjusted to be in close proximity (∼50 µm) to the droplet channel. A narrower neck (width, 50 µm; channel height, 80 µm) induces local elongation of the droplet, which facilitates absorbance measurements. The flow pattern in the device was designed so that droplets would naturally flow to the waste channel (Left) in the absence of an electric field. Whenever a transmittance below a set voltage threshold was detected, electric pulses (square 1 kVpp, 10 kHz, ∼5 ms long) were applied to attract droplets dielectrophoretically toward the sorting channel (Right). Salt water electrodes were used to conduct the electric field. (Scale bar, 125 µm.) (B) Calibration of the voltage (blue) required for successful sorting of droplets and the corresponding droplet sorting frequency (red) as a function of total flow rate (spacing oil plus emulsion). The ratio of the flow rate of emulsion to the spacing carrier phase was typically kept constant (1:15). Black lines are linear fits drawn to guide the eye.
The maximum droplet frequency at which stable and reliable sorting was achieved depended on the total flow rate (the sum of oil and emulsion flow rates): The faster the droplets moved through the sorting junction, the higher the electric field required for sorting had to be (Fig. 3B). Spacing oil was injected fast enough to bring about a large separation between droplets within the channel and ensure single droplet sorting (SI Appendix, A.2). With a square wave of 3 ms (1,000 Vpp, 10 kHz), a sorting frequency of over 300 Hz (at a total flow rate of 80 μL/min) could be achieved with droplets of 70 μm in diameter.
The current sorting algorithm simply compares real-time analog voltage to an arbitrary user-set voltage threshold. That implies that low dye concentrations (<100 μM) cannot be sorted, because they are effectively masked by signal spikes induced by refractive index mismatch between the phases (carrier oil, n = 1.29; dispersed aqueous phase, n = 1.34; see Fig. 2B and SI Appendix, Fig. S5 and A.4). To bypass this limitation, an offset of 1 mM WST-1 formazan was added in the single-cell sorting experiments to enable sorting with better sensitivity by masking the spikes (∼10 μM WST-1 formazan; see Fig. 2B).
Time Courses of Product Formation from Single-Cell Picoliter Lysates.
The required incubation time for single-cell lysate assays is dependent on the activity of the enzyme but also crucially on the droplet volume, because the amount of enzyme cannot exceed the quantity expressed by a single cell. To determine the incubation time necessary to detect product formation, time courses of the reaction with wild-type PheDH (wtPheDH) were performed. To this end, ∼105 cells expressing wtPheDH were encapsulated into ∼106 single emulsion droplets to ensure single-cell occupancy according to Poisson statistics, along with substrate l-Phe (10 mM; i.e., > KM, to test under conditions of maximal turnover rate) and lysis agents. The droplet absorbance was subsequently measured in the sorter at different incubation times to monitor the progress of the reaction (Fig. 4A and SI Appendix, Fig. S6).
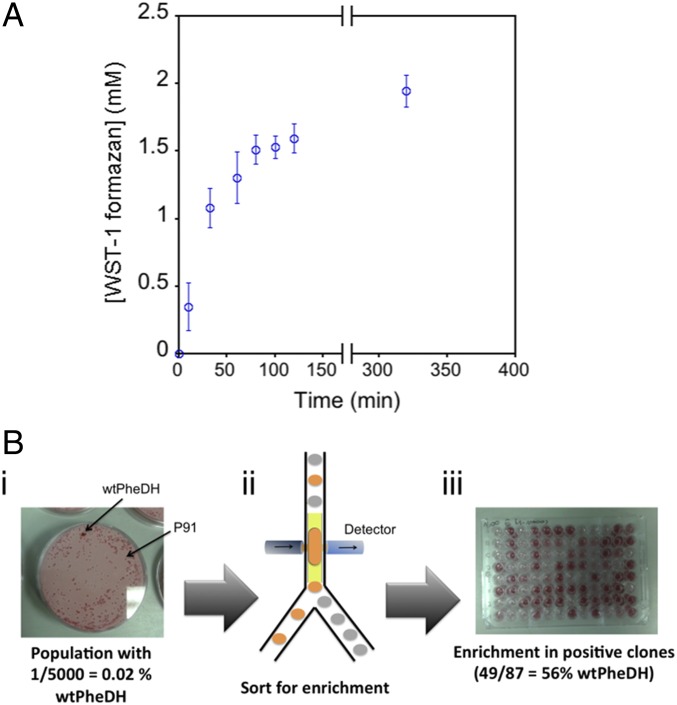
Fig. 4.
Detection and sorting of PheDH activity based on absorbance in droplets. (A) Time-dependent single-cell lysate in droplets for the reaction of wtPheDH. Conditions: [l-Phe], 10 mM; [NAD+], 10 mM; [WST-1], 5 mM; [mPMS], 5 μg/mL; in glycine–KOH buffer (100 mM), pH 10, 25 °C. Incubation time for library screening was chosen in order for wtPheDH to be close to the assay end point (>2 h). (B) Enrichment of cells by AADS based on PheDH activity. (i) E. coli cells expressing either wtPheDH or an enzyme not active on l-Phe (P91) were mixed at a 1:5,000 ratio before compartmentalization into single droplets; this population was analyzed by colony screening to detect PheDH activity (showing here the detection of one active colony out of ∼5,000 screened in total). (ii) After compartmentalization, droplets were sorted after 3 h of reaction (∼2 mM WST-1 formazan) to enrich for cells expressing wtPheDH. (iii) The sorted population was analyzed for PheDH activity by microplate cell lysate assays to determine enrichment ratios [here, 49 colonies (56%) were active out of 87 screened in total, corresponding to an enrichment value of 2,800].
Importantly, droplet-to-droplet cross-contamination of the reaction product was not observed (24), as droplets adjacent to positive droplets did not show detectable intake of dye over the course of the experiment (5 h; see SI Appendix, A.3). The signal reached saturation after 2 h of incubation, resulting in 2 mM WST-1 formazan production from a starting concentration of 10 mM l-Phe, indicating either depletion of or inhibition by at least one of the components within the droplets or enzyme inactivation.
Given the enzyme’s catalytic efficiency (kcat/KM ∼7 × 104 M−1·s−1; SI Appendix, Table S2) and the observation of ∼106 active enzymes produced per cell (SI Appendix, Text B, Table S1, and Fig. S14), the lower detection limit of the sorter (10 μM WST-1 formazan in a 180 pL droplet) is surpassed after 60 s, from which point onwards the reaction can be monitored and its signal used for sorting.
Sorting Efficiency Quantified by Enrichment Analysis.
The ability to isolate droplets containing an active enzyme that produces an absorbing downstream product (e.g., WST-1 formazan) was tested first by mimicking a library sorting experiment to enrich droplets containing wtPheDH from an overwhelming majority of droplets containing an enzyme not active on l-Phe (the phosphotriesterase P91) (15). After expression of both enzymes in separate liquid cultures, around 105 cells were mixed in a 1:5,000 ratio before Poisson-distributed single-cell compartimentalization with 10 mM substrate (i.e., l-Phe). After collection in a syringe and incubation (3 h), the droplets were injected into the absorbance sorting chip, and the ones containing active PheDH were collected based on WST-1 formazan product absorbance. After sorting, the plasmid DNA was recovered from droplets and transformed into Escherichia coli cells, which were subsequently grown on agar plates overnight. To estimate the enrichment ratio (defined as the percentage of positive variants after sorting divided by the same percentage before sorting), randomly picked colonies were screened for PheDH activity (Fig. 4B). The experiment yielded a 2,800-fold enrichment, corresponding to 56% positive hits in the sorted population. As previously shown (10), enrichment is mainly limited by accidental coencapsulation of negative and positive variants. Additional false-positives specific to absorbance detection include the presence of light-absorbing objects (e.g., dust particles, precipitates) inside droplets or the measurement of two droplets in close contact creating an enhanced scatter at their interface (see examples in Movie S2). In a typical model-sorting experiment, false-positive rates were found to be below 5% of sorted events. To further control the process, videos of sorting events were continuously and automatically acquired (via camera software) and saved, so that successful sorting could be ensured by inspection of videos during and after the experiment (see Movie S3 for examples of 10 short clips).
Directed Evolution of wtPheDH.
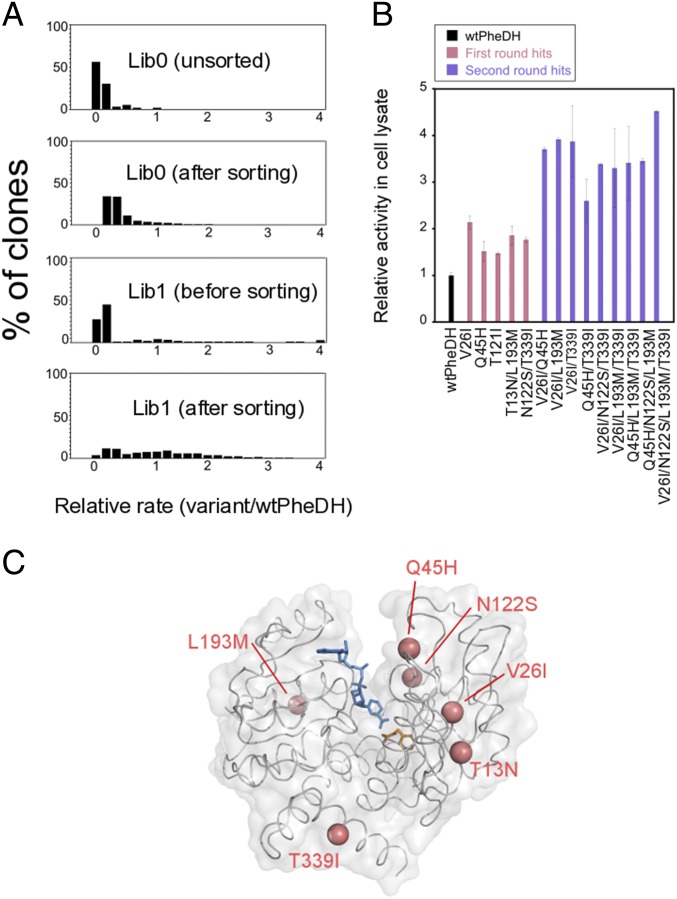
Next, the activity of wtPheDH toward its native substrate l-Phe was increased by directed evolution to further test whether the workflow outlined in Fig. 1A can yield improved enzymes. An error-prone PCR library of wtPheDH (Lib0; 106 transformants, 1.7 mutations per gene) was constructed and transformed into E. coli. Before sorting, randomly picked library members were assayed for cell lysate activity against l-Phe in 96-well plates to establish the activity profile of Lib0. This preliminary analysis (Fig. 5A) showed that the majority of variants (>90%; SI Appendix, Fig. S9) were highly deleterious (i.e., showing >twofold decreased activity relative to wtPheDH), whereas positive variants (showing >1.3-fold improvement relative to wtPheDH; i.e., significantly more than the SD) were less abundant (<1%; SI Appendix, Fig. S9). To enrich positive variants while maintaining high screening throughput, this library was sorted in droplets using a 1:2 (cell:droplet) ratio (forfeiting single-cell occupancy according to Poisson statistics for higher throughput). Around 5 × 105 cells (each of them expressing a single PheDH variant) were compartimentalized into single emulsions, along with 10 mM substrate. After incubation for 3 h at room temperature, these droplets were injected into the AADS device and sorted at ∼100 Hz with a selection threshold set to collect the 0.2% most absorbing droplets (SI Appendix, Fig. S8). This yielded ∼1,000 selected variants after DNA recovery and retransformation. Analysis of this sorted population in microwell plates (Fig. 5A) showed that the sorting experiment successfully enriched positives (∼6% of the sorted population; SI Appendix, Fig. S9) by at least sixfold, along with a decrease in the proportion of deleterious mutants (from >90% to 75%; SI Appendix, Fig. S9). Five mutants (containing mutations T13N, V26I, Q45H, T121I, N122S, L193M, and T339I; Fig. 5C) with up to 2.2-fold improved relative activity in cell lysates were identified (Fig. 5B), of which three showed improved soluble expression of the enzyme (up to twofold compared with wtPheDH; SI Appendix, Fig. S11). These results demonstrate that screening a higher number of variants under high droplet occupancy [e.g., using a 1:2 (cell:droplet) ratio] is an effective strategy to enrich a population for rare positive variants (e.g., from <1% before sorting to ∼6% after) that conventional plate screening approaches are unlikely to recover because of limited throughput (see SI Appendix, Fig. S9). For the second round of evolution, a shuffling library (Lib1; 106 transformants) was generated to identify beneficial combinations of the first-round hit mutations. As in the first round, the PheDH activity profile of the library was analyzed before screening by AADS (Fig. 5A and SI Appendix, Fig. S10). Around 10% of the assayed variants showed >twofold increased activity relative to wtPheDH (SI Appendix, Fig. S10). To enrich the library population for these potential hits, around 105 cells were compartimentalized into 106 single emulsions [i.e., corresponding to a 1:10 (cell:droplet) ratio for further AADS under single-cell occupancy conditions]. The selection threshold was set to collect the 1% most absorbing droplets (corresponding to the 10% most active variants; Fig. 5A and SI Appendix, Fig. S8), finally yielding ∼800 variants after DNA recovery and retransformation. The top 5% of the variants (with ≥ threefold improvements) were sequenced, leading to the identification of nine additional hits combining two to four mutations (Fig. 5B). Among these, the quadruple mutant V26I/N122S/L193M/T339I showed the highest improvement under our screening conditions, with a 4.6-fold improvement in lysate activity (relative to that of wtPheDH) (Fig. 5B). The kcat measured with purified protein was 2.7-fold improved (SI Appendix, Fig. S12 and Table S2), whereas no improvement in kcat/KM was observed, because KM and kcat showed compensatory behavior (SI Appendix, Table S2). Instead, this variant showed a 2.2-fold increase in soluble expression indicative of improved folding efficiency (SI Appendix, Fig. S11) as well as increased thermostability (Tm value increased by ∼12 °C at pH 8) and a half-life of inactivation at 50 °C increased by 7.5-fold (SI Appendix, Fig. S13 and Tables S3 and S4). Overall, this directed evolution experiment yielded 14 improved variants, demonstrating the validity of our approach. In single-cell lysate assays, the readout is a combination of the enzyme’s activity, solubility, stability, and its amount expressed within the cell, which is reflected in the enhanced properties of the selected enzyme variants. The mutations identified mainly affected solvent-exposed residues (V26I, Q45H, L193M, T339I; Fig. 5C), which may be indicative of their involvement in protein stabilization and folding efficiency (25–27). One mutation (N122S) is located close to the catalytic residue D118, in a region previously shown to be involved in substrate specificity in amino acid dehydrogenases (28).
Fig. 5.
Directed evolution of wtPheDH for increased activity against l-Phe. (A) Enrichment of positive hits over two rounds of droplet sorting. Shown are relative rates of 96 variants that were randomly picked from the original library (Lib0) and after each round of sorting (Lib0–Lib1). The data were normalized to the rate of wtPheDH (=1). (B) Activity changes among selected variants from the first and the second round of evolution. The activity for each variant is plotted relative to that of wtPheDH. Data are averages of triplicate values from three independent experiments, and error bars represent ±1 SEM. (C) The structure of wtPheDH (Protein Data Bank ID code 1BW9) with residues that were mutated in the two rounds of evolution shown in pink. The active site contains phenyl pyruvate (orange) and NAD+ (blue).
Discussion
A Sensitive Sorting Device Based on Absorbance Readouts.
The limited sensitivity of absorbance detection when working with small path lengths has generally been seen to preclude its use as a quantitative readout for droplet microfluidics assays. The design feature that makes our sorter useful for single-cell selections is the relatively large droplet volume (180 pL; diameter, 70 μm) that boosts sensitivity (by a larger path length) while keeping the sample/volume ratio high (1 million compartments generated from only 180 μL). This high sample/volume ratio and a screening rate of ∼300 per second exceed the throughput of assays in multiwell plates by 1,000-fold (14). In addition, building the system is relatively straightforward: Fiber-coupled LED sources and standard SI-photodiode detectors are inexpensive, integration of optical fibers on chip bypasses the need for optical tables and alignment procedures, and microcontroller actuation of the sorter is simple to implement (see SI Appendix, Fig. S4B). Further important design features of the microfluidic sorter are (i) the proximity of the detection area and the sorting junction (within 400 µm) to synchronize detection and sorting events; (ii) the slight squeezing of droplets in the detection area to form plugs that separate edge spikes from assay absorbance, ensuring reproducible measurements; (iii) the alignment of the detection point with the sorting channel to maximize the dielectrophoretic force acting on the droplets; and (iv) the use of a pressure equilibrator before the outlets to minimize hydrostatic effects on droplet flow, as reported previously (29). Previous absorbance measurements in droplets have lower analytical throughput [<10 Hz in published work (30–34); i.e., an order of magnitude below AADS] and crucially lack the connection to a sorting module, preventing their use for directed evolution.
Absorbance assays are intrinsically 3–4 orders of magnitude less sensitive compared with fluorescence assays: In platforms such as FACS or FADS, concentrations as low as 2.5 nM fluorescein can be readily detected (15, 18). Nevertheless, the ability to quantify micromolar concentrations of absorbing reaction product (∼10 μM) will make many established enzymatic assays accessible for droplet-based ultrahigh-throughput screening. The performance of AADS could be further improved by using monochromatic laser light with a wavelength centered at the absorption maximum of the dye, embedding optical cavities on chip, which can increase sensitivity about fivefold (35), or using a lock-in amplifier to extract the signal from electronic noise (36). Further increases in the sorting frequency may be achieved with smaller droplets, higher electric fields, or forcing droplets to flow closer to the central separation wall. Improvements to bypass the presence of spikes at the oil–aqueous interface could include algorithmic detection within the microcontroller, the use of refractive-index matching oils, or the use of electronic low-pass filters. The direct detection of NADH formation is possible but challenging due to its low extinction coefficient (6,220 M–1·cm−1 in water at 340 nm) as well as absorption losses within the fiber optics at this wavelength. However, the present setup allowed detection of 250 μM NADH (using a 365-nm LED light source).
Utility of Absorbance-Based Selections for Directed Evolution.
The power of in vitro compartmentalization for the screening of enzyme activities has been demonstrated previously with several formats based on fluorescence-based readouts (7, 12–14, 18, 37–39) The development of the AADS module further extends the range of applications toward enzymes for which no fluorescent assays are available or where fluorescent reporters are only accessible by complicated organic synthesis. Even when a fluorogenic assay is available, absorbance detection may offer advantages: Typically fluorogenic moieties are large and hydrophobic and render substrates and products often insoluble, so that solubility effects obscure the readout of enzymatic turnover. The size of the typically large fluorogenic moiety may also divert directed evolution away from the structure of the desired substrate. In the specific case of amino acid dehydrogenases, the substrate CTC–formazan cannot be used in assays in solution, because it is only fluorescent in the solid state (40, 41).
The sorting of improved PheDH variants was demonstrated by driving the absorbance detection sorter at high, practically useful rates (∼100 Hz). This throughput enables the analysis of 106 droplets within 3 h with a total reaction volume smaller than 200 μL, which offers a rapid and more economical strategy than any other conventional format [e.g., a more than half a million-fold smaller assay volume than microwell plate screening (14)]. Formation of absorbing product in the micromolar range can be readily detected given a high extinction coefficient (>30,000 M–1·cm−1) and a water-soluble dye. Although we conducted selections under conditions typical for biocatalytic applications (i.e., at saturating substrate concentration >KM, where a maximal turnover rate is achieved), experiments around KM are also possible (as suggested by the clear separation of positives and negatives at [S] = 1 mM; see SI Appendix, Fig. S7).
The sorter we describe can detect ∼109 molecules of the dye WST-1 formazan, which may be compared with close to a million enzyme copies produced in one E. coli cell (SI Appendix, Text B, Table S1, and Fig. S14) or >104 in in vitro expression (42). Based on this ratio, ∼1,300 turnovers per enzyme molecule in the single-cell lysate assays used in this work are necessary to reach the detection limit, which does not impose unreasonable demands on enzyme proficiency. In the case of PheDH, the high activity of the enzyme leads to a robust phenotype, producing ∼2 mM of product after 3 h. This means that enzymes with activities up to three orders of magnitude lower (kcat/KM ∼100 M−1·s−1) should still produce enough product to be amenable to selection by AADS within a reasonable time frame. Even conventional approaches that provide reasonable throughput (∼105 per day), such as colony screening, are an order of magnitude slower than AADS and additionally compromised by low assay quality.
Protein engineering efforts, including directed evolution, are often hampered by inefficient expression and lack of stability of the target proteins (43, 44), so identifying variants with both enhanced soluble expression levels (i.e., increased kinetic protein stability) and long-term endurance in ultrahigh throughput suggests that this system is a useful enabling module of the microfluidic toolkit for enzyme improvement. An improvement of these properties via this workflow sets the scene for step-by-step (45) accumulation of mutations that enhance evolvability (46, 47) and allow adaptation of substrate specificity or introduction of mechanistic switches (48–51). The more robust quadruple mutant PheDHV26I/N122S/L193M/T339I will be useful to buffer the effects of the growing mutational load that carries a stability cost in future evolution rounds. The ability to increase the activity of selected PheDH variants toward l-Phe substantially in two rounds of directed evolution validates AADS for an enzyme class with valuable applications in large-scale processes: NADH-dependent enzymes are widely used in industry for the production of chiral amines (52). The ultrahigh throughput that is achievable with droplet microfluidics can now be applied to a wider circle of reactions, specifically NADPH cofactor-dependent enzymes. The ease by which the proverbially vast sequence space can be explored for these types of reactions that so far could only be evolved with lower throughput will increase the chances of success in their directed evolution.
Materials and Methods
Flow-Focusing Droplet Generation.
A conventional droplet generator (height, 80 µm; width, 50 µm at the flow-focusing junction) was used to generate droplets at high rates (>1 kHz). Fluorinated oil HFE-7500 was purchased from 3M Novec and the surfactant (Picosurf1) from Dolomite Microfluidics. Chips were operated with syringe pumps (Nemesys, Cetoni) and gas-tight glass syringes (SGE). The flow rates for oil, cell preparation, and the lysis/substrate mix were 30 μL/min, 8 μL/min, and 8 μL/min, respectively.
Chip Fabrication.
The sorting chip was designed with CAD software (DraftSight, Dassault Systems) and the device fabricated following classical soft-lithography procedures by using a high-resolution acetate mask (Microlithography Services Ltd.) and SU-8–2025 photoresist patterning (53). The resulting microchannels obtained in polydimethylsiloxane (PDMS) were plasma bonded on a thin cured PDMS layer (5 g PDMS in a Petri dish; Ø, 9 cm), flushed with 2% (vol/vol) Trichloro(1H,1H,2H,2H-perfluorooctyl)silane in HFE-7500, put in a 65 °C oven for 30 min, and the PDMS–PDMS devices then bonded to a microscope glass slide. Electrodes were made by filling channels with salt solution (5 M NaCl) as a practical alternative to low-melting point metals (54). Optical fibers (SMA Fiber Patch Cable, 50 microns, 0.22 NA; Thorlabs) were stripped (so that only the cladding was left) and manually inserted into the chip under a microscope. Alignment was tested by connecting one fiber to the fiber-coupled LED light source (M455F1, royal blue, Thorlabs) and the other to the photodetector (PDA36A, Thorlabs). Once proper alignment was obtained, the fibers were fixed with epoxy glue, and freshly degassed PDMS was inserted into the fiber channels through specific inlets to remove air, preventing the scattering of light (see SI Appendix, Fig. S1). The PDMS was left overnight at room temperature inside the channels for curing. Design files will be deposited on the DropBase (openwetware.org/wiki/DropBase).
Sorting Electronics.
The voltage signal from the photodetector was split in two, recorded with a custom Labview program [using an Analog-to-Digital Converter (NI, USB-6009)], and at the same time sampled in 12-bit resolution via an analog-in pin of a 32-bit CortexM3 ARM-based microcontroller (Arduino Due). To match the voltage of the detector (max., 10 V) to the maximum voltage tolerable for the microcontroller board (3.3 V), a simple voltage divider (10 kΩ resistors) was used. On the microcontroller, the signal from the detector was compared with a threshold value for sorting. Below the threshold, a pin was set high to activate a pulse generator (TGP110, Thurlby Thandar Instruments), used to generate a 5-V pulse, and manually adjusted to be smaller than the droplet period (typically several milliseconds). The pulse generator controlled a function generator (20 MHz DDS Function Generator TG 2000, TTi) working in external gated mode, generating a 10-kHz square signal at an adjustable amplitude, which was then amplified 100 times with a voltage amplifier (TREK 601c) to actuate the sorter. This setup enabled convenient manual adjustment of all important sorting parameters.
Directed Evolution.
The wtPheDH gene and its variants were cloned into pASK-IBA63b-plus (IBA) and expressed in fusion with a C-terminal Strep-tag. Gene libraries were generated by error-prone PCR (Lib0; 106 transformants; 1.7 mutations per gene) and StEP PCR (55) (Lib1; 106 transformants) for the first and the second round of directed evolution, respectively. The library variants were screened by compartmentalization of single E. coli cells, each expressing an individual library variant, in single droplets and sorted by AADS. Briefly, E. coli BL21(DE3) cells were transformed with the plasmids carrying the wtPheDH library variants. The cells were grown in suspension culture (LB, 100 µg/mL ampicillin) for protein expression under the control of the Tet promoter (200 ng/mL anhydrotetracyclin) and subsequently compartmentalized in single emulsions (ratio 1:2 for Lib0 and 1:10 for Lib1) in 100 mM glycine–KOH buffer, pH 10, with substrates (10 mM l-Phe, 10 mM NAD+, 5 mM WST-1, and 5 µg/mL mPMS, and 1 mM of WST-1 formazan as offset) and cell lysis agents [120 KU/mL rLysozyme and 4% (vol/vol) CelLytic B]. After collection in a syringe and incubation at room temperature (90 min), the droplets were injected into the AADS chip and sorted based on their absorbance at 455 nm using a defined selection threshold (see SI Appendix, Fig. S8). Plasmid DNA from collected droplets was recovered by de-emulsification using 1H,1H,2H,2H-perfluorooctanol, purified, and transformed into E. coli E. cloni 10G cells (Lucigen) by electroporation (18). After overnight growth on solid media (LB agar, 100 µg/mL ampicillin), plasmid DNA was extracted from the colonies and retransformed into E. coli BL21(DE3) cells for further colony- or 96-well-plate–based screenings. Details of screening procedures, expression analysis, purification, and characterization of selected hits are available in SI Appendix.
Supplementary Material
Acknowledgments
This research was funded by the Engineering and Physical Sciences Research Council (studentship to R.H. and an Impact Acceleration Account Partnership Development Award), the Biological and Biotechnological Research Council (BBSRC), and Johnson Matthey. S.E. and M.F. were supported by postdoctoral Marie-Curie fellowships. F.H. is an ERC Investigator.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606927113/-/DCSupplemental.
References
- 1.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485(7397):185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 2.Turner NJ. Directed evolution drives the next generation of biocatalysts. Nat Chem Biol. 2009;5(8):567–573. doi: 10.1038/nchembio.203. [DOI] [PubMed] [Google Scholar]
- 3.Bershtein S, Tawfik DS. Advances in laboratory evolution of enzymes. Curr Opin Chem Biol. 2008;12(2):151–158. doi: 10.1016/j.cbpa.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 4.Park HS, et al. Design and evolution of new catalytic activity with an existing protein scaffold. Science. 2006;311(5760):535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]
- 5.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10(12):866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H, Cornish VW. Screening and selection methods for large-scale analysis of protein function. Angew Chem Int Ed Engl. 2002;41(23):4402–4425. doi: 10.1002/1521-3773(20021202)41:23<4402::AID-ANIE4402>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Colin PY, Zinchenko A, Hollfelder F. Enzyme engineering in biomimetic compartments. Curr Opin Struct Biol. 2015;33:42–51. doi: 10.1016/j.sbi.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Schaerli Y, Hollfelder F. The potential of microfluidic water-in-oil droplets in experimental biology. Mol Biosyst. 2009;5(12):1392–1404. doi: 10.1039/b907578j. [DOI] [PubMed] [Google Scholar]
- 9.Theberge AB, et al. Microdroplets in microfluidics: An evolving platform for discoveries in chemistry and biology. Angew Chem Int Ed Engl. 2010;49(34):5846–5868. doi: 10.1002/anie.200906653. [DOI] [PubMed] [Google Scholar]
- 10.Baret JC, et al. Fluorescence-activated droplet sorting (FADS): Efficient microfluidic cell sorting based on enzymatic activity. Lab Chip. 2009;9(13):1850–1858. doi: 10.1039/b902504a. [DOI] [PubMed] [Google Scholar]
- 11.Link DR, et al. Electric control of droplets in microfluidic devices. Angew Chem Int Ed Engl. 2006;45(16):2556–2560. doi: 10.1002/anie.200503540. [DOI] [PubMed] [Google Scholar]
- 12.Zinchenko A, et al. One in a million: Flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution. Anal Chem. 2014;86(5):2526–2533. doi: 10.1021/ac403585p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischlechner M, et al. Evolution of enzyme catalysts caged in biomimetic gel-shell beads. Nat Chem. 2014;6(9):791–796. doi: 10.1038/nchem.1996. [DOI] [PubMed] [Google Scholar]
- 14.Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA. 2010;107(9):4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colin P-Y, et al. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat Commun. 2015;6:10008. doi: 10.1038/ncomms10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjostrom SL, et al. High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab Chip. 2014;14(4):806–813. doi: 10.1039/c3lc51202a. [DOI] [PubMed] [Google Scholar]
- 17.Najah M, et al. Droplet-based microfluidics platform for ultra-high-throughput bioprospecting of cellulolytic microorganisms. Chem Biol. 2014;21(12):1722–1732. doi: 10.1016/j.chembiol.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Kintses B, et al. Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem Biol. 2012;19(8):1001–1009. doi: 10.1016/j.chembiol.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Chen B, et al. High-throughput analysis and protein engineering using microcapillary arrays. Nat Chem Biol. 2016;12(2):76–81. doi: 10.1038/nchembio.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bornscheuer UT. Protein engineering: Beating the odds. Nat Chem Biol. 2016;12(2):54–55. doi: 10.1038/nchembio.1989. [DOI] [PubMed] [Google Scholar]
- 21.Hall M, Bommarius AS. Enantioenriched compounds via enzyme-catalyzed redox reactions. Chem Rev. 2011;111(7):4088–4110. doi: 10.1021/cr200013n. [DOI] [PubMed] [Google Scholar]
- 22.Kohls H, Steffen-Munsberg F, Höhne M. Recent achievements in developing the biocatalytic toolbox for chiral amine synthesis. Curr Opin Chem Biol. 2014;19:180–192. doi: 10.1016/j.cbpa.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Ahn K, et al. Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl Phys Lett. 2006;88(2):024104. [Google Scholar]
- 24.Courtois F, et al. Controlling the retention of small molecules in emulsion microdroplets for use in cell-based assays. Anal Chem. 2009;81(8):3008–3016. doi: 10.1021/ac802658n. [DOI] [PubMed] [Google Scholar]
- 25.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369(5):1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 26.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci USA. 2009;106(29):11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer RM, Shende VR, Motl N, Pace CN, Scholtz JM. Toward a molecular understanding of protein solubility: Increased negative surface charge correlates with increased solubility. Biophys J. 2012;102(8):1907–1915. doi: 10.1016/j.bpj.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataoka K, et al. Site-directed mutagenesis of a hexapeptide segment involved in substrate recognition of phenylalanine dehydrogenase from Thermoactinomyces intermedius. J Biochem. 1993;114(1):69–75. doi: 10.1093/oxfordjournals.jbchem.a124142. [DOI] [PubMed] [Google Scholar]
- 29.Sciambi A, Abate AR. Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip. 2015;15(1):47–51. doi: 10.1039/c4lc01194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasan V, Pamula VK, Fair RB. An integrated digital microfluidic lab-on-a-chip for clinical diagnostics on human physiological fluids. Lab Chip. 2004;4(4):310–315. doi: 10.1039/b403341h. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi V, et al. A modular approach for the generation, storage, mixing, and detection of droplet libraries for high throughput screening. Lab Chip. 2010;10(18):2433–2442. doi: 10.1039/c004768f. [DOI] [PubMed] [Google Scholar]
- 32.Deal KS, Easley CJ. Self-regulated, droplet-based sample chopper for microfluidic absorbance detection. Anal Chem. 2012;84(3):1510–1516. doi: 10.1021/ac202791d. [DOI] [PubMed] [Google Scholar]
- 33.Gielen F, et al. A fully unsupervised compartment-on-demand platform for precise nanoliter assays of time-dependent steady-state enzyme kinetics and inhibition. Anal Chem. 2013;85(9):4761–4769. doi: 10.1021/ac400480z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi K, Mudrik JM, Wheeler AR. A guiding light: Spectroscopy on digital microfluidic devices using in-plane optical fibre waveguides. Anal Bioanal Chem. 2015;407(24):7467–7475. doi: 10.1007/s00216-015-8913-x. [DOI] [PubMed] [Google Scholar]
- 35.Rushworth CM, Jones G, Fischlechner M, Walton E, Morgan H. On-chip cavity-enhanced absorption spectroscopy using a white light-emitting diode and polymer mirrors. Lab Chip. 2015;15(3):711–717. doi: 10.1039/c4lc01264j. [DOI] [PubMed] [Google Scholar]
- 36.Novak L, Neuzil P, Pipper J, Zhang Y, Lee S. An integrated fluorescence detection system for lab-on-a-chip applications. Lab Chip. 2007;7(1):27–29. doi: 10.1039/b611745g. [DOI] [PubMed] [Google Scholar]
- 37.Griffiths AD, Tawfik DS. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003;22(1):24–35. doi: 10.1093/emboj/cdg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aharoni A, Amitai G, Bernath K, Magdassi S, Tawfik DS. High-throughput screening of enzyme libraries: Thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem Biol. 2005;12(12):1281–1289. doi: 10.1016/j.chembiol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Mastrobattista E, et al. High-throughput screening of enzyme libraries: In vitro evolution of a beta-galactosidase by fluorescence-activated sorting of double emulsions. Chem Biol. 2005;12(12):1291–1300. doi: 10.1016/j.chembiol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Huang Z, et al. Rapid detection of Zymomonas mobilis redox activity using 5-cyano-2,3-tolyl-tetrazolium chloride (CTC) Biotechniques. 2000;29(3):424–425, 428. doi: 10.2144/00293bm04. [DOI] [PubMed] [Google Scholar]
- 41.Gruden CL, Fevig S, Abu-Dalo M, Hernandez M. 5-Cyano-2,3-ditolyl tetrazolium chloride (CTC) reduction in a mesophilic anaerobic digester: Measuring redox behavior, differentiating abiotic reduction, and comparing FISH response as an activity indicator. J Microbiol Methods. 2003;52(1):59–68. doi: 10.1016/s0167-7012(02)00134-3. [DOI] [PubMed] [Google Scholar]
- 42.Courtois F, et al. An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem. 2008;9(3):439–446. doi: 10.1002/cbic.200700536. [DOI] [PubMed] [Google Scholar]
- 43.Bloom JD, Arnold FH. In the light of directed evolution: Pathways of adaptive protein evolution. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Socha RD, Tokuriki N. Modulating protein stability—Directed evolution strategies for improved protein function. FEBS J. 2013;280(22):5582–5595. doi: 10.1111/febs.12354. [DOI] [PubMed] [Google Scholar]
- 45.Tracewell CA, Arnold FH. Directed enzyme evolution: Climbing fitness peaks one amino acid at a time. Curr Opin Chem Biol. 2009;13(1):3–9. doi: 10.1016/j.cbpa.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103(15):5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLOS Comput Biol. 2008;4(2):e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6(4):R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 49.Renata H, Wang ZJ, Arnold FH. Expanding the enzyme universe: Accessing non-natural reactions by mechanism-guided directed evolution. Angew Chem Int Ed Engl. 2015;54(11):3351–3367. doi: 10.1002/anie.201409470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bornscheuer UT, Kazlauskas RJ. Catalytic promiscuity in biocatalysis: Using old enzymes to form new bonds and follow new pathways. Angew Chem Int Ed Engl. 2004;43(45):6032–6040. doi: 10.1002/anie.200460416. [DOI] [PubMed] [Google Scholar]
- 51.Toscano MD, Woycechowsky KJ, Hilvert D. Minimalist active-site redesign: Teaching old enzymes new tricks. Angew Chem Int Ed Engl. 2007;46(18):3212–3236. doi: 10.1002/anie.200604205. [DOI] [PubMed] [Google Scholar]
- 52.Kroutil W, et al. Asymmetric preparation of prim-, sec-, and tert-amines employing selected biocatalysts. Org Process Res Dev. 2013;17(5):751–759. doi: 10.1021/op4000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia YN, Whitesides GM. Soft lithography. Angew Chem. 1998;37(5):550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 54.Sciambi A, Abate AR. Generating electric fields in PDMS microfluidic devices with salt water electrodes. Lab Chip. 2014;14(15):2605–2609. doi: 10.1039/c4lc00078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol. 1998;16(3):258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.