Significance
Some bacteria contain tiny magnets, so-called magnetosomes, that allow them to swim oriented by the Earth’s magnetic field. An intracellular filamentous protein, MamK, has previously been shown to align the magnetosomes along a straight line to combine their magnetic moments. Here we show the structure the MamK filaments have at the atomic level. We also show what happens when individual MamK molecules come together to form the filaments, because it is known that filament formation triggers the hydrolysis of the ATP molecule bound to MamK. The structure of monomeric MamK was determined by X-ray crystallography and of the filament by electron cryomicroscopy, image processing, and helical reconstruction.
Keywords: magnetotactic bacteria, magnetosomes, filamentous proteins, bacterial cytoskeleton, cryo-EM
Abstract
Magnetotactic bacteria produce iron-rich magnetic nanoparticles that are enclosed by membrane invaginations to form magnetosomes so they are able to sense and act upon Earth’s magnetic field. In Magnetospirillum and other magnetotactic bacteria, to combine their magnetic moments, magnetosomes align along filaments formed by a bacterial actin homolog, MamK. Here, we present the crystal structure of a nonpolymerizing mutant of MamK from Magnetospirillum magneticum AMB-1 at 1.8-Å resolution, revealing its close similarity to actin and MreB. The crystals contain AMPPNP-bound monomeric MamK in two different conformations. To investigate conformational changes associated with polymerization, we used unmodified MamK protein and cryo-EM with helical 3D reconstruction in RELION to obtain a density map and a fully refined atomic model of MamK in filamentous form at 3.6-Å resolution. The filament is parallel (polar) double-helical, with a rise of 52.2 Å and a twist of 23.8°. As shown previously and unusually for actin-like filaments, the MamK subunits from each of the two strands are juxtaposed, creating an additional twofold axis along the filament. Compared with monomeric MamK, ADP-bound MamK in the filament undergoes a conformational change, rotating domains I and II against each other to further close the interdomain cleft between subdomains IB and IIB. The domain movement causes several loops to close around the nucleotide-binding pocket. Glu-143, a key residue for catalysis coordinating the magnesium ion, moves closer, presumably switching nucleotide hydrolysis upon polymerization—one of the hallmarks of cytomotive filaments of the actin type.
It was discovered in 1975 by Richard Blakemore that certain bacteria sense magnetic fields and swim according to the field’s direction (1). The cells contain specialized iron-rich, membrane-bounded organelles, magnetosomes (2, 3). Magnetosomes contain magnetic nanoparticles that either together confer a magnetic moment to the bacterium or produce a signal that is used by the bacterium’s chemotaxis machinery to swim in a particular direction.
Magnetosomes occur in many different bacteria, but have been studied in some detail in gram-negative Magnetospirillum gryphiswaldense and Magnetospirillum magneticum AMB-1. In these organisms, typically 10 to 20 magnetosomes form as invaginations of the inner membrane, meaning that the nanoparticles are outside of the inner membrane, in the periplasm (4). They align along a straight line, into a chain, along the inner perimeter of the helical cells. Biomineralization and membrane invagination are somewhat coupled, although first empty or almost empty membrane invaginations form (5).
In Magnetospirillum, magnetosome formation is encoded in gene clusters, included in a large magnetosome island. Many genes are required because the process of magnetosome formation is complex, requiring iron uptake, iron transport, biomineralization, membrane enclosure, magnetosome alignment, and regulation of chain formation, especially during cell division (6). Several of these functions have been mapped to particular genes, such as the mamGFDC operon, which seems involved in nanoparticle size regulation, although mechanistic insights are still missing for most processes (2, 3).
One process that is better understood is magnetosome alignment so that the individual magnetic moments combine. Electron cryotomography and genetics showed that MamK, an actin-like protein encoded in the mamAB cluster (Fig. 1A), forms filaments along the magnetosome chain (4, 7). The filaments coexist with the magnetosomes but rearrange during the cell cycle and during cell division, as do the magnetosomes (8). The mamK gene is immediately adjacent to mamJ, and it has been suggested that MamJ is either involved in magnetosome attachment of MamK filaments (7) or in the regulation of their dynamics (9). Previous work showed that MamK filaments are actin-like but double-helical and juxtaposed, but reliable atomic information was not obtained (10, 11).
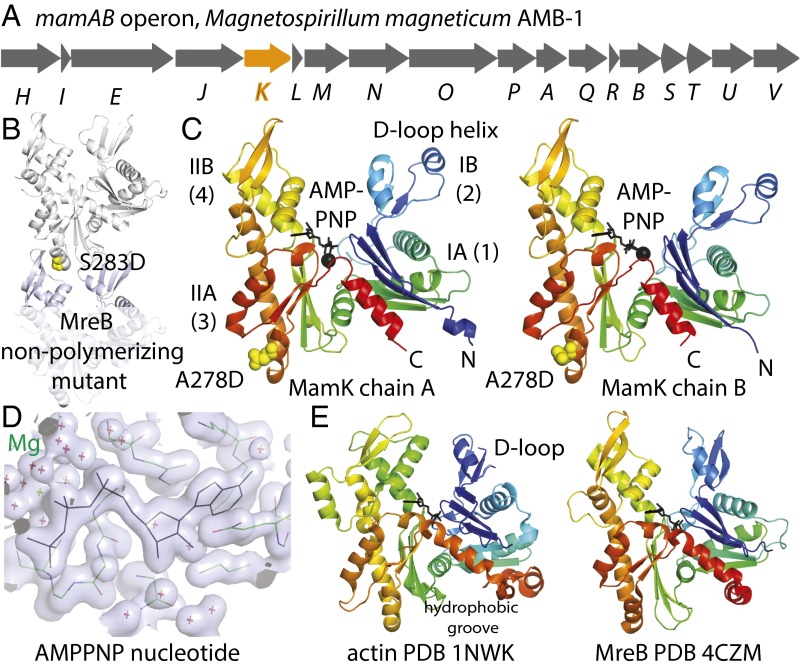
Fig. 1.
Crystal structure of MamK. (A) Schematic drawing of the M. magneticum magnetosome island mamAB operon, showing the position of the gene for the actin homolog MamK in orange. (B) Design of a nonpolymerizing MamK mutant protein (A278D) based on a previously reported nonpolymerizing mutant of C. crescentus MreB, located at the tip of subdomain IIA that inserts into the cleft formed by subdomains IIB and IB in the protofilament of MreB (12). (C) Crystal structure of M. magneticum AMB-1 MamK(A278D) at 1.8-Å resolution. The cocrystallized AMPPNP nucleotide is shown in black and the protein is colored from the N terminus to the C terminus like a rainbow. The asymmetric unit of the crystals contained two MamK molecules (chains A and B) with differing angles between domains I and II, changed by 13°, as determined by DynDom (27). (D) Close-up view of the electron density of the AMPPNP triphosphate nucleotide and magnesium coordination with the refined coordinates superimposed. (E) When comparing this MamK crystal structure with all structures in the PDB, it emerged that MamK is most closely related to eukaryotic actin and bacterial actin-like MreB. The D-loop in subdomain IB in actin (Left) is a similarly long insertion in MamK, where it forms an extra helix (C, Left).
To be able to investigate magnetosome alignment at the molecular level, we solved the crystal structure of MamK from M. magneticum AMB-1 at 1.8-Å resolution and its filament structure by cryo-EM and helical reconstruction at 3.6-Å resolution. A conformational change between monomer and polymer explains the allosteric regulation of ATP hydrolysis upon filament formation.
Results and Discussion
Crystal Structure of MamK from M. magneticum AMB-1.
MamK from M. magneticum AMB-1 (Fig. 1A) was expressed as a C-terminal truncation to improve the protein’s behavior, removing the last eight residues and adding a C-terminal GSHHHHH histidine tag for ease of purification. Crystallization trials produced many crystals but diffraction was very weak. Assuming that polymerization or at least the presence of a strong longitudinal protofilament (strand) contact caused a particular crystal form to emerge in a high percentage of crystallization conditions tried, we set out to use a nonpolymerizing mutant of MamK to solve its crystal structure. Previous work on the filaments of MreB, another bacterial actin-like protein, from Caulobacter crescentus highlighted that a mutation on the tip of subdomain IIA (Fig. 1B), MreB(S283D), was able to abrogate polymerization through the disruption of the longitudinal protofilament contact (12). In the canonical actin protofilament contact, the tip of subdomain IIA inserts into the cleft formed by subdomains IIB and IB (Fig. 1B).
Sequence alignments indicated that the equivalent residue in MamK should be A278 and the corresponding mutant protein, MamK(A278D), was produced. The mutant protein yielded well-diffracting crystals and the crystal structure was solved to 1.8-Å resolution using a mercury derivative and single isomorphous replacement with anomalous scattering (SIRAS) phasing (Materials and Methods and Table S1).
Table S1.
Crystallography and cryo-EM data
| Statistics | X-ray monomer phasing | X-ray monomer native | Cryo-EM filament |
| Sample | M. magneticum AMB-1 MamK | M. magneticum AMB-1 MamK | M. magneticum AMB-1 MamK |
| NCBI Database ID code | WP_041039444.1 | WP_041039444.1 | WP_041039444.1 |
| Constructs | 1–339–GSHHHHHH, A278D mutation | 1–339–GSHHHHHH, A278D mutation | 1–347, no tag |
| Method data collection | Crystallography 0.7 mM EMTS, SAD | Crystallography native | Cryo-EM with helical reconstruction in RELION |
| Beamline/microscope | In-house Rigaku FR-E | Diamond I04 | FEI Polara/Falcon III |
| Wavelength/energy | 1.5418 Å | 0.97941 Å | 300 kV |
| Crystal/helical | |||
| Space/point group | P21 | P21 | C2 along helical axis |
| Cell, Å | 73.2, 71.9, 79.5, 99.6° | 72.6, 71.8, 79.8, 98.4° | |
| Twist/rise | 23.77°, 52.15 Å | ||
| Data | |||
| Resolution, Å | 2.9 | 1.8 | 3.6 (0.143 FSC) |
| Completeness, %* | 98.0 (99.4) | 95.1 (97.4) | |
| Multiplicity* | 6.9 (6.5) | 3.4 (3.4) | |
| (I)/σ(I)* | 11.2 (2.7) | 14.5 (2.3) | |
| Rmerge* | 0.120 (0.654) | 0.083 (0.499) | |
| Rpim* | 0.074 (0.410) | 0.053 (0.315) | |
| CC1/2* | 0.996 (0.851) | 0.994 (0.810) | |
| Anomalous correlation* | 0.08 (−0.04) | ||
| Mercury sites | 6 | ||
| No. of images, pixel size | 1,665, 1.34 Å | ||
| Defocus range, dose | −1.1 to −3.0 µm, ∼40 e/Å2 | ||
| No. of helical segments | 596,427, 50 Å apart | ||
| Refinement | |||
| R/Rfree† | 0.178/0.206 | 0.250 | |
| FSC (REFMAC) | 0.920 | ||
| Models | Two chains per ASU: 4 to 334, AMPPNP, Mg, 536 waters | Six chains refined in P1: 10 to 334, ADP, Mg, no waters | |
| Bond length rmsd, Å | 0.013 | 0.030 | |
| Bond angle rmsd, ° | 1.406 | 2.865 | |
| Favored, %‡ | 99.4 | 99.9 | |
| Disallowed, %‡ | 0 | 0 | |
| MolProbity score | 99th percentile | 84th percentile | |
| ID codes | PDB 5LJW | PDB 5LJV, EMD-4062 |
ASU, asymmetric unit; SAD, single-wavelength anomalous diffraction.
Values in parentheses refer to the highest recorded resolution shell.
Five percent of reflections were randomly selected before refinement.
Percentage of residues in the Ramachandran plot (PROCHECK “most favored” and “additionally allowed” added together).
MamK(A278D) crystallized with two MamK molecules in the asymmetric unit, and they did not form filaments or protofilament contacts in the crystals. MamK’s structure shows the canonical actin fold (Fig. 1 C and E, Left), and also contains the ATP analog AMPPNP that had been added during purification to reach the high protein concentrations needed for crystallization.
The actin fold is composed of two domains, I and II, that can be further subdivided into four subdomains, IA (1), IB (2), IIA (3), and IIB (4). The two monomers (chains A and B) in the crystals display different interdomain angles between domains I and II (Fig. 1C, Left and Right), most likely facilitated by different crystal contacts for the two MamK molecules in the asymmetric unit of the crystals. Chain A is more closed, with subdomains IIB and IB closer together, and this is caused by a 13° rotation of the domains against each other. Related changes in conformation have been reported before when comparing monomeric and polymerized forms of actin (13, 14), MreB (12) and ParM (15), another bacterial actin-like protein involved in the segregation of plasmid R1 in Escherichia coli. It has been suggested that the formation of the protofilament leads to the closing of the subdomain IB–IIB cleft through removal of the propeller twist of domains I and II, and that this in turn may switch on nucleotide hydrolysis within the active site, which is not otherwise in direct contact with another subunit along the protofilament, as is the case for all tubulin-like polymers. In other words, it might constitute an allosteric mechanism that transmits information about the polymerization state to the active site in the middle of the molecule. To relate the conformational change seen between the two MamK monomers in the crystals to this mechanism, we then set out to determine the structure of MamK in the polymer state by cryo-EM as outlined below. This showed that in the filament the two domains of MamK (I and II) close further, but in the same direction.
Filament Structure of MamK.
For cryo-EM of MamK filaments, we used entirely unmodified protein from M. magneticum AMB-1, expressed in and purified from E. coli. As a change from previously published protocols (11), we introduced an ATP-induced polymerization/depolymerization step to select for polymerization-competent protein and to simplify the procedure. The protein was then polymerized in low-salt buffer in the presence of ATP and magnesium, without glycerol, because it interfered with the imaging by the electron microscope (Fig. 2C, Top). Helical reconstruction was performed with a RELION (16) version that we extended for this purpose (see also Materials and Methods). Briefly, this implementation performs single-particle-like processing of helical assemblies (17) in an empirical Bayesian framework, where a marginalized likelihood function is complemented with a prior on the reconstruction that effectively dampens high spatial-frequency terms in the absence of experimental data. Initial 2D classification of autopicked segments produced excellent 2D classes that showed both the unusual architecture of MamK double-helical filaments with juxtaposed subunits (Fig. 2C, Bottom) as well as some secondary structure within the subunits. For subsequent 3D refinement, we used a low-resolution reference model that was generated manually from the known helical parameters (11) and the crystal structure of MamK (this work; chain A). C2 symmetry was imposed during 3D refinement, and real-space optimization of the helical parameters at every cycle of the refinement yielded final values of 52.2 Å (rise) and 23.8° (twist). Notably, to prevent overfitting, we kept segments from each filament in the same half-set that was used for Fourier shell correlation (FSC). The final map showed an average resolution of 3.6 Å within the masked protein region around the central two subunits (Fig. 2E), and was easily interpretable (Fig. 2B and Movie S1). For example, it is clear from the map that the bound nucleotide is ADP (Fig. 2F and Movie S2) and the two β-sheets in MamK are well-resolved (Fig. 2G and Movie S2).
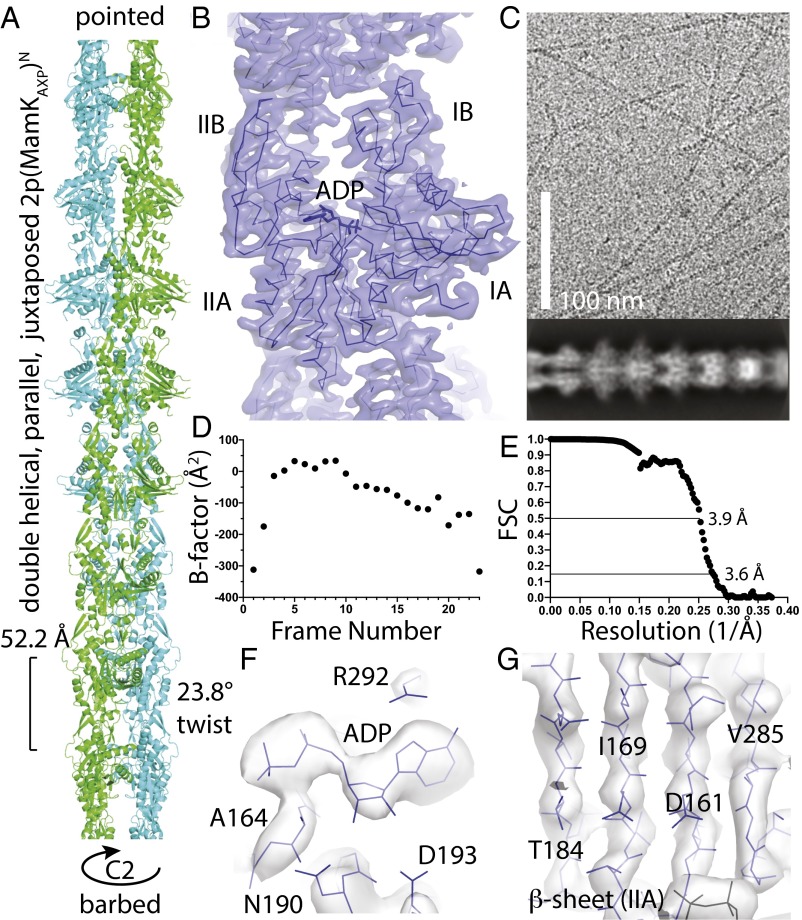
Fig. 2.
Filament structure of MamK. (A) Refined atomic model of the MamK filament. Unusual for actin-like filaments, the subunits from each strand are juxtaposed (in-register), creating an additional C2 symmetry axis along the filament axis. The filament is right-handed, juxtaposed, parallel (polar), and double-helical, 2p(MamKAXP)N (19), with a rise of 52.2 Å and a twist of 23.8°. (B) Cryo-EM density map at 3.6-Å resolution, showing one monomer of the fitted and refined MamK atomic model superimposed as a Cα trace in dark blue. See also Movie S1. (C, Top) Typical cryo-EM image as used for the helical reconstruction. Untagged MamK was polymerized at 1.5 mg/mL in low-salt buffer in the presence of ATP and magnesium. (C, Bottom) Two-dimensional classification in RELION clearly shows the juxtaposed subunits from each of the two strands along the double-helical filament and also some secondary structure within the monomers. (D) B-factor plot vs. frame number, resulting from particle polishing in RELION, indicating that the first two and the last few frames did not contain as much high-resolution information as the middle frames. Imaging conditions were 46 frames (averaged 2 frames for the analysis) over 1.5 s and ∼40 e/Å2 on a Falcon III direct electron detector prototype. (E) Gold standard FSC curve between two half-datasets as calculated by RELION, not separating segments from each filament into half-datasets. The 0.143 criterion indicates a resolution of the map for the central two subunits of 3.6 Å. (F) Cryo-EM density map around the nucleotide. The map clearly indicates that the nucleotide is ADP. (G) Cryo-EM density map showing the central β-sheet of subdomain IIA. The strands are well-separated. See also Movie S2.
The crystal structure of MamK (chain A; Fig. 1C, Left) was fitted into the cryo-EM density map and manually adjusted, especially accounting for the domain angle change (see below). The atomic model, comprising six subunits, three in each strand, was refined in reciprocal space with REFMAC (18), using additional geometric restraints based on the MamK crystal structure (chain A). Refinement in reciprocal space should be equivalent in performance to real-space approaches but has the advantage of providing an R factor that is very sensitive to small errors in the model compared with measures such as FSC or other correlative indicators that provide more linear quantities of agreement between the atomic model and the density map. The resulting model of MamK in the filament state has statistics comparable to good crystal structures (Table S1).
The MamK filament structure (Fig. 2A) shows a right-handed, double-helical, parallel (polar), juxtaposed filament with each monomer bound to ADP: 2p(MamKAXP)N (19). As has been deduced before (10, 11), the subunits from each strand in the filament are opposite each other, juxtaposed, in the filament, creating an additional twofold (C2) symmetry axis along the filament. Symmetry constraints mean that in a double-helical filament the subunits from each of the two strands can only be either staggered or juxtaposed (in-register) without creating chemically different strands. Actin and most other actin-like filaments show staggered filaments (20), possibly because such an arrangement produces ends that are not blunt, helping with filament elongation and nucleation. MreB also forms juxtaposed filaments but they are nonpolar, antiparallel, and not helical (12). Because MreB bends (21), one could suggest that the juxtaposed MreB architecture produces filaments that bend more easily because the longitudinal protofilament interfaces are lined up, creating points along the filament where bending could more easily occur. However, MamK filaments do not seem to bend more than actin or ParM (e.g., Fig. 2C), so the reason for juxtaposed subunit architecture awaits further investigation.
It seems to us that all actin-like filaments conserve the longitudinal protofilament contact architecture: Protofilaments are formed by the insertion of the previous subunit’s tip formed by subdomain IIA into a cleft formed between subdomains IB and IIB. Because of the large evolutionary distances involved, the contact surfaces have changed significantly at the level of amino acids (12, 22–24). One interesting feature to note is the D-loop helix (Fig. 1 C and E) that inserts into the hydrophobic groove region (25) of the preceding subunit (see also Movie S3). In contrast, lateral contacts, linking protofilaments into higher-order filament architectures, differ. The lateral interprotofilament contact in the MamK filament consists of a single contact per subunit (Fig. 3A). It comprises residues 81 to 84 and 191 to 198, and constitutes a small contact area of around 400 Å2 (assuming a 4-Å interaction radius per atom). Presumably, avidity, the repetition of this contact along the filament, results in stable protofilament association despite such a surprisingly small lateral contact. Small lateral interprotofilament contacts have also been reported for ParM (22) and microtubules (26), and to some extent for F-actin (24), where an additional “hydrophobic plug” between the protofilaments, now known to be mostly hydrophilic, yields a larger contact area (13, 24). Small and simple contacts mean that evolution will have little trouble diversifying and optimizing filament architectures, because only a few residues need changing.
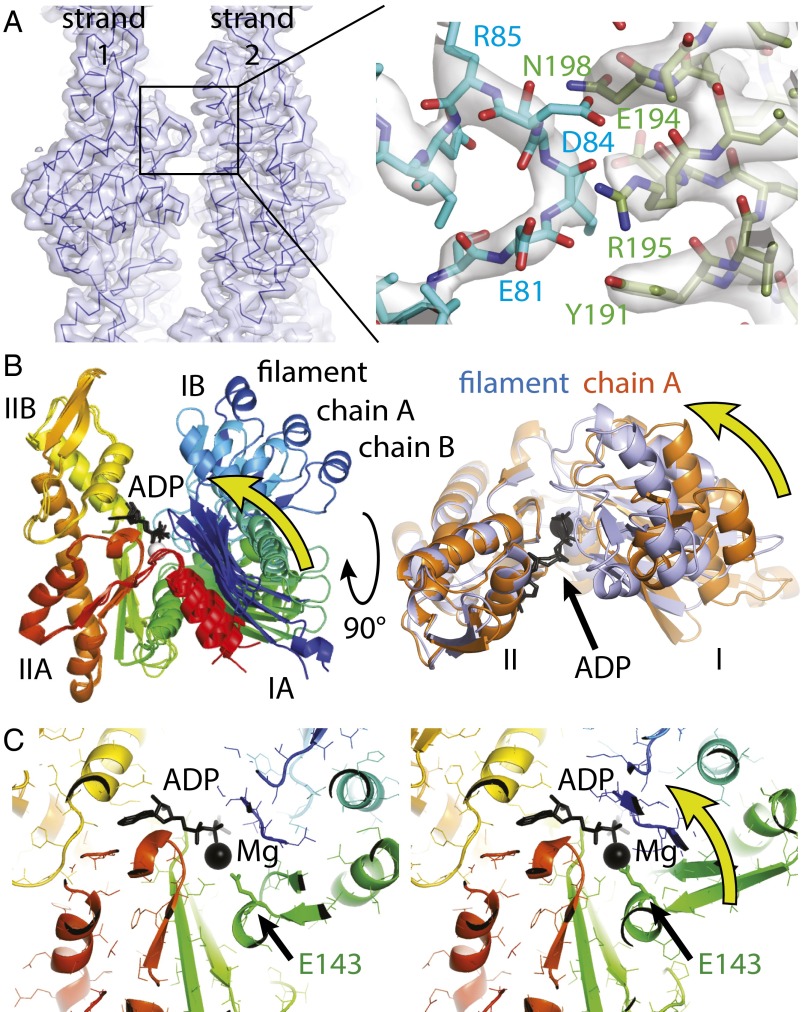
Fig. 3.
MamK lateral contacts and conformational changes upon polymerization. (A) The two protofilaments (strands) generating the double-helical MamK filament are held together by a single contact. The contact (detailed Right) is formed by two consecutive stretches of residues, 81 to 84 on one side and 191 to 198 on the other. Because this contact repeats exactly on the other side (through the C2 symmetry of the filament) and also along the filament between each pair of subunits, avidity keeps the protofilaments together despite such a small contact area. (B) The structure of MamK in the filament closes further, rotating domains I and II against each other by 19.8° such that the cleft between subdomains IB and IIB becomes smaller and also removing a propeller twist between domains I and II, as determined by DynDom (27). (B, Right) Removal of the propeller twist causes several loops (orange to purple) to close around the nucleotide (black). Directions of movements are indicated by yellow arrows. All structural alignments were performed superimposing domain II only (residues 156 to 314). See also Movies S3 and S4. (C) The conformational change upon polymerization moves a key residue, Glu-143, closer to the magnesium ion, presumably switching ATP hydrolysis during the polymerization cycle of MamK. As in B, the direction of the movement is indicated by a yellow arrow. See also Movie S5.
Conformational Changes upon Polymerization of MamK.
When comparing the atomic structures of MamK in the two monomeric forms (chains A and B; Fig. 1C, Left and Right) with the structure in the filament, a further domain angle closure becomes apparent (Fig. 3B and Movies S3 and S4, aligned on domain II only). The cleft between subdomains IIB and IB closes further by 19.8° because the tip of subdomain IIA of the preceding subunit in the protofilament inserts and binds to both the IIB and IB subdomains. Hence, extending the movement between the two conformations in the MamK crystals (Fig. 1C), the domains change to remove a propeller twist as indicated by the yellow arrows in Fig. 3B. The three MamK structures show different interdomain angles between domains I and II (Fig. 3B) but along a common trajectory, indicating a domain movement around a hinge. The hinge region is small, with significant changes limited to residues between domains I and II: 114 to 117, 142 and 143, 313 to 318, and 323 to 327, as determined by DynDom (27) after aligning on domain II only. A similar flattening has been previously reported for actin (13, 14), ParM (15), and to a lesser extent MreB (12). The MamK domain movement has many consequences at the residue level, but the overall effect is expected to be that polymerization enhances nucleotide hydrolysis, the hallmark of cytomotive filaments of the actin and tubulin type.
Domain movement closes several loops around the active site (Fig. 3B, Right and Movie S4), most importantly regions around Thr-20, Asp-76, Pro-116, and Glu-143 (all from domain I and all moving closer toward the phosphates of the nucleotide). With these changes, a residue key to catalysis, Glu-143, is moved closer to the magnesium ion, most likely enhancing the hydrolysis of ATP (11) (Fig. 3C and Movie S5). This means the structures provide important snapshots of an allosteric mechanism that regulates ATP hydrolysis upon polymerization in atomic detail.
In summary, we have determined the structure of MamK in the monomeric form and of MamK filaments at near-atomic resolution, also reiterating the power of cryo-EM for filamentous proteins that are often difficult or impossible to investigate by crystallography. The structures show glimpses of the allosteric mechanism of ATP activation through protofilament formation. In the future, the filament structure will enable detailed investigations of how MamK filaments are attached to magnetosome membranes, how they align magnetosomes, and how the filaments regulate their dynamics, for example the change from static to dynamic filaments during cell division.
Materials and Methods
Cloning, Expression, and Purification of C-Terminally Truncated and Tagged M. magneticum MamK and Its Nonpolymerizing Mutant.
The gene for mamK from M. magneticum (Mm) [National Center for Biotechnology Information (NCBI) Database ID code WP_041039444.1] was PCR-cloned from genomic DNA into vector pHis17 using Q5 polymerase (New England Biolabs) and the appropriate primers, enabling the production of MmMamK(1–339–GSHHHHHH) in E. coli C41(DE3) cells (Lucigen). MmMamK(1–339–GSHHHHHH, A278D) was expressed from the same plasmid, mutated with Q5 site-directed mutagenesis (New England Biolabs). Both proteins were expressed and purified the same way. Twelve liters of 2× TY (2× tryptone and yeast extract) media containing 100 µg/mL ampicillin was directly inoculated from 12 agar plates after overnight incubation. After reaching an OD600 of 0.6 to 1.0 at 36 °C, the culture was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 3 h at the same temperature and harvested. The entire pellet was resuspended in 300 mM buffer A [50 mM Tris⋅HCl, 300 mM NaCl, 10% (wt/vol) glycerol, 5 mM TCEP (tris(2-carboxyethyl)phosphine), pH 8.5], and some DNase I and RNase E were added. Cells were disrupted at 25 kpsi in a Constant Systems cell disruptor, and the resulting lysate was cleared in a Beckman 45 Ti rotor at 35,000 rpm and 4 °C for 30 min. The cleared lysate was loaded onto two 5-mL HisTrap HP columns (GE Healthcare), which were then washed with stepwise increases of imidazole in buffer A: 0, 20, 100, 300, and 1,000 mM. The protein eluted mostly at 100 mM imidazole, as determined by SDS/PAGE of the resulting fractions. Fractions were pooled and concentrated in Centriprep concentrators [10-kDa molecular weight cutoff (MWCO); Millipore]. The concentrate was loaded onto a Sephacryl S300 16/60 column (GE Healthcare) equilibrated in buffer A. The protein eluted as a single peak, and bound nucleotide was removed in this step as evident by a low-molecular-weight peak with high absorbance at 260 nm. To be able to concentrate the protein, 0.5 mM AMPPNP (Sigma) and 1 mM magnesium chloride were added and the pool was concentrated as before. Purity of the protein was confirmed by SDS/PAGE, and the protein was flash-frozen in liquid nitrogen in 100-µL aliquots at 7 mg/mL.
Crystallization and X-Ray Structure Determination of MmMamK.
Crystallization trials were performed using our in-house facility (28), using two-drop MRC crystallization plates (Swissci), C-terminally His-tagged MmMamK protein at 7 mg/mL (containing AMPPNP as described above), and 100 + 100 nL sitting drops. Refinement of one particular hit [reservoir solution: 50 mM Hepes, pH 7.3, 48% (wt/vol) PEG 1000] yielded large and well-diffracting crystals when using 200 + 200 nL sitting drops in the same crystallization plates. Crystals were flash-frozen in loops without further addition of cryoprotectant. A native dataset was collected at Diamond Light Source, beamline I04, to 1.8-Å resolution (see Table S1 for statistics), and a derivative dataset was collected in-house on a Rigaku FR-E rotating anode X-ray generator with MarDTB image plate detector (marXperts) from a crystal soaked for 1 h in 0.7 mM EMTS (thiomersal) to 2.9-Å resolution. autoSHARP (29) solved and built the structure with Buccaneer (30) using both isomorphous and anomalous differences (SIRAS phasing). The model was manually adjusted with MAIN (31) and refined with PHENIX.refine (32).
Cloning, Expression, and Purification of Untagged MmMamK.
The gene for mamK from M. magneticum (NCBI Database ID code WP_041039444.1) was codon-optimized for E. coli expression, synthesized (IDT), and PCR-cloned into vector pHis17 using Q5 polymerase (New England Biolabs), enabling the production of MmMamK(1–347) in E. coli C41(DE3) cells (Lucigen). Six liters of 2× TY media containing 100 µg/mL ampicillin was inoculated 1:100 from a preculture after overnight incubation. After reaching an OD600 of 0.6 to 1.0 at 36 °C, the culture was induced with 1 mM IPTG overnight at 20 °C and harvested. The entire pellet was resuspended in 300 mM buffer B [20 mM Tris⋅HCl, 10% (wt/vol) glycerol, 5 mM TCEP, 1 mM EDTA, 1 mM sodium azide, pH 8.5]. Cells were disrupted at 30 kpsi in a Constant Systems cell disruptor, and the resulting lysate was cleared in a Beckman 45 Ti rotor at 35,000 rpm and 4 °C for 1 h. The cleared lysate was adjusted to 20% (saturated) ammonium sulfate and centrifuged at 25,000 rpm (Beckman; JA-25.50 rotor) for 1 h at 4 °C. The resulting pellets were frozen before further processing. The pellet was resuspended in buffer B and adjusted to 10 mM ATP, 20 mM magnesium chloride from 100 and 200 mM stock solutions. After further centrifugation at 25,000 rpm (Beckman; JA-25.50 rotor) for 30 min at 4 °C, the resulting pellets were resuspended in buffer B plus 5 mM EDTA and incubated at 4 °C overnight. The solution almost cleared, and the protein was precipitated again by adjusting to 30% (saturated) ammonium sulfate. Centrifugation at 25,000 rpm (Beckman; JA-25.50 rotor) and 4 °C for 30 min yielded large white pellets. The pellets were dissolved in polymerization buffer C [10 mM Tris⋅HCl, 25 mM KCl, 10% (wt/vol) glycerol, 5 mM TCEP, pH 7.4 (11)]. The solution was loaded onto a Sephacryl S300 16/60 column (GE Healthcare) in polymerization buffer, and fractions were checked by SDS/PAGE. Pooled fractions were concentrated in Centriprep concentrators (10-kDa MWCO; Millipore) to 12 mg/mL and flash-frozen in 50-µL aliquots.
Cryo-EM of MmMamK Filaments and Filament Structure Determination by Helical Reconstruction.
For cryo-EM analysis, untagged MmMamK was diluted from 12 to 1.5 mg/mL in 10 mM Tris, 25 mM KCl, 5 mM TCEP (pH 7.4) (no glycerol) and 5 mM ATP and 10 mM magnesium chloride were added to start polymerization. After 5 to 25 min, 2.5 µL was vitrified on Quantifoil R2/2 Cu/Rh 200 EM grids in liquid ethane at below −160 °C using an FEI Vitrobot Mark IV. Images were taken using EPU automatic data collection software (FEI) on an FEI Polara G2 300-kV electron microscope at the temperature of liquid nitrogen on a Falcon III direct electron detector prototype (integrating mode, 30 frames per s) at a calibrated pixel size of 1.34 Å. Total dose was around 40 e/Å2, with 1.5-s exposures and 46 frames, and defocus values were varied between −3.0 and −1.1 µm. In total, 1,665 good movies were collected in three separate 24-h sessions, as judged after initial whole-image motion correction (MOTIONCORR) (33) and contrast transfer function (CTF) estimation (gctf) (34).
All further processing was performed with a modified version of RELION (16), extended for processing of helical specimens. A few hundred helical segments were picked manually in boxes of 280 pixels and 2D-classified. A few good classes were used for automatic, template-based picking in RELION, which was modified for efficient picking of helical segments, and the resulting particles were assessed by 2D classification. A modified autopicking algorithm was used to track from which filaments individual particles were derived, to be able to create two half-sets for FSC determination that separate only complete filaments. This yielded 596,427 good particles, and a 3D refinement with helical averaging was performed, using the published helical parameters of MamK filaments (11) as well as C2 symmetry along the filament axis, because previous work (10, 11) (and the 2D classes) showed that the subunits from each of the two strands in the filament are juxtaposed (in-register). For 3D refinement, a reference was constructed using the helical parameters and the MamK crystal structure (chain A), and the reference was low-pass-filtered to 30 Å before it was used in RELION. No mask was used during 3D refinement. Helical symmetry was applied after each but the last iteration in RELION. Subsequent movie refinement in RELION as well as particle polishing (35) yielded particles with higher signal-to-noise ratios because the final reconstruction and postprocessing produced the highest resolution map at 3.6 Å, as assessed by the gold standard FSC procedure implemented in RELION (0.143 FSC criterion) (36). Postprocessing used a mask around the map corresponding to the central two MamK subunits with an eight-pixel soft raised cosine edge. The FSC procedure was modified such that the two half-sets contained particles from complete filaments each, avoiding overfitting through the use of very similar particles in the two half-sets generated from overlapping filament segments.
For atomic fitting and refinement, the central portion covering six subunits of the cryo-EM filament density was cut out using REFMAC (18) and fitted with six MamK monomers (chain A of the crystal structure obtained in this work) in two strands (corresponding to three subunits along the filament). The atomic model was manually adjusted with MAIN (31) and subsequent refinement against the cryo-EM density was performed with REFMAC (37) in reciprocal space after back-transforming the cut-out density into structure factors (REFMAC SFCALC mode). To help with convergence of the refinement and to alleviate problems with low resolution, restraints were produced with ProSMART (38) from chain A of the crystal structure and added to REFMAC’s list of geometrical restraints and checked not to constrain differences through the density fit. Model quality was assessed using the standard R factor, as it is much more sensitive to small errors than FSC or other correlation methods, and further in MolProbity (39) for stereochemical plausibility.
Coordinates and Map Depositions.
Coordinates and structure factors of the M. magneticum AMB-1 MamK crystal structure were deposited in the Protein Data Bank (PDB) with ID code 5LJW. The refined filament structure was deposited in the PDB with ID code 5LJV, and the corresponding cryo-EM 3D map was deposited in the EMDataBank (EMDB) with ID code EMD-4062.
Supplementary Material
Acknowledgments
This work was funded by Medical Research Council U105184326 (to J.L.) and MC_UP_A025_1013 (to S.H.W.S.) and Wellcome Trust Award 095514/Z/11/Z (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors of the M. magneticum AMB-1 MamK crystal structure and refined filament structure reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5LJW and 5LJV, respectively). The corresponding cryo-EM 3D map of the latter has been deposited in the EMDataBank (ID code EMD-4062).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612034113/-/DCSupplemental.
References
- 1.Blakemore R. Magnetotactic bacteria. Science. 1975;190(4212):377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 2.Jogler C, Schüler D. Genomics, genetics, and cell biology of magnetosome formation. Annu Rev Microbiol. 2009;63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- 3.Komeili A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev. 2012;36(1):232–255. doi: 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311(5758):242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 5.Cornejo E, Subramanian P, Li Z, Jensen GJ, Komeili A. Dynamic remodeling of the magnetosome membrane is triggered by the initiation of biomineralization. MBio. 2016;7(1):e01898-15. doi: 10.1128/mBio.01898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin W, Pan Y. Snapping magnetosome chains by asymmetric cell division in magnetotactic bacteria. Mol Microbiol. 2011;82(6):1301–1304. doi: 10.1111/j.1365-2958.2011.07866.x. [DOI] [PubMed] [Google Scholar]
- 7.Scheffel A, et al. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440(7080):110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 8.Katzmann E, et al. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol. 2011;82(6):1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. [DOI] [PubMed] [Google Scholar]
- 9.Draper O, et al. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011;82(2):342–354. doi: 10.1111/j.1365-2958.2011.07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergeron JRC, et al. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci. August 19, 2016 doi: 10.1002/pro.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozyamak E, Kollman J, Agard DA, Komeili A. The bacterial actin MamK: In vitro assembly behavior and filament architecture. J Biol Chem. 2013;288(6):4265–4277. doi: 10.1074/jbc.M112.417030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Ent F, Izoré T, Bharat TA, Johnson CM, Löwe J. Bacterial actin MreB forms antiparallel double filaments. eLife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467(7316):724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 14.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457(7228):441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 15.Gayathri P, et al. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338(6112):1334–1337. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J Struct Biol. 2007;157(1):83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 1):136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosal D, Löwe J. Collaborative protein filaments. EMBO J. 2015;34(18):2312–2320. doi: 10.15252/embj.201591756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozyamak E, Kollman JM, Komeili A. Bacterial actins and their diversity. Biochemistry. 2013;52(40):6928–6939. doi: 10.1021/bi4010792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salje J, van den Ent F, de Boer P, Löwe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43(3):478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bharat TA, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523(7558):106–110. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31(10):2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von der Ecken J, et al. Structure of the F-actin-tropomyosin complex. Nature. 2015;519(7541):114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez R. Actin-binding proteins—A unifying hypothesis. Trends Biochem Sci. 2004;29(11):572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 2015;162(4):849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21(3):181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 28.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol. 2005;88(3):311–327. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 30.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 9):1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 31.Turk D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 8):1342–1357. doi: 10.1107/S0907444913008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193(1):1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333(4):721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.