Significance
Cells sense changes in environmental conditions and translate them into physiological responses that are mediated by a plethora of molecular intermediaries that converge at critical nodes. This is particularly relevant to multiple levels of biological organization in which the circadian clock and cell division are interlocked. Unexpectedly, the circadian rhythms of two critical components of the interlocked system, Period 2 (Per2) and p53, were in antiphase. This finding was analyzed using a systematic modeling approach, and predictions of the model were validated by experimental studies that revealed a distinct and complex scenario. Accordingly, we show that the spatiotemporal organization of Per2:p53 interactions and the nature of their chemical modifications are critical for their time-dependent subcellular redistribution and potential biological functions.
Keywords: tumor suppressor, circadian rhythms, p53, mathematical modeling, Period 2
Abstract
The circadian clock and cell cycle networks are interlocked on the molecular level, with the core clock loop exerting a multilevel regulatory role over cell cycle components. This is particularly relevant to the circadian factor Period 2 (Per2), which modulates the stability of the tumor suppressor p53 in unstressed cells and transcriptional activity in response to genotoxic stress. Per2 binding prevents Mdm2-mediated ubiquitination of p53 and, therefore, its degradation, and oscillations in the peaks of Per2 and p53 were expected to correspond. However, our findings showed that Per2 and p53 rhythms were significantly out-of-phase relative to each other in cell lysates and in purified cytoplasmic fractions. These seemingly conflicting experimental data motivated the use of a combined theoretical and experimental approach focusing on the role played by Per2 in dictating the phase of p53 oscillations. Systematic modeling of all possible regulatory scenarios predicted that the observed phase relationship between Per2 and p53 could be simulated if (i) p53 was more stable in the nucleus than in the cytoplasm, (ii) Per2 associates to various ubiquitinated forms of p53, and (iii) Per2 mediated p53 nuclear import. These predictions were supported by a sevenfold increase in p53’s half-life in the nucleus and by in vitro binding of Per2 to the various ubiquitinated forms of p53. Last, p53’s nuclear shuttling was significantly favored by ectopic expression of Per2 and reduced because of Per2 down-regulation. Our combined theoretical/mathematical approach reveals how clock regulatory nodes can be inferred from oscillating time course data.
The circadian clock is an autonomous molecular mechanism that controls biochemical, physiological, and behavioral processes with a periodicity of 24 h in living organisms and can be entrained by environmental cues (for review, see ref. 1). The clock is sustained by a coordinated interplay of positive and negative transcriptional–translational feedback loops driven by circadian factors, a core group of proteins that either possess intrinsic transcriptional activity or modulate gene expression (1). Disruption of the circadian clock by either desynchronizing the light/dark cycle entrainment, altering sleep/wake/feeding behavior, or manipulating the expression of core circadian genes result in an increased risk of development, progression, and exacerbation of a host of disease states, including proliferative disorders (for review, see refs. 2 and 3).
Circadian factor protein Period 2 (Per2) is a large and multifaceted protein with an incompletely defined domain architecture, whose functional relevance extends beyond its role as a circadian regulator (for review, see ref. 4). Loss of Per2 function predisposes genetically engineered mice to spontaneous lymphomagenesis, increases susceptibility to radiation, and influences cell growth, division, and cell death in various models of human cultured cells (for review, see ref. 5). Whereas the above findings are consistent with a role for PER2 in suppressing a cancer-prone phenotype, a note of caution is in order, as inconsistencies among in vitro and in vivo studies have been recently reported (6, 7).
At present, our understanding of the signaling events linking Per2 to arrest of proliferation and to cell death are just beginning to emerge. We now know that, in unstressed cells, Per2 transcriptionally modulates TP53 and directly associates with the C-terminus region of the p53 transcription factor (p53), which prevents murine double minute-2 (Mdm2)-mediated ubiquitination of p53 and further degradation by the proteasome pathway (8). As a result, low levels of p53 remain present at all times in the cell allowing for a “preconditioning” state to exist. This primes the cell for a rapid response to genotoxic stress, which is further sustained by a Per2-induced p53 transcriptional loop (8, 9). As Per2 blocks p53’s oligomeric domain, which is needed for the formation of tetrameric transcriptionally active p53, p53’s downstream response to genotoxic stress not only depends on its availability in the system but also on its dissociation from Per2 in the nucleus (9). Furthermore, it is the spatiotemporal distribution of Per2:p53 that, ultimately, provides the unifying framework with which to understand the link between the two components of our initial model. These findings place Per2 as a component of the checkpoint response by intersecting the p53 node, thus providing a mechanistic account for some of the Per2-associated growth inhibitory and proliferative phenotypes observed in cultured cells and in knockout animals, respectively, where endogenous levels of Per2 were experimentally modulated (10–12).
Using an approach that combines mathematical modeling of the protein interaction network with targeted experiments, we established the asymmetric distribution of Per2:p53 in the cell and its impact on the time-dependent regulatory mechanism that modulates p53’s rhythmic behavior, stability, and cellular distribution. Our models and data further revealed the relevance of posttranslational modification events that take place in separate cellular compartments and that contribute to the time-dependent phase shift in the accumulation of p53 proteins.
Results
Rhythmic Accumulation of p53 and Per2 Are Noticeably Out-of-Phase.
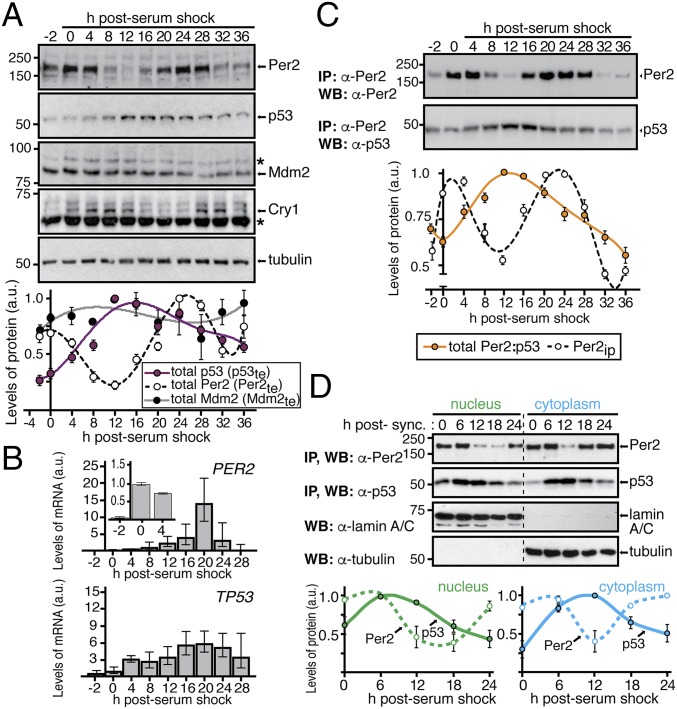
The observation that Per2 binding to p53 favors its stability by preventing Mdm2-mediated polyubiquitination of p53 prompted us to investigate whether p53 oscillation closely follows Per2 rhythms (Fig. 1A). Accordingly, extracts from circadian synchronized human colorectal carcinoma-116 (HCT116) cells were monitored for their level of expression of Per2, chryptochrome 1 (Cry1), Mdm2, and p53 proteins in a time-dependent fashion (Fig. 1A, Upper). As expected, Cry1 and Per2 undergo similar oscillations, albeit with a short delay, with a periodicity of about 24 h, following their mRNA by a few hours (Fig. 1B and Fig. S1A). Importantly, p53 was comparatively low at 0–4 h post serum shock, rose to a maximum at 12–16 h, and slowly decreased afterward (Fig. 1A). Thus, the peaks of Per2 and p53 expression in total cell extracts (named Per2te and p53te hereafter) is shifted by ∼8 h (out-of-phase), and the amplitude of the p53 peak is reduced compared with that of Per2 (Fig. 1A, Lower). Moreover, the out-of-phase relationship between Per2 and p53 remains a prevalent feature among cell-based systems and in response to different circadian stimuli as is the case, for example, in HeLa cells and with treatments such as dexamethasone (Fig. S1 B and C).
Fig. 1.
Distinct p53 and Per2 phases characterize their time-dependent subcellular distribution. (A, Upper) Extracts from circadian synchronized HCT116 cells were analyzed for the expression of endogenous Per2, p53, Mdm2, and Cry1 by immunoblotting. Asterisks indicate nonspecific bands. (Lower) Bands were quantified using ImageJ, and values were normalized to tubulin levels. Data are in arbitrary units (a.u.). (B) Samples from A were processed for qRT-PCR as described in SI Materials and Methods. Data for PER2 and TP53 gene expression are shown as the mean ± SEM from three independent experiments performed in triplicate. Bar graphs are fold increase normalized to the level of expression at t = 0 h. Inset indicates level of Per2 expression within the first 4 h. (C) HCT116 extracts from various times postcircadian synchronization (t = 0–36 h) were immunoprecipitated using α-Per2. Bound proteins were identified by immunoblotting and quantified as described in A. Relative amounts of Per2 and total Per2:p53 complex were plotted in arbitrary units relative to t = 0. (D) Nuclear and cytoplasmic fractions from circadian synchronized HCT116 cells were enriched for endogenous Per2 and p53 by immunoprecipitation and blotted using α-Per2 or -p53 antibodies, respectively. Tubulin and lamin A/C were used as controls. Bands were quantified and plotted as in A. In A, C, and D, immunoblot data were originated from a single experiment that was repeated three times with similar results. Error bars represent mean ± SEM.
Fig. S1.
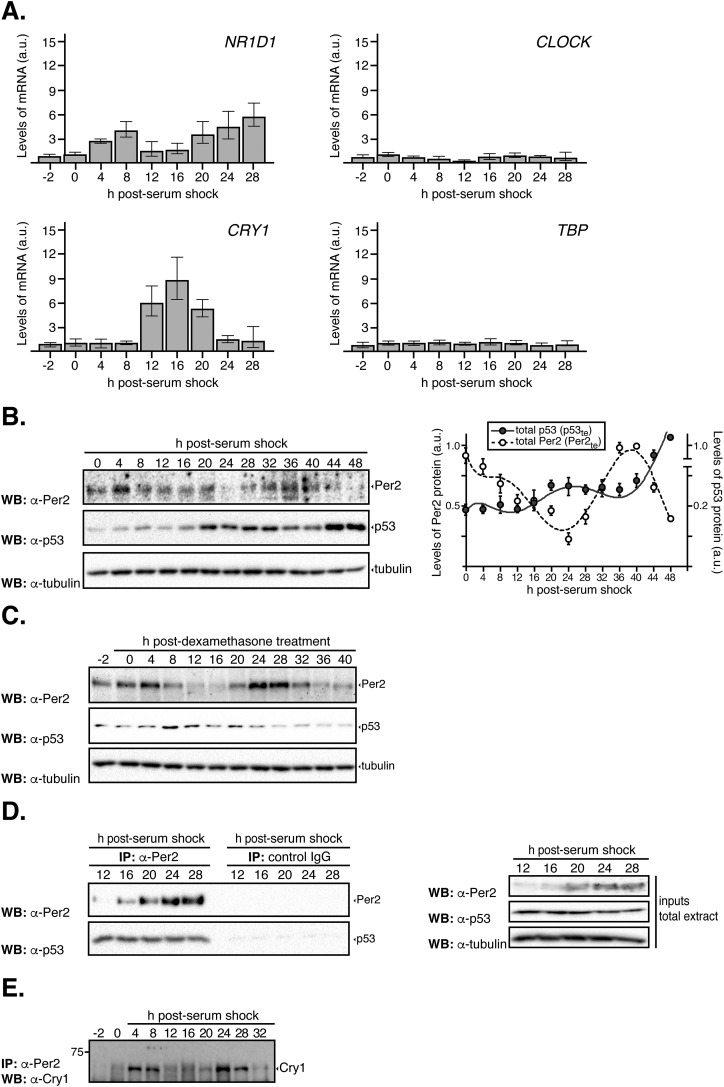
Expression of clock genes in circadian synchronized cells. (A) HCT116 cells were circadian synchronized by serum shock [50% serum for 2 h (t = −2–0)] after which they were maintained in serum-free media throughout the time course analyzed. Samples were collected at various times (t = 0–36 h) and processed for qRT-PCR analysis as described in Materials and Methods. Data for NR1D1 (encodes for Rev-erbα), circadian locomotor output cycles kaput (CLOCK), and CRY1 gene expression are shown as the mean ± SEM from three independent experiments performed in triplicate. The expression of TBP was monitored as a control of a noncircadian regulated gene. Bar graphs are fold increase normalized to the level of expression at t = 0 h. Primer sequences are shown in Table S4. (B) HeLa cells were circadian synchronized by serum shock and samples collected as described in the legend of Fig. 1A. Total extracts (5–10 μg) were analyzed for the expression of endogenous Per2 and p53 by immunoblotting using specific antibodies. Bands were quantified using Image Lab software/Gel Doc XR+ system, and values were normalized to tubulin levels. Data [in arbitrary units (a.u.)] are represented as the mean ± SEM. (C) HCT116 cells were circadian synchronized with dexamethasone [100 ng/mL for 2 h (t = −2–0) (42)], after which they were maintained in serum-free media throughout the time course analyzed. Samples were collected at various times and total extracts (10–100 μg) analyzed for expression of endogenous Per2, p53, and tubulin by immunoblotting using specific antibodies. (D) HCT116 extracts (1 mg) collected at various times postcircadian synchronization (t = 12–28 h) were immunoprecipitated using either α-Per2 antibody or rabbit IgG and protein A-beads, and bound proteins were identified by immunoblotting as indicated. For input, total cell extracts (50–100 μg) were loaded. Tubulin was used as a loading control. (E) HCT116 extracts from Fig. 1C were immunoprecipitated using α-Per2 antibody and protein A-beads, and Cry1 was identified by immunoblotting as indicated.
Next, we investigated whether p53 rhythms stem from the rhythmic transcription of TP53 or, instead, its negative regulator, the E3 ubiquitin protein ligase Mdm2. In contrast to the strong rhythmically expressed changes observed for PER2 mRNA (Fig. 1B), quantitative RT-PCRs (qRT-PCRs) exposed very weak rhythmic levels of the TP53 transcript. This was not unexpected as TP53’s transcript half-life is ∼9 h, which is much greater than that of Per2 (∼0.7 h) (13–15). Therefore, as the half-life of p53 protein is much shorter (∼2 h) than its mRNA (8), circadian modulation of p53 is likely to be more effective at the protein level than at the gene expression level. Furthermore, taking into account that the peak of the p53 protein precedes that of its transcript, it is unlikely that the rhythms in protein level result from rhythms in gene expression. Last, we examined whether antiphase expression of Mdm2 could drive the rhythmic accumulation of p53. Accordingly, detection of Mdm2 levels was by immunoblotting, and quantitative studies were carried out using JTK_CYCLE, a nonparametric algorithm testing rhythmicity (Fig. 1A and Table S1) (16). Whereas our results confirm the absence of rhythmic levels of Mdm2, a note of caution should be raised, as other scenarios involving Mdm2 cannot be excluded (e.g., posttranslational events, subcellular localization) that might contribute to the observed pattern of p53 expression.
Table S1.
Analysis of p53, Mdm2, and Per2 rhythms using JTK_CYCLE
| Protein | Benjamini–Hochberg q values (BH.Q)* | Permutation-based P values (ADJ.P)* | Phase | Amplitude |
| Per2 | 9.27E-06 | 4.64E-06 | 26 | 26.7781338 |
| p53 | 0.000784236 | 0.000784236 | 18 | 29.71262695 |
| Mdm2 | 0.341474333 | 0.341474333 | 14 | 3.60270905 |
For BH.Q, q < 0.05, and for ADJ.P, P < 0.001 to satisfy circadian rhythmicity (16).
These findings further motivate the search for the formation of the Per2:p53 complex in extracts from synchronized cells. Accordingly, we examined how Per2 and p53’s phase relationship impact the formation of the Per2:p53 complex over time. Extracts from circadian synchronized HCT116 cells were analyzed for the presence of total Per2 levels and endogenous Per2:p53 and Per2:Cry1 complexes by immunoprecipitation (Fig. 1C and Fig. S1 D and E). The Per2:p53 complex showed a peak at 8–16 h after synchronization (Fig. 1C, Lower), which is in clear contrast with the level of total Per2 detected in cell extracts (Fig. 1C, Upper) and is shifted from that of Per2:Cry1 (Fig. S1E), which is depicted in the quantitative profile. Because Per2 stabilizes p53 (8), the lack of correspondence between the Per2 peak and both the p53 (Fig. 1A) and Per2:p53 (Fig. 1C) peaks in total extracts implies the existence of an additional level of regulation (i.e., posttranslational) that needs to be factored into our initial model (9).
We then investigated whether the Per2 and p53 phase shifts identified in total extracts resulted from a complex redistribution of these proteins in different cellular compartments. The rationale behind this hypothesis is based on findings that Per2 shuttles between compartments following an oscillatory pattern that leads to an uneven distribution of the protein over time (17) and that p53 undergoes nuclear–cytoplasmic shuttling in a process that is both rapid and energy dependent (18, 19). We monitored Per2 and p53 protein levels in nuclear and cytoplasmic fractions purified from circadian synchronized HCT116 cells (Fig. 1D). Unexpectedly, whereas the rhythms of Per2 and p53 were in-phase in the nucleus, both proteins exhibited an out-of-phase relationship in the cytoplasm (Fig. 1D). In context with our previous findings (8), these results established that the phase relationship between p53 and Per2 observed in total extracts overlooks the role of compartmentalization.
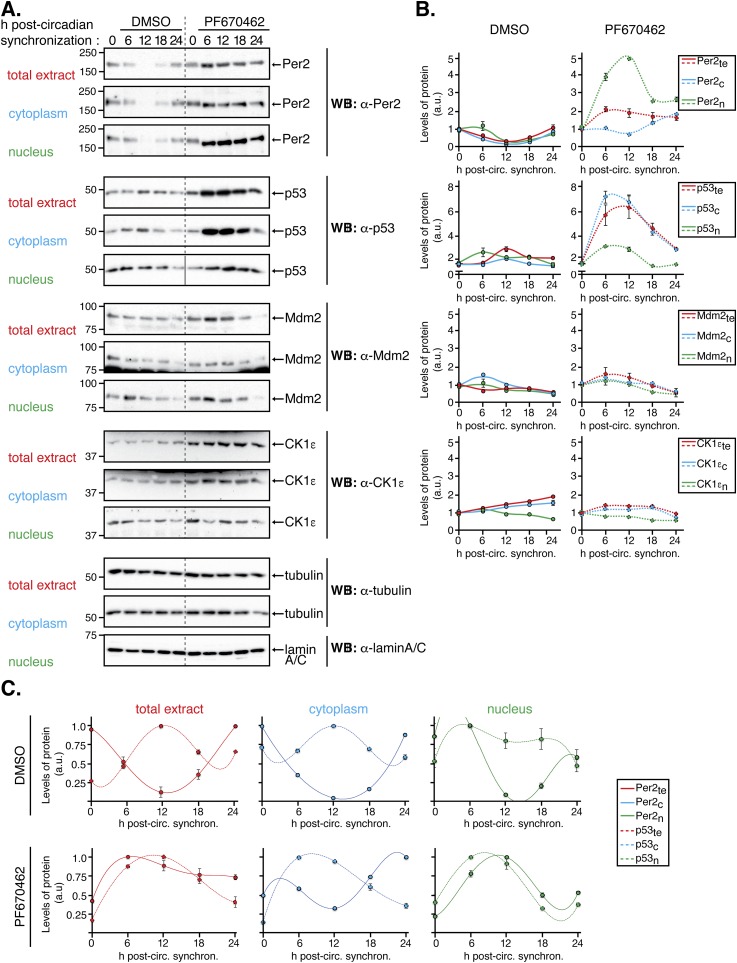
Next, we wanted to evaluate whether the phase relationships for Per2 and p53 in either compartment were maintained even when the circadian clock was perturbed (Fig. S2). Consequently, HCT116 cells were circadian synchronized by serum shock followed by addition of PF670462 (named PF670 hereafter), a potent dual pharmacological inhibitor of casein kinase 1 ε/δ (CK1ε/δ) that causes lengthening of clock oscillations by stabilizing Per2 (20). Lysates and subcellular fractions were quantitatively analyzed to evaluate the level of Per2, p53, Mdm2, and CK1ε expression (Fig. S2 A and B). As expected, addition of PF670 stabilized Per2 in both compartments (Fig. S2 A and B, Upper) and delayed the circadian phase (Fig. S2C). This is in agreement with the established role of CK1ε/δ-mediated phosphorylation of Per2 for its turnover and in sustaining the negative loop (21). Remarkably, and unlike Mdm2 and CK1 proteins (Fig. S2 A and B, Lower), the rhythms of Per2 and p53 were both delayed in PF670-treated samples in a manner that mimics the phase relationship observed in the absence of PF670 treatment (Fig. S2 A and B, Upper). Last, the activity of CK1ε/δ relevant to Per2’s stability and Per:Cry heterodimerization is not a contributing factor, at least under unstressed conditions, in regulating phase relationship between Per2 and p53. Instead, Per2 acts as the chief regulator (Fig. S2C).
Fig. S2.
The Per2 and p53 phase relationship remains unaltered despite inhibition of CK1δ/ε. HCT116 cells were circadian synchronized by serum shock after which they were collected (t = 0) or maintained in serum-free media with DMSO (control, 0.2%) or PF670462 (1 µM) throughout the time course analyzed (t = 6–24 h). Cells were collected at the indicated times and lysates were subjected to subcellular fractionation as indicated in Materials and Methods. Levels of endogenous proteins were detected in total (1.2 × 105 cells), cytoplasmic (0.63 × 105 cells), and nuclear (3.15 × 105 cells) extracts by immunoblotting using α-Per2, -p53, -Mdm2, and -CK1ε antibodies. Tubulin and lamin A/C were used as a loading and purity control for the cytoplasm and nuclear fractions, respectively. Immunoblot data originated from a single experiment that was repeated three times with similar results. (B) Protein bands from A were quantified using Image Lab software/Gel Doc XR+ system, and values were normalized to tubulin or lamin A/C levels (loading controls), and data [in arbitrary units (a.u.)] were represented as the mean ± SEM from three independent experiments. Plots were grouped by the protein’s class and expression in its various compartment in cells treated with either DMSO (Left) or PF670462 (Right) treatment. (C) Data from B was replotted to better show the conservation of the phase relationship between Per2 and p53 in the indicated compartments.
Mathematical Modeling Predicts Underlying Interactions Between Per2 and p53.
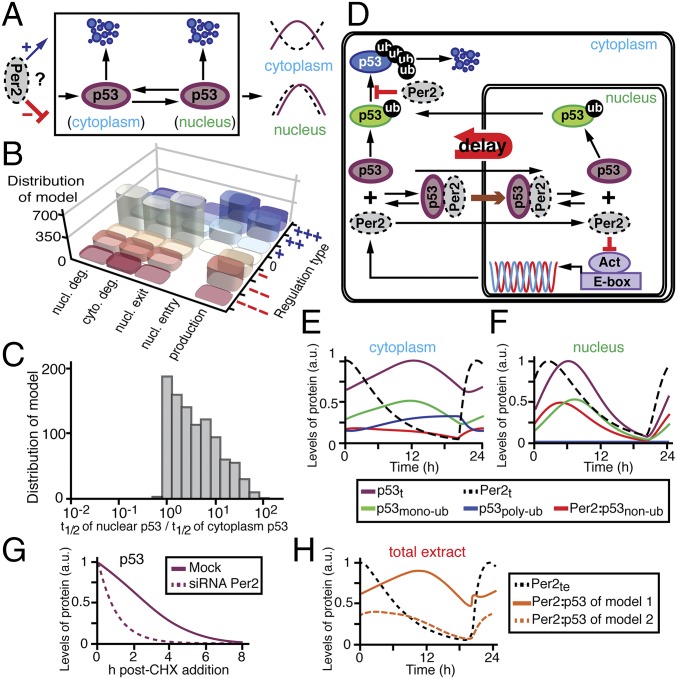
The inherent complexity of the interplay between Per2 and p53 begs for a combined theoretical and experimental approach to aid in the interpretation of experimental data, to generate testable predictions, and to develop a unified understanding of the phenomena (22, 23). Accordingly, the phase relationship between Per2 and p53 (Fig. 1) was analyzed in the context of all possible scenarios in which rhythmic Per2 could influence production, nucleus shuttling (entry/exit), and degradation of p53 (Fig. 2A). For each scenario, regulation types were randomly selected among positive (+), null (0), or negative (−) interactions during the model fitting process (Fig. 1D and SI Materials and Methods). As a result, we found 103 predicted models that successfully simulated the observed phase relationship between p53 and Per2. Interestingly, although the regulation types were randomly chosen, Per2-mediated entry of p53 to the nucleus was strongly skewed in the positive direction among all models, indicating that this event is essential to accurately simulate the rhythms of p53 (Fig. 2B). An additional unifying property among the 103 models predicted that the stability of nuclear p53 should be greater than that of the protein in the cytoplasmic compartment to successfully simulate p53 rhythms (Fig. 2C).
Fig. 2.
Mathematical modeling predicts unexpected mechanisms for Per2-mediated regulation of p53 phases. (A) Schematic representation of potential regulatory nodes for which Per2 can influence p53 phases. For each regulation, its type is randomly selected among positive (+)/null (0)/negative (−) throughout the process of fitting the model to the observed p53 and Per2 rhythms (see SI Materials and Methods for details). (B) Distribution of the 103 successful models according to their regulation type for p53. The strengths of regulation types are represented as the number of + and – (see SI Materials and Methods for details) (C) Distribution of the 103 successful models according to the relative half-life of nuclear vs. cytoplasmic p53. (D) Simplified representation of a comprehensive model for Per2 and p53 interaction based on the two essential properties predicted in B and C. (E and F) Simulated time course levels of Per2 and p53 species in both cytoplasm (E) and nuclear (F) compartments (see Fig. S3 for total extracts). (G) Prediction of p53 half-life in the presence or absence of Per2 in the system. (H) Phase predictions for the Per2:p53 complex differ depending on whether Per2 binding occurs regardless of p53 ubiquitination status (model 1) or is only associated to nonubiquitinated p53 (model 2).
Based on these initial two findings (Fig. 2 B and C), we developed a more detailed and realistic mathematical model (Fig. 2D, Tables S2 and S3, and SI Materials and Methods). First, we modified the Kim and Forger’s model used to represent the dynamics of the transcriptional negative arm generated by Per2 (24, 25). Second, we considered the ubiquitination status of p53 as it modulates p53 degradation and shuttling (26). Specifically, the dual role of monoubiquitination in p53 shuttling as a mode to impede nuclear import while facilitating p53 nuclear exit was addressed (18). In addition, the relevance of polyubiquitination in p53, which is associated with its rapid turnover (18), is reflected in the model by rapid clearance of p53 in the cytoplasm compared with the nucleus (Fig. 2C). Third, the model incorporates the interaction of Per2 with p53, which modulates p53 stability and function (8, 9). Accordingly, when p53 is prebound to Per2, both monoubiquitination and polyubiquitination of p53 are inhibited (8), thereby promoting p53 nuclear translocation (Fig. 2B) while blocking its proteasomal degradation.
Table S2.
Description of variables in the comprehensive mathematical model (Fig. 2D)
| Name | Description |
| The concentration of Per mRNA in the cytoplasm | |
| The concentration of Per protein in the cytoplasm | |
| The concentration of Per protein in the nucleus | |
| The concentration of nonubiquitinated p53 protein in the cytoplasm | |
| The concentration of monoubiquitinated p53 protein in the cytoplasm | |
| The concentration of polyubiquitinated p53 protein in the cytoplasm | |
| The concentration of Per and nonubiquitinated p53 heterodimer in the cytoplasm | |
| The concentration of Per and monoubiquitinated p53 heterodimer in the cytoplasm | |
| The concentration of Per and polyubiquitinated p53 heterodimer in the cytoplasm | |
| The concentration of nonubiquitinated p53 protein in the nucleus | |
| The concentration of monoubiquitinated p53 protein in the nucleus | |
| The concentration of Per and nonubiquitinated p53 heterodimer in the nucleus | |
| The concentration of Per and monoubiquitinated p53 heterodimer in the nucleus |
Subscript index indicates the subcellular location of proteins (c, cytoplasm, and n, nucleus).
Table S3.
Description of parameters in the comprehensive mathematical model (Fig. 2D)
| Name | Description | Value |
| α0 | Transcription rate constant for Per mRNA | 284 μM⋅h−1 |
| α1 | Translation rate constant for Per | 284 h−1 |
| α2 | Constitutive translation rate constant for p53 | 0.32 μM⋅h−1 |
| β0 | Degradation rate constant for Per mRNA | 0.17 h−1 |
| β1 | Degradation rate constant for Per in the nucleus | 0.5 h−1 |
| β2 | Degradation rate constant for polyubiquitinated p53 in the cytoplasm | 0.99 h−1 |
| δ1 | Ubiquitination rate constant for nonubiquitinated p53 in the cytoplasm | 46.5 μM⋅h−1 |
| δ2 | Ubiquitination rate constant for monoubiquitinated p53 in the cytoplasm | 0.464 μM⋅h−1 |
| δ3 | Ubiquitination rate constant for nonubiquitinated p53 in the nucleus | 12.35 μM⋅h−1 |
| Kd2 | Half-maximal constant of nonubiquitinated p53 for ubiquitination | 0.9 μM |
| Kd3 | Half-maximal constant of monoubiquitinated p53 for ubiquitination | 0.015 μM |
| kf | Binding rate constant for Per to p53 | 78 μM−1⋅h−1 |
| kb | Unbinding rate constant for Per to p53 | 370 h−1 |
| Kd1 | Dissociation constant between Per and activator protein | 2 × 10−5 μM |
| γnl | Nucleus localization rate constant for Per, nonubiquitinated p53, and their heterodimer | 0.525 h−1 |
| γne | Nucleus export rate constant for monoubiquitinated p53 | 22.5 h−1 |
| AT | Total concentration of activator protein in the nucleus | 14.8 μM |
| Nf | Ratio of cytoplasmic to nuclear compartment volume | 2 |
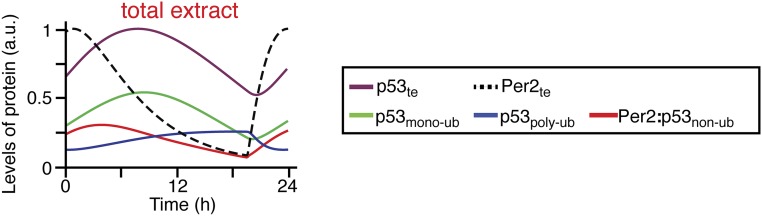
Next, we confirmed that the simulated time courses of Per2 and p53 (Fig. 2 E and F, and Fig. S3) matched the experimental data equally well (Fig. 1 A and D). In particular, the model predicted that the differences in phases observed between the p53 and Per2 peaks in the cytoplasm (Fig. 1D) were largely due to Per2-mediated shuttling of p53 into the nucleus, where p53 remains stable (Fig. 2 B and D). In this scenario, when Per2 levels are high (Fig. 2 E and F; Per2t, dashed black line), the cytoplasmic-to-nuclear translocation of nonubiquitinated p53 is promoted [Fig. 2D, brown arrow, and Fig. 2F; Per2:nonubiquitinated p53 (p53nonub), red solid line]. Once in the nucleus, the Per2:p53nonub complex dissociates allowing for free p53 to be monoubiquitinated [Fig. 2F; monoubiquitinated p53 (p53monoub), green solid line], stabilized, and ultimately exported to the cytoplasm (27). Consequently, export of p53monoub results in a peak delay of p53 levels in the cytoplasm (Fig. 2 D and E, green solid line). A corollary to that prediction is that p53 half-life should be shorter in a PER2−/− background (Fig. 2G) as demonstrated in ref. 8. Last, the model predicts that the phase of the Per2:p53 complex strongly depends on the ubiquitination status of p53 (nonubiquitinated, monoubiquitinated, or polyubiquitinated) that binds to Per2 and forms the complex (Fig. 2H). Accordingly, if binding of Per2 only occurs to nonubiquitinated p53, the phase of the Per2:p53 complex is predicted to be in-phase with that of total Per2 (Fig. 2H, orange dashed line) in clear contrast to the results shown in Fig. 1C. Alternatively, if Per2 binding occurs regardless of the ubiquitination status of p53, rhythms between the complex and total Per2 species are predicted to be antiphase (Fig. 2H, orange solid line) and in agreement with the experimental results shown in Fig. 1C. Overall, these predictions provide an initial framework for understanding what, a priori, was an unexpected observation (Fig. 1A). Therefore, we set out to test these specific predictions of the model experimentally.
Fig. S3.
Mathematical simulations of Per2 and p53 distribution in total extracts. Simulated total levels of Per2 (dashed black line), p53 (solid purple line), monoubiquitinated p53 (solid green line), polyubiquitinated p53 (solid blue line), and nonubiquitinated p53 associated to Per2 (solid red line) in total extracts. As shown, the rhythms of p53 and Per2 show the same phase relationship as the experimental data in Fig. 1A.
The Localization of p53 Influences the Kinetics of Its Degradation.
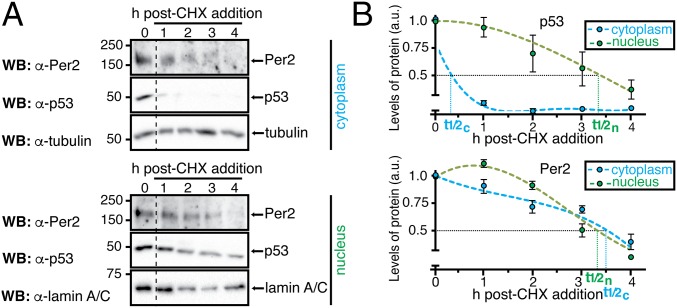
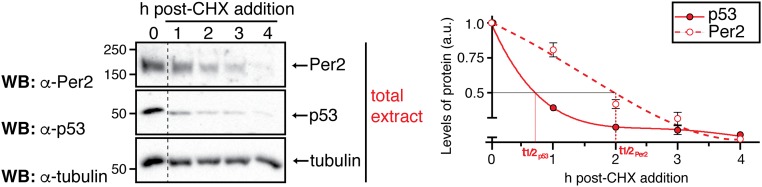
The model predicts that three essential properties simulate the correct rhythms of p53 (Fig. 2 B, C, and H). To test these predictions, initial experiments were devoted to evaluating whether p53 half-life differed in either the cytoplasmic or the nuclear compartment using cycloheximide (CHX). Whereas treatment of cells with CHX (an inhibitor of protein biosynthesis) can have a biasing effect on degradation rates of proteins in vitro and under normal growth conditions, and in different subcellular locations (28), this approach has been successfully used to compare the half-life of other core clock proteins in nuclear and cytoplasmic compartments (29). Therefore, HCT116 cells were treated with CHX (t = 0) and harvested at different times after treatment (Fig. 3; t = 1–4 h). The level of p53 was evaluated in both nuclear and cytoplasmic fractions as well as in whole-cell lysates by immunoblotting (Fig. 3 and Fig. S4). In agreement with the model’s prediction (Fig. 2C), the half-life of endogenous p53 in the nucleus was found to be considerably longer than that of p53 in the cytoplasmic compartment (Fig. 3B, Upper; t1/2n/t1/2c ∼ 7 times) where p53 is normally polyubiquitinated and targeted for degradation (30). Unlike p53, Per2 half-life remained unchanged in each compartment for the time course analyzed (Fig. 3B, Lower; t1/2n/t1/2c ∼ 1). These results further motivate the need to understand how Per2 shuttling influences the dynamic distribution of p53.
Fig. 3.
The half-life of p53, but not Per2, differs in nuclear and cytoplasmic compartments. (A) Samples were obtained from HCT116 cells in the absence (t = 0) or presence of CHX as indicated in SI Materials and Methods section. Extracts from cytoplasmic and nuclear fractions were analyzed by immunoblotting using α-Per2 and -p53 antibodies. Tubulin and lamin A/C were used as controls. (B) Protein levels were quantified as described in SI Materials and Methods section, values were normalized to tubulin or lamin A/C levels, and p53 and Per2 half-life (t1/2) calculated using Excel. Error bars represent mean ± SEM of three independent experiments.
Fig. S4.
Half-life of p53 and Per2 in total cell extracts. (Left) Samples (t = 1–4 h) were obtained from HCT116 cells maintained in complete medium in the absence (t = 0) or presence of cycloheximide (CHX) (100 μg/mL). Total extracts were analyzed by immunoblotting using α-Per2 and -p53 antibodies. Tubulin was used as a loading control. The immunoblot originated from a single experiment that was repeated three times with similar results. (Right) Protein levels were quantified using Image Lab software/Gel Doc XR+ system, and values were normalized to tubulin levels. Graphs indicate the level of remaining protein in arbitrary units (a.u.) plotted as a function of time. Curves were fitted and p53 and Per2 half-life (t1/2) calculated using Excel software. Error bars represent mean ± SEM of three independent experiments.
Qualitative Assessment of Per2 Association to the Various Ubiquitination Forms of p53.
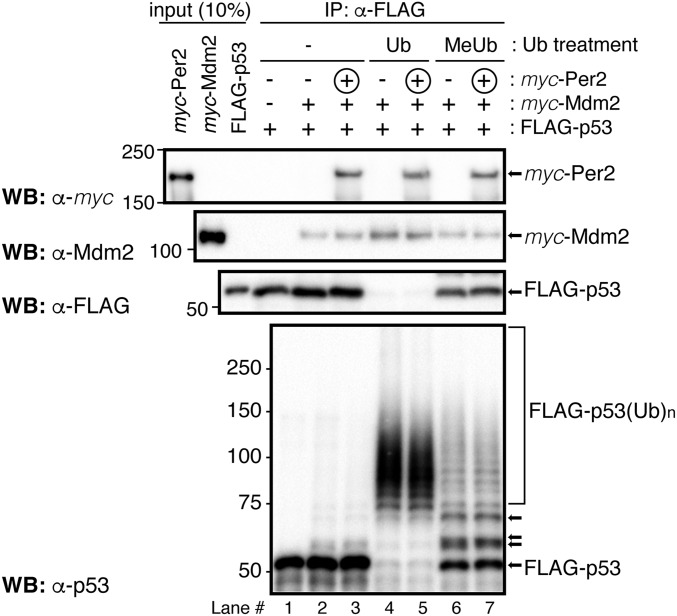
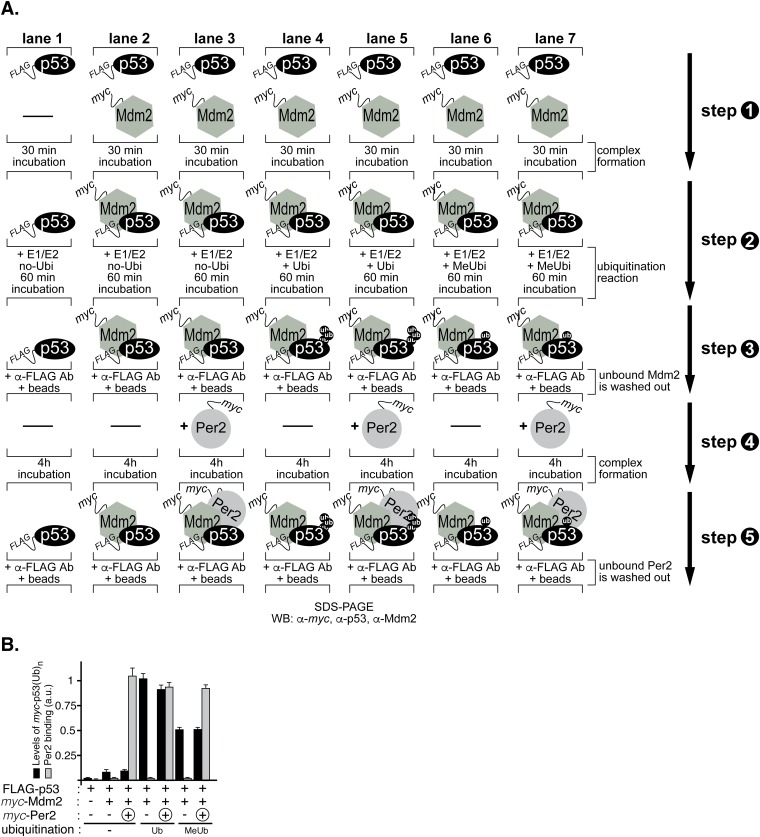
We previously showed that Per2 binding to p53 prevents subsequent p53 ubiquitination by Mdm2 (8). Whereas this interaction provides a means for enhancing p53’s stability and availability in the cell and for a rapid response to genotoxic stress (9), our model predicts that other ubiquitinated forms of p53 should be amenable to Per2 binding (Fig. 2H). To test this possibility in vitro, we performed a sequential ubiquitination and binding assay and monitored the presence of Per2 bound to the various ubiquitinated p53 forms (Fig. 4 and Fig. S5A). In vitro-transcribed and -translated myc-p53 was incubated with FLAG-Mdm2 in the presence (or absence) of a ubiquitin mixture and MG132, a proteasome inhibitor. Polyubiquitinated forms of p53 were purified and tested for Per2 binding by immunoprecipitation and blotting (Fig. 4 and Fig. S5A). As shown in Fig. 4 and Fig. S5B, p53 is efficiently polyubiquitinated by Mdm2 (lanes 2 and 3 vs. 4 and 5, Lower) and, despite this chemical modification, Per2 binding still occurs (lanes 3 vs. 5). When methyl-ubiquitin, which prevents elongation but not initiation of ubiquitin chains, was supplemented to the mixture instead, only monoubiquitinated p53 was generated. In accordance with our prediction, binding of Per2 was detected (Fig. 4 and Fig. S5B). Overall, these results, and those presented in Figs. 1C and 2H, support a model in which Per2 is capable of associating with p53 regardless of p53’s ubiquitination status and, thus, contributes to the pool of Per2:p53 complex whose distribution is antiphase to that of Per2. Still, much remains unknown regarding the underlying biochemistry that surrounds the association between Per2 and p53 as p53’s ubiquitination might influence, at multiple levels, the affinity of p53 for Per2 and/or that of its ternary partner Mdm2.
Fig. 4.
Per2 associates to ubiquitinated forms of p53. In vitro-transcribed and -translated FLAG-p53 was preincubated (+) with myc-Mdm2 and the complex added to a reaction mixture containing (+) or not (−) the ubiquitin substrate (ubiquitin, Ub, or methyl-ubiquitin, MeUb) as indicated in SI Materials and Methods and Fig. S5A. Circled “+” indicates myc-Per2 was added after complexes were formed and washed. Bound proteins were detected by immunoblotting. The FLAG-p53(Ub)n forms were detected using α-p53 antibody (Lower). Arrows indicate examples of monoubiquitinated forms of p53.
Fig. S5.
Analysis of Per2 association to various ubiquitinated forms of p53. (A) A schematic representation of the steps in the ubiquitination protocol detailed in Materials and Methods and used to generate the data summarized in Fig. 4 and Fig. S5B. (B) Bands from Fig. 4 were quantified as described and values plotted in arbitrary units (a.u.). In all cases, bars indicate levels of ubiquitinated p53 (black) and bound Per2 (gray) in the complex. Error bars represent mean ± SEM of three independent experiments.
Expression of Per2 Favors p53 Nuclear Translocation.
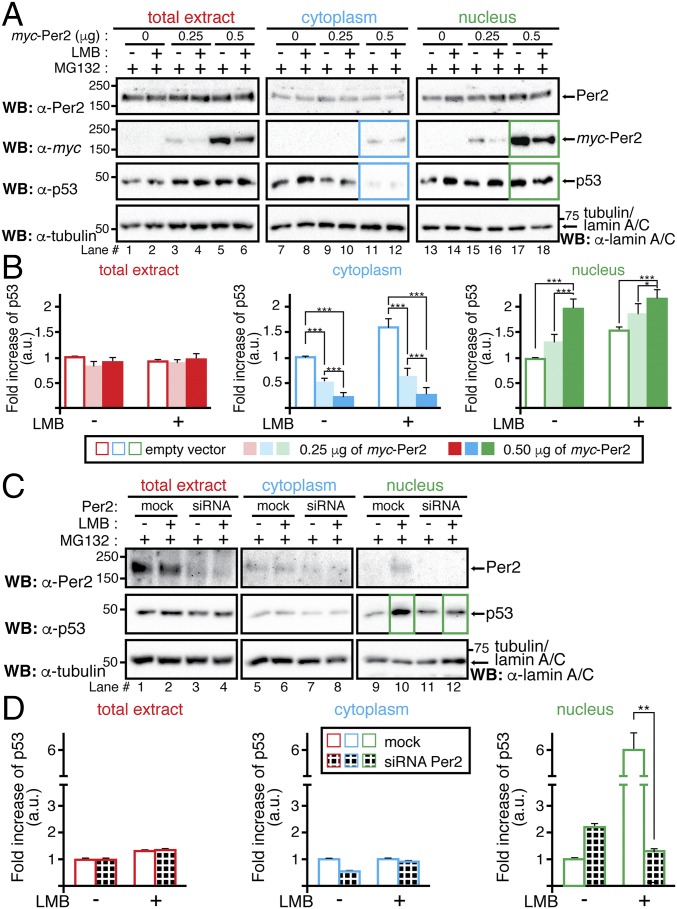
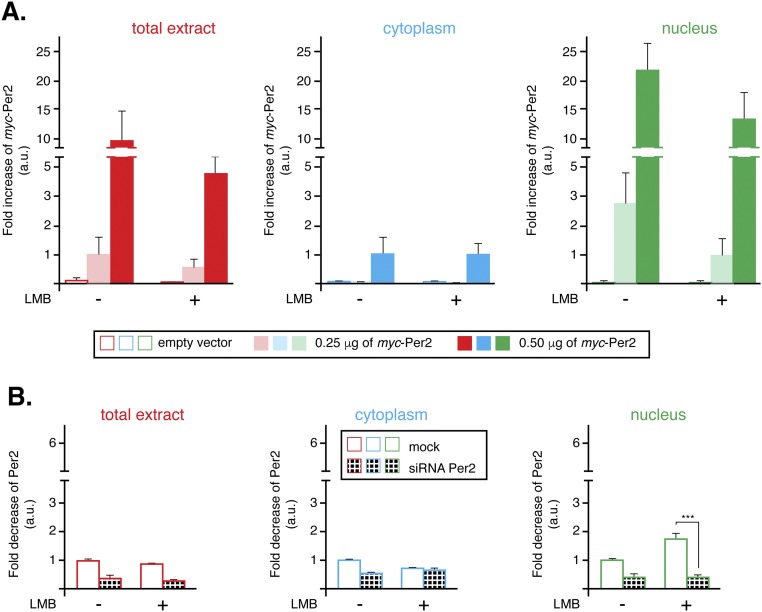
An essential property predicted by our model is that, to simulate the correct phase of p53 rhythm, Per2 should positively influence p53 nuclear translocation (Fig. 2B). Further experiments were done to confirm this prediction by evaluating the influence of Per2 expression levels on p53 trafficking. Initially, HCT116 cells were transfected to express trace levels of myc-Per2, low enough to keep total levels of Per2 relatively constant in the cell and avoiding potential overexpression artifacts (Fig. 5A, Upper). In fact, detection of the tagged form of Per2 was marginal to the point that only the addition of six tags in the construct allowed for its identification (Fig. 5A, Middle, and Fig. S6A). To better capture p53 distribution, experiments were performed in the presence of both MG132, a proteasome inhibitor, to block p53 degradation, and/or leptomycin B (LMB), which, by inhibiting the nuclear export factor CRM1, prevents p53 nuclear exit. As shown in Fig. 5A, augmented expression of myc-Per2 resulted in a concomitant decrease in p53 levels in the cytosol (lanes 5 and 6 vs. 11 and 12) and increased detection of both components in the nucleus (lanes 11 and 12 vs. 17 and 18), an observation that was further supported by quantitative analysis and statistical inference (Fig. 5B). Indeed, the converse experiment, where endogenous Per2 levels were down-regulated by small interfering RNA (siRNA) transfection (Fig. 5C and Fig. S6B), resulted in a sharp decrease of nuclear p53 accumulation (Fig. 5C, lanes 10 vs. 12 and Fig. 5D). Although existing shuttling mechanisms for p53 are in place in the cell (26), our results support a model in which partial translocation of p53 and its time-of-day cellular distribution are mediated by the availability and binding capability of Per2.
Fig. 5.
Per2 enhances p53 shuttling to the nucleus. (A) HCT116 cells were transfected with either myc-Per2 (0.25 or 0.5 μg) or empty vector (0) for 24 h before the addition of MG132 and/or leptomycin B (LMB). Lysates were subjected to subcellular fractionation, and total extract (Left), cytoplasmic (Middle), and nuclear (Right) fractions were analyzed for the presence of Per2, myc-Per2, and endogenous p53 by immunoblotting. Tubulin and lamin A/C were used as controls. (B) Endogenous p53 was quantified using Image Lab software/Gel Doc XR+ system and values normalized to tubulin or lamin A/C levels. (C) HCT116 cells were transfected with either scrambled siRNA (mock) or Per2 siRNA (siRNA) for 48 h before the addition of MG132 and/or LMB. Endogenous proteins were detected as indicated in A and quantified as in B. In B and D, data are represented as fold increase of p53 (in arbitrary units) compared with similar treatment in either vector-transfected or mock samples, respectively. Values are the mean ± SEM from three independent experiments. Statistical significance was determined by t test. *P ≤ 0.05; **P ≤ 0.005, ***P < 0.001.
Fig. S6.
Per2 distribution in cytoplasm and nucleus from LMB-treated cells. (A) From Fig. 5A, levels of myc-Per2 were quantified using Image Lab software/Gel Doc XR+ system and values normalized to tubulin or lamin A/C levels (loading controls). Data are represented as fold increases of myc-Per2 [in arbitrary units (a.u.)] compared with similar treatment in empty vector-transfected samples. Values are the mean ± SEM from three independent experiments. (B) From Fig. 5C, levels of endogenous Per2 were quantified as in A and values normalized to tubulin or lamin A/C levels (loading controls). Data are represented as fold increases of Per2 [in arbitrary units (a.u.)] compared with similar treatment in mock samples. Values are the mean ± SEM from three independent experiments. Statistical significance was determined by t test. ***P ≤ 0.001.
SI Materials and Methods
Cell Line.
The human colorectal carcinoma-116 (HCT116) (TP53+/+ and PER2+/+) cell line was purchased from the American Type Culture Collection (ATCC) and maintained according to manufacturer’s recommendations. We used HCT116 cells for our studies as they (i) are amenable to manipulation by gene transfer (9), (ii) can be circadian synchronized by serum shock (41) (Fig. 1A and Fig. S1), and (iii) have been used to study the contribution of Skp1-Cul1-F-Box-protein (SCF) ubiquitin E3 ligases in cell growth, expression of cell cycle genes, and turnover of circadian components (42).
Plasmid Constructs and siRNA Sequences.
The human Per2, p53, and mouse Mdm2 full-length cDNAs were cloned downstream from a tag encoding sequence into pCS2+3xFLAG- and (myc)6-tag vectors modified for ligation-independent cloning (EMD Millipore; named FLAG-Per2, FLAG-p53, FLAG-Mdm2, myc-Per2, myc-p53, and myc-Mdm2 throughout the text).
For small interfering RNA (siRNA)-mediated knockdown of Per2 (accession number NM_022817; National Center for Biotechnology Information), a 25-mer siRNA sequence (5′-CAACAAGGTGCTGAGAGTCAGCTTT-3′; Thermo Fisher Scientific) was designed using BLOCK-iT RNAi Designer (Thermo Fisher Scientific). A scrambled 25-mer siRNA sequence that did not show specificity to known cellular mRNAs was used as control.
Cell Culture and Transfections.
HCT116 cells were propagated in HyClone McCoy’s 5a medium (GE Healthcare), supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Corning), 50 IU/mL penicillin, and 50 μg/mL streptomycin (MP Biomedicals). For transfection experiments, cells were seeded into 6- or 12-well plates and maintained until reaching 50–80% confluence. For plasmid transfections, various DNA constructs were mixed with Lipofectamine LTX (Thermo Fisher Scientific) following manufacturer’s instructions. Transfections in HCT116 cells were in HyClone HyQ-RS without antibiotics for 4 h at 37 °C/5% CO2. Recombinant proteins were allowed to express in media containing 10% FBS (minus antibiotics) after which they were either collected or further circadian synchronized by serum shock treatment (41). When indicated, postsynchronized cells were incubated with the CK1δ/ε inhibitor PF670462 [1 µM (Cayman Chemical) (20)], MG132 (proteasome inhibitor; 50 μM), and/or leptomycin B (LMB) (nuclear export inhibitor; 50 ng/mL) throughout the time course analyzed. For siRNA experiments, cells (5 × 105) were transfected with either Per2 or scrambled siRNA using DharmaFECT2 (Thermo Scientific) following the manufacturer’s instructions. Cell extracts were prepared in “lysis buffer” containing 10 mM Tris⋅HCl (pH 7.5), 137 mM NaCl, 1 mM EDTA, 10% (vol/vol) glycerol, 0.5% Nonidet P-40, 80 mM β-glycerophosphate, 1 mM Na3VO4, 10 mM NaF, and protease inhibitors (10 μM leupeptin, 1 μM aprotinin A, and 0.4 μM pepstatin).
RNA Extraction and Quantitative RT-PCR.
Total RNA was purified from circadian synchronized HCT116 cell pellets using TRIzol (Invitrogen) following the manufacturer’s instructions. RNA was quantified by spectrophotometric reading at 260 nm and analyzed for quality assurance using an Agilent 2100 Bioanalyzer (Agilent Technologies) at the Biocomplexity Institute Proteomics Core Facility (Virginia Polytechnic Institute and State University, Blacksburg, VA). Reverse transcription was as described, and quantitative PCR assays were performed using IQ SYBR Green Supermix (Bio-Rad), 10 ng of cDNA, and the indicated primers for amplification (Table S4) (9). Real-time assays were performed in triplicate on a MyIQ single color Real-Time PCR Detection instrument (Bio-Rad). Data were collected and analyzed with Optical System Software, version 1.0. The GAPDH and β-ACTIN genes were used as internal controls to compute the relative expression level (ΔCT) for each sample. Fold change of gene expression in each sample was calculated as 2−ΔΔCt.
Table S4.
Primer sequences for qRT-PCR
| Gene | Encoding protein | Primer sequence, 5′ to 3′ | GenBank accession number | |
| ACTB | β-actin | Forward | TCAGAAGGATTCCTATGTGGGCGA | NM_001101.3 |
| Reverse | TTTCTCCATGTCGTCCCAGTTGGT | |||
| CLOCK | Clock | Forward | AGTTCAGCAACCATCTCAGGCTCA | NM_004898.3 |
| Reverse | TTGCTGGTGATGTGACTGAGGGAA | |||
| CRY1 | Cry1 | Forward | ATCATTGGTGTGGACTAC | NM_021117.3 |
| Reverse | TCTGCTTCATTCGTTCA | |||
| GAPDH | GAPDH | Forward | CTCTGGTAAAGTGGATATTGT | NM_002046.4 |
| Reverse | GGTGGAATCATATTGGAACA | |||
| PER2 | Per2 | Forward | TGAGAAGAAAGCTGTCCCTGCCAT | NM_022817.2 |
| Reverse | GACGTTTGCTGGGAACTCGCATTT | |||
| TP53 | p53 | Forward | GCGTGTGGAGTATTTGGATGA | NM_000546.5 |
| Reverse | AGTGTGATGATGGTGAGGATGG | |||
| NR1D1 | Rev-erbα | Forward | AGCATGACCAAGTCACCCTGCTTA | NM_021724.3 |
| Reverse | TGCGGCTTAGGAACATCACTGTCT | |||
| TBP | Tbp | Forward | CACGAACCACGGCACTGATT | NM_003194.4 |
| Reverse | TTTTCTTGCTGCCAGTCTGGAC | |||
Primers used throughout are summarized. Sequences were retrieved from their corresponding GenBank accession number and primers designed using Beacon Designer (Premierbiosoft) software.
In Vitro Binding Assays and Ubiquitination Reactions.
In vitro transcription and translation of pCS2+3xFLAG-p53 and pCS2+myc-Per2 and -Mdm2 were carried out using the SP6 high-yield TNT system (Promega) following manufacturer’s instructions. Aliquots of corresponding tagged protein (1–2 μL) were incubated at room temperature for 30 min to allow complex formation to happen (step 1 in Fig. S5A) before adding the reaction buffer containing 1× ubiquitination buffer (Enzo Biomol), 1 mM DTT, 20 U/mL inorganic pyrophosphate (Sigma-Aldrich), 20 μg/mL ubiquitin-aldehyde (Enzo Biomol), 200 μg/mL ubiquitin or 1 mg/mL methyl-ubiquitin, 1× ATP-energy regeneration system (5 mM ATP/Mg2+; Enzo Biomol), 40 μM MG132, 1× ubiquitin activating enzyme E1 (human, recombinant; Enzo Biomol), and 1× ubiquitin conjugating enzyme UbcH5c (human, recombinant; Enzo Biomol) to a final volume of 15 μL (step 2 in Fig. S5A). No ubiquitin treatment (indicated as “–” in Fig. 4 under “Ub treatment”) indicates the absence of ubiquitin-aldehyde, ubiquitin, and ATP-energy regeneration system in the reaction mixture (step 2 in Fig. S5A). Reactions were further incubated at 37 °C for 60 min before being terminated by the addition of binding buffer [20 mM Tris⋅HCl (pH 7.4), 100 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100]. Diluted samples were incubated twice at 4 °C for 1 h, first after addition of 2 μg of α-FLAG antibody (Sigma-Aldrich), and then after the addition of 8 μL of protein A–Sepharose 4B (50% slurry; Sigma-Aldrich) (step 3 in Fig. S5A). Then, bead-bound-(mono- or poly-)ubiquitinated p53 complexes were washed three times with binding buffer and resuspended in binding buffer (20 µL) containing 40 μM MG132, 20 μg/mL ubiquitin-aldehyde, and myc-Per2 (5 µL) (step 4 in Fig. S5A). Complex were allowed to form at 4 °C for 4 h before washing with binding buffer. Bound complexes were resolved by SDS/PAGE and proteins identified by immunoblotting using specific antibodies (step 5 in Fig. S5A). Goat α-mouse IgG (H+L), HRP-conjugated, cross-absorbed was used as secondary antibody (Thermo Scientific). The p53:Mdm2:Per2 ratio in the reaction was 1:2:5.
Subcellular Fractionation, Immunoblot, and Immunoprecipitation Assays.
Cells (HCT116) were harvested at different times after circadian synchronization and extracts (20–100 µg) were analyzed for endogenous expression of Per2, p53, Mdm2, Cry1, and CK1ε using specific primary antibodies [α-Per2 (clone 5C10 for immunoblotting; Sigma-Aldrich; and H-90 for immunoprecipitation; Santa Cruz Biotechnology), α-p53 (FL-393 for immunoprecipitation and DO-1 for immunoblotting; Santa Cruz Biotechnology), α-Mdm2 (Ab-5; Calbiochem), α-Cry1 (clone 4H4-1C4; Abnova), α-CK1ε (catalog no. 12448; Cell Signaling)]. Subcellular fractionation was performed essentially as described (39). Nuclear and cytoplasmic fractions from synchronized HCT116 cells were analyzed for the expression of endogenous proteins by immunoblotting. Tubulin and lamin A/C were used as loading controls and as positive markers for the cytoplasmic and nuclear fractions [α-tubulin (Sigma-Aldrich) and α-lamin A/C (Santa Cruz Biotechnology), respectively]. For immunoprecipitation of endogenous Per2:p53 complex, total extracts (∼1 mg) were incubated with α-Per2 in lysis buffer overnight at 4 °C before the addition of protein-A beads (50% slurry; Sigma-Aldrich). Samples were then kept for an additional 2 h at 4 °C with rotation and washed, and proteins were analyzed by immunoblotting following standard procedures (9). When indicated, immunoprecipitation of endogenous Per2 and p53 from nuclei and cytoplasmic fractions were from 8 × 106 and 2 × 106 cells, respectively.
Analysis of Protein Half-Life.
Total (1.2 × 105 cells), nuclear (3.15 × 105 cells), and cytoplasmic (0.63 × 105 cells) fractions were obtained from HCT116 cells collected at different times after cycloheximide (CHX) (100 μg/mL) addition (t = 0). Samples were analyzed for endogenous p53 and Per2 levels by immunoblotting. Protein levels were quantitated using the ImageJ software, version 1.45 [NIH software package (40)] or using Image Lab Software followed by detection using Gel Doc XR+ System (Bio-Rad). Values were normalized to tubulin or lamin A/C levels. Remaining protein levels [in arbitrary units (a.u.)] were calculated based on t = 0 and data fitted using Excel.
Systematic Search for Per2-Mediated Regulations of p53.
We used a mathematical model to investigate what type of Per2-mediated regulations of p53 are required to reproduce their phase relationship in the cytoplasmic and nuclear compartments (Fig. 1 A and D). The circadian rhythm of Per2 is simulated via the Kim and Forger model, a simple mathematical model of the intracellular mammalian circadian clock (24, 25). To simulate Per2’s regulation of p53 levels, we added the following equations to the Kim and Forger’s model (Per2 is referred as Per in all equations):
| [S1] |
where [p53c] and [p53n] represent the concentrations of p53 protein in the cytoplasm and nucleus, respectively. Similarly, [Perc] and [Pern] represent the concentrations of Per2 protein in the cytoplasm and nucleus, respectively. The functions f1, f2, f3, f4, and f5 in Eq. S1 describe the mode by which Per2 regulates production, nuclear import, cytosolic degradation, nuclear export, and nuclear degradation of p53, respectively (Fig. 2A). Specifically, the function fi can have one of the three forms shown below
| [S2] |
which represent positive regulation, negative regulation, or no regulation, respectively, via Per2. These equations (fi) have unknown parameters αi, and Ki. The parameters and Kdeg in Eq. S1 were estimated using a global stochastic parameter search algorithm (43) as described below.
Step 1: Initial parameter selection leading to sustained oscillation of p53.
We used a normalized version of Kim and Forger’s circadian clock model (24, 25), in which all reaction rate constants are equal to 1 to simulate circadian rhythms of [Per]. Thus, we assumed that new reaction rate constants (αi) would also take values close to 1, so we assigned initial parameter values for each αi and Kdeg to be a random number 10χ, where χ was chosen from a uniform distribution between −1 and 1. Regarding the regulatory functions fi, one of the three types in Eq. S2 was randomly selected. Then, for the positive regulation function, the parameter Ki was chosen to be 0.1, 1, or 10× the average concentration of [X], which represents weak, medium, and strong positive regulation, respectively (labeled as +, ++, and +++ in Fig. 2B). Conversely, for the negative regulation function, the chosen Ki with values of 0.1, 1, or 10× the average concentration of [X] represents strong, medium, and weak negative regulation, respectively (---, --, and - in Fig. 2B). We repeated this initial parameter selection process until we obtained a set of parameter values for which the model can simulate the sustained oscillations of p53 in both the cytoplasm and nucleus.
Step 2: Parameter calibration for calculating the correct phase relationship between p53 and Per2.
Using the simulated annealing method (42) with the initial parameter set obtained from step 1, we calibrated the parameters for which the model could simulate the correct phase and amplitude of p53 oscillations in both the cytoplasm and nucleus (Fig. 1D). During this parameter estimation process, the parameters determining the regulation types were fixed (i.e., only αi and Kdeg were perturbed) to test whether the initially selected regulation types (fi) could successfully simulate the correct phase and amplitude of p53 rhythms. The parameter estimation process stops after 104 perturbations of parameters from the initial parameter set. If the model with the newly estimated parameter set could reproduce the phase and amplitude of p53 rhythms in total extracts, cytoplasm, and nucleus (Fig. 1 A and D), we saved the parameter set as a potential parameter set for the model.
Step 3: Selection and analysis of 1,000 parameter sets.
We repeated steps 1 and 2 until we found 1,000 parameter sets for which the model successfully simulates the phase and amplitude of p53 rhythms. The distribution of parameters determining the regulation types in the 1,000 parameter sets is represented in Fig. 2B. Furthermore, the half-life of p53 in the cytoplasm and nucleus was estimated by simulating the time at which p53 reaches one-half of its concentration after the rate of production of p53 in the model was set to zero mimicking the results from the treatment with CHX (Fig. 2C).
Detailed Description of the Comprehensive Mathematical Model (Fig. 2D).
Based on the essential properties of the models that simulate the correct phase relationship between Per2 and p53 (Fig. 2 B and C), we developed a more detailed and realistic mathematical model (Fig. 2D). For this, we extended the Kim and Forger model (24, 25), to describe the interactions between p53 and Per. The model consists of two compartments, the cytoplasm and the nucleus. We used a subscript to denote which compartment a variable belongs to (c, cytoplasm, and n, nucleus) and a superscript to denote the ubiquitination status of p53 (0, nonubiquitinated; m, monoubiquitinated; and p, polyubiquitinated) (see Table S2 for a detailed description of variables). The model consists of a system of 13 differential equations (see Table S3 for a detailed description of the parameters). To consider the differences between the volume of the cytoplasm and the nucleus, the nucleus localization rate is multiplied by Nf, the ratio of cytoplasm to nuclear compartment volume. Similarly, the nuclear export rate in the cytoplasm is divided by Nf. The model largely consists of the transcriptional negative-feedback loop of the circadian clock, the interactions between Per and p53, and the ubiquitination modifications that p53 is subjected to in each compartment
Per transcriptional negative-feedback loop.
In the model, the rate of PER2 gene transcription is proportional to the activity of its promoter (E-box). The activity of the promoter is proportional to the fraction of free activator, that is, activator molecules that are not sequestered by the Per protein in the nucleus. Free activator concentration, A([Pern]), is described by the following function (24, 25):
A quasi–steady-state approximation is used to derive A([Pern]) under the assumption of fast binding and unbinding between the Per protein and the activator (24, 25). Per mRNA is translocated to the cytoplasm, where it is translated into Per protein, which returns to the nucleus where it binds to and inactivates its free transcriptional activator, A. As was the case with the previous model (24, 25), the total activator concentration in the nucleus (AT) is assumed to be a constant.
Ubiquitination and degradation of p53.
Because p53’s mRNA rhythm is weak (Fig. 1B), the model assumes that it is constitutively expressed and does not play a critical role in generating oscillations of the p53 protein. Hence, p53 protein is assumed to be constantly produced in the cytoplasm and ubiquitinated there by Mdm2. For simplicity, the ubiquitination process is described by a Michaelis–Menten function rather than explicit modeling of interactions between p53 and Mdm2. Monoubiquitination of p53 inhibits its nuclear translocation and promotes nuclear exit (18), and polyubiquitination of p53 leads to rapid proteasomal degradation. In the model, polyubiquitination occurs only in the cytoplasm, as suggested by experimental data (18). This leads to faster clearance of p53 in the cytoplasm than in the nucleus, which is required to simulate the correct rhythm of p53 (Fig. 2C).
Regulation of p53 by Per2.
When p53 is bound to Per, ubiquitination of p53 is inhibited (8). Because monoubiquitination is inhibited by Per, Per promotes translocation of p53 into the nucleus, a process that is also required, according to our simple mathematical model, to simulate the correct rhythm of p53 (Fig. 2B). Furthermore, Per also inhibits p53 polyubiquitination, which prevents proteasomal degradation of p53 and stabilizes p53 (8).
Computer Simulation of the Mathematical Model.
All simulations were performed using Mathematica 10.0 (Wolfram Research).
Discussion
Today, evidence points toward circadian factors controlling numerous processes that impact cell cycle transitions, growth, and death (for review, see ref. 31). However, we lack a unifying view about how those interactions operate under normal progression and how they adjust to stress conditions. To begin addressing this shortcoming, we focus on circadian components, specifically Per2, which play critical roles in controlling proliferation by interacting with cellular factors at key regulatory nodes. Accordingly, our previous findings placed Per2 at the core of the checkpoint response by directly interacting with p53, controlling p53’s stability in unstressed conditions and its transcriptional activity in response to genotoxic stimuli (8, 9); however, little is known about how Per2 and p53’s spatiotemporal distributions help to segregate signals in the cell.
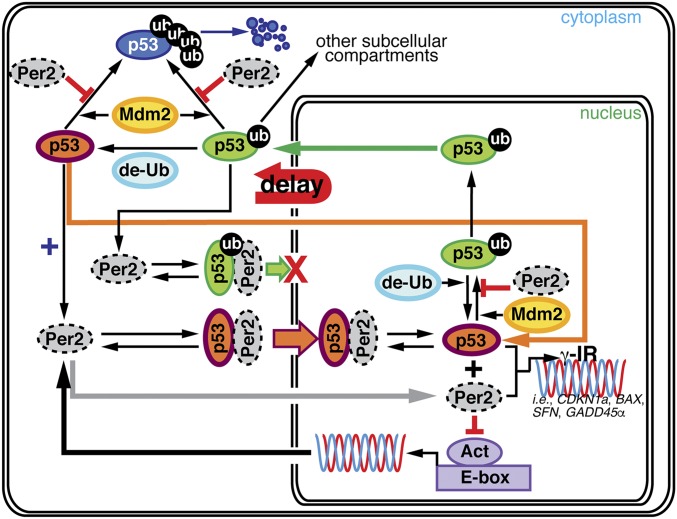
Our approach is simple in concept as it combines theoretical and experimental work and is motivated by an existing conundrum: how can discordant Per2 and p53 phases coexist in a model in which Per2 enhances p53’s stability (Fig. 1 and ref. 8)? To tackle this problem, we used a systematic investigation of models from which we could infer the interactions between Per2 and p53 using their time course data (Fig. 1 and refs. 22, 23, and 32). Three essential molecular mechanisms were predicted (Fig. 2 and Fig. S3) and validated experimentally: (i) p53 half-life is greater in the nucleus (Fig. 3 and Fig. S4), (ii) Per2 associates to various ubiquitinated forms of p53 (Fig. 4 and Fig. S5), and (iii) Per2 promotes p53’s nuclear entry (Fig. 5 and Fig. S6). Accordingly, the proposed model implies that the phase difference between Per2 and p53 in the cytoplasm may result from Per2:p53 binding and shuttling between compartments where posttranslational modifications in p53 take place (Fig. 6).
Fig. 6.
A unifying model for Per2-mediated regulation of p53 translocation and signaling. The schematic representation summarized in this section incorporates our previous and newly described findings. First, we focused on the Per2:p53 interaction as a mode of controlling the endogenous levels of p53 under physiological conditions. We proposed this to be a strategy to generate a stable pool of p53 that preconditions the cell for a rapid response to a stress situation (ref. 8; upper left corner of the model). Second, we demonstrated that Per2:p53 interaction is functionally relevant for modulating a p53-mediated transcriptional response in cells exposed to genotoxic stimuli (ref. 9; lower right corner). Last, we show that both processes are related and mediated by events that control the translocation and availability of p53 via Per2 in each subcellular compartment over the course of the circadian cycle (this publication).
Protein translocation dynamics and posttranslational events are recurrent themes when analyzing the importance of oscillations for modulating cell cycle transitions and for the generation of circadian rhythms. We know now that what makes the regulation of p53 by Per2 distinctive from others, besides Per2’s role as a p53 stabilizer, is its dual anchor/transporter function (depending on whether Per2 is bound to monoubiquitinated or nonubiquitinated p53) in the Per2:p53 complex, which takes place in a dynamic fashion throughout the day (Fig. 6). We first described the interplay between Per2, p53, and Mdm2, and how Per2:p53 interaction modulates the endogenous levels of p53 in unstressed conditions, thereby generating a readily available pool of p53 amenable for triggering a rapid response to, for example, genotoxic stress (ref. 8; Fig. 6, upper left corner). Then, we placed Per2 as a direct regulator of p53 expression and transcriptional activity under stress conditions, thus proving that Per2:p53 interaction could be functionally relevant in multiple scenarios (refs. 8 and 9; Fig. 6, lower right corner). Our findings bring both pieces of the Per2 and p53 story together by unveiling the role that Per2-mediated translocation of p53 plays in generating a time-dependent subcellular distribution of the p53’s pool (Fig. 6). In a larger context, and considering the functional significance of p53 in normal cell growth and carcinogenesis, understanding the underlying molecular mechanism by which circadian factors regulate p53 accumulation could help identify windows of opportunities for effective chronopharmaceutical treatments.
Last, whereas the focus of our work is in understanding how the circadian clock drives the circadian rhythms of p53 via Per2, it is worth mentioning that other p53 oscillations, with a period of ∼5 h, exist and are generated in response to specific stimuli. For example, in MCF7 cells, DNA damage triggers p53 pulses that are primarily driven by those occurring in upstream kinases (e.g., ATM and ChK) and are sustained as long as the DNA damage persists (33). The existence of p53 pulses impacts the expression of different subsets of p53 target genes and, consequently, influences the fate of the cell (34). Interestingly, even unstressed proliferating cells exhibit spontaneous pulses of p53 that are believed to be triggered by intrinsic and transient DNA damage that occurs during cell cycle transitions (35). Consequently, and in future studies, it would be of interest to investigate the existing interplay between stress-induced pulses of p53 (36) and the circadian rhythms oscillations of p53 (our work) as both the circadian and cell cycle oscillators are synchronized (37, 38).
Materials and Methods
Subcellular fractionation was performed essentially as described (39). Nuclear and cytoplasmic fractions from synchronized HCT116 cells were analyzed for expression of endogenous proteins by immunoblotting following standard procedures (8). When indicated, immunoprecipitation of endogenous Per2 and p53 from nuclei and cytoplasmic fractions were from 8 × 106 and 2 × 106 cells, respectively. To measure protein half-life, fractions were obtained from HCT116 cells collected after CHX (100 μg/mL) addition (t = 0). Endogenous protein levels were determined by immunoblotting and quantitated using the ImageJ software, version 1.45 [NIH software package (40)] or Image Lab Software followed by detection using Gel Doc XR+ System (Bio-Rad). For mathematical modeling, we extended a previous model of the intracellular mammalian circadian clock (24, 25) to describe interactions between p53 and Per2 (Fig. 2 A and D). Refer to SI Materials and Methods and Table S4 for technical details and Tables S2 and S3 for a list of variables and parameters used in modeling, respectively.
Acknowledgments
We thank Dr. D. Capelluto for critical reading of the manuscript and Dr. J. Webster for comments and manuscript editing. This work was supported by National Science Foundation Molecular and Cellular Biosciences Division Grant MCB-1517298 and Fralin Life Science Institute (to C.V.F.), and Korea Advanced Institute of Science and Technology Research Allowance Grant G04150020, National Research Foundation of Korea Grant N01160447, and T. J. Park Science Fellowship of POSCO (to J.K.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607984113/-/DCSupplemental.
References
- 1.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9(10):764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rana S, Mahmood S. Circadian rhythm and its role in malignancy. J Circadian Rhythms. 2010;8:3. doi: 10.1186/1740-3391-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht U, Bordon A, Schmutz I, Ripperger J. The multiple facets of Per2. Cold Spring Harb Symp Quant Biol. 2007;72:95–104. doi: 10.1101/sqb.2007.72.001. [DOI] [PubMed] [Google Scholar]
- 5.Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle. 2010;9(6):1097–1103. doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antoch MP, Toshkov I, Kuropatwinski KK, Jackson M. Deficiency in PER proteins has no effect on the rate of spontaneous and radiation-induced carcinogenesis. Cell Cycle. 2013;12(23):3673–3680. doi: 10.4161/cc.26614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sancar A, et al. Circadian clock, cancer, and chemotherapy. Biochemistry. 2015;54(2):110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh T, et al. The circadian factor Period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol Biol Cell. 2014;25(19):3081–3093. doi: 10.1091/mbc.E14-05-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gotoh T, Vila-Caballer M, Liu J, Schiffhauer S, Finkielstein CV. Association of the circadian factor Period 2 to p53 influences p53's function in DNA-damage signaling. Mol Biol Cell. 2015;26(2):359–372. doi: 10.1091/mbc.E14-05-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111(1):41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 11.Sun CM, et al. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol Oncol Res. 2010;16(3):403–411. doi: 10.1007/s12253-009-9227-0. [DOI] [PubMed] [Google Scholar]
- 12.Hua H, et al. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 2007;14(9):815–818. doi: 10.1038/sj.cgt.7701061. [DOI] [PubMed] [Google Scholar]
- 13.Lück S, Thurley K, Thaben PF, Westermark PO. Rhythmic degradation explains and unifies circadian transcriptome and proteome data. Cell Rep. 2014;9(2):741–751. doi: 10.1016/j.celrep.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Mazan-Mamczarz K, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100(14):8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129(5):1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamanini F, Yagita K, Okamura H, van der Horst GT. Nucleocytoplasmic shuttling of clock proteins. Methods Enzymol. 2005;393:418–435. doi: 10.1016/S0076-6879(05)93020-6. [DOI] [PubMed] [Google Scholar]
- 18.Marchenko ND, et al. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2010;17(2):255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middeler G, et al. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene. 1997;14(12):1407–1417. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- 20.Walton KM, et al. Selective inhibition of casein kinase 1 epsilon minimally alters circadian clock period. J Pharmacol Exp Ther. 2009;330(2):430–439. doi: 10.1124/jpet.109.151415. [DOI] [PubMed] [Google Scholar]
- 21.Vanselow K, Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:167–176. doi: 10.1101/sqb.2007.72.036. [DOI] [PubMed] [Google Scholar]
- 22.Clodong S, et al. Functioning and robustness of a bacterial circadian clock. Mol Syst Biol. 2007;3:90. doi: 10.1038/msb4100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locke JC, Westermark PO, Kramer A, Herzel H. Global parameter search reveals design principles of the mammalian circadian clock. BMC Syst Biol. 2008;2:22. doi: 10.1186/1752-0509-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JK, Forger DB. A mechanism for robust circadian timekeeping via stoichiometric balance. Mol Syst Biol. 2012;8:630. doi: 10.1038/msb.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JK. Protein sequestration versus Hill-type repression in circadian clock models. IET Syst Biol. 2016;10(4):125–135. doi: 10.1049/iet-syb.2015.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Brate A, Giannakakou P. The importance of p53 location: Nuclear or cytoplasmic zip code? Drug Resist Updat. 2003;6(6):313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA. 2006;103(35):13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoo SH, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci USA. 2005;102(7):2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, et al. Mono- versus polyubiquitination: Differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 31.Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol. 2013;23(5):724–731. doi: 10.1016/j.conb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JK, Forger DB. On the existence and uniqueness of biological clock models matching experimental data. SIAM J Appl Math. 2012;72(6):1842–1855. [Google Scholar]
- 33.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: A mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30(3):277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purvis JE, et al. p53 dynamics control cell fate. Science. 2012;336(6087):1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142(1):89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JK, Jackson TL. Mechanisms that enhance sustainability of p53 pulses. PLoS One. 2013;8(6):e65242. doi: 10.1371/journal.pone.0065242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bieler J, et al. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol. 2014;10:739. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feillet C, et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci USA. 2014;111(27):9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts”, prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 42.Yang X, et al. β-catenin induces β-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145(3):289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 43.Gonzalez OR, Küper C, Jung K, Naval PC, Jr, Mendoza E. Parameter estimation using Simulated Annealing for S-system models of biochemical networks. Bioinformatics. 2007;23(4):480–486. doi: 10.1093/bioinformatics/btl522. [DOI] [PubMed] [Google Scholar]