Significance
For DNA to be duplicated, it must first be unwound. Here, we examine the nature of the helicase engine that drives this unwinding process. In archaea and eukaryotes, the core helicase is the MCM complex. Our studies reveal that the active form of the archaeal replicative helicase is a complex of MCM with the accessory proteins Cdc45 and GINS. Our work reveals functional conservation of this architecture despite the 2 billion-year evolutionary gulf between archaea and eukaryotes.
Keywords: DNA replication, CMG, Archaea, MCM, helicase
Abstract
The regulated recruitment of Cdc45 and GINS is key to activating the eukaryotic MCM(2-7) replicative helicase. We demonstrate that the homohexameric archaeal MCM helicase associates with orthologs of GINS and Cdc45 in vivo and in vitro. Association of these factors with MCM robustly stimulates the MCM helicase activity. In contrast to the situation in eukaryotes, archaeal Cdc45 and GINS form an extremely stable complex before binding MCM. Further, the archaeal GINS•Cdc45 complex contains two copies of Cdc45. Our analyses give insight into the function and evolution of the conserved core of the archaeal/eukaryotic replisome.
The initiation of DNA replication is an important control point in the progression of the cell cycle. In eukaryotes, origins are defined by the initiator protein ORC complex that, via the actions of two additional factors, the helicase coloaders Cdc6 and Cdt1, directs loading of the MCM(2-7) replicative helicase onto double-stranded DNA. Activation of the MCM(2-7) replicative helicase occurs subsequent to recruitment, leading to DNA melting and assembly of the full replisome apparatus (1, 2). Key steps in MCM activation involve a series of phosphorylation-dependent events that promote the sequential association of Cdc45 and the GINS complex with the chromatin-associated MCM double hexamer. These recruitment events are regulated by the CDK and DDK kinases and require the additional accessory factors Sld3/7, Dpb11, and Sld2(3-6). The Cdc45•MCM(2-7)•GINS complex (CMG) forms the core of the eukaryotic replisome, and this 11-subunit assembly appears to be the functional helicase driving fork progression (3–6). The archaeal replication machinery resembles an ancestral form of its eukaryotic counterpart. Archaea possess a simple homohexameric MCM (5, 7). In addition, archaeal homologs of GINS and Cdc45 have been identified (8–14). In species of the genus Sulfolobus, we have previously demonstrated that the GINS complex interacts with the N-terminal domains of MCM (8). In Sulfolobus, GINS is a dimer of dimers: one subunit, Gins23, is related to the eukaryotic GINS components Psf2 and Psf3, and the second Sulfolobus subunit, Gins15, is related to the eukaryotic Sld5 and Psf1. These sequence relationships have been confirmed by structural studies of the Thermococcus kodakarensis GINS complex that have demonstrated the tetrameric assembly of archaeal (Gins15)2•(Gins23)2 and validated the organizational similarity of the archaeal and eukaryotic GINS complexes (15). Interestingly, Sulfolobus GINS copurifies over the course of eight steps with a further polypeptide that we initially named RecJdbh, based on its observed homology with the presumptive DNA binding domain of the bacterial exonuclease, RecJ (8). Subsequent sequence analyses have revealed a relationship between RecJ and eukaryotic Cdc45, and this has been elegantly confirmed by recent structural studies of eukaryotic Cdc45 (9, 11, 16, 17). We therefore propose renaming RecJdbh as Cdc45. As archaea lack orthologs of Sld2, Sld3, Sld7, and Dpb11, and do not possess counterparts of CDK or DDK, it appears that Cdc45 may form a constitutive complex with GINS in cells. In the current work, we investigate the role of the Cdc45•GINS (hereafter CG) complex in vivo and in vitro. We observe association of CG with the MCM complex at replication origins and during replication fork progression. We map the interactions between Cdc45 and GINS and between the CG complex and MCM. We reveal that although neither Cdc45 nor GINS individually stimulates MCM activity, the formation of the full CMG complex robustly enhances the basal helicase activity of MCM. Our data indicate that the CMG helicase is a conserved and central component of the replication fork in archaea and eukaryotes.
Results
Generation of a Strain Expressing an Epitope Tagged Cdc45.
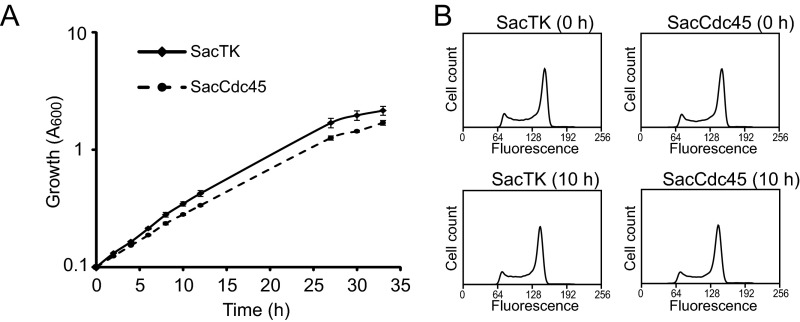
To investigate the in vivo role of Cdc45 in DNA replication, we modified the endogenous cdc45 gene of Sulfolobus acidocaldarius to encode a protein with a C-terminal dual affinity-tag (Fig. 1A). The addition of the C-terminal c-myc hexahistidine tag was confirmed by PCR amplification across the Saci_0177 gene (Fig. 1B), DNA sequencing, and Western blot analysis (Fig. 1C). The SacCdc45 strain displayed a similar growth rate compared with the parental strain (Fig. S1). Furthermore, analysis of the cell cycle profile by flow cytometry did not reveal any differences between the parental strain and SacCdc45 (Fig. S1). Thus, the C-terminal affinity tag to Cdc45 does not significantly perturb cell cycle progression.
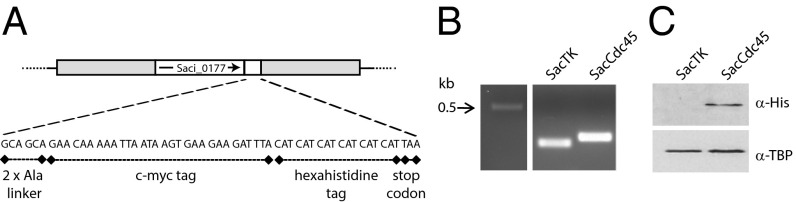
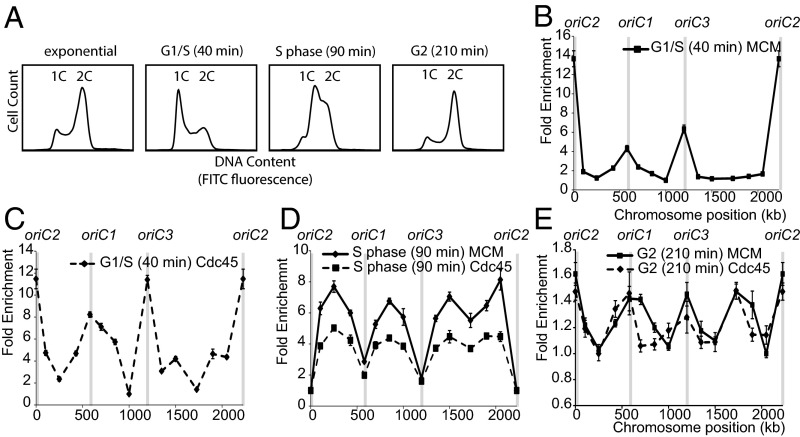
Fig. 1.
Construction of strain SacCdc45, encoding chromosomally expressed affinity-tagged Cdc45. (A) Schematic of the DNA sequence insertion in SacCdc45. A DNA sequence encoding a c-myc and hexahistine affinity tag was integrated into the 3′ end of gene Saci_0177, encoding Cdc45, using a “pop-in, pop-out” approach. (B) PCR amplification across the 3′ end of Saci_0177 using a genomic DNA template generated 188- and 245-bp products in SacTK and SacCdc45, respectively, indicating successful insertion of the affinity-tag encoding sequence. (C) SacCdc45 expresses C-terminal affinity-tagged Cdc45. Western blot analysis was performed using whole-cell extracts from exponential cultures of the parental strain, SacTK, and SacCdc45, using an anti-His-tag antibody, and anti-TBP antibody as a control.
Fig. S1.
Monitoring growth of SacCdc45 cells. (A) SacTK (solid line) and SacCdc45 (dashed line) cells were grown at 75 °C with shaking at 300 rpm, and growth was monitored by measuring absorbance at 600 nm. (B) Cell cycle analysis of SacCdc45. Flow cytometry analysis was performed throughout growth; shown are samples of SacTK and SacCdc45 taken at 0 and 10 h.
Cdc45 Interacts with GINS.
Cdc45 was initially identified in Sulfolobus solfataricus during purification of the archaeal GINS complex, as Cdc45 was stably associated with the GINS proteins (8). After partial purification of Cdc45•His6 from S. acidocaldarius using nickel affinity, gel filtration, and anion exchange chromatography, we detected two copurifying proteins (Fig. 2A). The coelution of Gins23 was confirmed by immunoblotting (Fig. 2B), and the proteins were identified by mass spectrometry as Gins15 (Saci_1278) and Gins23 (Saci_0901), both confirming the presence of a GINS•Cdc45 complex in S. acidocaldarius and demonstrating that the addition of the C-terminal tag had not disrupted formation of this complex.
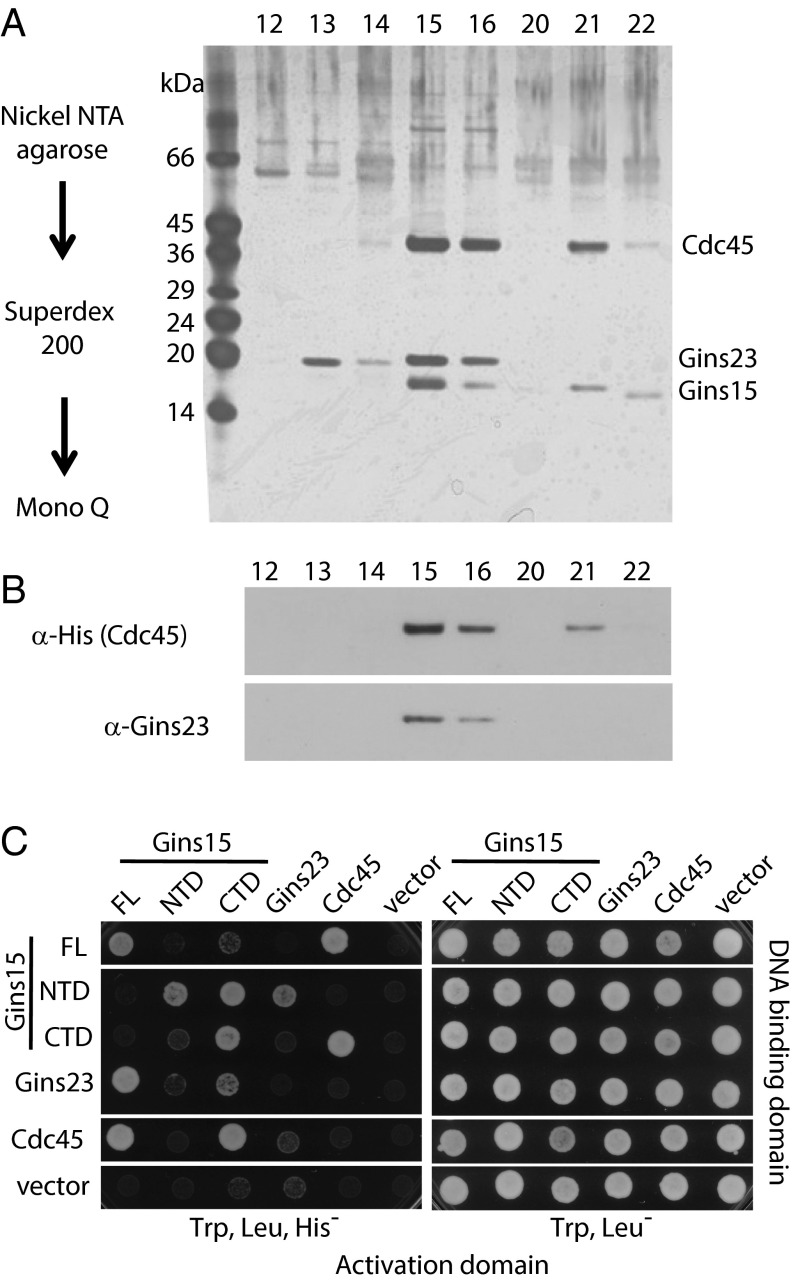
Fig. 2.
Cdc45 forms a complex with Gins15 and Gins23 in vivo. (A) Partial purification of the CG complex from extracts by successive affinity, size exclusion, and anion exchange chromatography; a silver-stained gel of the peak fractions from the final column is shown. (B) Western blotting was performed on the peak fractions from A, using the indicated antisera. (C) Yeast 2 hybrid analyses of interactions between the N- and C-terminal domains of Gins15 and full-length Gins23 and Cdc45. Growth on the selective plates (lacking histidine) is indicative of interaction.
We next performed yeast-two-hybrid analysis to examine the GINS–Cdc45 interaction in more detail (Fig. 2C). The Gins subunits contain an α-helical A domain and a β-strand rich B domain (14). The order of these domains is circularly permuted in Gins23 (BA) and Gins15 (AB) (Fig. S2). We could not observe any interaction between Gins23 and Cdc45, in agreement with our previous results (8). However, we confirmed interaction between Gin15 and Cdc45 and revealed that the C-terminal B domain of Gins15 was both necessary and sufficient for the interaction (Fig. 2C).
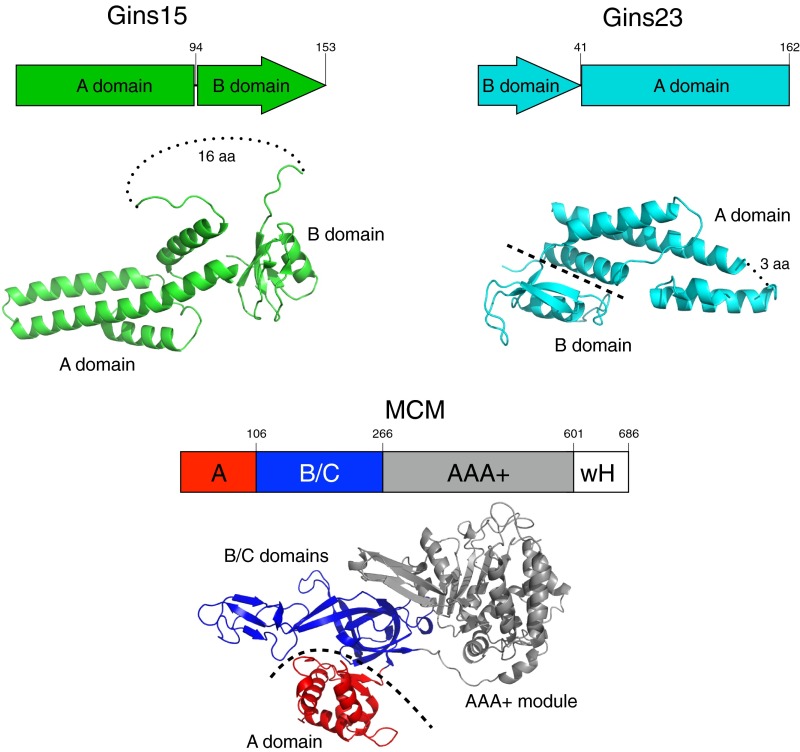
Fig. S2.
Representations and crystal structures of the domain organization of GINS subunits and MCM. Figures of the crystal structures were generated from PDB files 3ANW (T. kodakarensis GINS complex) and S. solfataricus MCM, the structure lacks the C-terminal wH domain. Dotted lines indicate missing density in the crystal structure, and dashed lines serve to highlight the boundaries between domains (15, 27).
Cdc45 Is a Component of the S. acidocaldarius Replisome.
Next we sought to determine whether Cdc45 is a component of the S. acidocaldarius replisome. To this end, we performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (qPCR) to determine the location of chromosomally associated Cdc45 and MCM throughout synchronous growth. qPCR was performed using 14 primer pairs for amplicons distributed across the SacCdc45 genome, including primers targeting the three origins of replication. MCM has previously been shown to be recruited to the S. acidocaldarius origins of replication at initiation of DNA replication (18), and as the presumed replicative helicase, MCM would be predicted to travel with the moving replication forks during S phase.
SacCdc45 strains were synchronized using the baby machine method (18). After formaldehyde-fixation, ChIP was performed using anti-MCM and anti-myc antibodies from cells fixed in G1/early S-phase, mid-S-phase and G2 phase (Fig. 3A). These experiments indicated recruitment of MCM and Cdc45 to the three origins of replication (oriC1, oriC2, and oriC3) in G1/S phase (Fig. 3 B and C). ChIP signals from the mid-S-phase samples reveal a relative reduction in Cdc45 and MCM at all three origins and relative increases in the interorigin regions. This is consistent with a model in which MCM and Cdc45 are associated with moving replication forks.
Fig. 3.
ChIP reactions using synchronized SacCdc45 cells were used to examine genomic localization of Cdc45 and MCM during the cell cycle. (A) Flow cytometry profiles of asynchronous, G1/early S-phase, S-phase, and G2-enriched synchronized cell populations are indicated. (B–E) ChIP profiles showing the enrichment of DNA in the immunoprecipitates at the various genomic loci at the indicated times.
This model is also supported by the observed occupancy of Cdc45 and MCM at the region midway between the two most separated origins (oriC2 and oriC3; ∼1,727 kb apart). This is the last region of S. acidocaldarius chromosome to be replicated (18). The ChIP signal from S phase cells indicates that replication forks have not yet progressed to this region in a significant number of cells (Fig. 3D). However, by the latest time (210 min), the moving forks, containing Cdc45 and MCM, have now reached this final region of DNA in a large number of cells (Fig. 3E). We note that flow cytometry reveals that some of the population has reentered G1 by 210 min; concomitant with this, we observe new enrichment of MCM and Cdc45 at the origins.
The CG Complex Is a Heterohexameric Assembly.
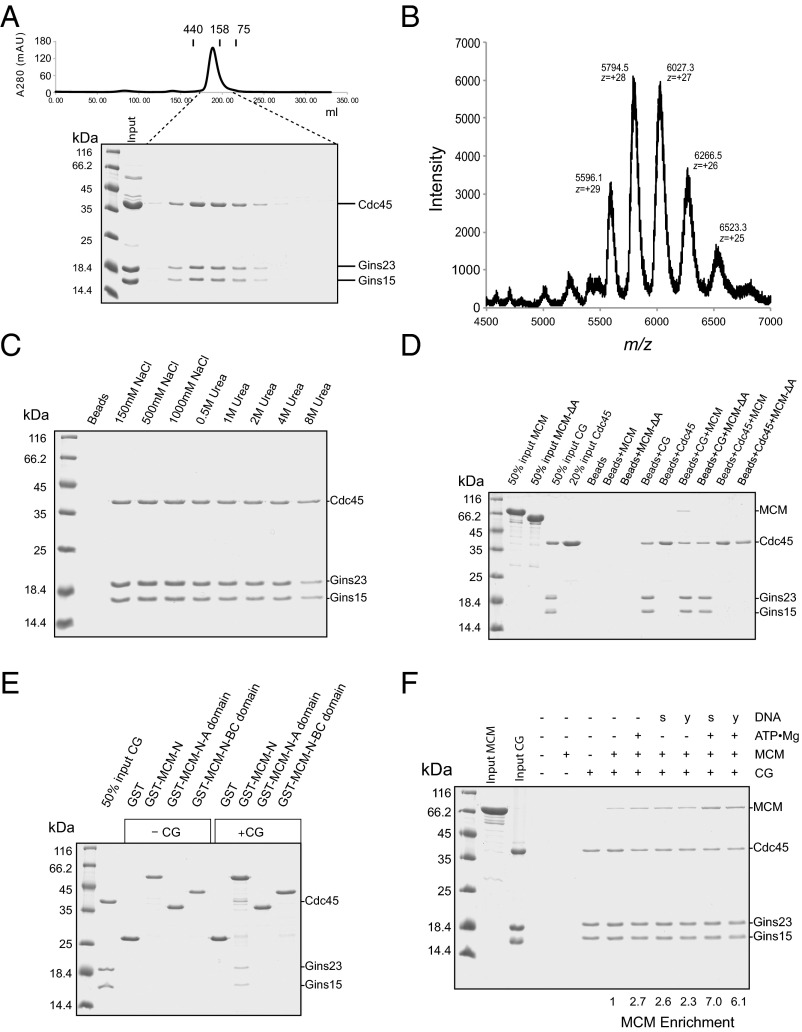
Next, we sought to determine the biochemical effect of CG association with MCM. Despite numerous attempts to express Cdc45 in a variety of bacterial and eukaryotic systems, we were unable to prepare the protein as recombinant from a heterologous system. Although it was possible to exploit the SacCdc45 strain to express a His-tagged version of Cdc45 from the endogenous locus, we could only obtain yields of a few hundred nanograms of protein from 6 L of culture. Accordingly, we switched to using the related strain Sulfolobus islandicus for our biochemical studies. Exploiting a strong and controllable promoter, we were able to express and purify milligram-scale amounts of C-terminally His-tagged Cdc45. We combined this with bacterially expressed GINS complex and recovered a reconstituted CG. Size exclusion chromatography of the reconstituted material reveals the majority of the CG complex has a mobility compatible with a 2:2:2 stoichiometry of Gins15:Gins23:Cdc45 (Fig. 4A). To further confirm the composition of the CG complex, we performed native electrospray ionization mass spectrometry (Fig. 4B). A 2:2:2 (Gins15:Gins23:Cdc45) complex has a predicted mass of 158,002 Da, whereas a 2:2:1 complex would have a mass of 117,346 Da, and the observed mass is 161,800, supporting the 2:2:2 stoichiometry. The excess mass that we detect is likely a result of tightly associated solvent molecules and ions.
Fig. 4.
(A) Gel filtration analysis of the reconstituted recombinant CG complex. The CG complex was analyzed by gel filtration on a Superdex 200 26/60 column. The UV absorption profile is shown with the positions of elution of 440, 158, and 75 kDa size standards (ferritin, aldolase, and conalbumin, respectively) indicated. (Bottom) SDS PAGE analysis of the corresponding fractions. (B) Native electrospray ionization mass spectrometry profile for the CG complex. The peak at m/z = ∼5,794.5 has a charge of +28. (C) Stability of the CG complex: 11 µg CG complex immobilized on Ni-NTA beads were washed four times with 500 µL TBS (10 mM Tris at pH 8.0, 150 mM) or TBS supplemented with NaCl or urea to the indicated concentration. The bead-retained material was analyzed by SDS/PAGE and proteins stained with Coomassie Brilliant Blue. (D) Pulldown assays to test for interaction between 13 µg Cdc45 or the CG complex immobilized on Ni-NTA via the His-tag on Cdc45 and 18 µg MCM or MCM-ΔA. (E) Interactions between the N-terminal domains of MCM and the CG complex. Glutathione sepharose beads bound to GST, GST-fused to MCM-N (residues 1–265), MCM-N-A domain (residues 1–104), or MCM-N-BC (residues 105–265) were incubated with 13 µg CG complex. After incubation and washing, bead-retained material was examined by SDS PAGE. (F) Ni-NTA pulldown assays with the indicated combination of CG complex, MCM, and ATP•Mg (2 mM final) and 20 µM single-stranded (s) or 10 µM Y-shaped DNA (y) (the distinct concentrations were to ensure same bulk amount of DNA per reaction). The relative enrichment of MCM was calculated by measuring band intensity of MCM, using ImageJ, and normalizing to the Cdc45 band intensity.
To test the stability of the CG complex, we immobilized it on Ni-NTA agarose beads. We then subjected it to washing in a variety of buffers, ranging in ionic concentration up to 1 M NaCl and with up to 8 M urea. Remarkably, the complex remained intact even when subjected to four washes containing 8M urea (Fig. 4C).
The CG Complex Interacts with MCM.
Next, we performed pulldown experiments, exploiting the His-tag on Cdc45, and could detect interaction between full-length MCM and the CG complex (Fig. 4D). No stable interaction was detectable with Cdc45 alone in the absence of GINS. Our previous work has demonstrated that Gins23 interacts with the N-terminal domains of MCM (8). The MCM N-terminal region can be further subdivided to A and B/C domains (19). Further, structural studies of eukaryal CMG have implicated the A domains of MCM2 and MCM5 as being of importance for CMG formation (3). We observe that deletion of the A domain of S. islandicus MCM abrogates association with CG (Fig. 4D). Interestingly, although the A domain is necessary for interaction, it is not sufficient. Using GST fusions of the N-terminal domains of MCM (domains A and B/C), and truncated derivatives thereof, we observe interaction mediated by the intact N-terminal domain, but this is abolished by deletion of either A or B/C domains (Fig. 4E). We also tested whether ATP•Mg or inclusion of single-stranded or Y-shaped oligonucleotides could influence the association of CG with MCM (Fig. 4F). Quantitation of the band intensity using ImageJ revealed a 2.5-fold enhancement of MCM retention on the CG beads by inclusion of fork-shaped DNA, single-stranded DNA, or nucleotide cofactor. The presence of either DNA with ATP resulted in a six- to sevenfold enrichment of MCM.
The Addition of CG Stimulates the Helicase Activity of MCM.
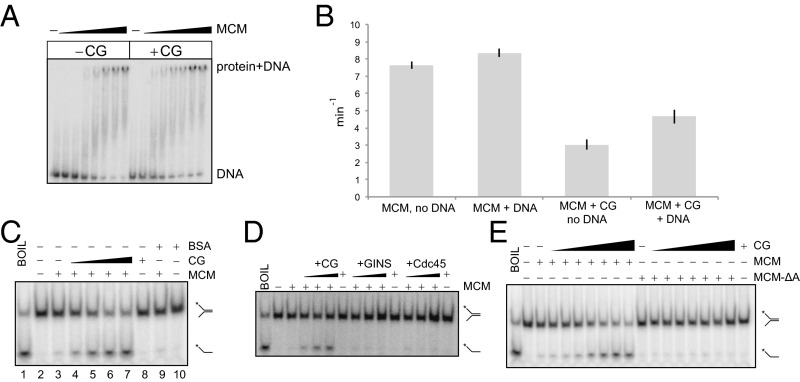
We sought to determine whether the CG complex has any effect on the ATPase or DNA binding activities of MCM. We examined DNA binding to a Y-shaped oligonucleotide substrate (Fig. 5A). Addition of CG resulted in a very slight (less than 1.5-fold) enhancement of the DNA binding activity of the MCM complex. We could not detect any DNA binding by the isolated CG complex. Next, we examined the influence of CG on the ATPase rate of MCM. The S. islandicus MCM has an intrinsic ATPase activity that is not affected by the addition of single-stranded DNA (Fig. 5B). Addition of CG lowers the basal ATPase rate by a factor of three, and addition of DNA now results in a ∼1.5-fold stimulation of ATPase activity.
Fig. 5.
Effect of CG on the biochemical properties of MCM. (A) EMSA analysis of MCM binding to 2 nM Y-shaped oligonucleotide DNA in the presence and absence of 500 nM CG. MCM concentrations were 5, 7.5, 10, 12.5, 15, 17.5, and 20 nM hexamer. (B) ATPase measurements of 100 nM MCM (hexamer) supplemented with 1 µM CG or corresponding buffer control. Where added, single-stranded DNA was at 1 µM. (C) DNA helicase assays with MCM and CG complex. DNA substrate was 1 nM. MCM was at 20 nM as hexamer; increasing amounts (125, 250, 500, 1,000 nM) of CG complex and 1 µM BSA as indicated were added. The control lane with CG complex alone had CG at 1 µM. (D) DNA helicase assays for MCM with CG complex, GINS, and Cdc45 separately. MCM was added 20 nM as hexamer; increasing amounts (125, 250, 500 nM) of CG complex, GINS, and Cdc45, as indicated, were added. Reactions with CG, GINS, or Cdc45 contained the indicated protein at 500 nM. (E) DNA helicase assays with MCM, MCM-ΔA, and CG complex. MCM and MCM-ΔA were added to 20 nM as hexamer; increasing amounts (15, 31, 62, 125, 250, 500, 1,000 nM) of CG complex, as indicated, were added. CG alone in a reaction without MCM or MCM-ΔA was at 1 µM.
Next, we determined whether CG, GINS, or Cdc45 could influence the helicase activity of MCM. At the concentrations of MCM we used in these experiments, we detect very low levels of activity by MCM alone (∼7% of template unwound). The addition of increasing concentrations of CG resulted in a clear stimulation of MCM helicase activity (Fig. 5C). Importantly, the CG complex alone had no helicase activity, and addition of a nonspecific control, BSA, failed to stimulate helicase activity. Next we tested whether GINS or Cdc45 could individually affect MCM’s helicase activity (Fig. 5D). Addition of GINS complex or Cdc45 individually had no detectable effect on the yield of product. Intriguingly, previous studies have yielded contrasting results on whether archaeal GINS can or cannot stimulate the helicase activity of MCM (8, 20–22). However, we observed a strong stimulation of helicase activity as we added the Cdc45•GINS complex to the reaction. Importantly, deletion of the A domain of MCM abolishes the stimulatory effect of CG on MCM’s helicase activity (Fig. 5E). We note that deletion of the A domain has no significant effect on the inherent helicase activity of MCM (23). Thus, association of the CG complex with MCM results in robust helicase activity.
Discussion
We provide a characterization of an archaeal CMG complex in vivo and in vitro. We demonstrate that Cdc45 and GINS form a stable complex in a range of species. Further, this complex associates with MCM at replication origins at the onset of replication and colocalizes with MCM during replication elongation. In agreement with our previous data, but in contrast with other reports, neither GINS nor Cdc45 individually affect MCM’s helicase activity (8). However, the CG complex promotes a robust stimulation of MCM’s helicase activity, and we can readily detect interaction between CG and MCM, using physical assays.
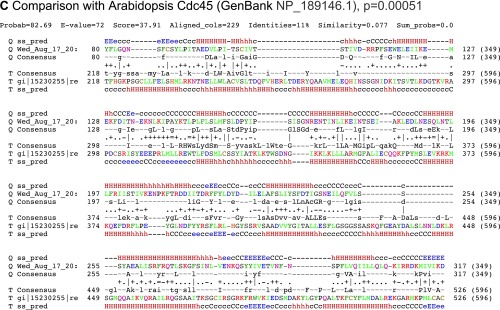
The proposed evolutionary relationship between RecJ and Cdc45 has been recently confirmed by structural studies. Cdc45 possesses the RecJ signature DHH and DHHA1 motifs, separated by a eukaryotic Cdc45-specific region termed the CMG interaction domain (CID) (24). We performed HHPred analysis and detected a highly significant homology (P = 3.1 × 10−11) between residues 20–335 of S. solfataricus Cdc45 and bacterial RecJ (Fig. S3A). However, to our considerable surprise, we also observed a significant relationship (P value = 0.00051 for relationship to Arabidopsis Cdc45) predicted by HHpred between residues 80–339 of S. solfataricus Cdc45 and the CID of eukaryal Cdc45s (Fig. S3 B and C).
Fig. S3.
HHPred analysis comparing Sulfolobus Cdc45 with its (A) bacterial and (B and C) eukaryotic counterparts. The analysis reveals an unanticipated relationship between the CID domain of eukaryotic Cdc45 and the archaeal protein.
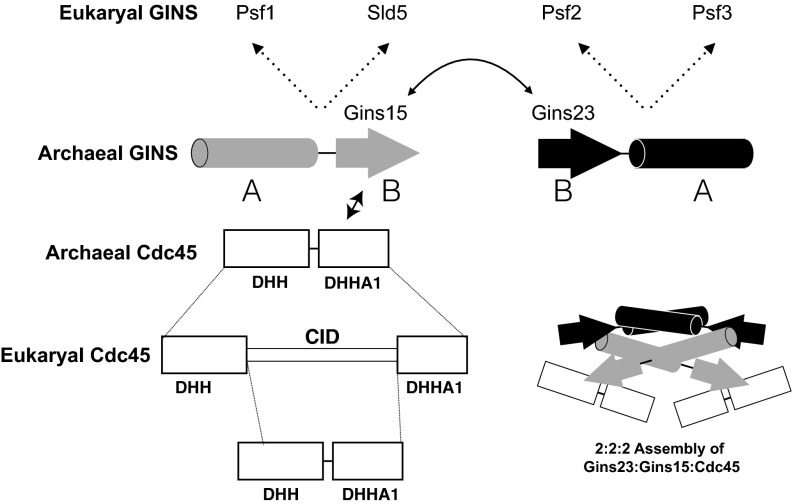
Thus, Sulfolobus Cdc45 is related both to the RecJ fold of eukaryotic Cdc45 and the CID (Fig. 6 and SI Materials and Methods). We speculate, therefore, that during the evolution of eukaryal Cdc45, a gene duplication and matryoshka-like internal fusion event occurred, with the CID subsequently diverging from the ancestral archaeal Cdc45 sequence. Thus, we suggest that the single eukaryal Cdc45 actually corresponds to two divergent copies of the archaeal ancestor. In agreement, we observe that the archaeal CG complex contains two copies of Cdc45, in contrast to the single Cdc45 in the eukaryotic assembly. The archaeal GINS complex is a dimer of dimers, and therefore is inherently more symmetric than the eukaryotic heterotetrameric GINS (8, 15). Archaeal GINS thus possesses two copies of the Gins15 C-terminal B domain we have demonstrated interacts with Cdc45. In eukaryotes, during the diversification of Psf1 and Sld5, presumably the progenitor ancestral Cdc45 interaction site on Sld5 has diverged and lost the ability to interact with Cdc45 (3). It is conceivable that this domain of Sld5 now plays a distinct and eukaryotic-specific role in orchestrating the replisome architecture. Indeed, we note that a yeast 2-hybrid study has provided evidence that Drosophila melanogaster Sld5 interacts with MCM10, a factor that lacks homologs in archaea. However, the region of Sld5 responsible for this interaction has not been mapped (25).
Fig. 6.
Evolutionary relationships and interactions between archaeal/eukaryotic GINS and Cdc45. Physical interactions between components are shown as double-headed solid arrows. Evolutionary relationships are shown as dotted lines. The diagram in the lower right provides a model for the 2:2:2 assembly of Gins23:Gins15:Cdc45, with the two copies of Cdc45 interacting with distinct Gins15 subunits via the latter’s B domain.
The greater symmetry in the archaeal CG complex is mirrored in the homohexameric MCM in archaea. In eukaryotes, CG appears to latch a gate between MCM2 and MCM5 (3). We have recently demonstrated that an open ring form of archaeal MCM is preferentially recruited to replication origins (26). We propose, therefore, that the archaeal CG complex will act similarly to its eukaryotic ortholog, conceivably facilitating closure of the gate in the archaeal MCM after its loading on replication origins.
Modeling of the human Cdc45 crystal structure into EM maps of CMG indicates that human Cdc45 makes multiple protein–protein contacts within the CMG assembly (24). More specifically, the DHH domain contacts the B domain of Psf1, whereas the CID mediates contacts with both Psf2 and MCM subunits. Our observations indicate that S. acidocaldarius Cdc45 is restricted to interactions with the Gins15 subunit. Thus, the eukaryotic Psf1–DHH interaction appears to be the key conserved feature of this assembly. Although we acknowledge that our binary interaction studies may have overlooked interactions important in the context of a higher-order CMG assembly, it is possible that during the evolution of the eukaryal CID, this region of Cdc45 acquired the ability to interact with Psf2 and its specific eukaryotic MCM subunit partners.
Our observations that Cdc45 and GINS form a remarkably stable complex both as recombinant proteins and in cell-free extracts highlight another key difference from the situation in eukaryotes. The sequential recruitment of eukaryal Cdc45 and GINS provide key control points for the cell cycle–dependent regulation of the activation of DNA replication (1, 2). This greater regulatory potential in the eukaryal assembly pathway presumably reflects the increased requirement for fidelity of control for the multiplicity of replication origins in eukaryotic chromosomes. It remains to be determined whether the recruitment of CG is a control point in the Sulfolobus cell cycle. Sulfolobus species have three replication origins per chromosome, and our work in S. islandicus has revealed that each is specified by a distinct initiator protein (23, 24). We have demonstrated control at the level of the initiator protein for one of these origins, oriC1, and this mechanism likely extends to oriC2. How initiation at oriC3, an orc1/cdc6-independent origin, is regulated is currently unknown. Given that the first common feature of all three origins is the recruitment of MCM, we speculate that MCM activation could be a key point for coordinate control of firing of all three origins. We note that we detect maximal stimulation of MCM’s helicase activity at a ratio of 25 molecules of GINS•Cdc45 per hexamer of MCM. Although this may simply reflect binding affinity in our in vitro assays conditions, it is conceivable that regulatory modifications to MCM and/or CG components in vivo could affect the strength of interaction we observe. Such a mode of regulation could provide a mechanism for the coordinate control of the three disparate origins present in the Sulfolobus chromosome (27).
SI Materials and Methods
Generation of a Strain of S. acidocaldarius Expressing Epitope-Tagged Cdc45.
DNA fragments corresponding to the 1 kb flanking the S. acidocaldarius cdc45 gene (Saci_0177) were amplified using primers TG75 and TG76 and primers TG77 and TG78, respectively (Table S1). These were then joined by overlap PCR, using primers TG75 and TG78, generating a fusion product encoding C-terminal hexahistidine and c-myc tags. This product was cloned into the KpnI and EcoRI sites of plasmid pEF (22) to generate plasmid pTG-Cdc45. S. acidocaldarius SacTK was transformed as described previously with methylated pTG-Cdc45 (22). Strains containing integrated plasmids were isolated on solid NZ-amine media, followed by liquid NZ-amine media. Unmarked mutants were then isolated by second selection, using solid NZ-amine media containing 0.25 mg/mL 5-fluoroorotic acid and 0.025 mg/mL uracil. PCR amplification across the cdc45 locus followed by DNA sequencing was used to confirm the insertion of the C-terminal tags. Western blotting was also used to detect production of the Cdc45-His-myc protein, using anti-His (1:1,000 dilution; Novagen) or anti-c-myc (1:500 dilution; Santa Cruz) primary antibodies and anti-IgG (mouse) HRP-coupled secondary antibody followed by standard chemiluminescence detection.
Table S1.
Oligonucleotides used in this study
| Primer name | Primer sequence 5′–3′ | Primer description |
| TG75 | TTTCTGCAGGTTAGTGAGATAATATCCTATCC | Saci_0177 upstream cloning F primer |
| TG76 | TTATTAATTTTTGTTCTGCTGCACTGAAATCTTTATTTTCAACTTTTATC | Saci_0177 upstream cloning R primer |
| TG77 | AGCAGAACAAAAATTAATAAGTGAAGAAGATTTACATCATCATCATCATCATTAAGATTTCCACTGAAGCTAG | Saci_0177 downstream cloning F primer |
| TG78 | TTTGGTACCTCTTCAGCGACATCACAAGC | Saci_0177 downstream cloning R primer |
| TG134 | AAAGTTATCCACGGACGCCG | qPCR F primer 1,727-kb region (ter) |
| TG135 | TGGCGACCAACAAATGGTTAGAT | qPCR R primer 1,727-kb region (ter) |
| TG136 | CCACGGTACTCCAAATCTTTATCA | qPCR F primer 429-kb region |
| TG137 | GTTGGAAGTTATCTGCGTTACCG | qPCR R primer 429-kb region |
| TG164 | CCCGATAACAGAGACTTTACTCG | qPCR F primer 579-kb region (oriC1) |
| TG165 | GATGAGATTGACGCACTTGTGAAA | qPCR R primer 579-kb region (oriC1) |
| TG172 | AGAAAGTCACCCAGGATATAGCG | qPCR F primer 0/2,226-kb region (oriC2) |
| TG173 | GAAGGAGTCCATTATTTGCACAAG | qPCR R primer 0/2,226-kb region (oriC2) |
| TG174 | TTGATAAGTGCCATCTTCCCG | qPCR F primer 1,197-kb region (oriC3) |
| TG175 | AGCCACAAATCTCAACCTGGATA | qPCR R primer 1,197-kb region (oriC3) |
| TG176 | CGTCTTGCCATATTTTAGTGCG | qPCR F primer 104-kb region |
| TG177 | AATTGCGTACTCTCCTTTAGGTCA | qPCR R primer 104-kb region |
| TG180 | TCCTGGAGGAGAAATAGGGAT | qPCR F primer 250-kb region |
| TG181 | TGTATACGCGCATGAATCAC | qPCR R primer 250-kb region |
| TG182 | TTGAATGGGCAATTTGGTAA | qPCR F primer 700-kb region |
| TG183 | CGTTAACGGTATACGTCACGA | qPCR R primer 700-kb region |
| TG186 | CACCAGTGGCACTTTACTCG | qPCR F primer 850-kb region |
| TG187 | TCAGGGCTACGCATTAGTGA | qPCR R primer 850-kb region |
| TG188 | AATGTACAAATTTCACCGACAGA | qPCR F primer 1,000-kb region |
| TG189 | CCTTAACTCCATTTGCCACA | qPCR R primer 1,000-kb region |
| TG190 | GACTATGCAGCTGGTCCTGA | qPCR F primer 1,350-kb region |
| TG191 | ATTTACGGAATTGGCGTAGG | qPCR R primer 1,350-kb region |
| TG192 | TCCATTTGTGGATTTCTTTGA | qPCR F primer 1,500-kb region |
| TG193 | GCACTTGGGAACATAAGGTG | qPCR R primer 1,500-kb region |
| TG194 | GGTGTTTCATGACGGAGAGA | qPCR F primer 1,900-kb region |
| TG195 | AGCCAAAGGAGCACCTAAAC | qPCR R primer 1,900-kb region |
| TG196 | GCAGACATAAGGAACGGGAT | qPCR F primer 2,050-kb region |
| TG197 | CCATCCTAATCACGCATTTG | qPCR R primer 2,050-kb region |
| Cdc45-F-NdeI | CGCCGCATATGGAACTATTCTTAGGAGAGCCCA | Amplification of SisCdc45 |
| Cdc45-R-SalI | CTAGTCGACTGTTCCGATAAGATTCTTAATTCTATC | Amplification of SisCdc45 |
| MCM-N-F-EcoRI | CCGAATTCCATGGAAATTCCTAGCAAACAGATTGAC | Construction of MCM-GST fusions |
| MCM-N-R-XhoI | TTCAATGCTCGAGTCATGAAACTTCTATGCTACTAACTTTCATA | Construction of MCM-GST fusions |
| MCM-N-A domain-R-XhoI | GCTAACCTCGAGTCAAGGTATTCCTACAATTCTAACGTGAA | Construction of MCM-GST fusions |
| MCM-N-BC domain-F-EcoRI | CCGGAATTCTAGAGTTATAGAACTTAGAAAAATAAGAAGTAC | Construction of MCM-GST fusions |
| SH TOP | CTCTATCTATACTCTATCTATATAAGTATTTAAACTTTACTCTATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | Substrate for EMSA and helicase assays |
| SH LOWER | TTTTTTTAGAGTAAAGTTTAAATACTTATATAGATAGAGTATAGATAGAG | Substrate for EMSA and helicase assays |
| ssDNA | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGAGAGTAAAGTTTAAATACTGGTATAGCTAGAGTCTAGATAGAG | ssDNA added to ATPase assays |
| ssDNA TOP | CTCTATCTAGACTCTAGCTATACCAGTATTTAAACTTTACTCTCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | For use in pulldown assay |
| ssDNA LOWER | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGAGAGTAAAGTTTAAATACTGGTATAGCTAGAGTCTAGATAGAG | For use in pulldown assay |
| MCM 5′ | GAATTCCCATGGAAATTCCTAGCAAACAGATTGACTATAG | For mcm gene amplification and cloning into pET33b |
| MCM delta A 5′ | GAATTCCCATGGAAATTCCTAGCAAACAGATTGACTATAG | For amplifying DNA corresponding to MCM(106-686), MCM-ΔA, and cloning into pET33b |
| MCM 3′ | GATCGTCGACTTAGACTTTTTTGTAACATTCTGGTTTTGCTTCGT | For mcm gene amplification and cloning into pET33b |
Protein Purification.
S. acidocaldarius CG complex was purified from 5 g cell mass of SacTK-Cdc45 harvested at OD600nm = 0.886. Cells were resuspended in 20 mL TBS (10 mM Tris at pH 8.0, 150 mM NaCl) and lysed in a French press. Extract was clarified by centrifugation and incubated with 1 mL Ni-NTA Agrose (Qiagen) resin at room temperature for 1 h. The matrix was transferred to a glass column and washed three times with 10 mL TBS + 10 mM imidazole before elution with TBS + 500 mM imidazole. The first five 1-mL elution fractions were combined and applied to Superdex 200 (26/60) column in TBS; 5-mL fractions were collected. Elution of Cdc45 was monitored by western blotting with an anti-His tag antibody. The 6 fractions containing the peak of Cdc45 were combined and diluted with 2 volumes (60 mL) of 10 mM Tris at pH 8.0. The resultant material was applied to a 1-mL MonoQ column and developed with a linear gradient to 10 mM Tris at pH 8.0, 500 mM NaCl. The CG complex eluted at ∼200 mM NaCl.
To express Cdc45 with C-terminal His tag in S. islandicus, cells containing the plasmid pSSR-Cdc45 were grown in 80 mL TSVY medium that contained mineral salts, 0.2% (wt/vol) sucrose, 0.1% (wt/vol) tryptone, 0.05% (wt/vol) yeast extract, and a mixed vitamin solution in the presence of 20 µg/mL uracil, 14 µM simvastatin at 78 °C. The culture was grown to optical density at 600 nm of 0.5–0.7; 60 mL of cells were diluted 1/100 in 6 L TVY medium (TSVY medium without sucrose) in the presence of 20 µg/mL uracil, 14 µM simvastatin. When the culture reached OD600nm = 0.2, expression was induced with 0.2% (wt/vol) L-arabinose; growth continues to OD600nm = 0.7 before being harvested by centrifugation. Cells were resuspended in TBS (10 mM Tris at pH 8.0, 150 mM NaCl) with Roche Minicomplete protease Inhibitors, lysed using a French press. After centrifugation (34,957 × g for 20 min at 4 °C), the supernatant was then passed over a Ni-NTA agarose column (Qiagen). The column was washed with wash buffer (10 mM Tris at pH 8.0, 150 mM NaCl, 30 mM imidazole), and then the bound proteins were eluted using elution buffer (10 mM Tris at pH 8.0, 150 mM NaCl, 300 mM imidazole). The 5 × 1-mL peak fractions were combined and applied to a HiLoad 26/600 Superdex 200 column (GE Healthcare) in TBS. The Cdc45 peak fractions were pooled and concentrated using a Vivaspin20 concentrator (10-kDa cutoff).
The Gins23 and Gins15 ORFs were synthesized and codon optimized for expression in Escherichia coli (Life Technologies) and then cloned into the pET-Duet vector. This was transformed into BL21 Rosetta. Cultures were grown in 1 L LB at 37 °C to an OD600nm = 0.6–0.8, and expression induced with 1 mM IPTG and then grown for an additional 3.5 h.
For CG complex purification, S. islandicus cells expressing Cdc45 and E. coli cells expressing GINS cells were lysed in 10 mM Hepes at pH 7.5, 100 mM NaCl, containing Roche Mini-Complete Protease Inhibitors, using a French press. The extracts were clarified by centrifugation (34,957 × g for 20 min at 4 °C). The GINS supernatant was then heated to 90 °C for 20 min. After further centrifugation, the heat-stable GINS-containing supernatant was combined with the Cdc45 supernatant and applied onto a Ni-NTA agarose column (Qiagen). The column was washed with 10 mM Hepes at pH 7.5, 100 mM NaCl, 10 mM imidazole, and then the bound proteins were eluted using 10 mM Hepes at pH 7.5, 100 mM NaCl, 300 mM imidazole. The 5-mL eluted fractions were then passed over a 1-mL Mono Q 5/50 GL column (GE Healthcare), using an AKTA purifier system (GE Healthcare). The column was developed with a linear gradient to 10 mM Hepes at pH 7.5, 1 M NaCl. CG was eluted at 0.28 M NaCl. Finally, CG complex was further purified over a HiLoad 26/600 Superdex 200 column (GE Healthcare) in 10 mM Hepes at pH 7.5, 150 mM NaCl. The CG peak fractions were pooled and concentrated in a Vivaspin concentrator (30 kDa molecular weight cut-off).
The ORFs for S. islandicus untagged MCM and MCM-ΔA (MCM residues 106–686) proteins were amplified by PCR, adding NcoI sites at the start codon and SalI sites immediately downstream of the native stop codon. The products were digested, with NcoI and SalI inserted at the NcoI and XhoI sites of pET33b. Proteins were expressed in E. coli by growth in LB at 37 °C to an OD600nm = 0.6–0.8 and induction with 1 mM IPTG for 16 h at 25 °C. Cells were resuspended in 10 mM Tris at pH 8.0, 150 mM NaCl, 1 mM DTT containing Roche Mini-Complete Protease Inhibitors, and lysed using a French press. Extract was clarified by centrifugation (34,957 × g for 20 min at 4 °C). The soluble lysate was heat treated for 20 min at 68 °C, recentrifuged as earlier, and the heat-stable soluble fraction was purified over a HiTrap heparin column (GE Healthcare). Proteins were further purified over a HiLoad 26/600 Superdex 200 column (GE Healthcare) in 20 mM Tris at pH 8.0, 300 mM NaCl, 1 mM DTT.
Pulldown Assays.
For GST pulldown assay, the indicated GST-fusion protein was immobilized on glutathione sepharose beads. Binding reactions were performed in a 100-µL reaction volume containing TBS and 10-µL bead volume with target protein, as indicated. After incubation at room temperature for 30 min with shaking at 800 rpm, beads were washed six times with TBS. After the final wash, samples were boiled in SDS PAGE loading buffer and subjected to SDS/PAGE.
For His-tag mediated pulldown assays, 100-µL reactions contained 40 µL Ni-NTA resin solution (∼20-µL beads) and 10–18 μg of each purified protein in TBS. Reactions were supplemented with 2 mM ATP+Mg2+, 20 µM ssDNA, 10 µM fork DNA, as indicated. Reactions were incubated at room temperature (65 °C was also used with no significant difference in outcome, not shown) for 30 min with shaking at 800 rpm, then washed six times with TBS. After the final wash, samples were boiled in SDS/PAGE loading buffer and analyzed by SDS/PAGE.
ChIP Experiments.
ChIP was performed using 100 mL stationary phase cells (OD600nm = 1.0), 100 mL exponentially growing cells (OD600nm = 0.15), or 200 mL synchronized cells (18). For synchronized samples, cells were collected simultaneously from two baby machines from 3 to 5 h elution, as described earlier, and then grown at 75 °C for the desired length of time. Fixation, quenching, sonication, and extract preparation were as described previously (23).
Immunoprecipitations (IPs) were performed using 1 μg of either affinity-purified sheep anti-Mcm antibody or anti-c-myc antibody (Santa Cruz), as described previously (18). The resulting purified DNA from a single anti-Mcm IP reaction or from three combined anti-c-myc IPs was resuspended in 250-μL TE for use in qPCR analysis. This approach was used because of the limited amount of total cell lysate that can be obtained from two combined baby machine runs. Input samples were treated as earlier, without the addition of antiserum and beads. Control experiments were performed using beads alone or using SacTK cultures that do not contain c-myc-tagged Cdc45, to ensure specificity of the IP reactions (not shown). Real-time qPCR was performed using the Brilliant III Ultra-Fast SYBR green qPCR mix (Agilent), with 5 µL ChIP DNA in a 25-µL reaction volume, using a Mastercycler qPCR instrument (Eppendorf). The qPCR reactions were performed in triplicate, using primers described in Table S1, and the resulting data were analyzed using the ∆∆Ct method.
ATPase, EMSA, and Helicase Assays.
Briefly, measurement of MCM ATPase activity was carried out in 100-µL reactions containing 50 mM potassium acetate, 20 mM Tris acetate at pH 7.9, 10 mM magnesium acetate, 1 mM DTT, 75 mM NaCl, 0.1 mg/mL BSA, 2 mM ATP. MCM (100 nM final concentration as hexamer), CG (1 µM final concentration), and ssDNA substrate (1 µM final concentration) were added when indicated. The reactions were preincubated at 55 °C for 1 h before the addition of 100 µL fresh reducing agents [one volume 6 N sulfuric acid, one volume 2.5% (wt/vol) ammonium molybdate, one volume 10% (wt/vol) ascorbic acid, and two volumes distilled water], mixed thoroughly, and then incubated at 37 °C for 30 min before measuring absorbance at 820 nm. The phosphate concentrations were determined according to standard curve.
EMSAs were performed in a 20-µL reaction volume containing 50 mM potassium acetate, 20 mM Tris acetate at pH 7.9, 10 mM magnesium acetate, 1 mM DTT, 75 mM NaCl, 0.1 mg/mL BSA, 5% (vol/vol) glycerol, 2 nM 32P-radiolabeled DNA substrate. MCM and CG were added when indicated. Reactions were incubated at 55 °C for 1 h and then loaded on a 5% (wt/vol) polyacrylamide gel in 1xTris borate EDTA (TBE). Gels were run for about 1 h at 175 V before drying and autoradiography.
Helicase assays were performed in a 20-µL reaction volume containing 50 mM potassium acetate, 20 mM Tris acetate at pH 7.9, 10 mM magnesium acetate, 1 mM DTT, 75 mM NaCl, 5 mM ATP, 0.1 mg/mL BSA, 2 nM 32P-radiolabeled DNA substrate. MCM, MCM-ΔA, CG, and BSA were added when indicated. Reactions were incubated at 55 °C for 1 h before the addition of 20 µL stop buffer [100 mM EDTA, 0.5% SDS, 0.1% bromophenol blue, and 50% (vol/vol) glycerol]. Then, 30-µL aliquots were loaded on an 8% (wt/vol) polyacrylamide gel in 1xTBE, 0.2% SDS. Gels were run for 1 h at 175 V before drying and autoradiography.
Methods
Cell Growth.
S. acidocaldarius SacTK, a pyrEF− TK+ strain derived from S. acidocaldarius DSM639 (28), was grown at 75 °C in Brock’s medium at pH 3.2 containing 0.1% (wt/vol) tryptone, 0.2% (wt/vol) xylose, and 0.025 mg/mL uracil. After transformations, SacTK cultures contained 0.1% NZ-amine as an alternative to tryptone. When required, Brock’s medium was solidified with 0.7% Gelrite (GmbH). Transformations with of S. islandicus with expression plasmid pSSR-Cdc45 and subsequent growth were as described (29). Yeast two-hybrid assays were performed in strain AH109, as described in the Matchmaker handbook (Clontech).
Genetic Manipulation of Sulfolobus.
The construct for tagging S. acidocaldarius cdc45 was generated by overlap PCR (Table S1) and transformation of S. acidocaldarius strain SacTK (28). Cdc45 was overexpressed in S. islandicus E223S, using the pSSR vector (23). For detailed information, see SI Materials and Methods.
Protein Purification.
For details of the purification methods, see SI Materials and Methods. Briefly, S. acidocaldarius CG complex was purified by metal affinity, size exclusion, and chromatography anion exchange. GST-fusion proteins (GST-MCM-N, GST-MCM-N-A domain, GST-MCM-N-BC domain) were purified using glutathione sepharose (for details, see SI Materials and Methods). Recombinant MCM and MCM ΔA were purified by chromatography over heparin sepharose and gel filtration matrices.
Intact Protein Complex Mass Spectrometry.
The CG complex was prepared in 50 mM ammonium acetate at a concentration of 4.4 μM. The sample was infused into a Synapt G2S mass spectrometer equipped with a nano ESI source at 0.5 μL/min. The source conditions were as follows: capillary voltage, 1.5 kV; source temperature, 100 °C; sampling cone, 80 V; source offset, 80 V; desolvation temperature, 150 °C; nanoflow gas pressure, 0.5 Bar. To promote declustering of solvent ions from the complex, the trap and transfer collision energies were varied between 70 and 100 V. The data were processed in MassLynx (Waters Corporation). Results from 5 min of scanning was averaged and smoothed, using the Savitzky Golay algorithm with four channels and 20 rounds of smoothing (30). To determine the mass of the intact complex, the data were processed using MaxEnt 1 (Waters Corporation), with a resolution of 10 Da/channel and damage model with a uniform Gaussian half height of 40 Da.
Pulldown Assays.
Pulldown assays used either beads with Ni-NTA–coupled protein or glutathione sepharose–coupled proteins.
ATPase, EMSA, and Helicase Assays.
These procedures are detailed in SI Materials and Methods. ATPase rate was assessed by colorimetry and EMSA, and helicase assays were essentially as described previously (20), with the exception of the inclusion of 75 mM NaCl in the binding and reaction buffers.
ChIP Experiments.
The procedure for ChIP assays is described in detail in SI Materials and Methods (26). ChIP was performed using anti-MCM or anti-c-myc antibodies, as described previously (18). The resulting purified DNA was quantified by qPCR.
Acknowledgments
We thank Luca Pellegrini for informative discussions about human Cdc45. We also thank Rachel Whitaker, University of Illinois, for sharing data before publication. We are indebted to Marleen van Wolferen with help with the S. acidocaldarius genetics. We thank the Biotechnology and Biological Sciences Research Council, Wellcome Trust, Max Planck Society, and Indiana University College of Arts and Sciences for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613825113/-/DCSupplemental.
References
- 1.Siddiqui K, On KF, Diffley JF. Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol. 2013;5(9):a012930. doi: 10.1101/cshperspect.a012930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5(12):a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18(4):471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SD, Botchan MR. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol. 2013;5(11):a012807. doi: 10.1101/cshperspect.a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103(27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelman LM, Kelman Z. Archaeal DNA replication. Annu Rev Genet. 2014;48:71–97. doi: 10.1146/annurev-genet-120213-092148. [DOI] [PubMed] [Google Scholar]
- 8.Marinsek N, et al. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7(5):539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Pulido L, Ponting CP. Cdc45: The missing RecJ ortholog in eukaryotes? Bioinformatics. 2011;27(14):1885–1888. doi: 10.1093/bioinformatics/btr332. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, et al. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 2011;39(14):6114–6123. doi: 10.1093/nar/gkr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Koonin EV, Kelman Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct. 2012;7:7. doi: 10.1186/1745-6150-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 2007;17(6):271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Onesti S, MacNeill SA. Structure and evolutionary origins of the CMG complex. Chromosoma. 2013;122(1-2):47–53. doi: 10.1007/s00412-013-0397-x. [DOI] [PubMed] [Google Scholar]
- 14.MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425(3):489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- 15.Oyama T, et al. Architectures of archaeal GINS complexes, essential DNA replication initiation factors. BMC Biol. 2011;9:28. doi: 10.1186/1741-7007-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23(3):217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2008;105(43):16737–16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher RJ, et al. The structure and function of MCM from archaeal M. Thermoautotrophicum. Nat Struct Biol. 2003;10(3):160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 20.Lang S, Huang L. The Sulfolobus solfataricus GINS complex stimulates DNA binding and processive DNA unwinding by minichromosome maintenance helicase. J Bacteriol. 2015;197(21):3409–3420. doi: 10.1128/JB.00496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino H, et al. Activation of the MCM helicase from the thermophilic archaeon, Thermoplasma acidophilum by interactions with GINS and Cdc6-2. Extremophiles. 2014;18(5):915–924. doi: 10.1007/s00792-014-0673-6. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y. The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J Biol Chem. 2008;283(3):1601–1609. doi: 10.1074/jbc.M707654200. [DOI] [PubMed] [Google Scholar]
- 23.Barry ER, McGeoch AT, Kelman Z, Bell SD. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 2007;35(3):988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon AC, Sannino V, Costanzo V, Pellegrini L. Structure of human Cdc45 and implications for CMG helicase function. Nat Commun. 2016;7:11638. doi: 10.1038/ncomms11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouge CA, Christensen TW. Drosophila Sld5 is essential for normal cell cycle progression and maintenance of genomic integrity. Biochem Biophys Res Commun. 2010;400(1):145–150. doi: 10.1016/j.bbrc.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samson RY, Abeyrathne PD, Bell SD. Mechanism of Archaeal MCM helicase recruitment to DNA replication origins. Mol Cell. 2016;61(2):287–296. doi: 10.1016/j.molcel.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samson RY, et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Reports. 2013;3(2):485–496. doi: 10.1016/j.celrep.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gristwood T, Duggin IG, Wagner M, Albers SV, Bell SD. The sub-cellular localization of Sulfolobus DNA replication. Nucleic Acids Res. 2012;40(12):5487–5496. doi: 10.1093/nar/gks217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng T, et al. Development of a simvastatin selection marker for a hyperthermophilic acidophile, Sulfolobus islandicus. Appl Environ Microbiol. 2012;78(2):568–574. doi: 10.1128/AEM.06095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savitzky A, Golay MJE. Smoothing and differentiation of data by simplified least squares procedures. Anal Chem. 1964;36(8):1627–1639. [Google Scholar]
- 31.Brewster AS, et al. Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci USA. 2008;105(51):20191–20196. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]