Abstract
Plant virus-based nanoparticles (VNPs) are a novel class of nanocarriers with unique potential for biomedical applications. VNPs have many advantageous properties such as ease of manufacture and high degree of quality control. Their biocompatibility and biodegradability make them an attractive alternative to synthetic nanoparticles (NPs). Nevertheless, as with synthetic NPs, to be successful in drug delivery or imaging, the carriers need to overcome several biological barriers including innate immune recognition. Plasma opsonization can tag (V)NPs for clearance by the mononuclear phagocyte system (MPS), resulting in shortened circulation half lives and non-specific sequestration in non-targeted organs. PEG coatings have been traditionally used to ‘shield’ nanocarriers from immune surveillance. However, due to broad use of PEG in cosmetics and other industries, the prevalence of anti-PEG antibodies has been reported, which may limit the utility of PEGylation in nanomedicine. Alternative strategies are needed to tailor the in vivo properties of (plant virus-based) nanocarriers. We demonstrate the use of serum albumin (SA) as a viable alternative. We demonstrate that SA conjugation to tobacco mosaic virus (TMV)-based nanocarriers results in a ‘camouflage’ effect more effective than PEG coatings. SA-’camouflaged’ TMV particles exhibit decreased antibody recognition, as well as enhanced pharmacokinetics in a Balb/C mouse model. Therefore, SA-coatings may provide an alternative and improved coating technique to yield (plant virus-based) NPs with improved in vivo properties enhancing drug delivery and molecular imaging.
Keywords: nanomedicine, drug delivery, tobacco mosaic virus (TMV), stealth and camouflage, pharmacokinetics
INTRODUCTION
Nanoparticle (NP)-based biomedical approaches allow for delivery of contrast agents and therapeutics. Tissue-targeted delivery of these payloads decreases dose-limiting systemic side effects, and increases therapeutic efficacy and/or site-specific accumulation of contrast enhancement agents [1–5]. Various NP-based formulations carrying chemotherapeutic agents [6,7], photothermal therapeutics [6,8], or magnetic resonance imaging (MRI) contrast agents [7,9–12] are currently in the development pipeline. A few formulations have been approved for clinical applications; these include superparamagnetic iron oxide (SPIO) NPs, which are used in MRI prognosis [13] as well as liposomal doxorubicin [7] and micellar or albumin-bound paclitaxel [7]. NPs can solubilize and carry large payloads of medical cargo and provide a size regime ideal for navigating circulation, tissues, and cells. Nevertheless, to reach their target site, nanocarriers must overcome various biological barriers. Some NPs show poor dispersion properties in biological media. In blood, NPs are opsonized with antibodies and complement proteins, which can tag them for recognition by the mononuclear phagocyte system (MPS). Therefore, stealth coatings must be applied to overcome non-specific uptake by the MPS to enhance pharmacokinetic and biodistribution profiles.
The most common ‘stealth’ technique used is the coating of nanocarriers with polyethylene glycol (PEG); PEG coatings ‘shield’ the nanocarrier surface from opsonization and therefore reduce innate immune surveillance leading to sequestration in the MPS [14]. Nevertheless, recent research indicates that up to 25% of the human population (estimated based on blood titers from 44 healthy donors) have developed PEG-specific antibodies [15,16]. This is a 124 fold increase compared to a study reported over 20 years ago, where it was shown that only 0.2% of humans tested presented with anti-PEG antibodies [17]. This increase may be due to improved assays or due to immune response caused by increased use of PEG in industry, medicine and cosmetics [15]. Furthermore, in patients previously treated/immunized with PEG-containing formulations, higher prevalence of anti-PEG antibodies has been reported [15,16], resulting in enhanced blood clearance after repeat administration of PEGylated NPs [18]. These studies raise concerns about the applicability of PEG containing formulations in the clinic. A paradigm shift is needed.
Toward this goal, we turned toward a bio-mimicry approach where the NP surface is coated with serum albumin (SA), the most abundant plasma protein. SA is an ellipsoidal-shaped molecule of 15 nm x 3.8 nm x 3.8 nm dimensions. Its main function is to maintain oncotic pressure and to transport hydrophobic molecules in the blood. SA makes up for approximately 55% of the total protein content in plasma; we therefore reasoned that it would be a suitable candidate for the proposed ‘camouflage’ strategy. In fact, certain strains of pathogenic bacteria (e.g. streptococci) employ a strategy of binding SA upon entry into mammalian organisms via cell surface-expressed HSA-binding protein G. The SA coating allows the bacteria to hide from immune surveillance [19].
Protein ligands have been applied in nanomedicine, although they have been most typically employed as a targeting strategy: for example, NP-transferrin conjugates have been used to target cancer cells [20–22]. In addition to conferring tissue-specificity, the protein-coating also has been shown to reduce protein corona formation, therefore reducing innate immune recognition [23]. Furthermore, recent reports reveal that conjugation of minimal “self” peptides (fragment derivatives of CD47 protein) can efficiently inhibit the clearance of NPs by phagocytic cells [24]. Together these studies indicate that protein camouflage coatings may be a viable strategy for nanomedicine.
In this work, we compared the effectiveness of SA vs. PEG coatings using the NPs formed by the nucleoprotein components of tobacco mosaic virus (TMV). TMV is a plant virus-based nanoparticle (VNP); the TMV capsids forms 300x18 nm-sized hollow nanotubes composed of 2,130 identical copies of a ~17-kDa coat protein. TMV, like other VNPs have many advantageous properties, such as ease of manufacture and high degree of quality control. Their biocompatibility and biodegradability make them an attractive alternative to synthetic NPs. Indeed VNPs are undergoing development for use in drug delivery, immunotherapy, and molecular imaging [25–31].
Due to its high aspect ratio shape of TMV, the platform offers advantages over spherical nanomaterials. High aspect ratio materials evade MPS clearance, show enhanced vessel wall margination and tissue penetration [32,33]. For example, we demonstrated that based on its enhanced tumbling and margination properties, TMV effectively targets solid tumors [34] and thrombosis [35]. The shape-mediated enhanced disease-targeting properties also align with reduced macrophage clearance [34] and translate to high potential for molecular imaging [10], and drug delivery (the latter will be reported elsewhere). As a model NP, TMV serves as an excellent platform because its structure is known to atomic resolution [36,37] and genetic and chemical methods are available to impart new functionalities[25]. For these reasons, TMV serves as a good candidate material to assess the proposed ‘camouflage’ strategy.
In this work, we investigated SA vs. PEG surface coatings on TMV. Structure-function studies were performed to assess immune-recognition by TMV-specific antibodies, clearance by cells of the MPS, and in vivo pharmacokinetics (PK) profiles.
RESULTS AND DISCUSSION
Serum albumin (SA) was conjugated to the external surface of a TMV lysine-added chimera (TMV-lys) [38]. The side-chain of lysine residue contains an amine group susceptible to conjugation using NHS ester chemistry. Three strategies were tested to cross-link human and mouse SA to TMV-lys (Supplementary Figure S1): (a) carbodiimide-based condensation reaction between the carboxyl groups of SA and the surface anime groups of TMV-lys were used; (b) homobifunctional NHS-PEG5-NHS to cross-link NH2 groups of SA and TMV-lys was explored; and (c) a three-stage process was developed:
NHS-to-NH2 conjugation of NHS-PEG4-MAL to the TMV-lys surface;
NHS-to-NH2 conjugation between NHS-PEG4-SH and SA;
MAL-to-SH conjugation of product (i) to (ii) to achieve TMV-PEG8-SA (see Materials and Methods and Figure 1A).
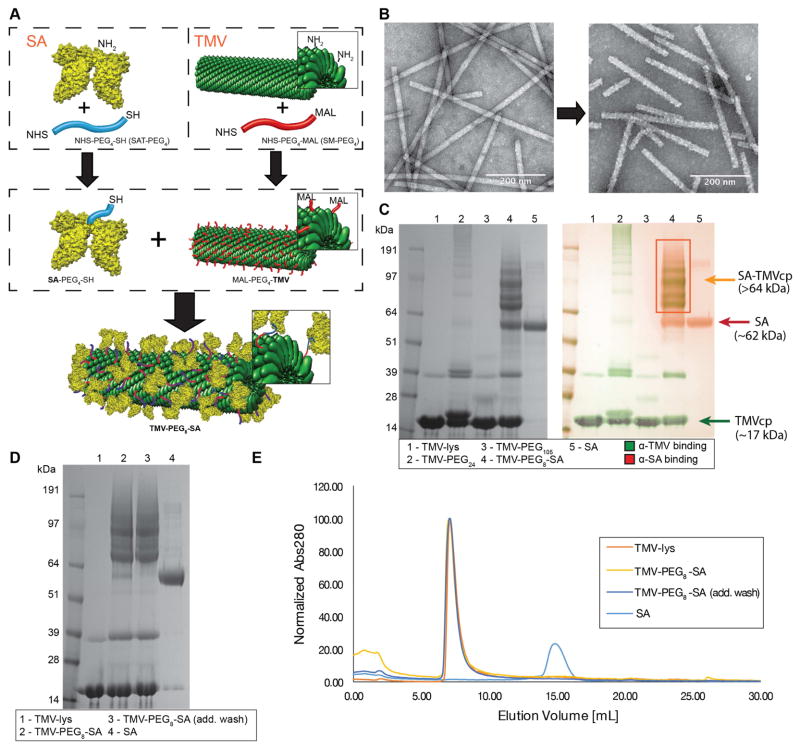
Figure 1. Coating TMV surface with SA.
A, schematic representation of the SA conjugation to TMV. Step one: Conjugation of SAT(PEG)4 and SM(PEG)4 to the lysine residues of SA protein and TMV-lys respectively (-NHS ester to -NH2 reaction). Step two: conjugation of SA-PEG4-SH to TMV-PEG4-maleimide (-SH to -maleimide reaction). Human serum albumin (HSA) was used to prepare the TMV-PEG8-SA particles. B, TEM images of VNPs before (TMV-lys, left) and after SA conjugation (TMV-PEG8-SA, right). The morphology of TMV-lys surface changes from smooth to ‘patchy’ upon SA conjugation. C, SDS-PAGE and Western Blot (WB) analysis of TMV-lys particles before and after PEGylation and SA conjugation. Free SA was used as reference. Successful conjugation of PEG is indicated by presence of bands with apparent molecular weights (>17 kDa) higher in respect to TMVcp band (~17 kDa). SA conjugation was also indicated by presence of multiple protein bands corresponding to SA-TMVcp (multiple bands of apparent Mw > 64kDa; theoretical molecular weight of 1:1 SA:TMVcp monomer = 83 kDa;) as shown by WB immune recognition. Additional WB images are shown Supplementary Figure S2. D, SDS-PAGE analysis of TMV-PEG8-SA before and after additional wash in PBS. Reduction of non-coupled SA band (apparent Mw of ~62 kDa) can be observed. E, Size exclusion chromatography (SEC) analysis of TMV-PEG8-SA before and after additional washes. 100 μg of TMV conjugates were analyzed. No significant amount of free SA has been detected thus the non-coupled SA (visible in panels C + D) is expected to be physically adsorbed on the surface of TMV-PEG8-SA particles. SA was used as reference.
Both (a) and (b) were low yielding and/or resulted in extensive particle aggregation (for details see Supplementary Figure S1). Strategy (c), using a combination of two heterobifunctional PEG cross-linkers, allowed for efficient conjugation of SA to TMV while avoiding aggregation (see Materials and Methods and Figure 1A). TMV was also modified with PEG chains only to yield TMV-PEG24 (MwPEG24=1394.55 Da) and TMV-PEG105 (MwPEG105=1394.55 Da). These samples served as controls and were prepared using procedure (i) with longer PEG chains and subsequent quenching of the maleimide groups of PEG with use of excess L-cysteine.
We confirmed that the TMV formulations maintained their structural integrity upon SA and PEG conjugation. Irregular surface morphology of the TMV-PEG8-SA particles was observed using TEM (Figure 1B), indicating successful protein coating. The particles were further characterized by SDS-PAGE and westerns blotting (WB) to analyze the SA-to-TMV ratios (Figure 1C). SA conjugated to the TMV coat protein (TMVcp) was detectable as multiple protein bands of high molecular weight (>64 kDa) not present in either TMV-lys or SA controls. Based on densitometric analysis, approximately 0.3–0.4 mg SA were conjugated per 1 mg of TMV particles, or ~180–240 SA proteins per TMV particle. This corresponds to 1 SA molecule conjugated to every 9th–12th TMVcp. Assuming the exposed surface area of TMVcp ATMVcp=~8nm2 and surface area of SA cross section ASA=~11–45nm2 (approximating SA as an 15 nm x 3.8 nm x 3.8 nm ellipsoid, and depending on its orientation), the theoretical coverage of TMV surface by SA could be as high as ~60%.
WB against SA and TMV provided further insight into the particle composition and make up of the protein bands. WB analysis confirmed that both SA and TMVcp were present in the >64-kDa bands detected by SDS-PAGE. This suggests that SA binds as a multimer, or anchors to multiple TMVcps, rather than forming a uniform single layer coating.
Although samples have been extensively washed and dialyzed against PBS buffer (see Materials and Methods), a small amount of non-covalently attached or unbound SA has been detected in TMV-PEG8-SA sample and quantified as approximately 0.02–0.1 mg (~12–58 SA proteins) per TMV particle, or ~6–20% of total SA present in the sample. Extensive washing procedures with PBS allowed the removal of free SA (Figure 1D + 1E). However, removal of the bound SA may not be a requirement or it may not even be desirable, because the bound (but non-coupled) SA, if it is stably adsorbed to TMV in plasma (not exchanging with other proteins) [23,39], could seal the gaps in the SA coating to enhance the camouflage and stealth effect.
The degree of surface coverage with PEG for the TMV-PEG24 and TMV-PEG105 samples was estimated as ~45% and ~18%, respectively (i.e. the % of total TMVcp conjugated to PEG). Lower conjugation efficiency of PEG105 is expected due to longer chain length and consequential increase in steric hindrance of the PEG chains interacting with the TMV surface. Depending on its density, PEG can adopt a brush conformation (for high density) or mushroom conformation (for low-to-medium density) [40]. The theoretical PEG conformation can be estimated by comparison of distance between adjacent PEG chains (D) and the Flory dimension (RF) [40,41], with RF < D resulting in a mushroom conformation, and when RF > D, then a brush conformation is more likely. For the TMV-PEG24 particles we find that RF=~2.4 < D=~6.3, and for TMV-PEG105 particles RF=~5.7 < D=~15.9. Therefore, for both a mushroom conformation is assumed.
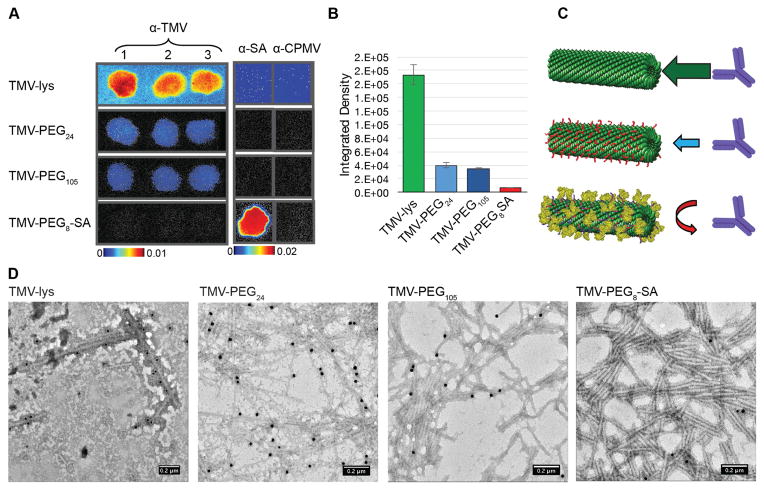
TMV and other plant viruses are wide-spread in nature; TMV in particular has been detected in agricultural products (e.g. vegetables) [42,43] and tobacco [44]. Antibodies against TMV, therefore, may be prevalent in the human population. Indeed, we have previously found binding of immunoglobulins to TMV upon incubation in human plasma collected from random donors [45]. If antibodies are not already present, these are developed after repeat administration of TMV-based therapeutics. Therefore, effective stealth coatings are needed to overcome antibody recognition and clearance by MPS. To investigate the stealth efficacy of SA vs. PEG coatings on TMV, immune-recognition experiments have been performed. Immuno dot-blots were prepared by spotting α-TMV (polyclonal antibodies produced through immunization of rabbits) or α-SA antibodies on a nitrocellulose membrane, followed by addition of ‘naked’ TMV-lys or coated TMV-SA and TMV-PEG. Fluorescently-labeled TMV formulations were used in these experiments where Cy5 dyes were conjugated to glutamic acids on the interior channel (see Materials and Methods). Antibody-recognition was analyzed based on fluorescence readout quantified using a Maestro Imaging system. While recognition of bare TMV-lys by α-TMV antibodies was high (as anticipated), it decreased for PEGylated TMV and was minimal for TMV-PEG8-SA (Figure 2A + 2B). We also note that bare TMV-lys were non-specifically adsorbed across the entire membrane, most likely reflecting the propensity of proteins to adsorb onto nitrocellulose membranes. Non-specific membrane adsorption was not observed for either the TMV-PEG or TMV-SA formulations. But most importantly, the PEG/SA-coated TMV VNPs showed decreased antibody recognition with SA coatings outperforming the PEG-coating by 5–6 times. The difference may be explained by the difference in the nature of the coating: SA is a relatively high molecular weight protein (~66 kDa) with a globular and ‘rigid’ structure, while PEG is a flexible polymer with low molecular weight (~1.4 kDa and 5 kDa for PEG24 and PEG105 respectively). Those structural differences are a possible reason for better stealth properties of TMV-PEG8-SA; it is expected that SA provides improved steric hindrance compared to PEG.
Figure 2. Immune recognition of TMV-PEG8-SA.
A, Immune recognition of fluorescent ‘naked’ and ‘stealth’ TMV particles by α-TMV and α-SA antibodies using dot blots. The binding of particles to α-TMV antibody spotted on the membrane is decreased with PEG coatings and was effectively prevented using SA coatings. B, Quantitative densitometric analysis of the dot-blots (A). C, Schematic representation of the antibody-recognition of the various TMV-based particles. D, TEM images of TMV formulations after immunogold staining using α-TMV and gold-labeled secondary antibodies. HSA was used to prepare the TMV-PEG8-SA particles.
These results were in good agreement with immunogold TEM studies (see Materials and Methods). We observed high recognition of ‘naked’ TMV-lys particles by α-TMV antibodies; antibody recognition was significantly reduced upon conjugation of stealth PEG and SA coatings, again with SA outperforming the PEG coatings (Figure 2D).
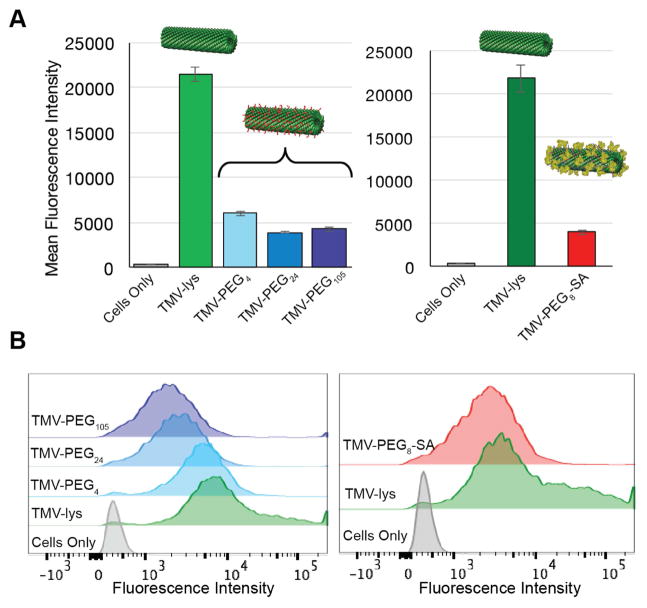
While antibodies can trigger recognition and clearance of (V)NPs by the MPS, another route is direct recognition by macrophages’ membrane receptors. Therefore, we have studied in vitro interactions between stealth TMV-PEG and TMV-SA particles and RAW264.7 cells; quantitative data were obtained using the flow cytometry. The conjugation of PEGs and SA decreased the interactions between TMV and macrophages (Figure 3) but did not eliminate uptake. Overall the uptake rates of PEGylated and SA-coated TMV particles were comparable resulting in a 4-fold reduction of macrophage cell interactions. It appears that the SA-coating does not confer additional advantages over the PEG coatings in terms of macrophage clearance in vitro. Even a comparison between TMV particles coated with a short PEG4 linker vs. SA-coated particles yielded no difference in this experiment (Figure 3A), indicating the low density coating with high-molecular weight PEG, medium density coatings with low-molecular weight PEGs, or addition of SA provides similar levels of evasion from macrophage uptake tested in vitro.
Figure 3. In vitro recognition of SA- and PEG-coated TMV vs. ‘naked’ TMV by RAW264.7 macrophages.
A, Quantitative FACS analysis of the interactions between ‘naked’ and ‘stealth’ TMV formulations and RAW264.7 cells. B, FACS histograms of the same. Mouse serum albumin (MSA) was used to prepare the TMV-PEG8-SA particles (RAW264.7 is a mouse-derived macrophage cell line).
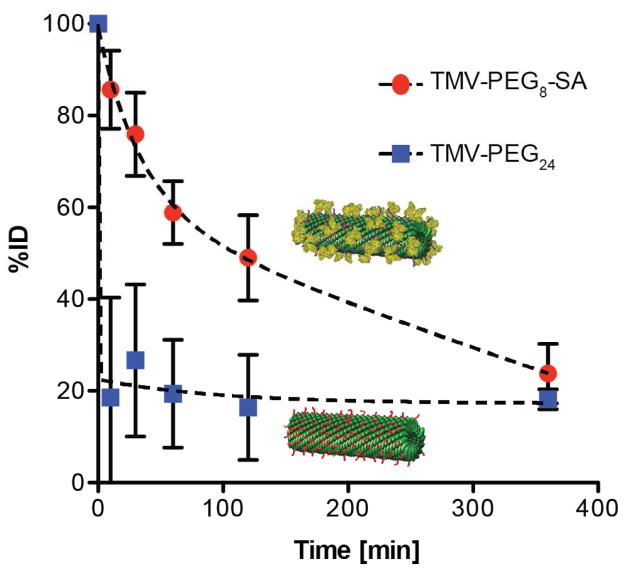
Finally, we performed in vivo pharmacokinetic studies (PK) to evaluate the circulation half-lives of PEG- and SA-coated TMV particles. Fluorescently-labeled TMV-PEG and TMV-SA formulations and Balb/C mice were used; PK was evaluated after bolus administration of 0.4 mg of TMV formulation in the tail vein. Blood was collected via retro-orbital bleeds, and after removal of cellular components, the percent injected dose (%ID) of TMV was determined based on fluorescence measurements and use of standard curve prepared by spiking mouse serum with known concentrations of TMV. Although PEGylation of VNPs has been shown to reduce the antibody recognition and reduce interactions with macrophages, our results indicate fast clearance of the PEGylated VNPs (t1/2~10 mins; Figure 4), consistent with previous observations[46]. The SA-coated TMV exhibited much slower clearance from blood stream with average half-life of ~100 minutes, therefore increasing the circulation time by one order of magnitude.
Figure 4. Pharmacokinetics of the TMV-PEG24 and TMV-PEG8-SA particles in Balb/C mouse model.
The particles were administered intravenously at the amount of 400 μg/mouse. Blood was collected before injection at t=0 and after injection at t=10 min, t=30 min, t=60 min, t=120 min, and t=360 min (the experiments were completed at an n=3 per group).
CONCLUSIONS
We have investigated the ‘stealth’ properties of SA coatings vs. PEGylation using TMV as a candidate material. Firstly, we have established chemistries that enable high-density conjugation of SA to the surface of TMV-lys; we envision that these methods could be expended to other protein coatings. Secondly, we demonstrate that SA coatings overcome recognition of the carrier by TMV-specific antibodies, decrease TMV clearance by macrophages, and improve its pharmacokinetic profile. In particular, SA coatings outperformed PEG coatings in terms of antibody evasion and pharmacokinetics, resulting in effective evasion from antibody recognition and 10-times increased circulation half-life of t1/2 ~ 100 mins vs. the PEGylated TMV formulation. Macrophage uptake was reduced compared to ‘naked’ TMV but no differences were observed comparing SA- vs. PEG-coated TMV, indicating that antibody-mediated clearance is the dominating factor for the rapid plasma clearance of TMV (we have previously shown that the protein corona formed on ‘naked’ TMV in plasma consists of immunoglobulins [45]). To be effective for systemic and repeat administration, evasion of antibody surveillance is an important goal. Prevalence of neutralizing antibodies has limited the use of viral vectors for localized treatment, an example are the oncolytic virus therapies, including T-VEC developed and commercialized by Amgen. The incorporation of serum proteins or other self-coating strategies may provide a powerful strategy to overcome the immunogenicity of virus-based and nanoparticle-based therapeutics. If the carrier is protein-based, fusion could be achieved through genetic engineering and overcoat expression; in the case of synthetic materials bio-conjugate ligation strategies as described in this article could be applied. Therefore, SA-coatings could be widely applied as a coating technique to yield (V)NPs with improved in vivo properties enhancing drug delivery and molecular imaging.
MATERIALS AND METHODS
Virus propagation and purification
Viruses were propagated by mechanical inoculation using 5–10 μg of virus per leaf. Wild-type TMV and TMV-lys mutants were propagated in Nicotiana benthamiana. The isolation of VNPs using established procedures yielded approximately 1 mg of virus per gram of infected leaf material [47].
TMV sCy5 labeling
First, alkynes were attached to internal TMV carboxylic acids 100 e.q. of propargylamine (P50900; Sigma Aldrich) per capsid protein and 50 e.q. of EDC (25 equivalents added at 0 and 18 h; E6383; Sigma Aldrich) in 100 mM HEPES buffer, pH 7.4; the reaction was allowed to proceed for 24 h. Second, an alkyne-azide click reaction was performed by adding 1 e.q. of sCy5-azide (B3330; Lumiprobe) per coat protein using 2 mg/mL TMV in the presence of 1 mM CuSO4 (AC423615000; Fisher), 2 mM AMG (AC36891025; Fisher), and 2 mM Asc (AC352681000; Fisher) in 10 mM potassium phosphate (KP) buffer (pH 7.4) on ice for 30 min. TMV was purified by ultracentrifugation at 42,000 rpm for 3 h on a 40% (w/v) sucrose cushion.
TMV conjugation with SA
Human serum albumin (HSA, A9511; Sigma Aldrich) or mouse serum albumin (MSA, 22070104–1; Bioworld) was conjugated to the external surface of TMV-lys using a three-step reaction: 1) The NHS ester-to-lysine binding between SA and SAT(PEG)4 (NHS-(PEG)4–SAT, 26099; Thermo Fisher) was performed by mixing SA (80 mg/mL final conc.) and SAT(PEG)4 using the linker at a 1 e.q. per SA protein. The reaction was carried out in 0.01 M phosphate buffer 0.125 M saline (PBS; pH 7.4) containing 10% (v/v) DMSO overnight at room temperature (RT). To de-protect the -SH group de-acetylation solution (0.5 M hydroxylamine, 25 mM EDTA in PBS, pH 7.2–7.5) was added at a final concentration of 10% by volume. 2) The NHS ester-to-lysine binding between TMV-lys and SM(PEG)4 (NHS-(PEG)4–mal, 22104; Thermo Fisher) was performed by mixing TMV-lys (at 2 mg/mL final conc.) and SM(PEG)4, the linker was used at a 10 e.q. per TMV-lys coat protein. The reaction was carried out in 0.01 M potassium phosphate buffer pH 7.4 containing 10% (v/v) DMSO for 2 h at RT. Both (1) and (2) products were purified using PD MiniTrap G-25 desalting columns (28-9180-08; GE) and combined in step 3. 3) The maleimide-to-thiol coupling between TMV-(PEG)4-mal and SA-(PEG)4-SH (6 e.q. of SA per TMV-lys coat protein) was carried out overnight at RT and quenched by addition of excess glycine/L-cysteine. The TMV-(PEG)8-SA particles were then purified by ultracentrifugation over a 40% (w/v) sucrose cushion.
TMV conjugation with PEG
TMV-PEG particles were obtained by conjugation of SM- (PEG)24 (NHS-(PEG)24-mal, 22114; Thermo Fisher) or SM-(PEG)105 (NHS-(PEG)105-mal, PG2-MLNS-5k; Nanocs) to the external surface of TMV-lys. The reaction was performed by mixing TMV-lys (at 2 mg/mL final conc.) and SM-(PEG)24/105 at 10 e.q. per TMVcp in 0.01 M potassium phosphate buffer pH 7.4 containing 10% (v/v) DMSO overnight at RT. The reaction was quenched by addition of excess glycine/L-cysteine, quenching was allowed to proceed for 1 h at RT. The products were then purified by ultracentrifugation over a 40% (w/v) sucrose cushion.
SDS-PAGE and Western blotting (WB)
HSA was used to prepare the TMV-PEG8-SA particles for SDS-PAGE and WB. The TMV samples (40 μg) were denatured by boiling at 100°C for 7 minutes in gel lading buffer (62.5 mM Tris-HCl pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 0.01% (w/v) bromophenol blue, 10% (v/v) 2-mercaptoethanol). Denatured protein samples were then separated on 4–12% NuPAGE polyacrylamide gels in 1x MOPS running (Invitrogen) buffer at 200 V for 50 min. The gels were stained with Coomassie Brilliant Blue and visualized using an AlphaImager imaging system (Biosciences).
For WB, samples separated by SDS-PAGE were transferred from the gel onto nitro-cellulose membranes under a constant voltage of 30 V for 1 h. The membranes were then incubated at RT for 1 h in blocking solution using 5% (w/v) skimmed milk in TBST (150 mM NaCl, 10 mM Tris HCl, 0.1% (v/v) Tween-20, pH 7.5). Then, blots were incubated with: a) 0.5 μg/mL rabbit polyclonal antibody against human serum albumin (NBP1-32458; Novus Biologicals), or b) 0.5 μg/mL rabbit anti-TMV antibody (custom made; Pacific Immunology) in blocking solution and subsequently washed 3x for 5 minutes in TBST. After washing, membranes were incubated with 1 μg/mL of alkaline phosphatase goat anti-rabbit antibody in blocking solution for 1 h at RT and washed 3x for 15 minutes in TBST, and 1x for 5 minutes in Millipore water. Specific antibody binding was visualized using Novex AP Chromogenic Substrate (BCIP/NBT) (WP20001; Invitrogen).
Transmission electron microscopy and immunogold staining
HSA was used to prepare the TMV-PEG8-SA particles for TEM and immunogold staining. Particles were adsorbed to carbon-coated copper grids at a concentration of 0.1 mg/mL (2 μl per grid), rinsed with deionized water, and negatively stained with 2% (w/v) uranyl acetate for 2 min before analysis with a Zeiss Libra 200FE TEM at 200 kV. For immunogold labeling experiments, prior the uranyl acetate stain, the grids were blocked with 1% (w/v) BSA/0.1% (v/v)Tween-20 for 30 min, incubated with 0.5 μg/mL rabbit α-TMV antibody (polyclonal antibody from rabbits, Pacific Immunology) in 10 mM PBS for 1.5 h at RT, washed 4 x 3 min with 0.01% (v/v) Tween-20 in 10 mM PBS, and incubated with goat anti-rabbit gold conjugated antibody (1:10 dilution in 10 mM PBS) for 1.5 h at RT, washed 4 x 3 min with 0.01% Tween-20 in PBS and washed by 3 x 20 dips in DI water.
Size exclusion chromatography (SEC)
The samples (100 μL of 1.0 mg/mL solution) were analyzed by SEC using a Superose6 column on the ÄKTA Explorer chromatography system (GE Healthcare), at a flow rate of 0.5 mL/min using either KP or PBS buffer (pH 7.4) for TMV-lys and TMV-PEG8-SA particles respectively. The absorbance at wavelengths of 260 nm and 280 nm was registered.
Immuno dot-blots
The dot-blots were prepared by spotting 1 μL of α-TMV (polyclonal antibody from rabbits, Pacific Immunology), α-HSA (ab10241; AbCam) and α-CPMV control (polyclonal antibody from rabbits, Pacific Immunology) antibodies either at 3 different concentrations each (100 μg/mL, 20 μg/mL and 10 μg/mL in 10 mM PBS) or using only the highest concentration (100 μg/mL in 10 mM PBS) on nitrocellulose membrane, previously equilibrated in 10 mM PBS. The prepared blots were then blocked in 5% (w/v) skimmed milk solution in 10 mM PBS for 1 h in RT, washed three times for 5 min in 10 mM PBS and incubated in 40 μg/mL TMV solution in PBS for 2.5 h at RT. HSA was used to prepare the TMV-PEG8-SA particles for immune recognition experiments. After subsequent 3x 5 min washes in PBS, blots were dried and imaged for fluorescence using Maestro imaging system with yellow excitation (576–621 nm) and emission (635 nm longpass) filters and with automatically determined optimal exposure times. Cy5-labeled TMV was used for these experiments and therefore read out of binding was quantified based on the fluorescence signals.
Flow cytometry
RAW264.7 cells were grown to confluence in DMEM medium, supplemented with 10% (v/v) FBS (to make complete medium) and 1% (v/v) penicillin/streptomycin, at 37°C and 5% CO2. The cells were washed with PBS, collected in enzyme-free Hank’s-based Cell Dissociation Buffer (Fisher), washed in PBS again and resuspended in the complete medium. Cells were then added to 96-well v-bottom plates (200,000 cells in 200 μL per well) and incubated with 13 μg of VNPs per well in triplicate for 6 h at 37°C and 5% CO2. MSA was used to prepare the TMV-PEG8-SA particles for the uptake studies in RAW264.7 cells (a mouse-derived cell line). Cells were washed twice in FACS buffer (1 mM EDTA, 25 mM HEPES, 1% (v/v) FBS in PBS, pH 7.0) and fixed in 2% (v/v) paraformaldehyde in FACS buffer for 10 min at RT. After fixation, cells were washed twice in FACS buffer, resuspended in 300 μL FACS buffer, and stored at 4°C. Cells were analyzed using a BD LSR II Flow Cytometer and 10,000 gated events were recorded. Data were analyzed using FlowJo v8.6.3 software.
In vivo pharmacokinetics experiments
MSA was used to prepare the TMV-PEG8-SA particles for PK study. All animal procedures were performed using approved protocols from the Institutional Animal Care and Use Committee at Case Western Reserve University. Particles (4 mg/mL stock in PBS or KP buffer) at a dose of 0.40 mg/mice were administered by tail vein injection using Balb/C mice (Charles River). Blood was collected pre- and post-injection via retro-orbital bleed (at t=0, 10, 30, 60, 120, and 360 minutes); an n=3 was assigned for each time-point. Heparin coated capillaries and collection tubes have been used; and collected blood was stored on ice and storage and handling time was kept to minimum. The collected blood was centrifuged at 2000 rcf for 10 minutes to separate the blood plasma (supernatant) from the cells (pellet). The fluorescence (λex=600, λem=650) of the plasma samples was then analyzed with use of Tecan Infinite M200 plate reader. The fluorescence reading was correlated to the standard curves prepared for each of used particles to determine the particle concentration at each timepoint. %ID was determined as ratio between initial particle concentration (determined using injected dose and total mouse blood volume) and the concentration at each time point. The calculations were made with the assumption of a total mouse blood volume of 8mL/100g. The mice weights were measured before injections.
Protein corona preparation
HSA was used to prepare the TMV-PEG8-SA particles for protein corona analysis. Hard protein coronas from TMV rods were prepared by incubating the particles in 2 mL ~100% human plasma (70039.6; Stemcell Technologies) at concentration of 0.3 mg/mL TMV at RT for 1 h. The samples were then diluted in 25 mL PBS, purified by ultracentrifugation on a 40% (v/v) sucrose cushion, and washed twice with 25 mL PBS to remove loosely-bound proteins. The final pellet was resuspended in 0.1 mL gel loading buffer (62.5 mM Tris-HCl pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 0.01% (w/v) bromophenol blue, 10% (v/v) 2-mercaptoethanol) and denatured by boiling at 100°C for 7 min. Subsequently 10–15 μl (~40 μg) of each sample was analyzed by SDS-PAGE (see Supplementary Information and Figure S3).
Supplementary Material
Figure S1. Comparison of different strategies of TMV-lys functionalization. A, Chemical schemes of different TMV-lys surface functionalization strategies. Abbreviations: EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; BS(PEG)5, PEGylated bis(sulfosuccinimidyl)suberate; SAT(PEG)4, PEGylated N-succinimidyl S-acetylthioacetate; SM(PEG)4, PEGylated succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate crosslinker. B, SDS-PAGE analysis of SA coated TMV prepared using strategies presented in panel (A). Absence of >65 kDa SA-TMVcp conjugate bands indicates no detectable SA conjugation to TMV-lys using EDC coupling. Conjugation with use of BS(PEG)5 promoted extensive TMV-crosslinking evident by presence of a 38-kDa band corresponding to crosslinked TMVcps and sedimentation of particles due to lack of colloidal stability. Conjugation using two different heterobifunctional PEG linkers SAT(PEG)4 and SM(PEG)4 was most successful yielding ~ 60% coverage of SA per TMVcps. Presence of >67kD high molecular weight bands corresponded to SA bound to TMVcp, which was confirmed by WB stained positively for both SA and TMVcp (see Figure 1 + S2).
Figure S2. SDS-PAGE and WB analysis of TMV-lys particles before and after PEGylation and SA conjugation – the primary images used in Figure 1. A, SDS-PAGE; Coomassie stained. B, α-TMV WB. C, α-SA WB.
Figure S3. Protein corona formation using ‘naked’ and ‘stealth’ TMV. SDS-PAGE analysis of TMV after protein corona formation (1 h incubation in ~100% human plasma, followed by removal of the soft corona). ‘Bare’ (not exposed to human plasma) TMV-lys and 1:100 diluted human plasma were used as control.
Acknowledgments
FUNDING SOURCES
This work was supported in part by a grant from the National Science Foundation CAREER DMR 1452257 (to NFS) and grants from the National Institute of Health (NIH): NHLBI R21 HL121130 (to NFS) and a pilot grant from Case-Coulter Translational Research Partnership and the Harrington Heart & Vascular Institute.
We thank Prof. Christina Wege and her team from the University of Stuttgart, Germany for the TMV-lys mutant. We thank CWRU Farm for help with the scaled-up production and growth of N. benthamiana plants. Selected molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
ABBREVIATIONS
- TMV
Tobacco Mosaic Virus
- TMVcp
Tobacco Mosaic Virus capsid protein
- VNP
Viral Nanoparticle
- NP
nanoparticle
- MPS
Mononuclear Phagocyte System
- SA
Serum Albumin
- HSA
Human Serum Albumin
- MSA
Mouse Serum Albumin
- PEG
poly ethylene glycol
- MRI
Magnetic Resonance Imaging
- PK
pharmacokinetics
- TEM
Transmission Electron Microscopy
- SDS-PAGE
sodium dodecyl sulphate poly-acrylamide electrophoresis
- WB
Western Blot
Footnotes
Author Contributions
A.S.P. prepared and characterized nanoparticle formulations, performed in vitro binding assays and in vivo pharmacokinetics study, analyzed and interpreted the data; S.A.J and F.A.V. purified TMV, assisted in synthesis and characterization of nanoparticle formulations, analyzed and interpreted the data; S.S. assisted in in vivo pharmacokinetics study; A.S.P and N.F.S. conceived and designed the experiments and wrote the paper; all authors edited and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could aFFect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moghimi SM, Hunter AC, Murray JC. Nanomedicine: current status and future prospects. Faseb J. 2005;19:311–330. doi: 10.1096/fj.04-2747rev. [DOI] [PubMed] [Google Scholar]
- 2.Zamboni WC, Torchilin V, Patri AK, Hrkach J, Stern S, Lee R, et al. Best practices in cancer nanotechnology: perspective from NCI nanotechnology alliance. Clin Cancer Res. 2012;18:3229–3241. doi: 10.1158/1078-0432.CCR-11-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 4.Godin B, Sakamoto JH, Serda RE, Grattoni A, Bouamrani A, Ferrari M. Emerging applications of nanomedicine for the diagnosis and treatment of cardiovascular diseases. Trends Pharmacol Sci. 2010;31:199–205. doi: 10.1016/j.tips.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iverson N, Plourde N, Chnari E, Nackman GB, Moghe PV. Convergence of nanotechnology and cardiovascular medicine : progress and emerging prospects. BioDrugs. 2008;22:1–10. doi: 10.2165/00063030-200822010-00001. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Xu Z, Yu H, Chen X, Feng B, Cui Z, et al. Treatment of metastatic breast cancer by combination of chemotherapy and photothermal ablation using doxorubicin-loaded DNA wrapped gold nanorods. Biomaterials. 2014;35:8374–8384. doi: 10.1016/j.biomaterials.2014.05.094. [DOI] [PubMed] [Google Scholar]
- 7.Cole JT, Holland NB. Multifunctional nanoparticles for use in theranostic applications. Drug Deliv Transl Res. 2015;5:295–309. doi: 10.1007/s13346-015-0218-2. [DOI] [PubMed] [Google Scholar]
- 8.Huang P, Gao Y, Lin J, Hu H, Liao H-S, Yan X, et al. Tumor-Specific Formation of Enzyme-Instructed Supramolecular Self-Assemblies as Cancer Theranostics. ACS Nano. 2015;9:9517–9527. doi: 10.1021/acsnano.5b03874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruckman MA, Randolph LN, Gulati NM, Stewart PL, Steinmetz NF. Silica-coated Gd (DOTA)-loaded protein nanoparticles enable magnetic resonance imaging of macrophages. Journal of Materials Chemisstry B. 2015;3:7503–7510. doi: 10.1039/C5TB01014D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruckman MA, Jiang K, Simpson EJ, Randolph LN, Luyt LG, Yu X, et al. Dual-modal magnetic resonance and fluorescence imaging of atherosclerotic plaques in vivo using VCAM-1 targeted tobacco mosaic virus. Nano Lett. 2014;14:1551–1558. doi: 10.1021/nl404816m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shukla S, Steinmetz NF. Virus-based nanomaterials as positron emission tomography and magnetic resonance contrast agents: from technology development to translational medicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2015;7:708–721. doi: 10.1002/wnan.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikhaylov G, Mikac U, Magaeva AA, Itin VI, Naiden EP, Psakhye I, et al. Ferri-liposomes as an MRI-visible drug-delivery system for targeting tumours and their microenvironment. Nature Nanotechnology. 2011;6:594–602. doi: 10.1038/nnano.2011.112. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y-XJ. Superparamagnetic iron oxide based MRI contrast agents: Current status of clinical application. Quant Imaging Med Surg. 2011;1:35–40. doi: 10.3978/j.issn.2223-4292.2011.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Garay RP, El-Gewely R, Armstrong JK, Garratty G, Richette P. Antibodies against polyethylene glycol in healthy subjects and in patients treated with PEG-conjugated agents. Expert Opin Drug Deliv. 2012;9:1319–1323. doi: 10.1517/17425247.2012.720969. [DOI] [PubMed] [Google Scholar]
- 16.Garratty G. Modulating the red cell membrane to produce universal/stealth donor red cells suitable for transfusion. Vox Sang. 2008;94:87–95. doi: 10.1111/j.1423-0410.2007.01003.x. [DOI] [PubMed] [Google Scholar]
- 17.Richter AW, Akerblom E. Polyethylene glycol reactive antibodies in man: titer distribution in allergic patients treated with monomethoxy polyethylene glycol modified allergens or placebo, and in healthy blood donors. Int Arch Allergy Appl Immunol. 1984;74:36–39. doi: 10.1159/000233512. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Ishida T, Kiwada H. Anti-PEG IgM elicited by injection of liposomes is involved in the enhanced blood clearance of a subsequent dose of PEGylated liposomes. J Control Release. 2007;119:236–244. doi: 10.1016/j.jconrel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Egesten A, Frick I-M, Mörgelin M, Olin AI, Björck L. Binding of albumin promotes bacterial survival at the epithelial surface. J Biol Chem. 2011;286:2469–2476. doi: 10.1074/jbc.M110.148171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews NC. Iron homeostasis: insights from genetics and animal models. Nat Rev Genet. 2000;1:208–217. doi: 10.1038/35042073. [DOI] [PubMed] [Google Scholar]
- 21.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Baldelli Bombelli F, Hristov DR, et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nature Nanotechnology. 2013;8:137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 22.Huang RK, Steinmetz NF, Fu C-Y, Manchester M, Johnson JE. Transferrin-mediated targeting of bacteriophage HK97 nanoparticles into tumor cells. Nanomedicine (Lond) 2011;6:55–68. doi: 10.2217/nnm.10.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pitek AS, O’Connell D, Mahon E, Monopoli MP, Baldelli Bombelli F, Dawson KA. Transferrin coated nanoparticles: study of the bionano interface in human plasma. PLoS ONE. 2012;7:e40685. doi: 10.1371/journal.pone.0040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez PL, Harada T, Christian DA, Pantano DA, Tsai RK, Discher DE. Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science. 2013;339:971–975. doi: 10.1126/science.1229568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koudelka KJ, Pitek AS, Manchester M, Steinmetz NF. Virus-Based Nanoparticles as Versatile Nanomachines. Annual Review of Virology. 2015;2:379–401. doi: 10.1146/annurev-virology-100114-055141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmetz NF. Viral nanoparticles in drug delivery and imaging. Mol Pharmaceutics. 2013;10:1–2. doi: 10.1021/mp300658j. [DOI] [PubMed] [Google Scholar]
- 27.Cho C-F, Shukla S, Simpson EJ, Steinmetz NF, Luyt LG, Lewis JD. Molecular targeted viral nanoparticles as tools for imaging cancer. Methods Mol Biol. 2014;1108:211–230. doi: 10.1007/978-1-62703-751-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasch M, de la Escosura A, Ma Y, Uetrecht C, Heck AJR, Torres T, et al. Encapsulation of phthalocyanine supramolecular stacks into virus-like particles. J Am Chem Soc. 2011;133:6878–6881. doi: 10.1021/ja110752u. [DOI] [PubMed] [Google Scholar]
- 29.Douglas T, Young M. Host|[ndash]|guest encapsulation of materials by assembled virus protein cages. Nature. 1998;393:152–155. doi: 10.1038/30211. [DOI] [Google Scholar]
- 30.Steinmetz NF, Hong V, Spoerke ED, Lu P, Breitenkamp K, Finn MG, et al. Buckyballs meet viral nanoparticles: candidates for biomedicine. J Am Chem Soc. 2009;131:17093–17095. doi: 10.1021/ja902293w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lizotte PH, Wen AM, Sheen MR, Fields J, Rojanasopondist P, Steinmetz NF, et al. In situ vaccination with cowpea mosaic virus nanoparticles suppresses metastatic cancer. Nature Nanotechnology. 2015 doi: 10.1038/nnano.2015.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caldorera-Moore M, Guimard N, Shi L, Roy K. Designer nanoparticles: incorporating size, shape and triggered release into nanoscale drug carriers. Expert Opin Drug Deliv. 2010;7:479–495. doi: 10.1517/17425240903579971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S-Y, Ferrari M, Decuzzi P. Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 34.Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, et al. The Impact of Aspect Ratio on the Biodistribution and Tumor Homing of Rigid Soft-Matter Nanorods. Adv Healthc Mater. 2015;4:874–882. doi: 10.1002/adhm.201400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen AM, Wang Y, Jiang K, Hsu GC, Gao H, Lee KL, et al. Shaping bio-inspired nanotechnologies to target thrombosis for dual optical-magnetic resonance imaging. J Mater Chem B Mater Biol Med. 2015;3:6037–6045. doi: 10.1039/C5TB00879D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CASPAR DLD. Structure of Tobacco Mosaic Virus: Radial Density Distribution in the Tobacco Mosaic Virus Particle. Nature. 1956;177:928–928. doi: 10.1038/177928a0. [DOI] [Google Scholar]
- 37.Sachse C, Chen JZ, Coureux P-D, Stroupe ME, Fändrich M, Grigorieff N. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol. 2007;371:812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, et al. TMV nanorods with programmed longitudinal domains of differently addressable coat proteins. Nanoscale. 2013;5:3808–3816. doi: 10.1039/C3NR33724C. [DOI] [PubMed] [Google Scholar]
- 39.Milani S, Bombelli Bombelli F, Pitek AS, Dawson KA, Rädler J. Reversible versus irreversible binding of transferrin to polystyrene nanoparticles: soft and hard corona. ACS Nano. 2012;6:2532–2541. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 40.de Gennes PG. Polymers at an interface; a simplified view. Advances in Colloid and Interface Science. 1987;27:189–209. doi: 10.1016/0001-8686(87)85003-0. [DOI] [Google Scholar]
- 41.Lee KL, Shukla S, Wu M, Ayat NR, El Sanadi CE, Wen AM, et al. Stealth filaments: Polymer chain length and conformation affect the in vivo fate of PEGylated potato virus X. Acta Biomater. 2015;19:166–179. doi: 10.1016/j.actbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva RMD, de Souto ER, Pedroso JC, Arakava R, Almeida ÁMR, Barboza AAL, et al. Detection and identification of TMV infecting tomato under protected cultivation in Paraná State. Brazilian Archives of Biology and Technology. 2008;51:903–909. doi: 10.1590/S1516-89132008000500005. [DOI] [Google Scholar]
- 43.Hu Q, Niu Y, Zhang K, Liu Y, Zhou X. Virus-derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virology Journal. 2011;8:41. doi: 10.1186/1743-422X-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wetter C. Tobacco mosaic virus and para-tobacco mosaic virus in cigarettes. Naturwissenschaften. 1975;62:533. doi: 10.1007/BF00609072. [DOI] [PubMed] [Google Scholar]
- 45.Pitek AS, Wen AM, Shukla S, Steinmetz NF. The Protein Corona of Plant Virus Nanoparticles Influences their Dispersion Properties, Cellular Interactions, and In Vivo Fates. Small. 2016 doi: 10.1002/smll.201502458. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, et al. Biodistribution, pharmacokinetics, and blood compatibility of native and PEGylated tobacco mosaic virus nano-rods and -spheres in mice. Virology. 2014;449:163–173. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruckman MA, Steinmetz NF. Chemical modification of the inner and outer surfaces of Tobacco Mosaic Virus (TMV) Methods Mol Biol. 2014;1108:173–185. doi: 10.1007/978-1-62703-751-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of different strategies of TMV-lys functionalization. A, Chemical schemes of different TMV-lys surface functionalization strategies. Abbreviations: EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; BS(PEG)5, PEGylated bis(sulfosuccinimidyl)suberate; SAT(PEG)4, PEGylated N-succinimidyl S-acetylthioacetate; SM(PEG)4, PEGylated succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate crosslinker. B, SDS-PAGE analysis of SA coated TMV prepared using strategies presented in panel (A). Absence of >65 kDa SA-TMVcp conjugate bands indicates no detectable SA conjugation to TMV-lys using EDC coupling. Conjugation with use of BS(PEG)5 promoted extensive TMV-crosslinking evident by presence of a 38-kDa band corresponding to crosslinked TMVcps and sedimentation of particles due to lack of colloidal stability. Conjugation using two different heterobifunctional PEG linkers SAT(PEG)4 and SM(PEG)4 was most successful yielding ~ 60% coverage of SA per TMVcps. Presence of >67kD high molecular weight bands corresponded to SA bound to TMVcp, which was confirmed by WB stained positively for both SA and TMVcp (see Figure 1 + S2).
Figure S2. SDS-PAGE and WB analysis of TMV-lys particles before and after PEGylation and SA conjugation – the primary images used in Figure 1. A, SDS-PAGE; Coomassie stained. B, α-TMV WB. C, α-SA WB.
Figure S3. Protein corona formation using ‘naked’ and ‘stealth’ TMV. SDS-PAGE analysis of TMV after protein corona formation (1 h incubation in ~100% human plasma, followed by removal of the soft corona). ‘Bare’ (not exposed to human plasma) TMV-lys and 1:100 diluted human plasma were used as control.