Abstract
The network structure of biological systems suggests that effective therapeutic intervention may require combinations of agents that act synergistically. However, a dearth of systematic chemical combination datasets have limited the development of predictive algorithms for chemical synergism. Here, we report two large datasets of linked chemical-genetic and chemical-chemical interactions in the budding yeast Saccharomyces cerevisiae. We screened 5,518 unique compounds against 242 diverse yeast gene deletion strains to generate an extended chemical-genetic matrix (CGM) of 492,126 chemical-gene interaction measurements. This CGM dataset contained 1,434 genotype-specific inhibitors, termed cryptagens. We selected 128 structurally diverse cryptagens and tested all pairwise combinations to generate a benchmark dataset of 8,128 pairwise chemical-chemical interaction tests for synergy prediction, termed the cryptagen matrix (CM). An accompanying database resource called ChemGRID was developed to enable analysis, visualisation and downloads of all data. The CGM and CM datasets will facilitate the benchmarking of computational approaches for synergy prediction, as well as chemical structure-activity relationship models for anti-fungal drug discovery.
Subject terms: High-throughput screening, Chemical genetics, Networks and systems biology, Small molecules, Screening
Background & Summary
The network-based organization of biological systems suggests that combinations of small molecules will be needed to achieve therapeutic efficacy and selectivity for infectious disease, cancer and many other disorders1,2. Cellular networks are strongly buffered against loss of gene function, manifest as synthetic lethal genetic interactions3. A genetic interaction occurs when a phenotype caused by a mutation in one gene is exacerbated (or suppressed) by a mutation in another gene4. Genome-wide screens in the budding yeast S. cerevisiae have uncovered over 200,000 genetic interactions, in contrast to the ~1,000 essential genes3,5. Analogous to genetic interactions, combinations of chemicals that individually cause minimal phenotypes may exhibit greater than additive effects, termed synergism2,6–9. Notably, compounds that phenocopy the effect of mutations in non-essential genes may have no discernable effect on wild-type cells, but might inhibit growth in a given genetic context2. By definition, the biological activity of such compounds would not be detected in many high-throughput screens used in modern drug discovery. Compounds with such latent activities have been termed cryptagens or dark chemical matter10,11.
We recently generated a systematic chemical-genetic dataset in S. cerevisiae to allow the discovery and prediction of synergistic interactions between cryptagens that do not have obvious effects on cell proliferation on their own11. Various algorithmic approaches have been developed to predict synergistic compound combinations1,12,13. However, in most cases such predictions have been made on focused datasets and/or known chemical activities, which inherently constrains the development of general methods14. The dearth of fully factorial drug combination data matrices has hampered the systematic testing and comparisons of different predictive approaches1. To address this shortfall, we generated two large-scale data sets: a chemical-genetic matrix (CGM) of 356,500 pairwise chemical-gene interaction tests and a derived cryptagen matrix (CM) of 8,128 chemical-chemical interaction tests11. Based on this data, we developed a machine learning approach that integrates structural features of compounds with chemical-genetic interactions to predict compound synergism11,15. This systematic approach identified many novel synergistic anti-fungal combinations, many of which also exhibited species-selective effects against clinical isolates of pathogenic fungi11. The CM represents a benchmark dataset for the development and refinement of synergy prediction algorithms.
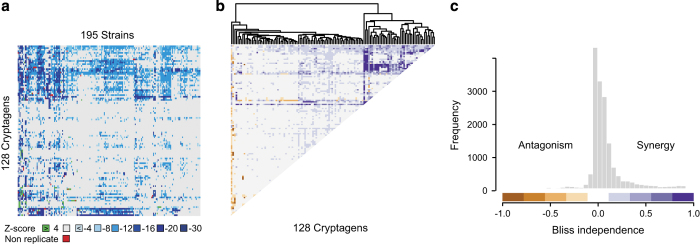
Here, we describe the CGM and CM datasets in detail to facilitate use of this data for synergy prediction by computational approaches. The original CGM was generated by screening 4,915 compounds drawn from four different chemical libraries (LOPAC, Maybridge Hitskit 1000, Spectrum Collection and an in-house collection called Bioactive 1). These libraries were screened against 195 diverse S. cerevisiae deletion strains, which we termed sentinel strains for their ability to detect otherwise hidden chemical activities11. The updated CGM described here is an extended version of the dataset reported previously: the number of sentinels has been increased from 195 to 242 yeast deletion strains and the cohort of chemical libraries has been expanded to include a second in-house collection of 892 compounds with bioactivity in yeast, termed Bioactive 2. This extended CGM dataset contains data for 5,518 unique compounds, 242 sentinel strains and duplicate measurements for 492,126 pairwise chemical-gene interaction tests, which represent an additional 135,626 duplicate interaction tests compared to the original CGM dataset (Figs 1,2; Table 1 (available online only)). As previously, we defined cryptagens as compounds that were active against more than 4 and less than 2/3 of tested sentinel strains. Out of the 5,518 compounds in the expanded CGM, 1,434 compounds were categorized as cryptagens (Table 2). From the original CGM dataset11, we selected a subset of 128 cryptagens that were used to generate a complete single concentration combination matrix, termed the cryptagen matrix (CM) (Fig. 3a). All 8,128 possible combinations between the 128 cryptagens were tested for synergy at 10 μM concentration for each compound in a drug pump-deficient S. cerevisiae strain (Fig. 3b). Bliss independence values were calculated for each compound pair in the CM dataset (see Methods for details). Independent dose-response surface (checkerboard) assays demonstrated a 65% confirmation rate of synergistic compound interactions from the CM dataset. The full CGM and CM datasets can be accessed at ChemGRID, a web portal that also houses a suite of tools that enable the interrogation and visualization of the chemical interaction datasets (Fig. 4). The CGM dataset and a detailed accompanying description of the yeast cell growth assay have been deposited at NCBI PubChem BioAssay (Data citations 1,2).
Figure 1. Schematic overview of experimental workflow for CGM and CM dataset generation and data deposition.
Figure 2. CGM heatmaps for activity (Z-score) of cryptagens contained in each of the five different compound libraries screened against the sentinel deletion strains.
Corresponding histograms with Z-score distributions are shown below.
Table 1. Strains screened against each library in the CGM.
| ORF | Gene Name | LOPAC | Maybridge HitsKit 1000 | Spectrum | Bioactive 1 | Bioactive 2 | Use in CM | Synonym | Description * | Phenotype information * | SGD ID * |
|---|---|---|---|---|---|---|---|---|---|---|---|
| YLR027C | AAT2 | + | ASP5 | Cytosolic aspartate aminotransferase, involved in nitrogen metabolism; localizes to peroxisomes in oleate-grown cells | Null mutant is viable in S288C, inviable in Sigma1278b | S000004017 | |||||
| YCR088W | ABP1 | + | Actin-binding protein of the cortical actin cytoskeleton, important for activation of the Arp2/3 complex that plays a key role actin in cytoskeleton organization | Null mutant is viable, has abnormalities in actin cytoskeleton morphology | S000000684 | ||||||
| YDR226W | ADK1† | + | AKY1|AKY2 | Adenylate kinase, required for purine metabolism; localized to the cytoplasm and the mitochondria; lacks cleavable signal sequence | Null mutant is viable. displays slow growth and inability to utilize non-fermentable carbon sources | S000002634 | |||||
| YNR030W | ALG12 | + | ECM39 | Alpha-1,6-mannosyltransferase localized to the ER; responsible for the addition of the alpha-1,6 mannose to dolichol-linked Man7GlcNAc2, acts in the dolichol pathway for N-glycosylation | Null mutant is viable, confers increased resistance to oxidative stress | S000005313 | |||||
| YLR102C | APC9 | + | Subunit of the Anaphase-Promoting Complex/Cyclosome (APC/C), which is a ubiquitin-protein ligase required for degradation of anaphase inhibitors, including mitotic cyclins, during the metaphase/anaphase transition | Null mutant is viable, shows delay in entry into anaphase at 37 C | S000004092 | ||||||
| YLR370C | ARC18 | + | Subunit of the ARP2/3 complex, which is required for the motility and integrity of cortical actin patches | Null mutant is viable, fails to localize actin patches, accumulates high amounts of chitin in cell wall, and shows defects in mitochondrial movement | S000004362 | ||||||
| YHR013C | ARD1 | + | Subunit of the N-terminal acetyltransferase NatA (Nat1p, Ard1p, Nat5p); N-terminally acetylates many proteins, which influences multiple processes such as the cell cycle, heat-shock resistance, mating, sporulation, and telomeric silencing | Null mutant is viable, displays decreased pheromone sensitivity, reduced mating efficiency and sporulation | S000001055 | ||||||
| YDL192W | ARF1 | + | ADP-ribosylation factor, GTPase of the Ras superfamily involved in regulation of coated formation vesicles in intracellular trafficking within the Golgi; functionally interchangeable with Arf2p | Null mutant is viable, shows slow growth, cold sensitivity and sensitivity to normally sublethal concentrations of fluoride ion in the medium. | S000002351 | ||||||
| YNL020C | ARK1 | + | + | Serine/threonine protein kinase involved in regulation of the cortical actin cytoskeleton; involved in control of endocytosis | Null mutant is viable, shows slight delocalisation of actin cytoskeleton | S000004965 | |||||
| YPL051W | ARL3 | + | GTPase of the Ras superfamily required to recruit Arl1p to the Golgi; similar to ADP-ribosylation factor | Null mutant is viable, displays cold-sensitive growth | S000005972 | ||||||
| YHR129C | ARP1 | + | + | ACT5 | Actin-related protein of the dynactin complex; required for spindle orientation and nuclear migration; putative ortholog of mammalian centractin | Null mutant is viable, both null mutations and overexpression lead to defects in spindle orientation and nuclear migration | S000001171 | ||||
| YLR085C | ARP6 | + | + | Nuclear actin-related protein involved in chromatin remodeling, component of chromatin-remodeling enzyme complexes | Null mutant is viable, disorganized actin filaments and abnormal vacuolar morphology | S000004075 | |||||
| YDR101C | ARX1 | + | Protein associated with the ribosomal export complex | Null mutant is viable, modest nuclear retention of 60S ribosomal subunits | S000002508 | ||||||
| YJL115W | ASF1 | + | CIA1 | Nucleosome assembly factor, involved in chromatin assembly after DNA replication, anti-silencing protein that causes derepression of silent loci when overexpressed | Null mutant is viable, delayed cell cycle progression through metaphase, defect in telomere localization, decreased silencing, shortened lifespan | S000003651 | |||||
| YPL078C | ATP4 | + | LPF7 | Subunit b of the stator stalk of mitochondrial F1F0 ATP synthase, which is a large, evolutionarily conserved enzyme complex required for ATP synthesis | Null mutant is viable, oxidative phosphorylation deficient, unable to grow on glycerol | S000005999 | |||||
| YOL078W | AVO1† | + | Component of a membrane-bound complex containing the Tor2p kinase and other proteins, which may have a role in regulation of cell growth | Null mutant is inviable, depolarized actin cytoskeleton | S000005438 | ||||||
| YJL095W | BCK1 | +++ | + | + | + | LAS3|SAP3|SLK1|SSP31 | Mitogen-activated protein (MAP) kinase kinase kinase acting in the protein kinase C signaling pathway, which controls cell integrity; upon activation by Pkc1p phosphorylates downstream kinases Mkk1p and Mkk2p | Null mutant is viable, temperature-sensitive cell wall defect | S000003631 | ||
| YER167W | BCK2 | + | + | CTR7 | Protein rich in serine and threonine residues, overproduction suppresses pkc1 mutation, activates G1/S transcription | Null mutant is viable, increased cell size and pheromone sensitivity | S000000969 | ||||
| YER155C | BEM2 | ++ | + | IPL2|SUP9|TSL1 | Rho GTPase activating protein (RhoGAP) involved in the control of cytoskeleton organization and cellular morphogenesis; required for bud emergence | Null mutant is viable, randomized bud-site selection and defective bud emergence | S000000957 | ||||
| YCL029C | BIK1 | + | ARM5|PAC14 | Microtubule-associated protein, component of the interface between microtubules and kinetochore, involved in sister chromatid separation; essential in polyploid cells but not in haploid or diploid cells; ortholog of mammalian CLIP-170 | Null mutant is viable, bilateral defects in karyogamy | S000000534 | |||||
| YER016W | BIM1 | + | EB1|YEB1 | Microtubule-binding protein that together with Kar9p makes up the cortical microtubule capture site and delays the exit from mitosis when the spindle is oriented abnormally | Null mutant is viable, cold sensitivie, benomyl supersensitive, aberrant microtubule morphology | S000000818 | |||||
| YNL271C | BNI1 | + | PPF3|SHE5 | Formin, nucleates the formation of linear actin filaments, involved in cell processes such as budding and mitotic spindle orientation which require the formation of polarized actin cables, functionally redundant with BNR1 | Null mutant is viable, bni1 bnr1 double deletion mutant is temperature sensitive, deficient in bud emergence, random distribution of cortical actin patches | S000005215 | |||||
| YNL233W | BNI4 | + | Targeting subunit for Glc7p protein phosphatase, localized to the bud neck, required for localization of chitin synthase III to the bud neck via interaction with the chitin synthase III regulatory subunit Skt5p | Null mutant is viable, shows delocalized chitin, elongated buds, enlarged bud necks | S000005177 | ||||||
| YDL074C | BRE1 | + | + | E3 ubiquitin ligase for Rad6p, required for the ubiquitination of histone H2B, recruitment of Rad6p to promoter chromatin and subsequent methylation of histone H3 (on L4 and L79), contains RING finger domain | Null mutant is viable, sensitive to brefeldin A | S000002232 | |||||
| YFL029C | CAK1† | + | + | CIV1 | Cyclin-dependent kinase-activating kinase required for passage through the cell cycle, phosphorylates and activates Cdc28p; nucleotide-binding pocket differs significantly from those of most other protein kinases | Null mutant is inviable, temperature-sensitive mutant confers a G2 delay | S000001865 | ||||
| YNL161W | CBK1† | + | Serine/threonine protein kinase that regulates cell morphogenesis pathways; involved in cell wall biosynthesis, apical growth, proper mating projection morphology, bipolar bud site selection in diploid cells, and cell separation | Null mutation is inviable, defect in morphogenesis and cell separation | S000005105 | ||||||
| YPR025C | CCL1† | + | Cyclin associated with protein kinase Kin28p, which is the TFIIH-associated carboxy-terminal domain (CTD) kinase involved in transcription initiation at RNA polymerase II promoters | Null mutant is inviable, reduced function mutants show increased competitive fitness | S000006229 | ||||||
| YAL021C | CCR4 | + | FUN27|NUT21 | Component of the CCR4-NOT transcriptional complex, which is involved in regulation of gene expression; component of the major cytoplasmic deadenylase, which is involved in mRNA poly(A) tail shortening | Null mutant is viable, temperature sensitive growth on nonfermentative medium, sensitive to many drugs | S000000019 | |||||
| YFR028C | CDC14† | + | OAF3 | Protein phosphatase required for mitotic exit; located in the nucleolus until liberated by the FEAR and Mitotic Exit Network in anaphase, enabling it to act on key substrates to effect a decrease in CDK/B-cyclin activity and mitotic exit | Null mutant is inviable; temperature sensitive mutant arrests at late anaphase | S000001924 | |||||
| YAR019C | CDC15† | + | LYT1 | Protein kinase of the Mitotic Exit Network that is localized to the spindle pole bodies at late anaphase; promotes mitotic exit by directly switching on the kinase activity of Dbf2p | Null mutant is inviable, temperature sensitive mutant arrests at late anaphase | S000000072 | |||||
| YBR160W | CDC28† | + | CDK1|HSL5|SRM5 | Catalytic subunit of the main cell cycle cyclin-dependent kinase (CDK); alternately associates with G1 cyclins (CLNs) and G2/M cyclins (CLBs) which direct the CDK to specific substrates | Null mutant is inviable, temperature sensitive mutants arrest at G1/S and G2/M | S000000364 | |||||
| YDR054C | CDC34† | + | DNA6|UBC3 | Ubiquitin-conjugating enzyme or E2; together with Skp1p, Rbx1p, Cdc53p, and an F-box protein, forms a ubiquitin-protein ligase called the SCF complex which regulates cell cycle progression by targeting key substrates for degradation | Null mutant is inviable, temperature sensitive mutant arrests at G1/S with multiple buds | S000002461 | |||||
| YFL009W | CDC4† | + | F-box protein required for G1/S and G2/M transition, associates with Skp1p and Cdc53p to form a complex, SCFCdc4, which acts as ubiquitin-protein ligase directing ubiquitination of the phosphorylated CDK inhibitor Sic1p | Null mutant is inviable, temperature sensitive mutant arrests at G1/S with multiple buds | S000001885 | ||||||
| YMR001C | CDC5† | + | MSD2|PKX2 | Polo-like kinase with similarity to Xenopus Plx1 and S. pombe Plo1p; found at bud neck, nucleus and SPBs; has multiple functions in mitosis and cytokinesis through phosphorylation of substrates; may be a Cdc28p substrate | Null mutant is inviable, temperature sensitive mutant arrests in late anaphase | S000004603 | |||||
| YDL132W | CDC53† | + | Cullin, structural protein of SCF complexes (which also contain Skp1p, Cdc34p, and an F-box protein) involved in ubiquitination; SCF promotes the G1-S transition by targeting G1 cyclins and the Cln-CDK inhibitor Sic1p for degradation | Null mutant is inviable, temperature sensitive mutant arrests at G1/S with multiple buds | S000002290 | ||||||
| YDL017W | CDC7† | + | LSD6|SAS1 | DDK (Dbf4-dependent kinase) catalytic subunit required for firing origins and replication fork progression in mitosis through phosphorylation of Mcm2-7p complexes and Cdc45p; kinase activity correlates with cyclical DBF4 expression | Null mutant is inviable, temperature sensitive mutant arrests at G1/S | S000002175 | |||||
| YLR418C | CDC73 | + | Constituent of Paf1 complex with RNA polymerase II, Paf1p, Hpr1p, Ctr9, Leo1, Rtf1 and Ccr4p, distinct from Srb-containing Pol II complexes; required for expression of certain genes, modification of some histones, and telomere maintenance | Null mutant is viable, temperature sensitive and cold sensitive | S000004410 | ||||||
| YBR023C | CHS3 | + | CAL1|CSD2|DIT101|KTI2 | Chitin synthase III, catalyzes the transfer of N-acetylglucosamine (GlcNAc) to chitin; required for synthesis of the majority of cell wall chitin, the chitin ring during bud emergence, and spore wall chitosan | Null mutant is viable, exhibits cell wall defects | S000000227 | |||||
| YLR330W | CHS5 | + | + | CAL3 | Protein of unknown function, involved in chitin biosynthesis by regulating Chs3p localization, also involved in cell fusion during mating | Null mutant is viable, exhibits cell wall defects | S000004322 | ||||
| YHR142W | CHS7 | + | Protein of unknown function, involved in chitin biosynthesis by regulating Chs3p export from the ER | Null mutant is viable, exhibits cell wall defects | S000001184 | ||||||
| YPL241C | CIN2 | + | + | + | + | Tubulin folding factor C (putative) involved in beta-tubulin (Tub2p) folding; isolated as mutant with increased chromosome loss and sensitivity to benomyl | Null mutant is viable, sensitive to benomyl, cold sensitive, increased rate of chromosome loss | S000006162 | |||
| YEL061C | CIN8 | + | ++ | + | KSL2|SDS15 | Kinesin motor protein involved in mitotic spindle assembly and chromosome segregation | Null mutant is viable, sensitive to benomyl, increased rate of chromosome loss | S000000787 | |||
| YOR061W | CKA2 | + | + | + | YOR29-12 | Alpha- catalytic subunit of casein kinase 2, a Ser/Thr protein kinase with roles in cell growth and proliferation; the holoenzyme also contains CKA1, CKB1 and CKB2, the many substrates include transcription factors and all RNA polymerases | Null mutant is viable, resistant to many drugs, increased lifespan | S000005587 | |||
| YLR133W | CKI1 | + | Choline kinase, catalyzes the first step in the CDP-choline pathway phosphatidylcholine synthesis (Kennedy pathway); mRNA expression is regulated by inositol and choline, enzyme activity is stimulated by phosphorylation by protein kinase | Null mutant is viable, resistant to many drugs, increased lifespan | S000004123 | ||||||
| YBR135W | CKS1† | + | Subunit of the Cdc28 protein kinase, required for mitotic proteolysis, may also be involved in the proteolysis of the G1 cyclins | Null mutant is inviable, temperature sensitive mutant arrests at G1/S | S000000339 | ||||||
| YNL298W | CLA4 | + | + | ERC10 | Cdc42p activated signal transducing kinase of the PAK (p21-activated kinase) family, involved in septin ring assembly and cytokinesis; directly phosphorylates septins Cdc3p and Cdc10p; other yeast PAK family members are Ste20p and Skm1p | Null mutant is viable, cytokinesis defect | S000005242 | ||||
| YPR119W | CLB2 | + | B-type cyclin involved in cell cycle progression; activates Cdc28p to promote the transition from G2 to M phase; accumulates during G2 and M, then targeted via a destruction box motif for ubiquitin-mediated degradation by the proteasome | Null mutant is viable, delayed G2/M progression, increased cell size | S000006323 | ||||||
| YPL256C | CLN2 | ++ | + | + | G1 cyclin involved in regulation of the cell cycle; activates Cdc28p kinase to promote the G1 to S phase transition; late G1 specific expression depends on transcription factor complexes, MBF (Swi6p-Mbp1p) and SBF (Swi6p-Swi4p) | Null mutant is viable, delayed G1/S progression, increased cell size | S000006177 | ||||
| YAL040C | CLN3 | + | + | DAF1|FUN10|WHI1 | G1 cyclin involved in cell cycle progression; activates Cdc28p kinase to promote the G1 to S phase transition; plays a role in regulating transcription of the other G1 cyclins, CLN1 and CLN2; regulated by phosphorylation and proteolysis | Null mutant is viable, delayed G1/S transition, increased cell size | S000000038 | ||||
| YKL190W | CNB1 | + | + | + | CRV1|YCN2 | Calcineurin B; the regulatory subunit of calcineurin, a Ca++/calmodulin-regulated protein phosphatase which regulates Crz1p (a stress-response transcription factor), the other calcineurin subunit is encoded by CNA1 and/or CMP1 | Null mutant is viable, Li+ and Na+ sensitive | S000001673 | |||
| YOR316C | COT1 | + | Vacuolar transporter that mediates zinc transport into the vacuole; overexpression confers resistance to cobalt and rhodium | Null mutant is viable, increased sensitivity to cobalt | S000005843 | ||||||
| YMR078C | CTF18 | + | CHL12 | Subunit of a complex with Ctf8p that shares some subunits with Replication Factor C and is required for sister chromatid cohesion; may have overlapping functions with Rad24p in the DNA damage replication checkpoint | Null mutant is viable, increased mitotic recombination, slow growth, cold sensitivity | S000004683 | |||||
| YPR135W | CTF4 | + | CHL15|POB1 | Chromatin-associated protein, required for sister chromatid cohesion; interacts with DNA polymerase alpha (Pol1p) and may link DNA synthesis to sister chromatid cohesion | Null mutant is viable, increased chromosome loss rate, and mitotic recombination | S000006339 | |||||
| YML070W | DAK1 | + | Dihydroxyacetone kinase, required for detoxification of dihydroxyacetone (DHA); involved in stress adaptation | Null mutant is viable, sensitive to dihydroxyacetone | S000004535 | ||||||
| YGR092W | DBF2 | + | + | Ser/Thr kinase involved in transcription and stress response; functions as part of a network of genes in exit from mitosis; localization is cell cycle regulated; activated by Cdc15p during the exit from mitosis | Null mutant is viable, dbf1 dbf20 double mutant arrests in late telophase | S000003324 | |||||
| YPR111W | DBF20 | + | Ser/Thr kinase involved in late nuclear division, one of the mitotic exit network (MEN) proteins; necessary for the execution of cytokinesis | Null mutant is viable, dbf1 dbf20 double mutant arrests in late telophase | S000006315 | ||||||
| YDR052C | DBF4† | + | DNA52|LSD7 | Regulatory subunit of Cdc7p-Dbf4p kinase complex, required for Cdc7p kinase activity and initiation of DNA replication; phosphorylates the Mcm2-7 family of proteins; cell cycle regulated | Null mutant is inviable; temperature sensitive mutant arrests at G1/S | S000002459 | |||||
| YGL078C | DBP3 | + | Putative ATP-dependent RNA helicase of the DEAD-box family involved in ribosomal biogenesis | Null mutant is viable, severe growth defect, thermotolerant | S000003046 | ||||||
| YCL016C | DCC1 | + | Subunit of a complex with Ctf8p and Ctf18p that shares some components with Replication Factor C, required for sister chromatid cohesion and telomere length maintenance | Null mutant is viable, benomyl sensitive, increased chromosome loss rate | S000000521 | ||||||
| YIR030C | DCG1 | + | Protein of unknown function, expression is sensitive to nitrogen catabolite repression and regulated by Dal80p; contains transmembrane domain | Null mutant is viable, abnormal vaculoar morphology | S000001469 | ||||||
| YAL013W | DEP1 | + | + | + | + | FUN54 | Transcriptional modulator involved in regulation of structural phospholipid biosynthesis genes and metabolically unrelated genes, as well as maintenance of telomeres, mating efficiency, and sporulation | Null mutant is viable, increased silencing, increased cell size | S000000011 | ||
| YOR080W | DIA2 | + | YOR29-31 | Origin-binding F-box protein that forms an SCF ubiquitin ligase complex with Skp1p and Cdc53p; plays a role in DNA replication, involved in invasive and pseudohyphal growth | Null mutant is viable, sensitive to DNA damage, increased chromsome loss rate, anaphase delay | S000005606 | |||||
| YLL001W | DNM1 | + | Dynamin-related GTPase required for mitochondrial fission and the maintenance of mitochondrial morphology, assembles on the cytoplasmic face of mitochondrial tubules at sites at which division will occur; also participates in endocytosis | Null mutant is viable, reduced autophagy, increased lifespan; aberrant mitochondrial and peroxisomal morphology | S000003924 | ||||||
| YDR069C | DOA4 | + | DOS1|MUT4|NPI2|SSV7|UBP4 | Ubiquitin hydrolase, required for recycling ubiquitin from proteasome-bound ubiquitinated intermediates, acts at the late endosome/prevacuolar compartment to recover ubiquitin from ubiquitinated membrane proteins en route to the vacuole | Null mutant is viable, sensitive to many drugs | S000002476 | |||||
| YGL240W | DOC1 | + | APC10 | Processivity factor required for the ubiquitination activity of the anaphase promoting complex (APC), mediates the activity of the APC by contributing to substrate recognition; involved in cyclin proteolysis | Null mutant is viable, delayed metaphase to anaphase transition, increased chromsome loss rate | S000003209 | |||||
| YAL026C | DRS2 | + | FUN38|SWA3 | Integral membrane Ca(2+)-ATPase involved in aminophospholipid translocation; required to form a specific class of secretory vesicles that accumulate upon actin cytoskeleton disruption; mutation affects maturation of the 18S rRNA | Null mutant is viable, sensitive to cell wall-damaging agents, defective protein transport through the Golgi | S000000024 | |||||
| YDL101C | DUN1 | + | + | + | + | Cell-cycle checkpoint serine-threonine kinase required for DNA damage-induced transcription of certain target genes, phosphorylation of Rad55p and Sml1p, and transient G2/M arrest after DNA damage; also regulates postreplicative DNA repair | Null mutant is viable, defective in DNA damage response | S000002259 | |||
| YKL160W | ELF1 | + | Transcription elongation factor that contains a conserved zinc finger domain; implicated in the maintenance of proper chromatin structure in actively transcribed regions; deletion inhibits Brome mosaic virus (BMV) gene expression | Null mutant is viable, decreased stress resistance | S000001643 | ||||||
| YOR144C | ELG1 | + | RTT110 | Protein required for S phase progression and telomere homeostasis, forms an alternative replication factor C complex important for DNA replication and genome integrity; mutants are sensitive to DNA damage | Null mutant is viable, increased chromosome loss rate, defective DNA damage response | S000005670 | |||||
| YKL048C | ELM1 | + | LDB9 | Serine/threonine protein kinase that regulates cellular morphogenesis, septin behavior, and cytokinesis; required for the regulation of other kinases; forms part of the bud neck ring | Null mutant is viable, abnormal morphology, delayed G2/M transition | S000001531 | |||||
| YMR219W | ESC1 | + | Protein localized to the nuclear periphery, involved in telomeric silencing; interacts with PAD4-domain of Sir4p | Null mutant is viable, defective telomeric silencing | S000004832 | ||||||
| YDR363W | ESC2 | + | + | Protein involved in mating-type locus silencing, interacts with Sir2p; probably functions to recruit or stabilize Sir proteins | Null mutant is viable, defective telomeric silencing, increased chromosome loss rate | S000002771 | |||||
| YER015W | FAA2 | + | FAM1 | Long chain fatty acyl-CoA synthetase; accepts a wider range of acyl chain lengths than Faa1p, preferring C9:0-C13:0; involved in the activation of endogenous pools of fatty acids | Null mutant is viable | S000000817 | |||||
| YFR019W | FAB1 | + | SVL7 | 1-phosphatidylinositol-3-phosphate 5-kinase; vacuolar membrane kinase that generates phosphatidylinositol (3,5)P2, which is involved in vacuolar sorting and homeostasis | Null mutant is viable, temperature sensitive, defective vacuolar transport and morphology | S000001915 | |||||
| YJL157C | FAR1 | + | Cyclin-dependent kinase inhibitor that mediates cell cycle arrest in response to pheromone; also forms a complex with Cdc24p, Ste4p, and Ste18p that may specify the direction of polarized growth during mating; potential Cdc28p substrate | Null mutant is viable, fails to arrest in G1 upon pheromone exposure | S000003693 | ||||||
| YMR277W | FCP1† | + | Carboxy-terminal domain (CTD) phosphatase, essential for dephosphorylation of the repeated C-terminal domain of the RNA polymerase II large subunit (Rpo21p) | Null mutant is inviable | S000004890 | ||||||
| YMR058W | FET3 | + | Ferro-O2-oxidoreductase required for high-affinity iron uptake and involved in mediating resistance to copper ion toxicity, belongs to class of integral membrane multicopper oxidases | Null mutant is viable, defective for high affinity Fe(II) uptake | S000004662 | ||||||
| YLR342W | FKS1 | ++ | CND1|CWH53|ETG1|GSC1|PBR1 | Catalytic subunit of 1,3-beta-D-glucan synthase, functionally redundant with alternate catalytic subunit Gsc2p; binds to regulatory subunit Rho1p; involved in cell wall synthesis and maintenance; localizes to sites of cell wall remodeling | Null mutant is viable, growth defect, sensitive to FK506, cyclosporin A, and echinocandin | S000004334 | |||||
| YGR052W | FMP48 | + | + | Localized to the mitochondrion, interacts with TORC1 complex | Null mutant is viable, abnormal vaculoar morphology | S000003284 | |||||
| YBR021W | FUR4 | + | + | Uracil permease, localized to the plasma membrane; expression is tightly regulated by uracil levels and environmental cues | Null mutant is viable, abnormal bud morphology | S000000225 | |||||
| YMR307W | GAS1 | + | + | + | + | CWH52|GGP1 | Beta-1.3-glucanosyltransferase, required for cell wall assembly; localizes to the cell surface via a glycosylphosphatidylinositol (GPI) anchor | Null mutant is viable, cell wall defect, decreased growth rate, abnormal vacuolar morphology | S000004924 | ||
| YCL011C | GBP2 | + | RLF6 | Poly(A+) RNA-binding protein, involved in the export of mRNAs from the nucleus to the cytoplasm; similar to Hrb1p and Npl3p; also binds single-stranded telomeric repeat sequence in vitro | Null mutant is viable, thermotolerant | S000000517 | |||||
| YKR026C | GCN3 | + | AAS2 | Alpha subunit of the translation initiation factor eIF2B, the guanine-nucleotide exchange factor for eIF2; activity subsequently regulated by phosphorylated eIF2; first identified as a positive regulator of GCN4 expression | Null mutant is viable, fails to derepress amino acid-regulated genes | S000001734 | |||||
| YGR252W | GCN5 | + | ADA4|SWI9 | Histone acetyltransferase, acetylates lysine 14 on histone H3; catalytic subunit of the ADA and SAGA histone acetyltransferase complexes; founding member of the Gcn5p-related N-acetyltransferase superfamily | Null mutant is viable, grows poorly on minimal media, sensitive to many drugs | S000003484 | |||||
| YNL153C | GIM3 | + | + | + | + | PFD4 | Subunit of the heterohexameric cochaperone prefoldin complex which binds specifically to cytosolic chaperonin and transfers target proteins to it | Null mutant is viable, cold sensitive, benomyl sensitive, increased chromsome loss rate | S000005097 | ||
| YEL003W | GIM4 | + | PFD2 | Subunit of the heterohexameric cochaperone prefoldin complex which binds specifically to cytosolic chaperonin and transfers target proteins to it | Null mutant is viable, cold sensitive, benomyl sensitive, increased chromsome loss rate | S000000729 | |||||
| YML094W | GIM5 | + | PFD5 | Subunit of the heterohexameric cochaperone prefoldin complex which binds specifically to cytosolic chaperonin and transfers target proteins to it | Null mutant is viable, cold sensitive, benomyl sensitive, increased chromsome loss rate | S000004559 | |||||
| YER133W | GLC7† | + | + | CID1|DIS2|DIS2S1|PP1 | Catalytic subunit of type 1 serine/threonine protein phosphatase, involved in many processes including glycogen metabolism, sporulation, and mitosis; interacts with multiple regulatory subunits; predominantly isolated with Sds22p | Null mutant is inviable, defective actin cytoskeleton and endocytosis, heat and cold sensitive, increased chromosome loss rate | S000000935 | ||||
| YDL035C | GPR1 | + | + | Plasma membrane G protein coupled receptor (GPCR) that interacts with the heterotrimeric G protein alpha subunit, Gpa2p, and with Plc1p; sensor that integrates nutritional signals with the modulation of cell fate via PKA and cAMP synthesis | Null mutant is viable, decreased cell size, increased lifespan | S000002193 | |||||
| YLR258W | GSY2 | + | Glycogen synthase, similar to Gsy1p; expression induced by glucose limitation, nitrogen starvation, heat shock, and stationary phase; activity regulated by cAMP-dependent, Snf1p and Pho85p kinases as well as by the Gac1p-Glc7p phosphatase | Null mutant is viable, gsy1 gsy2 double mutant defective in glycogen deposition | S000004248 | ||||||
| YOR070C | GYP1 | + | YOR29-21 | Cis-golgi GTPase-activating protein (GAP) for the Rab family members Ypt1p (in vivo) and for Ypt1p, Sec4p, Ypt7p, and Ypt51p (in vitro); involved in vesicle docking and fusion | Null mutant is viable, sensitive to many drugs | S000005596 | |||||
| YBR009C | HHF1 | + | One of two identical histone H4 proteins (see also HHF2); core histone required for chromatin assembly and chromosome function; contributes to telomeric silencing; N-terminal domain involved in maintaining genomic integrity | Null mutant is viable, increased chromosome loss rate, decreased telomere length | S000000213 | ||||||
| YJR075W | HOC1 | + | + | + | + | Alpha-1,6-mannosyltransferase involved in cell wall mannan biosynthesis; subunit of a Golgi-localized complex that also contains Anp1p, Mnn9p, Mnn11p, and Mnn10p; identified as a suppressor of a cell lysis sensitive pkc1-371 allele | Null mutant is viable, sensitive to many drugs | S000003836 | |||
| YLR113W | HOG1 | + | + | + | + | SSK3 | Mitogen-activated protein kinase involved in osmoregulation via three independent osmosensors; mediates the recruitment and activation of RNA Pol II at Hot1p-dependent promoters; localization regulated by Ptp2p and Ptp3p | Null mutant is viable, unable to grow in high osmolarity media | S000004103 | ||
| YPL204W | HRR25† | + | Protein kinase involved in regulating diverse events including vesicular trafficking, gene expression, DNA repair, and chromosome segregation; binds the CTD of RNA pol II; homolog of mammalian casein kinase 1delta (CK1delta) | Null mutant is viable, slow growth, sensitive to DNA damage | S000006125 | ||||||
| YKL101W | HSL1 | + | NIK1 | Nim1p-related protein kinase that regulates the morphogenesis and septin checkpoints; associates with the assembled septin filament; required along with Hsl7p for bud neck recruitment, phosphorylation, and degradation of Swe1p | Null mutant is viable, abnormal bud morphology, G2/M delay | S000001584 | |||||
| YLL026W | HSP104 | + | Heat shock protein that cooperates with Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously denatured, aggregated proteins; responsive to stresses including: heat, ethanol, and sodium arsenite; involved in [PSI+] propagation | Null mutant is viable, defective in induced thermotolerance | S000003949 | ||||||
| YPL240C | HSP82 | + | HSP83|HSP90 | Cytoplasmic chaperone (Hsp90 family) required for pheromone signaling and negative regulation of Hsf1p; docks with the mitochondrial import receptor Tom70p for preprotein delivery; interacts with co-chaperones Cns1p, Cpr6p, Cpr7p, and Sti1p | Null mutant is viable, temperature sensitive | S000006161 | |||||
| YOL012C | HTZ1 | + | H2A.F/Z|H2AZ|HTA3 | Histone variant H2AZ, exchanged for histone H2A in nucleosomes by the SWR1 complex; involved in transcriptional regulation through prevention of the spread of silent heterochromatin | Null mutant is viable, loss of silencing at telomeres, sensitive to many drugs | S000005372 | |||||
| YGL253W | HXK2 | + | HEX1|HKB|SCI2 | Hexokinase isoenzyme 2 that catalyzes phosphorylation of glucose in the cytosol; predominant hexokinase during growth on glucose; functions in the nucleus to repress expression of HXK1 and GLK1 and to induce expression of its own gene | Null mutant is viable, fails to show glucose repression | S000003222 | |||||
| YHR094C | HXT1 | + | HOR4 | Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting | Null mutant is viable, decreased transport of glucose | S000001136 | |||||
| YOR136W | IDH2 | + | Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase, which catalyzes the oxidation of isocitrate to alpha-ketoglutarate in the TCA cycle | Null mutant is viable, G1 delay, increased thermotolerance | S000005662 | ||||||
| YLR384C | IKI3 | + | ELP1|TOT1 | Subunit of RNA polymerase II elongator histone acetyltransferase complex, involved in maintaining its structural integrity; negatively regulates exocytosis independent of transcription, homolog of human familial dysautonomia (FD) protein | Null mutant is viable, insensitive tokiller toxin, G1 delay | S000004376 | |||||
| YJR016C | ILV3† | + | Dihydroxyacid dehydratase, catalyzes third step in the common pathway leading to biosynthesis of branched-chain amino acids | Null mutant is viable, isoleucine and valine auxotroph | S000003777 | ||||||
| YPL209C | IPL1† | + | PAC15 | Aurora kinase involved in regulating kinetochore-microtubule attachments, associates with Sli15p, which stimulates Ipl1p kinase activity and promotes its association with the mitotic spindle, potential Cdc28p substrate | Null mutant is inviable, temperature sensitive mutant has incrwased chromosome loss rate | S000006130 | |||||
| YHR079C | IRE1 | ++ | + | + | ERN1 | Serine-threonine kinase and endoribonuclease; transmembrane protein that initiates the unfolded protein response signal by regulating synthesis of Hac1p through HAC1 mRNA splicing | Null mutant is viable, sensitive to ER stress ER | S000001121 | |||
| YPR106W | ISR1 | + | Predicted protein kinase, overexpression causes sensitivity to staurosporine, which is a potent inhibitor of protein kinase C | Null mutant is viable, exacerbates phenotype of a temperature-sensitive allele of PKC1 | S000006310 | ||||||
| YER110C | KAP123 | + | YRB4 | Karyopherin beta, mediates nuclear import of ribosomal proteins prior to assembly into ribosomes and import of histones H3 and H4; localizes to the nuclear pore, nucleus, and cytoplasm; exhibits genetic interactions with RAI1 | Null mutant is viable, defective nuclear import | S000000912 | |||||
| YPR141C | KAR3 | + | OSR11 | Minus-end-directed microtubule motor that functions in mitosis and meiosis, localizes to the spindle pole body and localization is dependent on functional Cik1p, required for nuclear fusion during mating; potential Cdc28p substrate | Null mutant is viable, defective in nuclear fusion | S000006345 | |||||
| YGL173C | KEM1 | + | DST2|RAR5|SEP1|SKI1|XRN1 | Evolutionarily-conserved 5--3- exonuclease component of cytoplasmic processing (P) bodies involved in mRNA decay; plays a role in microtubule-mediated processes, filamentous growth, ribosomal RNA maturation, and telomere maintenance | Null mutant is viable, abnormal vaculoar morphology, reduced lifespan, increased chromosome loss rate | S000003141 | |||||
| YIL125W | KGD1 | + | OGD1 | Component of the mitochondrial alpha-ketoglutarate dehydrogenase complex, which catalyzes a key step in the tricarboxylic acid (TCA) cycle, the oxidative decarboxylation of alpha-ketoglutarate to form succinyl-CoA | Null mutant is viable, respiration deficient | S000001387 | |||||
| YDL108W | KIN28† | + | + | Serine/threonine protein kinase, subunit of the transcription factor TFIIH; involved in transcription initiation at RNA polymerase II promoters | Null mutant is inviable | S000002266 | |||||
| YNL322C | KRE1 | + | + | ++ | + | Cell wall glycoprotein involved in beta-glucan assembly; serves as a K1 killer toxin membrane receptor | Null mutant is viable, reduced cell wall biosynthesis | S000005266 | |||
| YHR082C | KSP1 | + | + | Serine/threonine protein kinase; associates with TORC1; overproduction causes allele-specific suppression of the prp20-10 mutation | Null mutant is viable, increased autophagy | S000001124 | |||||
| YGR040W | KSS1† | + | Mitogen-activated protein kinase (MAPK) involved in signal transduction pathways that control filamentous growth and pheromone response | Null mutant is viable, decreased filamentous and invasive growth | S000003272 | ||||||
| YNL071W | LAT1 | + | ODP2|PDA2 | Dihydrolipoamide acetyltransferase component (E2) of pyruvate dehydrogenase complex, which catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA | Null mutant is viable, increased rate of petite formation | S000005015 | |||||
| YJL134W | LCB3 | + | + | LBP1|YSR2 | Long-chain base-1-phosphate phosphatase, regulates ceramide and long-chain base phosphates levels, involved in incorporation of exogenous long chain bases in sphingolipids | Null mutant is viable, reduced rate of exogenous long chain base incorporation into sphingolipids | S000003670 | ||||
| YPL055C | LGE1 | + | + | + | + | Protein of unknown function; null mutant forms abnormally large cells | Null mutant is viable, increased cell size | S000005976 | |||
| YNL006W | LST8† | + | Protein required for the transport of amino acid permease Gap1p from the Golgi to the cell surface; component of the TOR signaling pathway; associates with both Tor1p and Tor2p; contains a WD-repeat | Null mutant is inviable, reduction-of-function mutations cause sensitivity to rapamycin | S000004951 | ||||||
| YGL086W | MAD1 | + | Coiled-coil protein involved in the spindle-assembly checkpoint; phosphorylated by Mps1p upon checkpoint activation which leads to inhibition of the activity of the anaphase promoting complex; forms a complex with Mad2p | Null mutant is viable, benomyl sensitive, increased chromosome loss rate | S000003054 | ||||||
| YNL307C | MCK1 | + | + | YPK1 | Protein serine/threonine/tyrosine (dual-specificity) kinase involved in control of chromosome segregation and in regulating entry into meiosis; related to mammalian glycogen synthase kinases of the GSK-3 family | Null mutant is viable, benomyl sensitive, increased chromosome loss rate, thermosensitive | S000005251 | ||||
| YDR318W | MCM21 | + | CTF5 | Protein involved in minichromosome maintenance; component of the COMA complex (Ctf19p, Okp1p, Mcm21p, Ame1p) that bridges kinetochore subunits that are in contact with centromeric DNA and the subunits bound to microtubules | Null mutant is viable, benomyl sensitive, increased chromosome loss rate | S000002726 | |||||
| YKL085W | MDH1 | + | + | Mitochondrial malate dehydrogenase, catalyzes interconversion of malate and oxaloacetate; involved in the tricarboxylic acid (TCA) cycle | Null mutant is viable, severe respiratory growth defect | S000001568 | |||||
| YBR136W | MEC1† | + | ESR1|SAD3 | Genome integrity checkpoint protein and PI kinase superfamily member; signal transducer required for cell cycle arrest and transcriptional responses prompted by damaged or unreplicated DNA; monitors and participates in meiotic recombination | Null mutant is inviable, reduction-of-function mutant has elevated mitotic recombination and decreased telomere length | S000000340 | |||||
| YDL005C | MED2 | + | Subunit of the RNA polymerase II mediator complex; associates with core polymerase subunits to form the RNA polymerase II holoenzyme; essential for transcriptional regulation | Null mutant is viable, unable to grow on galactose | S000002163 | ||||||
| YPR167C | MET16 | + | 3--phosphoadenylsulfate reductase, reduces 3--phosphoadenylyl sulfate to adenosine-3-,5--bisphosphate and free sulfite using reduced thioredoxin as cosubstrate, involved in sulfate assimilation and methionine metabolism | Null mutant is viable, methionine auxotroph | S000006371 | ||||||
| YOR231W | MKK1 | + | + | + | SSP32 | Mitogen-activated kinase kinase involved in protein kinase C signaling pathway that controls cell integrity; upon activation by Bck1p phosphorylates downstream target, Slt2p; functionally redundant with Mkk2p | Null mutant is viable, cannot grow on glycerol, sensitive to nitrogen starvation, temperature sensitive cell wall defect | S000005757 | |||
| YLR320W | MMS22 | + | SLM2 | Protein involved in resistance to ionizing radiation; acts with Mms1p in a repair pathway that may be involved in resolving replication intermediates or preventing the damage caused by blocked replication forks | Null mutant is viable, sensitive DNA damage | S000004312 | |||||
| YDL028C | MPS1† | + | RPK1 | Dual-specificity kinase required for spindle pole body (SPB) duplication and spindle checkpoint function; substrates include SPB proteins Spc42p, Spc110p, and Spc98p, mitotic exit network protein Mob1p, and checkpoint protein Mad1p | Null mutant is inviable, accumulates cells with less than 1N DNA content, required for sporulation | S000002186 | |||||
| YCL061C | MRC1 | + | + | + | + | YCL060C | S-phase checkpoint protein found at replication forks, required for DNA replication; also required for Rad53p activation during DNA replication stress, where it forms a replication-pausing complex with Tof1p and is phosphorylated by Mec1p; protein involved in replication checkpoint | Null mutant is viable, sensitive to DNA damage, increased chromsome loss rate, anaphase delay | S000000566 | ||
| YKL009W | MRT4 | + | Protein involved in mRNA turnover and ribosome assembly, localizes to the nucleolus | Null mutant is viable, decreased mRNA decay rates | S000001492 | ||||||
| YDR335W | MSN5 | + | KAP142|STE21 | Karyopherin involved in nuclear import and export; shown to be responsible for nuclear import of replication protein A and for export of several proteins including Swi6p, Far1p, and Pho4p; cargo dissociation involves binding to RanGTP | Null mutant is viable, defective nuclear import | S000002743 | |||||
| YNR049C | MSO1 | + | Probable component of the secretory vesicle docking complex, acts at a late step in secretion; shows genetic and physical interactions with Sec1p and is enriched in microsomal membrane fractions; required for sporulation | Null mutant is viable, accumulates secretory vesicles in bud | S000005332 | ||||||
| YDL040C | NAT1 | + | AAA1 | Subunit of the N-terminal acetyltransferase NatA (Nat1p, Ard1p, Nat5p); N-terminally acetylates many proteins, which influences multiple processes such as the cell cycle, heat-shock resistance, mating, sporulation, and telomeric silencing | Null mutant is viable, reduced acetyltransferase activity, defective mating type locus silencing, fails to enter G0 | S000002198 | |||||
| YDR162C | NBP2 | + | + | Protein involved in the HOG (high osmolarity glycerol) pathway, negatively regulates Hog1p by recruitment of phosphatase Ptc1p the Pbs2p-Hog1p complex, found in the nucleus and cytoplasm, contains an SH3 domain that binds Pbs2p | Null mutant is viable | S000002569 | |||||
| YML120C | NDI1 | + | NADH:ubiquinone oxidoreductase, transfers electrons from NADH to ubiquinone in the respiratory chain but does not pump protons, in contrast to the higher eukaryotic multisubunit respiratory complex I; homolog of human AMID | Null mutant is viable, decreased apoptosis, replicative lifespan and respiratory growth | S000004589 | ||||||
| YKL171W | NNK1 | + | + | Nitrogen Network Kinase - Protein kinase | Null mutant is viable, overexpression causes hypersensitivity to rapamycin | S000001654 | |||||
| YNL175C | NOP13 | + | + | Protein of unknown function, localizes to the nucleolus and nucleoplasm; contains an RNA recognition motif (RRM) and has similarity to Nop12p, which is required for processing of pre-18S rRNA | Null mutant is viable | S000005119 | |||||
| YER002W | NOP16 | + | + | Constituent of 66S pre-ribosomal particles, involved in 60S ribosomal subunit biogenesis | Null mutant is viable, defect in 60S ribosomal subunit biogenesis | S000000804 | |||||
| YNL183C | NPR1 | + | + | + | Protein kinase that stabilizes several plasma membrane amino acid transporters by antagonizing their ubiquitin-mediated degradation | Null mutant is viable, defect in ammonia-sensitive amino acid permeases | S000005127 | ||||
| YDR001C | NTH1 | + | Neutral trehalase, degrades trehalose; required for thermotolerance and may mediate resistance to other cellular stresses; may be phosphorylated by Cdc28p | Null mutant is viable | S000002408 | ||||||
| YDR150W | NUM1 | + | + | + | + | PAC12 | Protein required for nuclear migration, localizes to the mother cell cortex and the bud tip; may mediate interactions of dynein and cytoplasmic microtubules with the cell cortex | Null mutant is viable, defective nuclear segregation | S000002557 | ||
| YKL068W | NUP100 | + | NSP100 | Subunit of the nuclear pore complex (NPC) that is localized to both sides of the pore; contains a repetitive GLFG motif that interacts with mRNA export factor Mex67p and with karyopherin Kap95p; homologous to Nup116p | Null mutant is viable, defective nucelar transport | S000001551 | |||||
| YGR078C | PAC10 | + | + | + | + | GIM2|PFD3|RKS2 | Part of the heteromeric co-chaperone GimC/prefoldin complex, which promotes efficient protein folding | Null mutant is viable, benomyl sensitive, cold sensitive | S000003310 | ||
| YBR279W | PAF1 | + | RNA polymerase II-associated protein, defines a large complex that is biochemically and functionally distinct from the Srb-Mediator form of Pol II holoenzyme and is required for full expression of a subset of cell cycle-regulated genes | Null mutant is viable, sensitive to many drugs | S000000483 | ||||||
| YJL128C | PBS2 | + | HOG4|SFS4|SSK4 | MAP kinase kinase that plays a pivotal role in the osmosensing signal-transduction pathway, activated under severe osmotic stress | Null mutant is viable, sensitive to osmotic stress | S000003664 | |||||
| YGL248W | PDE1 | + | Low-affinity cyclic AMP phosphodiesterase, controls glucose and intracellular acidification-induced cAMP signaling, target of the cAMP-protein kinase A (PKA) pathway; glucose induces transcription and inhibits translation | Null mutant is viable, accumulates cAMP, sensitive to rapamycin | S000003217 | ||||||
| YMR231W | PEP5 | + | END1|VAM1|VPL9|VPS11|VPT11 | Peripheral vacuolar membrane protein required for protein trafficking and vacuole biogenesis; forms complex with Pep3p that promotes vesicular docking/fusion reactions in conjunction with SNARE proteins, also interacts with Pep7p | Null mutant is viable, abnormal vaculoar morphology | S000004844 | |||||
| YOL147C | PEX11 | + | PMP24|PMP27 | Peroxisomal membrane protein required for peroxisome proliferation and medium-chain fatty acid oxidation, most abundant protein in the peroxisomal membrane, regulated by Adr1p and Pip2p-Oaf1p, promoter contains ORE and UAS1-like elements | Null mutant is viable, abnormal peroxisomal morphology | S000005507 | |||||
| YMR205C | PFK2 | + | + | Beta subunit of heterooctameric phosphofructokinase involved in glycolysis, indispensable for anaerobic growth, activated by fructose-2,6-bisphosphate and AMP, mutation inhibits glucose induction of cell cycle-related genes | Null mutant is viable, slow growth, decreased glucose utilization | S000004818 | |||||
| YOL136C | PFK27 | + | PFK-2 | 6-phosphofructo-2-kinase, has negligible fructose-2,6-bisphosphatase activity, inhibited by phosphoenolpyruvate and sn-glycerol 3-phosphate, expression induced by glucose and sucrose, transcriptional regulation involves protein kinase A | Null mutant is viable, abnormal vacuolar morphology, resistant to killer toxin | S000005496 | |||||
| YPL031C | PHO85† | + | LDB15 | Cyclin-dependent kinase, with ten cyclin partners; involved in environmental stress response; in phosphate-rich conditions, Pho85p-Pho80p complex phosphorylates Pho4p which in turn represses PHO5 | Null mutant is viable, abnormal morphology, increased chromosome loss rate, increased lifespan, sensitivity to many drugs | S000005952 | |||||
| YBL105C | PKC1† | + | + | CLY15|HPO2|STT1 | Protein serine/threonine kinase essential for cell wall remodeling during growth; localized to sites of polarized growth and the mother-daughter bud neck; homolog of the alpha, beta, and gamma isoforms of mammalian protein kinase C (PKC) | Null mutant is inviable, arrests growth with small buds due to cell wall lysis | S000000201 | ||||
| YGL167C | PMR1 | + | BSD1|LDB1 | High affinity Ca2+/Mn2+ P-type ATPase required for Ca2+ and Mn2+ transport into Golgi; involved in Ca2+ dependent protein sorting and processing; mutations in human homolog ATP2C1 cause acantholytic skin condition Hailey-Hailey disease | Null mutant is viable, inositol auxotroph, abnormal vacuolar morphology, decreased lifespan | S000003135 | |||||
| YJR043C | POL32 | + | + | Third subunit of DNA polymerase delta, involved in chromosomal DNA replication; required for error-prone DNA synthesis in the presence of DNA damage and processivity; interacts with Hys2p, PCNA (Pol30p), and Pol1p | Null mutant is viable, increased chromosome loss rate, cold sensitive, DNA damage sensitive | S000003804 | |||||
| YMR129W | POM152 | + | Nuclear pore membrane glycoprotein; may be involved in duplication of nuclear pores and nuclear pore complexes during S-phase; | Null mutant is viable, defective nuclear transport | S000004736 | ||||||
| YNR052C | POP2 | + | CAF1 | RNase of the DEDD superfamily, subunit of the Ccr4-Not complex that mediates 3- to 5- mRNA deadenylation | Null mutant is viable, increased chromosome loss rate, sensitive to many drugs | S000005335 | |||||
| YDL134C | PPH21 | + | + | + | PPH1 | Catalytic subunit of protein phosphatase 2A, functionally redundant with Pph22p; methylated at C terminus; forms alternate complexes with several regulatory subunits; involved in signal transduction and regulation of mitosis | Null mutant is viable, increased chromosome loss rate, abnormal lipid particles | S000002292 | |||
| YML016C | PPZ1 | + | + | Serine/threonine protein phosphatase Z, isoform of Ppz2p; involved in regulation of potassium transport, which affects osmotic stability, cell cycle progression, and halotolerance | Null mutant is viable, increased cell size, resistant to ionic stress | S000004478 | |||||
| YGR135W | PRE9 | + | 20S proteasome beta-type subunit; the only nonessential 20S subunit | Null mutant is viable | S000003367 | ||||||
| YDL214C | PRR2 | + | Protein kinase with a possible role in MAP kinase signaling in the pheromone response pathway | Null mutant is viable, increased filamentous growth, decreased pheromone response | S000002373 | ||||||
| YJL166W | QCR8 | + | COR5 | Subunit 8 of ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; oriented facing the intermembrane space; expression is regulated by Abf1p and Cpf1p | Null mutant is viable, respiration deficient | S000003702 | |||||
| YEL037C | RAD23 | + | Protein with ubiquitin-like N terminus, recognizes and binds damaged DNA (with Rad4p) during nucleotide excision repair; regulates Rad4p levels, subunit of Nuclear Excision Repair Factor 2 (NEF2); homolog of human HR23A and HR23B proteins | Null mutant is viable, increased chromosome loss rate, defective DNA damage response | S000000763 | ||||||
| YNL250W | RAD50 | + | Subunit of MRX complex, with Mre11p and Xrs2p, involved in processing double-strand DNA breaks in vegetative cells, initiation of meiotic DSBs, telomere maintenance, and nonhomologous end joining | Null mutant is viable, increased chromosome loss rate, defective DNA damage response | S000005194 | ||||||
| YML032C | RAD52 | + | Protein that stimulates strand exchange by facilitating Rad51p binding to single-stranded DNA; anneals complementary single-stranded DNA; involved in the repair of double-strand breaks in DNA during vegetative growth and meiosis | Null mutant is viable, deficient in homologous recombination | S000004494 | ||||||
| YPL153C | RAD53† | + | + | LSD1|MEC2|SPK1 | Protein kinase, required for cell-cycle arrest in response to DNA damage; activated by trans autophosphorylation when interacting with hyperphosphorylated Rad9p | Null mutant is inviable, point mutants sensitive to DNA-damaging agents | S000006074 | ||||
| YOR265W | RBL2 | + | Protein involved in microtubule morphogenesis, required for protection from excess free beta-tubulin; proposed to be involved the folding of beta-tubulin | Null mutant is viable, sensitive to many stresses | S000005791 | ||||||
| YDR195W | REF2 | + | RNA-binding protein involved in the cleavage step of mRNA 3--end formation prior to polyadenylation, and in snoRNA maturation; part of holo-CPF subcomplex APT, which associates with 3--ends of snoRNA- and mRNA-encoding genes | Null mutant is viable, sensitive to many stresses | S000002603 | ||||||
| YDR137W | RGP1 | + | Subunit of a Golgi membrane exchange factor (Ric1p-Rgp1p) that catalyzes nucleotide exchange on Ypt6p | Null mutant is viable, sensitive to many stresses | S000002544 | ||||||
| YLR039C | RIC1 | + | + | + | Protein involved in retrograde transport to the cis-Golgi network; forms heterodimer with Rgp1p that acts as a GTP exchange factor for Ypt6p; involved in transcription of rRNA and ribosomal protein genes | Null mutant is viable, sensitive to many stresses | S000004029 | ||||
| YOR119C | RIO1† | + | + | RRP10 | Essential serine kinase involved in cell cycle progression and processing of the 20S pre-rRNA into mature 18S rRNA | Null mutant is inviable, defective in ribosome biogenesis | S000005645 | ||||
| YNL207W | RIO2† | + | + | Essential serine kinase involved in the processing of the 20S pre-rRNA into mature 18S rRNA; has similarity to Rio1p | Null mutant is inviable, defective in ribosome biogenesis | S000005151 | |||||
| YIL066C | RNR3 | + | + | DIN1|RIR3 | Ribonucleotide-diphosphate reductase (RNR), large subunit; the RNR complex catalyzes the rate-limiting step in dNTP synthesis and is regulated by DNA replication and DNA damage checkpoint pathways via localization of the small subunits | Null mutant is viable, sensitive to dNTP pool depletion | S000001328 | ||||
| YLR371W | ROM2 | + | GDP/GTP exchange protein (GEP) for Rho1p and Rho2p; mutations are synthetically lethal with mutations in rom1, which also encodes a GEP | Null mutant is viable, temperature and cold sensitive, increased cell size, abnormal bud morphology, sensitive to benomyl | S000004363 | ||||||
| YBR229C | ROT2 | + | + | + | GLS2 | Glucosidase II catalytic subunit required for normal cell wall synthesis; mutations in rot2 suppress tor2 mutations, and are synthetically lethal with rot1 mutations | Null mutant is viable, abnormal vacuollar morphology, rot 1 rot2 double mutant is inviable | S000000433 | |||
| YNL248C | RPA49 | + | A49 | RNA polymerase I subunit A49 | Null mutant is viable, temperature and cold sensitive, abnormal nuclear morphology | S000005192 | |||||
| YNL067W | RPL9B | + | + | Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl9Ap and has similarity to E. coli L6 and rat L9 ribosomal proteins | Null mutant is viable | S000005011 | |||||
| YHR027C | RPN1† | + | HRD2|NAS1 | Non-ATPase base subunit of the 19S regulatory particle of the 26S proteasome; may participate in the recognition of several ligands of the proteasome; contains a leucine-rich repeat (LRR) domain, a site for protein?protein interactions | Null mutant is inviable; point mutants are sensitive to canavanine, global accumulation of ubiquitin-conjugates | S000001069 | |||||
| YHR200W | RPN10 | + | MCB1|SUN1 | Non-ATPase base subunit of the 19S regulatory particle (RP) of the 26S proteasome; N-terminus plays a role in maintaining the structural integrity of the RP; binds selectively to polyubiquitin chains; homolog of the mammalian S5a protein | Null mutant is viable, sensitive to amino acid analogs, increased ubiquitin conjugates | S000001243 | |||||
| YOR001W | RRP6 | + | Exonuclease component of the nuclear exosome; contributes to the quality-control system that retains and degrades aberrant mRNAs in the nucleus | Null mutant is viable, accumulates incompletely processed mRNA, rRNA, snRNA, snoRNA species | S000005527 | ||||||
| YLR221C | RSA3 | + | Protein with a likely role in ribosomal maturation, required for accumulation of wild-type levels of large (60S) ribosomal subunits; binds to the helicase Dbp6p in pre-60S ribosomal particles in the nucleolus | Null mutant is viable, required for 60S ribosomal subunit biogenesis | S000004211 | ||||||
| YLR357W | RSC2 | + | One of 15 subunits of the -Remodel the Structure of Chromatin- (RSC) complex; required for expression of mid-late sporulation-specific genes; involved in telomere maintenance | Null mutant is viable, increased chromosome loss rate, sensitive to many drugs | S000004349 | ||||||
| YDR388W | RVS167 | + | Actin-associated protein, subunit of a complex (Rvs161p-Rvs167p) involved in regulation of actin cytoskeleton, endocytosis, and viability following starvation or osmotic stress; homolog of mammalian amphiphysin | Null mutant is viable, reduced viability upon starvation | S000002796 | ||||||
| YDR159W | SAC3 | + | LEP1 | Nuclear pore-associated protein, forms a complex with Thp1p that is involved in transcription and in mRNA export from the nucleus | Null mutant is viable, increased cell size, sensitive to many drugs, cold and temperature sensitive | S000002566 | |||||
| YDR502C | SAM2 | + | ETH2 | S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) | Null mutant is viable, sam1 sam2 double mutant is AdoMet auxotroph | S000002910 | |||||
| YMR263W | SAP30 | + | + | + | + | Subunit of a histone deacetylase complex, along with Rpd3p and Sin3p, that is involved in silencing at telomeres, rDNA, and silent mating-type loci; involved in telomere maintenance | Null mutant is viable, decreased mating efficiency and telomere length | S000004876 | |||
| YOR329C | SCD5† | + | FTB1 | Protein required for normal cortical actin organization and endocytosis; multicopy suppressor of clathrin deficiency; acts as a targeting subunit for protein phosphatase type 1 | Null mutant is inviable, point mutant has abnormal morphology and decreased endocytosis | S000005856 | |||||
| YHR205W | SCH9† | + | + | KOM1 | Protein kinase that regulates signal transduction activity and G1 progression, controls cAPK activity, required for nitrogen activation of the FGM pathway, involved in life span regulation, homologous to mammalian Akt/PKB | Null mutant is viable, grows slowly, increased lifespan | S000001248 | ||||
| YLL041C | SDH2 | + | ACN17 | Iron-sulfur protein subunit of succinate dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation of succinate to the transfer of electrons to ubiquinone | Null mutant is viable, fails to grow on nonfermentable carbon sources, increased lifespan | S000003964 | |||||
| YKL193C | SDS22† | + | EGP1 | Conserved nuclear regulatory subunit of Glc7p type 1 protein serine-threonine phosphatase (PP1), functions positively with Glc7p to promote dephosphorylation of nuclear substrates required for chromosome transmission during mitosis | Null mutant is inviable, frameshift mutation causes heat and cold sensitivity, and increased chromosome loss rate | S000001676 | |||||
| YLR268W | SEC22 | + | SLY2|TSL26 | R-SNARE protein; assembles into SNARE complex with Bet1p, Bos1p and Sed5p; cycles between the ER and Golgi complex; involved in anterograde and retrograde transport between the ER and Golgi; synaptobrevin homolog | Null mutant is viable, cold and heat sensitive, abnormal ER morphology, cell wall defects | S000004258 | |||||
| YBR171W | SEC66 | + | HSS1|SEC71 | Non-essential subunit of Sec63 complex (Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP and Lhs1p forms a channel competent for SRP-dependent and post-translational SRP-independent protein targeting and import into the ER | Null mutant is viable, defective in filamentous growth, sensistive to many drugs | S000000375 | |||||
| pdr1Δpdr3Δ | Sensitized strain | + | + | + | + | + | |||||
| YOR184W | SER1 | + | ADE9 | 3-phosphoserine aminotransferase, catalyzes the formation of phosphoserine from 3-phosphohydroxypyruvate, required for serine and glycine biosynthesis; regulated by the general control of amino acid biosynthesis mediated by Gcn4p | Null mutant is viable, serine auxotroph | S000005710 | |||||
| YJL168C | SET2 | + | EZL1 | Histone methyltransferase with a role in transcriptional elongation, methylates a lysine residue of histone H3; associates with the C-terminal domain of Rpo21p; histone methylation activity is regulated by phosphorylation status of Rpo21p | Null mutant is viable, lacks methylation of histone H3 at Lys36 | S000003704 | |||||
| YLR403W | SFP1† | + | Transcription factor that controls expression of many ribosome biogenesis genes in response to nutrients and stress, regulates G2/M transitions during mitotic cell cycle and DNA-damage response, involved in cell size modulation | Null mutant is viable, grows slowly, defective in ribosome biogenesis gene expression | S000004395 | ||||||
| YLR058C | SHM2 | + | + | SHMT2 | Cytosolic serine hydroxymethyltransferase, involved in one-carbon metabolism | Null mutant is viable, shm2 srp40 ade3 triple mutnat is inviable | S000004048 | ||||
| YBL061C | SKT5 | + | CAL2|CHS4|CSD4 | Activator of Chs3p (chitin synthase III), recruits Chs3p to the bud neck via interaction with Bni4p; has similarity to Shc1p, which activates Chs3p during sporulation | Null mutant is viable, reduced cell wall chitin deposition | S000000157 | |||||
| YIL147C | SLN1† | + | + | YPD2 | Histidine kinase osmosensor that regulates a MAP kinase cascade; transmembrane protein with an intracellular kinase domain that signals to Ypd1p and Ssk1p, thereby forming a phosphorelay system similar to bacterial two-component regulators | Null mutant is inviable, constitutive activation of the HOG1 MAPK cascade | S000001409 | ||||
| YHR030C | SLT2 | + | ++ | + | + | + | BYC2|MPK1|SLK2 | Serine/threonine MAP kinase involved in regulating the maintenance of cell wall integrity and progression through the cell cycle; regulated by the PKC1-mediated signaling pathway | Null mutant is viable, temperature sensitive cell lysis defect | S000001072 | |
| YGR229C | SMI1 | + | + | + | + | KNR4 | Protein involved in the regulation of cell wall synthesis; proposed to be involved in coordinating cell cycle progression with cell wall integrity | Null mutant is viable, sensitive to osmotic stress, sensitive to many chemicals | S000003461 | ||
| YAL030W | SNC1 | + | Vesicle membrane receptor protein (v-SNARE) involved in the fusion between Golgi-derived secretory vesicles with the plasma membrane; proposed to be involved in endocytosis; member of the synaptobrevin/VAMP family of R-type v-SNARE proteins | Null mutant is viable; snc1 snc2 double mutant is deficient in bulk secretion | S000000028 | ||||||
| YDR477W | SNF1† | + | CAT1|CCR1|GLC2|HAF3|PAS14 | AMP-activated serine/threonine protein kinase found in a complex containing Snf4p and members of the Sip1p/Sip2p/Gal83p family; required for transcription of glucose-repressed genes, thermotolerance, sporulation, and peroxisome biogenesis | Null mutant is viable, fails to accumulate glycogen; sucrose nonfermenting | S000002885 | |||||
| YOR290C | SNF2 | + | GAM1|HAF1|SWI2|TYE3 | Catalytic subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; contains DNA-stimulated ATPase activity; functions interdependently in transcriptional activation with Snf5p and Snf6p | Null mutant is viable, defects in chromatin remodeling and transcriptional regulation, inability to switch mating type, increased sensitivity to DNA damage, defective cell wall | S000005816 | |||||
| YIL073C | SPO22 | + | + | Meiosis-specific protein with similarity to phospholipase A2, involved in completion of nuclear divisions during meiosis; induced early in meiosis | Null mutant is viable, defective spore production | S000001335 | |||||
| YDR392W | SPT3 | + | Subunit of the SAGA and SAGA-like transcriptional regulatory complexes, interacts with Spt15p to activate transcription of some RNA polymerase II-dependent genes, also functions to inhibit transcription at some promoters | Null mutant is viable, defective in mating and sporulation, Ty transcription | S000002800 | ||||||
| YNL224C | SQS1 | + | + | SQuelch of Splicing suppression | Null mutant is viable, facilitates 18S rRNA maturation | S000005168 | |||||
| YKR091W | SRL3 | + | Cytoplasmic protein that, when overexpressed, suppresses the lethality of a rad53 null mutation; potential Cdc28p substrate | Null mutant is viable | S000001799 | ||||||
| YJL092W | SRS2 | + | RADH|RADH1|HPR5 | DNA helicase and DNA-dependent ATPase involved in DNA repair, required for proper timing of commitment to meiotic recombination and the transition from Meiosis I to Meiosis II; potential Cdc28p substrate | Null mutant is viable, causes RAD52-dependent hyperrecombination | S000003628 | |||||
| YNL209W | SSB2 | + | YG103 | Cytoplasmic ATPase that is a ribosome-associated molecular chaperone, functions with J-protein partner Zuo1p; may be involved in the folding of newly-synthesized polypeptide chains; member of the HSP70 family; homolog of SSB1 | Null mutant is viable, ssb1 ssb2 double mutant has growth defect, hypersensitivity to protein synthesis inhibitors | S000005153 | |||||
| YPL106C | SSE1 | + | LPG3|MSI3 | ATPase that is a component of the heat shock protein Hsp90 chaperone complex; binds unfolded proteins; member of the heat shock protein 70 (HSP70) family; localized to the cytoplasm | Null mutant is viable, slow growth, decreased glucose utilization | S000006027 | |||||
| YNL222W | SSU72† | + | Transcription/RNA-processing factor that associates with TFIIB and cleavage/polyadenylation factor Pta1p; exhibits phosphatase activity on serine-5 of the RNA polymerase II C-terminal domain; affects start site selection in vivo | Null mutant is inviable, increased chromosome loss rate | S000005166 | ||||||
| YMR125W | STO1 | + | CBC1|CBP80|GCR3|SUT1 | Large subunit of the nuclear mRNA cap-binding protein complex, interacts with Npl3p to carry nuclear poly(A)+ mRNA to cytoplasm; also involved in nuclear mRNA degradation and telomere maintenance; orthologous to mammalian CBP80 | Null mutant is viable, defective growth on fermentable carbon sources | S000004732 | |||||
| YDR297W | SUR2 | + | SYR2 | Sphinganine C4-hydroxylase, catalyses the conversion of sphinganine to phytosphingosine in sphingolipid biosyntheis | Null mutant is viable, altered sphingolipid levels | S000002705 | |||||
| YBR231C | SWC5 | + | AOR1 | Protein of unknown function, component of the Swr1p complex that incorporates Htz1p into chromatin | Null mutant is viable, increased lifespan, abnormal bud and vacuolar morphology | S000000435 | |||||
| YJL187C | SWE1 | + | ++ | WEE1 | Protein kinase that regulates the G2/M transition by inhibition of Cdc28p kinase activity; localizes to the nucleus and to the daughter side of the mother-bud neck; homolog of S. pombe Wee1p; potential Cdc28p substrate | Null mutant is viable, defective for morphogenesis checkpoint | S000003723 | ||||
| YER111C | SWI4 | + | ART1 | DNA binding component of the SBF complex (Swi4p-Swi6p), a transcriptional activator that in concert with MBF (Mbp1-Swi6p) regulates late G1-specific transcription of targets including cyclins and genes required for DNA synthesis and repair | Null mutant is viable, defect in G1/S transcription causes G1/S delay | S000000913 | |||||
| YDR334W | SWR1 | + | Swi2/Snf2-related ATPase, component of the SWR1 complex; required for the incorporation of Htz1p into chromatin | Null mutant is viable, increased lifespan and thermotolerance | S000002742 | ||||||
| YNL128W | TEP1 | + | + | + | + | Homolog of human tumor suppressor gene PTEN/MMAC1/TEP1 that has lipid phosphatase activity and is linked to the phosphatidylinositol signaling pathway; plays a role in normal sporulation | Null mutant is viable, abnormal cell wall morphology | S000005072 | |||
| YDL185W | TFP1 | + | CLS8|VMA1 | Vacuolar ATPase V1 domain subunit A containing the catalytic nucleotide binding sites; protein precursor undergoes self-catalyzed splicing to yield the extein Tfp1p and the intein Vde (PI-SceI), which is a site-specific endonuclease | Null mutant is viable, abnormal vacuolar and mitochondrial morphology, reduced lipid droplets | S000002344 | |||||
| YPR163C | TIF3 | + | RBL3|STM1 | Translation initiation factor eIF-4B, has RNA annealing activity; contains an RNA recognition motif and binds to single-stranded RNA | Null mutant is viable, abnormal bud morphology, increased cell size | S000006367 | |||||
| YOL006C | TOP1 | + | + | + | MAK1|MAK17 | Topoisomerase I, nuclear enzyme that relieves torsional strain in DNA by cleaving and re-sealing the phosphodiester backbone; relaxes both positively and negatively supercoiled DNA; functions in replication, transcription, and recombination | Null mutant is viable, increased chromosome loss rate | S000005366 | |||
| YJR066W | TOR1 | + | + | + | DRR1 | PIK-related protein kinase and rapamycin target; subunit of TORC1, a complex that controls growth in response to nutrients by regulating translation, transcription, ribosome biogenesis, nutrient transport and autophagy; involved in meiosis | Null mutant is viable, grows slowly, increased lifespan, thermotolerance and oxidative stress resistance, increased sensitivity to rapamycin, tor1 tor2 double mutant is inviable | S000003827 | |||
| YKL203C | TOR2† | + | + | DRR2 | PIK-related protein kinase and rapamycin target; subunit of TORC1, a complex that regulates growth in response to nutrients and TORC2, a complex that regulates cell-cycle dependent polarization of the actin cytoskeleton; involved in meiosis | Null mutant is inviable, defective actin cytoskeleton and endocytosis, heat and cold sensitive, increased chromosome loss rate | S000001686 | ||||
| YJL164C | TPK1 | + | PKA1|SRA3 | Subunit of cytoplasmic cAMP-dependent protein kinase, which contains redundant catalytic subunits Tpk1p, Tpk2p, and Tpk3p and regulatory subunit Bcy1p; promotes vegetative growth in response to nutrients; inhibits filamentous growth | Null mutant is viable, sensitive to desiccation, multicopy suppressor of ras mutant | S000003700 | |||||
| YJL129C | TRK1 | + | Component of the Trk1p-Trk2p potassium transport system; 180 kDa high affinity potassium transporter | Null mutant is viable, requires added potassium | S000003665 | ||||||
| YML124C | TUB3 | + | Alpha-tubulin; associates with beta-tubulin (Tub2p) to form tubulin dimer, which polymerizes to form microtubules; expressed at lower level than Tub1p | Null mutant is viable, sensitive to benomyl, poor spore viability | S000004593 | ||||||
| YFR010W | UBP6 | + | Ubiquitin-specific protease situated in the base subcomplex of the 26S proteasome, releases free ubiquitin from branched polyubiquitin chains; deletion causes hypersensitivity to cycloheximide and other toxic compounds | Null mutant is viable, decreased lifespan, increased [Psi+] prion formation | S000001906 | ||||||
| YMR223W | UBP8 | + | Ubiquitin-specific protease that is a component of the SAGA (Spt-Ada-Gcn5-Acetyltransferase) acetylation complex; required for SAGA-mediated deubiquitination of histone H2B | Null mutant is viable, increased lifespan, sensitive to DNA damage and desiccation, defective in sporulation | S000004836 | ||||||
| YOR106W | VAM3 | + | PTH1 | Syntaxin-related protein required for vacuolar assembly; functions with Vam7p in vacuolar protein trafficking; member of the syntaxin family of proteins | Null mutant is viable, defective processing of vacuolar hydrolases | S000005632 | |||||
| YCL069W | VBA3 | + | Permease of basic amino acids in the vacuolar membrane | Null mutant is viable, abnormal vacuolar morphology | S000000574 | ||||||
| YGL227W | VID30 | + | GID1 | Protein involved in proteasome-dependent catabolite degradation of fructose-1,6-bisphosphatase (FBPase); shifts the balance of nitrogen metabolism toward the production of glutamate; localized to the nucleus and the cytoplasm | Null mutant is viable, vacuolar degradation of cytosolic proteins, sensitive to starvation | S000003196 | |||||
| YOR270C | VPH1 | + | Subunit of vacuolar-ATPase V0 domain, one of two isoforms (Vph1p and Stv1p); Vph1p is located in V-ATPase complexes of the vacuole while Stv1p is located in V-ATPase complexes of the Golgi and endosomes | Null mutant is viable, deficient in acidification of the vacuole | S000005796 | ||||||
| YKR001C | VPS1 | + | GRD1|LAM1|SPO15|VPL1|VPT26 | GTPase required for vacuolar protein sorting, functions in actin cytoskeleton organization via its interaction with Sla1p; required for late Golgi-retention of some proteins including Kex2p; involved in regulating peroxisome biogenesis | Null mutant is viable, sporulation defective, abnormal organization of intracellular membranes | S000001709 | |||||
| YJL154C | VPS35 | + | GRD9|VPT7 | Endosomal protein that is a subunit of the membrane-associated retromer complex essential for endosome-to-Golgi retrograde transport; forms a subcomplex with Vps26p and Vps29p that selects cargo proteins for endosome-to-Golgi retrieval | Null mutant is viable, defects in vacuolar sorting | S000003690 | |||||
| YOR069W | VPS5 | + | GRD2|PEP10|VPT5|YOR29-20 | Nexin-1 homolog required for localizing membrane proteins from a prevacuolar/late endosomal compartment back to the late Golgi apparatus; structural component of the retromer membrane coat complex; forms a retromer subcomplex with Vps17p | Null mutant is viable, defects in vacuolar sorting | S000005595 | |||||
| YNL197C | WHI3 | + | + | RNA binding protein that binds to and sequesters the G1 cyclin CLN3 mRNA; regulates cell fate and dose-dependently inhibits passage through Start by regulating the critical cell size requirement necessary for cell cycle progression | Null mutant is viable, defective in filamentous growth | S000005141 | |||||
| YOR083W | WHI5 | + | + | + | Protein that regulates the critical cell size required for passage through Start and commitment to cell division; may act upstream of SCB binding factor (SBF) and MCB binding factor (MBF); periodically expressed in G1 | Null mutant is viable, reduced cell size | S000005609 | ||||
| BY4741 | Wild type | + | + | + | + | + | + | ||||
| YER123W | YCK3 | + | + | CKI3 | Palmitoylated, vacuolar membrane-localized casein kinase I isoform; negatively regulates vacuole fusion during hypertonic stress via phosphorylation of the HOPS complex subunit, Vps41p; shares overlapping essential functions with Hrr25p | Null mutant is viable; abnormal vacuolar and endomembrane system morphology | S000000925 | ||||
| YHR087W | YHR087W | + | HGI1|RTC3 | Protein of unknown function involved in RNA metabolism; has structural similarity to SBDS, the human protein mutated in Shwachman-Diamond Syndrome (the yeast SBDS ortholog = SDO1); null mutation suppresses cdc13-1 temperature sensitivity; restriction of telomere capping | Null mutation is viable | S000001129 | |||||
| YKL067W | YNK1 | + | NDK1 | Nucleoside diphosphate kinase, catalyzes the transfer of gamma phosphates from nucleoside triphosphates, usually ATP, to nucleoside diphosphates by a mechanism that involves formation of an autophosphorylated enzyme intermediate | Null mutation is viable | S000001550 | |||||
| YFR003C | YPI1† | + | Inhibitor of the type I protein phosphatase Glc7p, which is involved in regulation of a variety of metabolic processes; overproduction causes decreased cellular content of glycogen | Null mutation is inviable, G2/M phase defect | S000001899 | ||||||
| YLR262C | YPT6 | + | ++ | + | GTPase, Ras-like GTP binding protein involved in the secretory pathway, required for fusion of endosome-derived vesicles with the late Golgi, maturation of the vacuolar carboxypeptidase Y; has similarity to the human GTPase, Rab6 | Null mutant is viable, increased lifespan, abnormal vacuolar morphology, impaired utilization of nitrogen sources, impaired endocytosis | S000004252 | ||||
| Total Strains Screened | 22 | 34 | 175 | 71 | 16 | 58 |
*taken from Saccharomyces Genome Database.
†screen performed on heterozygous diploid deletion strain.
Table 2. Library screen strains and hits that comprise the chemical genetic matrix.
| Library | Compounds | Unique cryptagen hits | Sentinel strains | Description |
|---|---|---|---|---|
| Spectrum03 | 2000 | 394 | 89 | Microsource Spectrum Collections consisting of 60% drug-like compounds, 25% natural products and 15% other bioactive molecules |
| Spectrum05 | 2000 | 54 | 87 | |
| Spectrum08 | 2000 | 31 | 7 | |
| LOPAC | 1267 | 67 | 20 | Library Of Pharmalogically Active Compounds |
| Maybridge | 1000 | 254 | 32 | Maybridge Hitskit 1000 selection of synthetic compounds representative of Maybridge Hitfinder library |
| Bioactive 1 | 678 | 421 | 69 | Selected from Maybridge Hitfinder 50,000 compound library for growth inhibtory activity in yeast |
| Bioactive 2 | 892 | 213 | 14 | Selected from Maybridge Hitfinder 50,000 compound library for growth inhibtory activity in yeast |
| Total | 5518 | 1434 | 242 |
Figure 3. Representations of CM dataset.
(a) Heatmap of growth inhibition for the 128 cryptagen compounds from the Microsource Spectrum library against 195 sentinel strains. (b) Heatmap of Bliss scores for all pairwise combinations of 128 cryptagen compounds. (c) Histogram of the Bliss score distribution.
Figure 4. Screenshots of ChemGRID web portal functions.
(a) Heatmap view of representative screens in ChemGRID (left) and accompanying data plot for a single screen (right). Mouseover functions allow interactive exploration of small molecule activities across different screens. (b) Overview page for the LOPAC screen component of the CGM. (c) The compound view page summarizes chemical properties, structure, and activity across all screens in the CGM, with automated internal links to similar compounds.
Methods
The methods detailed below are expanded versions of descriptions in our related work11.
Compound libraries used to generate the CGM
Compound libraries used were the LOPAC (Sigma), Maybridge Hitskit 1000 (Ryan Scientific) and the Spectrum Collection (MicroSource Discovery Systems Inc). We also screened two custom Yeast Bioactive Collections, termed Bioactive 1 and Bioactive 2, both of which were derived from screens of a 53,000 compound synthetic library (Ryan Scientific) in an S. cerevisiae cell proliferation assay at 10 μM (refs 16,17). The Bioactive 1 library contained 678 compounds that inhibited growth of a pdr1Δ̣pdr3Δ strain between 20% and 80%. Bioactive 2 contained 892 compounds that inhibited growth of a pdr1Δ̣pdr3Δ strain by at least 80%. Both of the Bioactive 1 and Bioactive 2 collections were selected to maximize chemical structural diversity compared to approved drugs listed by the World Health Organization (WHO). The approved drug list contains about 1,500 compounds that represent 50 Bemis-Murcko Fragments18. In contrast, the Bioactive 1 and Bioactive 2 collections contain 78 novel Bemis-Murcko scaffolds with uncharacterized modes of action. For all screens in the CGM, compound library stocks of 10 mM were diluted to 1 mM working stocks in DMSO in 96 well plates. Over the course of the study, the Spectrum Collection was re-purchased twice: the original library was called Spectrum03, and the two repurchased versions were called Spectrum05 and Spectrum08. As the composition of each release of this library differed somewhat, this non-redundancy resulted in 2,300 unique compounds in the combined Spectrum Collection used in this study. Library compositions and chemical structures are available at www.chemgrid.org.
Yeast strains used to generate the CGM and the CM
The 242 different S. cerevisiae deletion strains used as sentinels to generate the CGM (Table 1 (available online only)) were obtained from the Euroscarf deletion collection and are isogenic to BY4741, which has the genotype MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0. For the CM, an isogenic pdr1Δ̣pdr3Δ; strain (MT2481) was generated from BY4741 using PDR1::nat and PDR3::URA3 deletion cassettes.
Generation of the chemical genetic matrix (CGM)
All screens were conducted in synthetic complete (SC) medium with 2% glucose. Yeast deletion strains (Table 1 (available online only)) were seeded at 50,000 cells per well from fresh overnight cultures in a screening volume of 100 μl in 96 well plates. 2 μl of 1 mM compound stock was added to each well for a final compound concentration of 20 μM. Screens were conducted in technical duplicates using a Biomek FX liquid handling workstation with an integrated stacker carousel. 10 μM cycloheximide positive controls and DMSO solvent-only controls were set up in columns 1 and 12 of each 96 well plate. All plates were incubated at 30 °C without shaking for approximately 18 h or until solvent-treated control cultures were saturated. Cultures were then resuspended by shaking on the robotic platform before reading OD600 values on either Tecan Sunrise or Tecan M1000 plate readers19. Biological repeats were generated every 8 months over the course of the study for both the pdr1Δpdr3Δ strain and a wild-type S. cerevisiae strain (BY4741) to ensure consistency of compound activity and data reproducibility.
Statistical analysis of the CGM data
All screening data was subjected to the following analysis workflow:
LOWESS regression was used to correct spatial effects on growth across all plates for all screens performed with the LOPAC, Maybridge Hitskit 1000 and Spectrum Collection libraries, and for all but seven screens with the Bioactive 1 library. An empirically estimated sliding window of 1/3 was used and data normalization was based on the plate median. The LOWESS normalization method effectively removed variable plate edge effects within the non-active fraction of compounds, which sometimes occurred when plates were read at late time points.
For the seven Bioactive 1 screens with higher hit rates, and for all Bioactive 2 screens, the data was not LOWESS corrected but was instead normalized to DMSO controls. The Bioactive 1 library was selected for enrichment of compounds with moderate bioactivity against yeast, whereas the Bioactive 2 library was selected for compounds with strong bioactivity.
Median-normalization was applied to all plates and experiments.
Z-scores for growth inhibition were calculated based on the median and the interquartile range (IQR) by fitting a normal distribution with N(1,IQR) to the experimental data. We note that the IQR was intentionally chosen as a conservative estimate of variance to reduce the risk of false positives among the weakly active compounds in the screen. This approach slightly underestimates the significance of the Z-scores. In addition, Z-scores, per cent inhibition and normalized OD values were calculated for manual data validation.
Data points with high variation between replicates with growth inhibition up to 30% (>3 MAD) were flagged as inconsistent outliers and removed from further analysis.
Classification of compounds into ‘active’ and ‘enhanced growth’ was based on Z-score cut-offs. Compounds with Z-scores<−4 were classified as ‘active’, i.e., with reduced OD values compared to the negative solvent-only control. Positive Z-scores >4 were classified as ‘enhanced growth’, i.e., with OD values greater than the negative control.
All raw and processed CGM data are available online at http://chemgrid.org/cgm and from NCBI PubChem BioAssay (Data citation 1).
Selection of cryptagen compounds for the cryptagen matrix (CM)
From the CGM dataset, we identified 1,434 cryptagen compounds. These compounds inhibited growth of at least four and less than two-thirds of the yeast deletion strains. Each of the four chemical libraries yielded cryptagen compounds: the LOPAC at a hit rate of 5%, the Maybridge Hitskit at 27%, the Spectrum Collection at 18%, Bioactive 1 at 10% and Bioactive 2 at 23%. For the generation of the CM, cryptagens were selected from the Microsource Spectrum Collection based on activities against 58 different sentinel strains (Table 1 (available online only)). All cryptagens from the Spectrum Collection were clustered (average linkage hierarchical clustering) based on chemical structure and a structurally diverse set of 128 compounds with diverse chemical-genetic profiles was selected for the generation of the CM.
Generation of the CM
The 128 Spectrum compounds used for the CM were resupplied from MicroSource Discovery Systems Inc. (Groton, CT). Compounds were diluted to a stock concentration of 0.5 mM and arrayed in two 96 well plates. The 128×128 matrix was generated at 10 μM per compound in duplicate experiments (i.e, biological replicates) using a pdr1Δ̣pdr3Δ S. cerevisiae strain in 96 well plates. Yeast cultures were seeded at 50,000 cells per well in synthetic complete (SC) medium with 2% glucose at a screening volume of 100 μl. For the combination screens, 2 μl of 0.5 mM compound stock was added for both compounds for a final concentration of 10 μM per compound. The 128 compounds were also screened in combination with 2 μl DMSO to obtain growth inhibition data for each compound alone for compound interaction calculations. DMSO solvent-only controls were set up in columns 1 and 12 as well as four wells with a 10 μM cycloheximide positive control for complete growth inhibition in column 12. Plates were incubated for approximately 18 h or until solvent control cultures were saturated at 30 °C without shaking. Cultures were then resuspended by shaking on the robotic platform before reading OD600 values on either Tecan Sunrise or Tecan M1000 plate readers19.
Analysis of CM data
OD600 measurements for the CM were normalized to DMSO controls and data was averaged between the biological replicates. Bliss independence20 for compounds X and Y was calculated using the equation I xy =I x +I y −(I x ×I y ) where I x and I y correspond to growth inhibition in the presence of 10 μM compound X and Y, respectively. The expected growth inhibition for combination treatment with X and Y was compared to the actual growth inhibition observed in the CM to obtain the Bliss independence values. Bliss independence values within 90% density kernel fit represented additive effects. Based on the density kernel density estimation, Bliss independence values larger than 0.25 represented synergism and values below −0.18, antagonism. All raw and processed CM data are available online at http://chemgrid.org/cgm and from NCBI PubChem BioAssay (Data citation 2).
Note about the Bliss independence model
The Bliss independence model is based on testing two compounds and the pairwise combination at single concentrations20. The Bliss model does not account for possible non-linear concentration effects of either drug, which would instead require assessment over a two dimensional dose-response surface to determine Loewe additivity21, also sometimes calculated as the fractional inhibitory concentration index or FICI6. The Loewe additivity model requires extensive single and combination drug inhibition measurements that are not practical for large-scale surveys of drug combinations. Therefore, our initial estimates for synergism in the CM relied on the Bliss independence model, as calculated from single concentrations for each drug and the pairwise combination, using the equation I xy =I x +I y −(I x I y )6. The Bliss independence model for growth inhibition is equivalent to the multiplicative fitness model used to quantify genetic interactions. If two genes A and B do not interact, the expected fitness F of the double deletion strain is F ΔAΔB =F ΔA ×F ΔB where F ΔA and F ΔB represent the fitness defects of the two single deletion strains. Replacing fitness F with growth inhibition (F=1−I) in the genetic model yields: 1−I ΔAΔB =(1−I ΔA )×(1−I ΔB ) which can be simplified to 1−I ΔAΔB =1−I ΔB −I ΔA +I ΔA ×I ΔB . Subtracting ‘1’ and multiplication with (–1) yields the Bliss independence formula for growth inhibition: I ΔAΔB =I ΔA +I ΔB −I ΔA ×I ΔB . For example, if two single deletion strains, ΔA and ΔB, have fitness values of 0.6 and 0.8, compared to a wild type fitness of 1.0, the expected fitness of the double deletion strain ΔAΔB is FΔAΔB=0.6 * 0.8=0.48, corresponding to a growth inhibition of 52%. Similarly, if two small molecules A and B inhibit growth by 40 and 20%, their expected combined growth inhibition based on the Bliss independence model is IAB=0.4+0.2−(0.5×0.2)=0.6–0.08=0.52. Thus, if compounds A and B do not interact (i.e., have an additive effect), we expect a growth inhibition of 52% for this compound combination. Two genes are said to display a negative genetic interaction when the observed fitness of the double deletion strain is lower than expected. Similarly, synergy between two small molecules is observed when growth inhibition in response to the combination treatment is higher than expected.
Code availability
The data analysis procedures for the CGM were implemented in R using the additional packages RMySQL, outliers, matlab, amap, RSvgDevice and RSvgTipsDevice. We developed ChemGRID (http://chemgrid.org/cgm) as a webportal for the upload, processing and visualisation of chemical-genetic screen data (Fig. 4a,b) using PHP, PEAR, Perl and MySQL. The cheminformatics functionalities to register structural information were implemented with Python and FROWNS, PerlMol and MolDB4 (ref. 22). All code for data analysis is available at https://github.com/jwildenhain/chemical-genetic-matrix.
Data Records
Data record 1—chemical genetic matrix (CGM)
The chemical genetic data set of five different compound libraries screened against different subsets of 242 different S. cerevisiae deletion strains. In total, 492,126 chemical-genetic interaction tests (i.e., 984,252 independent measurements since each test was performed in duplicate) between 5,518 compounds and 242 S. cerevisiae deletion strains are represented in the CGM dataset. All raw and processed CGM data can be downloaded and visualized using ChemGRID (http://chemgrid.org/cgm) and are available from NCBI PubChem BioAssay (Data citation 1).
Data record 2—cryptagen matrix (CM)
The raw data as well as the Bliss independence values for all 8,128 pairwise combinations of 128 cryptagen compounds are available from NCBI PubChem BioAssay (Data citation 2). Bliss independence value calculations are described in detail in the Method section above.
Technical Validation
In-plate controls
For the CGM and the CM, each screening plate contained DMSO solvent-only controls as well as cycloheximide positive controls to ensure that inoculum preparation, compound pipetting and screening conditions were not compromised. DMSO solvent-only controls were used for data normalization to account for plate-to-plate variation within each screen and for variation between different screens.
Technical and biological replicates
During generation of the CGM, each S. cerevisiae deletion strain was screened in a technical or biological replicate. In addition, we repeatedly screened the compound collections against the pdr1Δpdr3Δ strain and a wild-type S. cerevisiae strain (BY4741) to ensure data reproducibility (Fig. 5a,b).
Figure 5. Technical validation of CGM data.
(a) Technical replicates of S. cerevisiae wild type screen data. (b) Scatter plot of S. cerevisiae wild type versus rbl9Δ screen data. (c) Z-factor distributions for each individual compound library screened in the CGM.
Z-factor
For each library screen, Z-factors were calculated to ensure quality of the screening data (Fig. 5c).
Quality filter
Small molecules that did not reproduce between replicate screens were filtered out and not used for further analysis of the CGM data.
Biological replicates
Every compound combination was tested in biological replicate on different days.
Surface dose-response (checkerboard) assays
A set of 75 combinations from the CM was tested in 4×4 checkerboard assays. 40 of these combinations initially showed synergistic interactions and 35 did not. This checkerboard analysis confirmed 23 interactions as true positives and 26 as true negatives, with 9 false negatives and 17 false positives. Overall, these confirmatory assays resulted in a 65% confirmation rate of synergistic compound interactions from the CM dataset.
Usage Notes
Dataset applications
The linked CGM chemical-genetic interaction and CM chemical-chemical interaction datasets were designed to serve as resources for computational predictions of chemical synergism, and were used in our original study to predict synergistic antifungal combinations11. To our knowledge, the CM is the first unbiased large-scale benchmark dataset for chemical-chemical interactions. The CGM and CM datasets can also be used to derive chemical structure-activity relationships for anti-fungal drug discovery. Furthermore, the CGM data can be used to infer compound mode of action11,13,23,24.
ChemGRID functionality
The ChemGRID webportal is designed to allow the upload, processing and visualisation of chemical-genetic screen data (Fig. 4a,b). All of the raw and processed CGM data is available on ChemGRID and the dataset for each individual chemical library can be browsed, visualized and downloaded. Screens against different S. cerevisiae deletion strains with the same compound collection can be compared and a mouse-over functionality allows for interactive data exploration, including chemical structures and properties. Scatterplots can be generated to compare data from different screens, for example between a wild type and a specific deletion strain. Compounds can also be searched by name and chemical structure. Data for single compounds across different screens can be retrieved and viewed on a summary page together with chemical properties, structures, and links to similar compounds (Fig. 4c). ChemGRID also provides linkouts to other webportals for drug target and compound activity information, including Drugbank25, PubChem26, MolClass15 and BioGRID5.
Additional Information
How to cite this article: Wildenhain, J. et al. Systematic chemical-genetic and chemical-chemical interaction datasets for prediction of compound synergism. Sci. Data 3:160095 doi: 10.1038/sdata.2016.95 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank S. Giguere, G. Giaever and L. Fischer for discussions. This work was supported by Canada Research Chairs in Molecular Studies of Antibiotics (G.D.W.) and in Systems and Synthetic Biology (M.T.), a Howard Hughes Medical Institute International Research Scholar Award (M.T.), a Royal Society Wolfson Research Merit Award (M.T.), a Scottish Universities Life Sciences Alliance Research Chair (M.T.), and by grants from the Canadian Institutes for Health Research (MOP 119572 to G.D.W. and M.T.), the European Research Council (SCG-233457 to M.T.), the Wellcome Trust (085178 to M.T.), the National Institutes of Health (R01RR024031 to M.T.), an award from the Ministère de l’enseignement supérieur, de la recherche, de la science et de la technologie du Québec through Génome Québec (M.T.), and the US Office of the Assistant Secretary of Defence for Health Affairs through the Breast Cancer Research Program (DAMD17-03-1-0471 to D.B.).
Footnotes
The authors declare no competing financial interests.
Data Citations
References
- Feala J. D. et al. Systems approaches and algorithms for discovery of combinatorial therapies. Wiley Interdiscip Rev Syst Biol Med 2, 181–193 (2010). [DOI] [PubMed] [Google Scholar]
- Sharom J. R., Bellows D. S. & Tyers M. From large networks to small molecules. Curr Opin Chem Biol 8, 81–90 (2004). [DOI] [PubMed] [Google Scholar]
- Costanzo M. et al. The genetic landscape of a cell. Science 327, 425–431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R. St Onge R. P., Hartman J. L., Giaever G. & Roth F. P. Defining genetic interaction. Proc Natl Acad Sci USA 105, 3461–3466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A. et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res 43, D470–D478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco W. R., Bravo G. & Parsons J. C. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev 47, 331–385 (1995). [PubMed] [Google Scholar]
- Fitzgerald J. B., Schoeberl B., Nielsen U. B. & Sorger P. K. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2, 458–466 (2006). [DOI] [PubMed] [Google Scholar]
- Kitano H. A robustness-based approach to systems-oriented drug design. Nat Rev Drug Discov 6, 202–210 (2007). [DOI] [PubMed] [Google Scholar]
- Roemer T. & Boone C. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol 9, 222–231 (2013). [DOI] [PubMed] [Google Scholar]
- Wassermann A. M. et al. Dark chemical matter as a promising starting point for drug lead discovery. Nat Chem Biol 11, 958–966 (2015). [DOI] [PubMed] [Google Scholar]
- Wildenhain J. et al. Prediction of synergism from chemical-genetic interactions by machine learning. Cell Systems 1, 383–395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M. et al. Systematic exploration of synergistic drug pairs. Mol Syst Biol 7, 544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M. et al. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol Syst Biol 7, 499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M. et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol 32, 1213–1222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenhain J., Fitzgerald N. & Tyers M. MolClass: a web portal to interrogate diverse small molecule screen datasets with different computational models. Bioinformatics 28, 2200–2201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki H. et al. Combined zebrafish-yeast chemical-genetic screens reveal gene-copper-nutrition interactions that modulate melanocyte pigmentation. Dis Model Mech 3, 639–651 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. et al. ALDH2 mediates 5-nitrofuran activity in multiple species. Chem Biol 19, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis G. W. & Murcko M. A. The properties of known drugs. 1. Molecular frameworks. J Med Chem 39, 2887–2893 (1996). [DOI] [PubMed] [Google Scholar]
- Wong L. H. et al. A yeast chemical genetic screen identifies inhibitors of human telomerase. Chem Biol 20, 333–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss C. I. The toxicity of poisons applied jointly. Ann Appl Biol 26, 585–615 (1939). [Google Scholar]
- Loewe S. Die quantitativen Probleme der Pharmakologie. Ergebnisse der Physiologie 27, 47–187 (1928). [Google Scholar]
- Haider N. Functionality pattern matching as an efficient complementary structure/reaction search tool: an open-source approach. Molecules 15, 5079–5092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M. E. et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons A. B. et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol 22, 62–69 (2004). [DOI] [PubMed] [Google Scholar]
- Knox C. et al. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res 39, D1035–D1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. PubChem Substance and Compound databases. Nucleic Acids Res 44, D1202–D1213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.