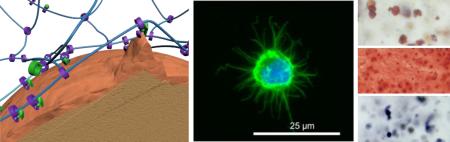
Abstract
Here we report the development of sliding hydrogel with mobile crosslinks and biochemical ligands as a 3D stem cell niche. The molecular mobility of this sliding hydrogel allows stem cells to reorganize the surrounding ligands and change their morphology in 3D. Without changing matrix stiffness, sliding hydrogels support efficient stem cell differentiation toward multiple lineages including adipogenesis, chondrogenesis and osteogenesis.
Keywords: Hydrogels, sliding, cell niche, cyclodextrin, mobile
Graphical abstract
Stem cells have been used as promising recourses in cell-based therapies and tissue regeneration due to their capacity for self-renewal and for differentiation. To guide stem cells to differentiate into desired lineages, efforts have been dedicated to identifying optimal soluble factors and insoluble niche cues, including cell-cell interactions and cell-niche interactions.[1, 2] To harness niche properties in order to regulate stem-cell differentiation, particularly in 3D, various artificial cell niches have been developed and utilized. For example, hydrogels have been widely employed as artificial matrices to enhance stem cell-based therapy and to serve as artificial cell niches for mechanistic studies. [3-5] Hydrogels are usually categorized according to the crosslinking mechanisms used to construct them: as physical hydrogels or as chemical hydrogels. Physical hydrogels are crosslinked via physical interactions such as calcium ion bonding, which is used to crosslink alginate hydrogels. [4, 6] Unlike chemical bonds, physical interactions are usually dynamic and flexible, allowing physical hydrogels to easily respond to morphological changes and matrix reorganization by cells that exert traction forces in response to niche cues, which in turn promote stem cell differentiation. [6, 7] However, this flexibility also inherently decreases the stability of the hydrogel. On the other hand, chemical hydrogels, which are crosslinked by chemical covalent bonds, are much more stable, but at the cost of with significantly reduced flexibility.[3, 5] As a result, cells are often restricted by the nano-sized meshes of chemical hydrogel networks and cannot change their morphology or reorganize the matrix, which may inhibit stem cells from differentiating and from performing critical functions. To overcome these limitations of chemical hydrogels, degradation has been used to enlarge the mesh size, giving cells the freedom to function and differentiate. [5] However, the extent and timing of degradation can be variable depending on cell type and enzymatic activity[8], and may simultaneously impact hydrogel diffusivity and loss of mechanical strength.
To address these challenges, here we report the development of a sliding hydrogel with mobile crosslinks and biochemical ligands that is suitable for use as a 3D stem-cell niche. A sliding hydrogel has a supramolecular architecture with topological characteristics that makes it highly mobile but stable (Figure 1a). [9] However, since the synthesis and crosslinking procedures of previously reported sliding hydrogels are usually not cell-friendly [9, 10], sliding hydrogels have only been used as strengthening materials [11]. The goal of this study is to apply the concept of sliding hydrogels for constructing synthetic stem cell niche. Our sliding hydrogel is chemically crosslinked, exhibiting stability comparable to that of chemical hydrogels, but it displays molecular mobility due to its sliding/mobile crosslinks and biochemical ligands. The hydrogel is crosslinked under cell-friendly, physiological conditions, rendering it appropriate for cell encapsulation and culture. The molecular mobility of this sliding hydrogel allows stem cells to reorganize the surrounding ligands and change their morphology in 3D, supporting more efficient stem-cell differentiation toward multiple lineages. This sliding hydrogel thus constitutes a novel material platform as a stem-cell niche, and is a useful tool for elucidating the molecular mobility of niche cues for the regulation of stem-cell fate in 3D.
Figure 1. Engineering sliding hydrogels as stem-cell niches.
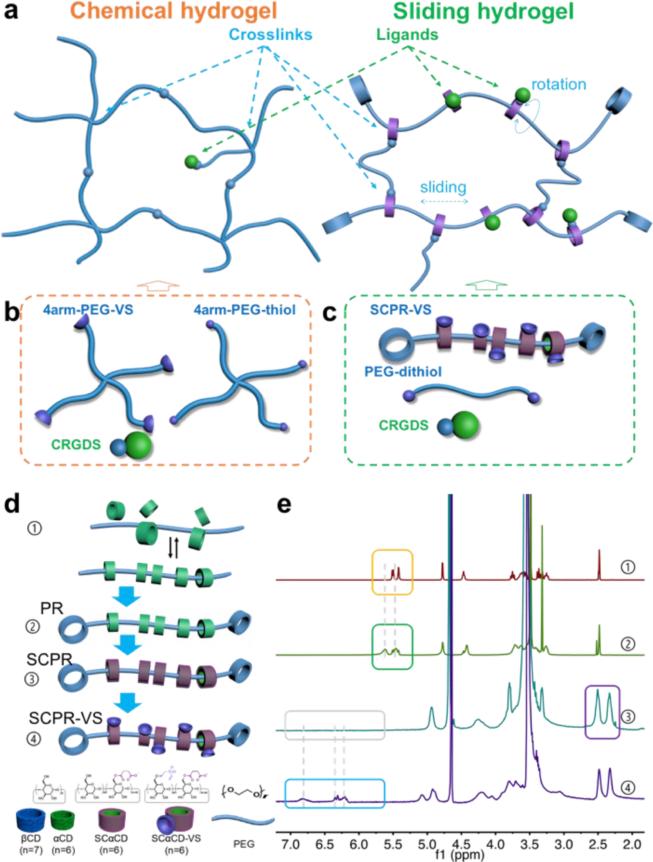
(a) Chemical hydrogels (left) are crosslinked by chemical covalent bonds, are stable but lacks flexibility. In contrast, sliding hydrogels (right) can be crosslinked from polyrotaxane, a supramolecular precursor with a topological architecture in which α-cyclodextrins (αCDs) are threaded on a linear polyethylene glycol (PEG) chain and trapped with capping agents with large spatial hindrance. αCDs are confined to the PEG chains but can slide and rotate, yielding crosslinks and ligands that are more mobile than those in conventional chemical hydrogels. However, the crosslinks attached to αCDs are stably confined in the hydrogel network, rendering sliding hydrogels more stable than physical hydrogels. (b) Polymer precursor to be crosslinked into control chemical hydrogels. 4arm-PEG-VS, 4-arm PEG with vinyl sulfone groups; 4arm-PEG-thiol, 4-arm PEG with thiol groups; CRGDS, a small peptide that promotes cell adhesion and various cell functions. (c) Polymer precursor to be crosslinked into sliding hydrogels. SCPR VS, succinic-polyrotaxane with vinyl sulfone groups; PEG-dithiol, dithiol-functionalized PEG. (d) Synthesis strategy for the sliding hydrogel precursor in (c). βCD, β-cyclodextrin; αCD, α-cyclodextrin; PR, polyrotaxane; SCαCD, succinic-α-cyclodextrin; SCPR, succinic-polyrotaxane; SCαCD-VS, succinic-α-cyclodextrin with vinyl sulfone groups; SCPR-VS, succinic-polyrotaxane with vinyl sulfone groups. (e) 1H NMR spectra of each step for synthesizing the sliding hydrogel precursor in (c); proton peaks on hydroxyl of 2-, 3- position on αCD (yellow box) shifted and broadened (green box) due to the inclusion of polyrotaxane; the peaks of protons on succinic ethylenes (purple box) indicates the successful succinic modification of α-cyclodextrins; peaks of protons on vinyl sulfone (from grey box to blue box) confirms successful incorporation of vinyl sulfone group.
To harness the mobility and stability of sliding hydrogels for applications as a 3D stem-cell niche, we first synthesized an aqueous-soluble supramolecular precursor for crosslinking and ligand incorporation under physiological conditions. We synthesized conventional polyrotaxane by mixing α-cyclodextrins (αCDs) with linear polyethylene glycol (PEG) in aqueous solution to form the inclusion complex (Figure 1d-1), followed by capping with β-cyclodextrins, which are larger than αCDs (Figure 1d-2). Polyrotaxane is insoluble in neutral aqueous solutions due to the crystal structure of the densely packed αCDs on the PEG chain[12, 13], making it difficult to fabricate hydrogels under physiological conditions. We therefore converted the αCDs of polyrotaxane into succinic-αCDs (Figure 1d-3). The resulting succinic-polyrotaxane was soluble in aqueous buffer because the succinic incorporation loosened the packing in the crystal structure of αCDs and allowed ionization in water. Finally, we obtained the sliding hydrogel precursor by introducing vinyl sulfone groups onto the succinic-polyrotaxane to allow crosslinking and ligand incorporation (SCPR-VS, Figure 1d-4). The synthesis and structure of the obtained sliding hydrogel precursor were all confirmed by 1H NMR spectra (Figure 1e).
Next, we crosslinked the supramolecular precursor SCPR-VS with dithiol-functionalized PEG and incorporated the thiol-containing biochemical ligand CRGDS (Cys-Arg-Gly-Asp-Ser), which is a widely used peptide that promotes cell adhesion and various cell functions.[14] We used 4-arm PEG with vinyl sulfone groups and with thiol groups to crosslink a conventional chemical hydrogel as a control (Figure 1b).
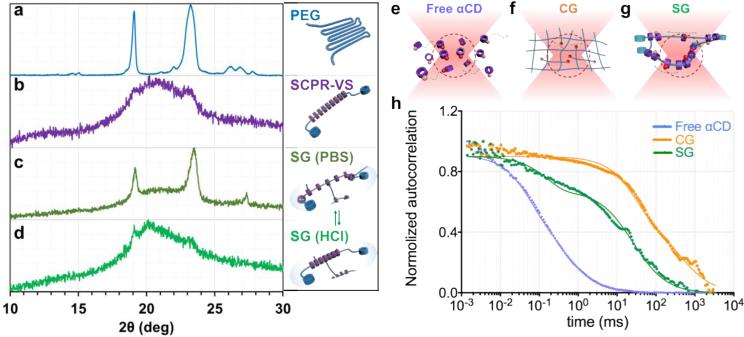
To assess the molecular mobility of the αCDs on the embedded PEG chains in our sliding hydrogel, we studied the static distribution and arrangement of αCDs using X-ray diffraction. We first examined the ability of αCDs to slide and change their distribution on the PEG chains. According to previous reports, the αCDs threaded on the PEG chains of polyrotaxane exhibit an oriented and densely packed channel-like crystal structure that yielded strong X-ray diffraction at 19.8°, which was assigned a (210) reflection for the hexagonal lattice [12, 15]. Consistently, our supramolecular precursor (SCPR-VS) presented an X-ray diffraction pattern with a broadened peak ranging from 19.9° to 23.3°, which covered the representative diffraction of polyrotaxane and PEG (Figure 2a,b). This pattern suggests that modified αCDs still pack closely in the sliding hydrogel, although not as neatly as free αCDs due to the spatial effect of the functional groups. When SCPR-VS was crosslinked with dithiol-functionalized PEG in phosphate-buffered saline to form a sliding hydrogel, only the diffraction of PEG was apparent (Figure 2c), suggesting that ionization in aqueous buffer disrupts the packing structure of succinic-αCDs, leading to their dispersal on the chains. However, when the sliding hydrogel was placed into an HCl solution (0.1 M) to deionize the succinic-αCDs, the diffraction of the channel-type crystal structure returned (Figure 2d), indicating that decreasing the pH deionized the succinic-αCDs and induced their re-packing. At the lower pH, the sliding hydrogel changed from clear to slightly opaque due to crystal formation (data not shown). Taken together, these observations of reversible changes in the distribution of αCDs on the PEG chains indicates that the αCDs can slide in the sliding hydrogel.
Figure 2. Sliding hydrogels (SG), but not chemical hydrogels (CG), demonstrate reversible ligand distribution and dynamic molecular ligand mobility, driven by sliding αCDs along the PEG backbone in 3D.
(a-d) X-ray diffraction (XRD) pattern confirms reversible distribution of αCDs in sliding hydrogel. a: Baseline pattern of PEG backbone without αCD; b: Crystal structure formation confirms packing of sliding CDs along PEG backbone; c. Sliding hydrogels in PBS; αCDs are ionized in crosslinked SG, reverting from packed pattern to more dispersed distribution, exposing structure of PEG backbone similar to a; d. Upon switching to HCL solution, αCDs in sliding hydrogels are deionized, reverting to packed state similar to pattern in b. (e-g) Scheme of movement of tetramethylrhodamine-labeled ligands (red spheres) toward and away from the focal volume (red dotted region) of confocal microscopy when they are tethered on (e) free αCDs, (f) chemical hydrogel (CG), and (g) sliding hydrogel (SG). (h) Normalized autocorrelation curve from fluorescence correlation spectroscopy showed fastest decay of free αCDs (blue) due to their free diffusion. The sliding hydrogel (SG; green) exhibited a faster decay of auto-correlation than the chemical hydrogel (CG; orange), indicating higher mobility, but more stable than freely diffusive αCDs.
Since X-ray diffraction only yields a snapshot of the αCD distribution, we assessed the dynamic mobility of these units in real time using fluorescence correlation spectroscopy (FCS). We used tetramethylrhodamine (TRITC)-labeled CRGDS as model ligand and incorporated it into free αCDs, chemical hydrogels, and sliding hydrogels. The real-time movement of the TRITC-labeled ligand in and out of the focal volume results in a fluctuation of the fluorescence intensity; higher mobility causes faster movement of the TRITC-labeled ligand and thus faster fluctuations in fluorescence intensity, which manifest as a faster decay of the auto-correlation in the FCS spectrum. Due to their free diffusion (Figure 2e), free αCDs presented the fastest decay of auto-correlation (Figure 2h). This diffusive mobility was also confirmed by fluorescence recovery after photobleaching (Figure S2), where recovery happened within minutes. Neither the chemical hydrogel nor the sliding hydrogel, which both lack freedom of diffusion across the hydrogel, displayed recovery of fluorescence after photobleaching (Figure S2), suggesting that covalent immobilization substantially enhanced the retention and stability of ligands in these hydrogels. In the chemical hydrogel, TRITC-labeled ligands were covalently immobilized and bonded to the hydrogel network, rendering them unable to diffuse or move (Figure 2f). In contrast, although TRITC-labeled ligands were immobilized on αCDs that were confined to the network in sliding hydrogels, these ligands retained enough mobility to slide along the chain (Figure 2g). This scenario was confirmed via FCS: the sliding hydrogel exhibited a faster decay of auto-correlation than the chemical hydrogel (Figure 2h), indicating higher mobility (albeit less than that of free αCDs).
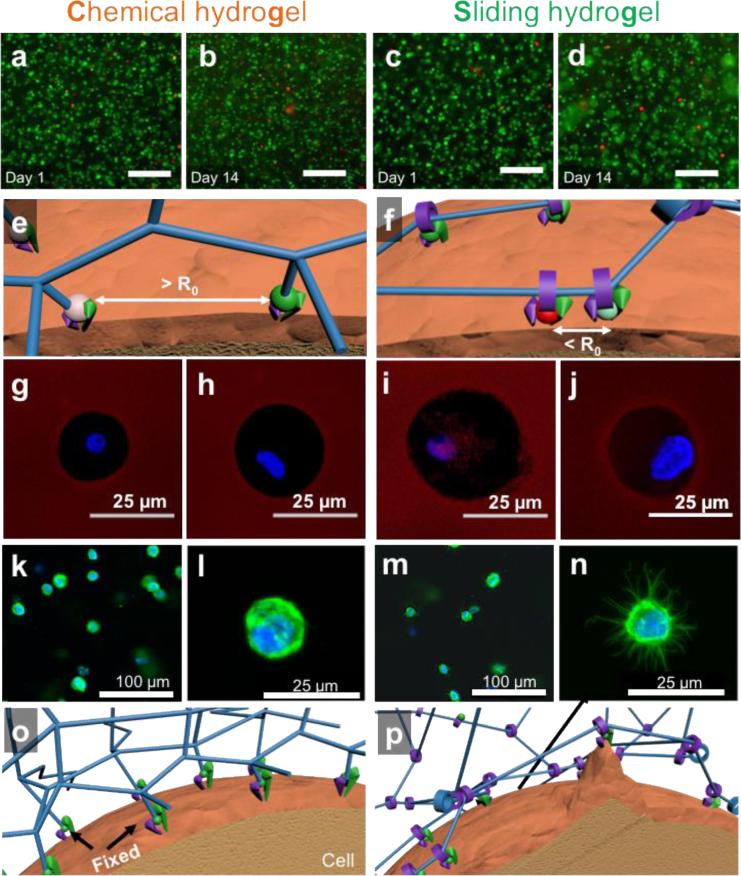
To assess the cellular response to molecular mobility of our sliding hydrogel, we studied the ability of human mesenchymal stem cells (hMSCs) to reorganize ligands and crosslinks in hydrogels in 3D. We have chosen hMSCs as a model cell type given their promise as an autologous cell source for tissue regeneration. Importantly, the sliding hydrogel developed here is crosslinked under physiological conditions, allowing the encapsulation of live cells in 3D. In this study, both chemical hydrogels and sliding hydrogels supported >95% cell survival after encapsulation and even after 14 days (Figure 3a-d), indicating that both hydrogels are cell friendly.
Figure 3. Sliding hydrogels, but not chemical hydrogels, facilitate cells to reorganize ligands and cytoskeleton in in 3D.
(a-d) Fluorescent microscopy images of encapsulated human mesenchymal stem cells 1 day and 14 days after encapsulation in chemical hydrogel or sliding hydrogel stained with calcium-AM (green; highlights live cells) and ethidium homodimer (red; highlights dead cells). The scale bar is 100 μm in panels (a) to (d). (e) Fluorescein isothiocyanate- (green ball) and tetramethylrhodamine- (red ball) labeled ligands are immobilized in chemical hydrogels with a distance greater than the Förster distance (R0). (f) Fluorescein isothiocyanate and tetramethylrhodamine-labeled ligands in sliding hydrogels cluster together by sliding along the hydrogel network, resulting in a distance less than R0. (g-j) Assessment of the ability of cells to reorganize ligands in the hydrogels via fluorescence resonance energy transfer, by taking confocal microscopy images of tetramethylrhodamine emission from chemical hydrogels (g) or sliding hydrogels (i) with cells encapsulated excited with a 488-nm laser. Cells treated with blebbistatin to inhibit myosin contraction were used as controls in chemical hydrogels (h) and sliding hydrogels (j). FRET signals were only observed in sliding hydrogels, suggesting molecular ligand mobility in sliding hydrogels but not in chemical hydrogels. Blebbistatin treatment abolished the ability of cells to pull on mobile ligands, with no changes in FRET signals (h, j). (k-n) Assessment of the ability of cells to change morphology in hydrogels, by taking confocal microscopy images of cells in chemical hydrogels (k-l) and sliding hydrogels (m-n) stained for F-actin (green) and the nucleus (blue). Higher-resolution images of cell morphology are shown in (l,n). (o,p) Schematic illustrations of the difference in molecular ligand mobility on cell morphology in hydrogels by reorganizing ligands and crosslinks in 3D.
Next we assessed the ability of cells to reorganize ligands in the hydrogels via fluorescence resonance energy transfer[16]. For these analyses, we labeled the cell adhesion ligand CRGDS with fluorescein isothiocyanate (FITC; green fluorescence) and with TRITC (red fluorescence), which served as donor and acceptor fluorophores, respectively. In chemical hydrogels, due to the static immobilization of ligands on hydrogel network, the distance of these ligands was far greater than the Förster distance (R0) (Figure 3e), prohibiting energy transfer from FITC to TRITC and minimizing red fluorescence across the cell area (Figure 3g). In contrast, in sliding hydrogels, due to the sliding mobility of the ligands along the PEG chains, the cells reorganized the ligands to be closer to each other (Figure 3f), decreasing the distance between the donor and acceptor fluorophores and transferring energy from FITC to TRITC, increasing the red fluorescence across the cell area (Figure 3i). To further confirm the molecular mobility of sliding hydrogels, cells in chemical hydrogels (Figure. 3h) or sliding hydrogels (Figure. 3j) were also treated with blebbistatin, a small molecule to inhibit myosin contraction, as additional controls. Our results showed blebbistatin treatment abolished the FRET signals observed in sliding hydrogels. These data indicate that hMSCs in sliding hydrogels are sense the flexibility of the hydrogel and exploit this molecular mobility to carry out particular functions, as the clustering of adhesion ligands is usually associated with behavior such as the exertion of traction forces. [4]
Since the exertion of traction forces is also associated with cell-morphology changes [5, 17], we next evaluated these changes in hMSCs cultured in hydrogels. We predicted that the sliding crosslinks in sliding hydrogels would render the hydrogel network amenable to cell-induced forces and morphology changes. Note that unlike degradable hydrogels, sliding hydrogels do not undergo cleavage or breakdown of the network. We observed that at the micron scale and larger, hMSCs cultured in sliding hydrogels did not spread out (as is commonly seen in degradable hydrogels [5, 18]), but instead displayed a spherical shape (Figure 3m). At the nano-scale, hMSCs induced nano-scale cavities or channels, allowing them to form protrusions (Figure 3n), likely due to their ability to exploit the molecular mobility of the sliding hydrogel to rearrange the ligands and crosslinks by sliding them along the network chains (Figure 3p). In contrast, hMSCs cultured in chemical hydrogels lacked protrusions (Figure 3k,l), consistent with a failure to reorganize the surrounding hydrogel network (Figure 3o). Thus, molecular mobility is an important hydrogel characteristic that enables cells to reorganize ligands and crosslinks in order to change their morphology.
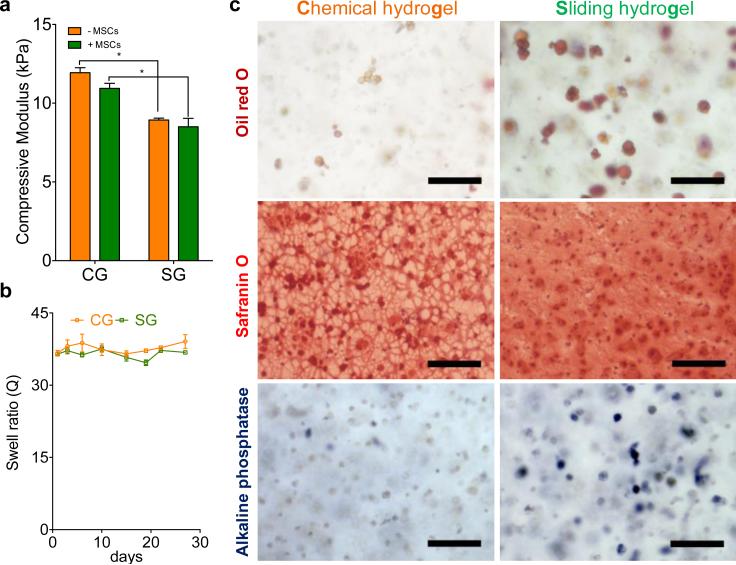
The morphologies displayed by hMSCs cultured in sliding hydrogels led us to expect the cells to undergo related downstream cellular functions, such as differentiation toward specific lineages. [5, 19] We therefore evaluated how hydrogel molecular mobility affected the differentiation of hMSCs toward three lineages: adipogenesis, chondrogenesis and osteogenesis. These three lineages have been reported to favor differing levels of niche stiffness and degradation.[3, 5, 20] To isolate the effect of mobility from effects due to mechanical stiffness and degradation, we fabricated sliding hydrogels and chemical hydrogels with comparable mechanical stiffness, both are around 10 kPa which has been reported to favor MSCs differentiation towards muscle lineage (Figure 4a), and minimum degradation, which was verified by stable swelling ratios over 30 days (Figure 4b). To further characterize the mechanical properties of chemical hydrogels and sliding hydrogels, we performed additional rheological measurements for both hydrogels with and without cells. Storage and loss moduli of chemical hydrogels and sliding hydrogels are close (Figure S3), and is within a range that has been reported to favor MSC differentiation towards tissues with intermediate stiffness such as muscle. To compare the diffusion of soluble factors in chemical hydrogels and sliding hydrogels, we performed FRAP measurements using FITC labeled bovine serine albumin (FITC-BSA) as a model molecule with molecular weight of 65 kDa, which is larger than most of the nutrients and growth factors. Our results confirmed that the diffusion of BSA in chemical hydrogels and sliding hydrogels is very comparable, with no significant difference (Figure S4). Inclusion of cells did not significantly change the diffusion (Figure S4). Thus, the differences of cell responses in chemical hydrogels and sliding hydrogels are not due to the differences in diffusion. Adipogenesis was previously reported to be favored by less spatial restriction [5] and rounded cell morphology [19]. We detected oil accumulation, a functional marker of adipogenesis, in hMSCs cultured in chemical hydrogels and in sliding hydrogels, with sliding hydrogels inducing more oil production than chemical hydrogels (Figure 4c, Figure S5). Chondrogenesis requires fewer morphological changes of hMSCs, which usually retain a spherical morphology [21]; both chemical hydrogels and sliding hydrogels allowed chondrogenesis, as indicated by high glycosaminoglycan deposition (Figure 4c). Note that the distribution of glycosaminoglycan was more even and interconnected in sliding hydrogels than in chemical hydrogels (Figure 4c). This distribution may be beneficial for the regeneration and function of cartilage tissues.[22] Unlike adipogenesis and chondrogenesis, osteogenesis requires more cell-niche interactions, reorganization of ligands, and morphology changes. As expected, there was minimal expression of alkaline phosphatase, an osteogenic biomarker, in chemical hydrogels (Figure 4c) due to the static and nano-size mesh of chemical hydrogels. In sliding hydrogels, however, hMSCs presented enhanced alkaline phosphatase expression (Figure 4c, Figure S5), suggesting that the molecular mobility possible in sliding hydrogels facilitates traction forces and downstream osteogenesis in response to the soluble induction factors. We also performed quantitative analyses of differentiation marker staining of oil droplets or ALP positive cells in chemical hydrogels (CG) versus sliding hydrogels (SG). Sliding hydrogels induced significantly higher level of differentiation than chemical hydrogels for both markers (**: p<0.01) (Figure S5). The observed trend was also consistent with gene expression by hMSCs (Figure S6).
Figure 4. Sliding hydrogels enhanced differentiation of human mesenchymal stem cells toward adipogenesis, chondrogenesis, and osteogenesis in 3D versus chemical hydrogels with similar stiffness and degradation.
(a) Comparable compressive moduli of chemical hydrogels (CGs) and sliding hydrogel (SGs), with (−MSCs) and without (−MSCs) hMSCs encapsulated.( * indicates p value < 0.05.) (b) Minimum degradation of both SG and CG verified by stable equilibrium swell ratios over 30 days. (c) Representative images of hMSCs in a CG and a SG stained for neutral lipid accumulation (Oil Red O; a marker of adipogenesis), glycoaminoglycan deposition (Safranin O) and ALP activity (Fast Blue; osteogenic biomarker, blue) after culture in adipogenic, chondrogenic and osteogenic medium. Data were presented as average ± standard deviations with 3 samples per group in (a) and (b). Scale bar is 100 μm in panel (c).
Taken together, our investigations demonstrate that without changes in matrix stiffness and degradation (Figure 4a,b), sliding hydrogels facilitated the differentiation of hMSCs toward all three lineages in response to soluble induction factors (Figure 4c, Figure S6). In contrast, non-degradable chemical hydrogels favored chondrogenesis more than adipogenesis and osteogenesis (Figure 4, Figure S6). These data suggest that the molecular mobility of sliding hydrogels renders the hydrogel network dynamic and more adaptable, giving stem cells the freedom to respond to soluble factors and to adjust their behavior correspondingly. For example, with comparable stiffness of chemical and sliding hydrogels (Figure 4a,b), expression of the gene encoding ROCK1, an important mechanosensing protein associated with cell differentiation[2, 23], was 3-fold higher when hMSCs were cultured in sliding hydrogels in osteogenesis medium versus adipogenesis medium (#: p<0.05) (Figure S7); this difference was not evident with cells grown in chemical hydrogels (Figure S7). Regardless of the type of induction medium, the expression of both ROCK1 and RhoA of hMSCs in sliding hydrogels are lower than those in chemical hydrogels (Figure S7). This suggests that the sliding hydrogel constitutes a less restrictive and more permissive environment than chemical hydrogels, which may also enhance the stem-cell differentiation.
In conclusion, we have developed a sliding hydrogel for use as a niche for the culture and differentiation of stem cells in 3D. These sliding hydrogels endow ligands and crosslinks with molecular mobility, enhancing stem-cell differentiation toward multiple lineages. Here, we specifically confirmed the reversible distribution and the dynamic movement of αCDs tethered with ligands and crosslinks to the sliding hydrogel network (Figure 2). This mobility allows cells to reorganize ligands and to rearrange crosslinks to change the network structure of the hydrogel, which in turn allows them to change their morphology and to form protrusions (Figure 3). We detected an enhanced differentiation of stem cells toward adipogenesis, chondrogenesis, and osteogenesis in sliding versus chemical hydrogels (Figure 4c), with comparable stiffness and degradation of the hydrogels (Figure 4a,b). We therefore conclude that our sliding hydrogel constitutes a versatile material for supporting differentiating stem cells toward desired lineages for specific applications in stem cell therapy and tissue regeneration. These sliding hydrogels can also serve as a useful tool for elucidating the effects of the molecular mobility of niche cues on stem cell-fate regulation in 3D.
Supplementary Material
Acknowledgements
This work was supported by the following grants: NIH R01DE024772 (F.Y.), NSF CAREER award (CBET-1351289) (F.Y.), and California Institute for Regenerative Medicine Tools and Technologies Award (RT3-07804) (F.Y.). The authors also acknowledge funding from the Stanford Chem-H Institute (F.Y.), Stanford Bio-X Interdisciplinary Initiative Program (F.Y.), the Stanford Child Health Research Institute Faculty Scholar Award (F.Y.), and Alliance for Cancer Gene Therapy Young Investigator award grant (F.Y.).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Xinming Tong, Department of Orthopaedic Surgery, Stanford University School of Medicine, CA, 94305, United States..
Prof. Fan Yang, Department of Orthopaedic Surgery and Bioengineering, Stanford University School of Medicine, 300 Pasteur Dr., Edwards R105, CA, 94305, United States..
References
- 1.Pittenger MF. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]; Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Cell stem cell. 2009;5:17. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Celiz AD, Smith JGW, Langer R, Anderson DG, Winkler DA, Barrett DA, Davies MC, Young LE, Denning C, Alexander MR. Nature Materials. 2014;13:570. doi: 10.1038/nmat3972. [DOI] [PubMed] [Google Scholar]
- 2.Murphy WL, McDevitt TC, Engler AJ. Nat Mater. 2014;13:547. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12:458. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo CK, Ma PX. Biomaterials. 2001;22:511. doi: 10.1016/s0142-9612(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee H.-p., Lippens E, Duda GN, Mooney DJ. Nat Mater. 2015 doi: 10.1038/nmat4489. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson J, Hubbell JA. Biomaterials. 2011;32:1301. doi: 10.1016/j.biomaterials.2010.10.016. [DOI] [PubMed] [Google Scholar]; Patterson J, Hubbell JA. Biomaterials. 2010;31:7836. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 9.Okumura Y, Ito K. Adv. Mater. 2001;13:485. [Google Scholar]
- 10.Araki J, Ito K. Soft Matter. 2007;3:1456. doi: 10.1039/b705688e. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka Y, Gong JP, Osada Y. Progress in Polymer Science. 2005;30:1. [Google Scholar]; Ito K. Current Opinion in Solid State and Materials Science Polymers. 2010;14:28. [Google Scholar]
- 12.Harada A, Kamachi M. Macromolecules. 1990;23:2821. [Google Scholar]
- 13.Harada A, Li J, Kamachi M. Nature. 1992;356:325. [Google Scholar]
- 14.Pierschbacher MD, Ruoslahti E. Nature. 1984;309:30. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]; Shin H, Jo S, Mikos AG. Biomaterials. 2003;24:4353. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]; Jonker AM, Löwik DWPM, van Hest JCM. Chemistry of Materials. 2011 [Google Scholar]; Tong X, Yang F. Biomaterials. 2014;35:1807. doi: 10.1016/j.biomaterials.2013.11.064. [DOI] [PubMed] [Google Scholar]
- 15.Kataoka T, Kidowaki M, Zhao C, Minamikawa H, Shimizu T, Ito K. J Phys Chem B. 2006;110:24377. doi: 10.1021/jp0649246. [DOI] [PubMed] [Google Scholar]
- 16.Kong HJ, Polte TR, Alsberg E, Mooney DJ. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4300. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nature methods. 2010;7:733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]; Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Nature methods. 2010;7:969. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Adv Mater. 2003;15:888. [Google Scholar]
- 19.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Developmental cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 20.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]; Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Nat Mater. 2014 doi: 10.1038/nmat4051. advance online publication, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]; Guvendiren M, Burdick JA. Nat Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 21.Huey DJ, Hu JC, Athanasiou KA. Science. 2012;338:917. doi: 10.1126/science.1222454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bian L, Hou C, Tous E, Rai R, Mauck RL, Burdick JA. Biomaterials. 2013;34:413. doi: 10.1016/j.biomaterials.2012.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sridhar BV, Dailing EA, Brock JL, Stansbury JW, Randolph MA, Anseth KS. Regenerative Engineering and Translational Medicine. 2015;1:11. doi: 10.1007/s40883-015-0002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ. Nat Mater. 2014;13:970. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.