Abstract
Poly(ethylene glycol) (PEG) hydrogels are widely used to deliver therapeutic biomolecules, due to high hydrophilicity, tunable physicochemical properties, and anti-fouling properties. Although different hydrogel crosslinking mechanisms are known to result in distinct network structures, it is still unknown how these various mechanisms influence biomolecule release. Here we compared the effects of chain-growth and step-growth polymerization for hydrogel crosslinking on the efficiency of protein release and diffusivity. For chain-growth-polymerized PEG hydrogels, while decreasing PEG concentration increased both the protein release efficiency and diffusivity, it was unexpected to find out that increasing PEG molecular weight did not significantly change either parameter. In contrast, for step-growth-polymerized PEG hydrogels, both decreasing PEG concentration and increasing PEG molecular weight resulted in an increase in the protein release efficiency and diffusivity. For step-growth-polymerized hydrogels, the protein release efficiency and diffusivity were further decreased by increasing crosslink functionality (4-arm to 8-arm) of the chosen monomer. Altogether, our results demonstrate that the crosslinking mechanism has a differential effect on controlling protein release, and this study provides valuable information for the rational design of hydrogels for sophisticated drug delivery.
Introduction
Poly(ethylene glycol) (PEG) hydrogels have been widely used for the delivery of therapeutic proteins due to their tissue-like water content, tunable physicochemical properties, and resistance to non-specific protein adsorption.1–4 To construct chemically stable PEG hydrogels as delivery vehicles, the two most commonly used hydrogel crosslinking mechanisms are chain-growth and step-growth polymerization.5 For either crosslinking mechanism, the hydrogel mesh size can be modulated to control protein release.6 The mesh size is commonly controlled by varying the monomer molecular weight and concentration.5,7 As an example, hydrogels with a mesh size smaller than the hydrodynamic radius of encapsulated protein will result in sustained release of the proteins. Previous studies demonstrated that increasing the PEG molecular weight or lowering the PEG concentration increased the diffusivity of various small molecules and proteins from both chain-growth-polymerized8–10 and step-growth-polymerized PEG hydrogels.11 However, no direct comparisons of protein release have been performed between these two types of hydrogels.
It is known that chain-growth polymerization can create a more heterogeneous network structure than step-growth polymerization.5 Chain-growth-polymerized hydrogels are formed by rapid propagation of active centers through monomers containing multiple carbon–carbon double bonds (e.g.PEG diacrylate (PEGDA) and PEG dimethacrylate (PEGDMA)), which forms a high-molecular-weight kinetic chain, serving as a crosslinking point (Fig. S1†). Due to the random nature of radical propagation and termination in chain-growth-polymerization, crosslink functionality or the number of arms per crosslinking point is not fixed across the hydrogel. On the other hand, step-growth-polymerized hydrogels are more homogeneous because they are formed by reacting at least two multifunctional monomers with mutually reactive groups, and each monomer with defined functionality serves as a crosslinking point.12 Since these two crosslinking mechanisms form different hydrogel network structures, varying the monomer molecular weight or concentration may have a differential effect on varying the hydrogel mesh size, which can ultimately influence protein release from the hydrogels. Previously, we have shown that varying the monomer molecular weight or concentration can differentially influence protein accumulation in the three-dimensional PEG hydrogel formed by different crosslinking mechanisms.13 However, the effect of the crosslinking mechanism on protein release is still unknown because the hydrogel crosslinking process can be altered in the presence of encapsulated proteins or therapeutics. Therefore, the goal of the study was to investigate how chain-growth and step-growth polymerization influence the effect of varying the PEG molecular weight or concentration on the efficiency of encapsulated protein release and protein diffusivity. Since distinct crosslinking mechanisms yield different hydrogel network structures, we hypothesize that varying the molecular weight and concentration of PEG for generating chain-growth- or step-growth-polymerized hydrogels would have differential effects on protein release efficiency and diffusivity.
To verify this, we prepared chain-growth- and step-growth-polymerized hydrogels with different PEG molecular weights (2, 2.5, 4, 5 kDa), concentrations (10%, 15%, 20% (w/v)), and functionalities (linear, 4-arm, 8-arm) (Fig. 1). Bovine serum albumin (BSA), a model protein, was encapsulated in chain-growth- or step-growth-polymerized PEG hydrogels, and BSA release was monitored over periods of hours to weeks. To characterize the efficiency of BSA release from these hydrogels, we evaluated the percentage of total protein release. Protein diffusivity was calculated by fitting the fractional BSA release profile to a three-dimensional Fickian model.14 We believe this study helps deepen our understanding of how the hydrogel crosslinking mechanism influences protein release. We anticipate that our observations will guide the rational design and evaluation of hydrogels for sophisticated drug delivery.
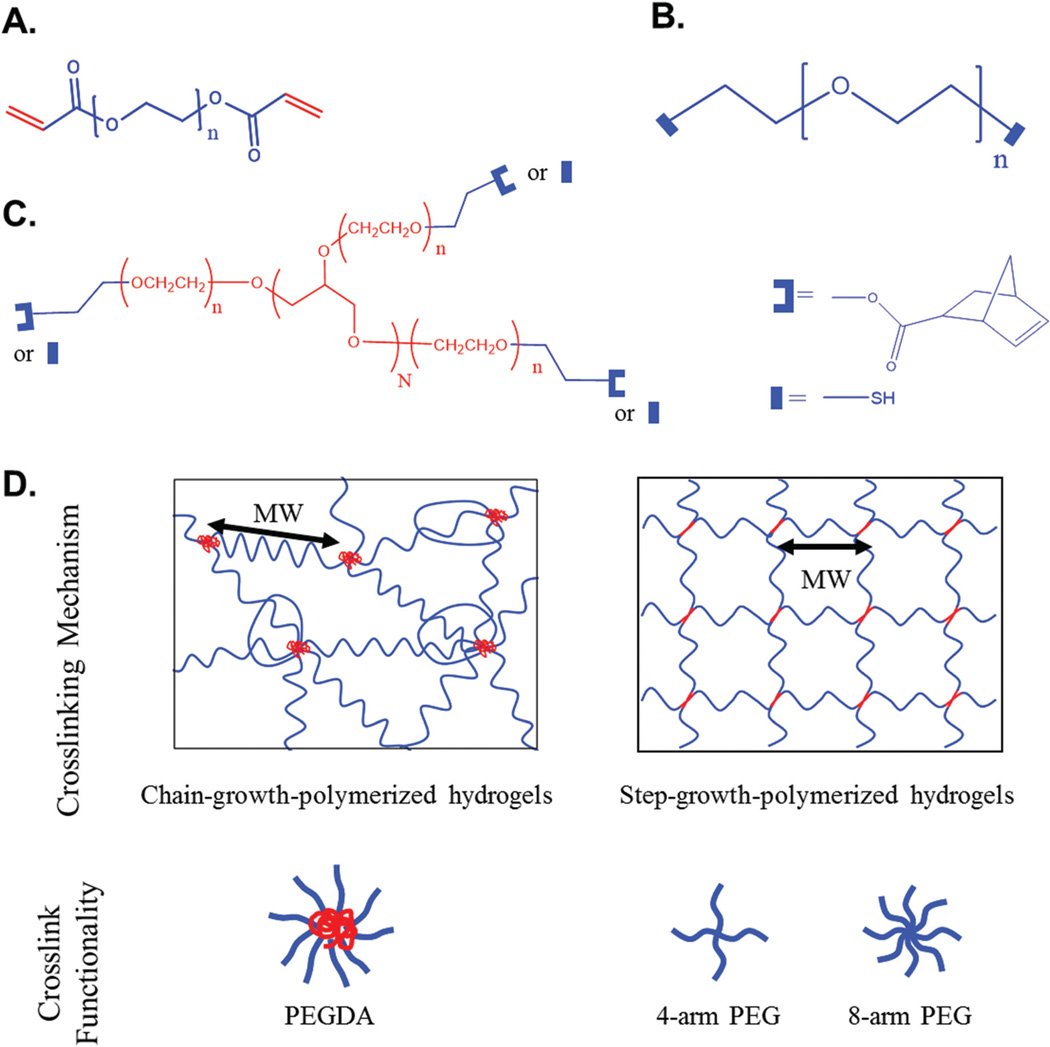
Fig. 1.
Polymer chemical structures and hydrogel networks formed by chain-growth and step-growth polymerization. (A) Chemical structure of PEGDA. (B) Chemical structure of PEG-dithiol. (C) Chemical structures of 4-arm (N = 2) and 8-arm (N = 6) PEG-norbornene (U-shape) and PEG-thiol (rectangle). (D) The networks of chain-growth-polymerized hydrogels and step-growth-polymerized hydrogels and their crosslink functionality. Crosslink functionality is defined as the number of arms per crosslinking point (red). In chain-growth-polymerized hydrogels, the crosslinking point is a high-molecular weight-kinetic chain and crosslink functionality is not constant because it is determined by the degree of polymerization (Fig. S1†). In step-growth-polymerized hydrogels, the crosslinking point is the core of a multi-arm PEG monomer and crosslink functionality is determined by the number of arms in a PEG monomer. MW is defined as the molecular weight between adjacent crosslinking points.
Experimental section
Materials
PEG (molecular weight (MW) 2 kDa, 4 kDa), K2CO3, dichloromethane, acryloyl chloride, KI, Celite® 521, 4-(dimethylamino)-pyridine, N,N′-diisopropylcarbodiimide, 5-norbornene-2-carboxylic acid, NaH, allyl bromide, 2,2-dimethoxy-1,2-diphenylethan-1-one, dithiothreitol, tetrahydrofuran, and thioacetic acid were purchased from Sigma-Aldrich (St Louis, MO, USA). 4-(2-Hydroxyethoxy)phenyl-(2-hydroxy-2-propyl)ketone (Irgacure 2959) was purchased from BASF (Florham Park, NJ, USA). PEGDA (MW 5 kDa) was purchased from Laysan Bio (Arab, AL, USA). Four-arm PEG (MW 5 kDa), 4-arm PEG-thiol (MW 5 kDa), 8-arm PEG (MW 10 kDa), and 8-arm PEG-thiol (MW 10 kDa) were purchased from JenKem Technology (Allen, TX, USA). PEGDA (MW 2 kDa, 4 kDa), 8-arm PEG-norbornene (MW 10 kDa), 4-arm PEG-norbornene (MW 5 kDa), and PEG-dithiol (MW 1.5 kDa) were synthesized in house. BSA was purchased from Fisher Scientific (Pittsburg, PA, USA). The BioRad Protein Assay was purchased from BioRad (Hercules, CA, USA).
Polymer synthesis
To synthesize PEGDA (2 kDa), linear PEG (2 kDa) was dissolved in dichloromethane with 3 eq. (mole equivalents with respect to hydroxyls) K2CO3. Three eq. acryloyl chloride and 0.1 eq. KI were added to the solution. The solution was stirred at 0–4 °C overnight and filtered with Celite® 521 to remove insoluble materials. The product was collected through evaporation of the solvent and precipitation in cold ether. The product was purified by dialysis against deionized water (MCO 1 kDa) for 2 days, followed by lyophilization for 2–3 days to collect the purified product. PEGDA (4 kDa) was synthesized via the same procedure. Conversion ratios of >95% were confirmed with proton nuclear magnetic resonance (1H NMR; data not shown).
Four-arm and eight-arm PEG-norbornene (PEG-NB) were synthesized as previously reported.15,16 Four-arm PEG (5 kDa) was dissolved in dichloromethane plus 0.2 eq. 4-(dimethylamino) pyridine and 3 eq. 5-norbornene-2-carboxylic acid. After cooling the solution in an ice bath, 3 eq. N,N′-diisopropylcarbodiimide were added to the solution. After overnight stirring, the reaction mixture was filtered and concentrated by evaporating most of the solvent. The concentrated solution was then poured into ice-cold diethyl ether to precipitate the product. Eight-arm PEG-norbornene (10 kDa) was synthesized via the same procedure. Conversion ratios of >95% were confirmed with1H NMR (data not shown).
PEG-dithiol was synthesized as previously reported.15,16 Linear PEG (1.5 kDa) was dissolved in tetrahydrofuran followed by the addition of 5 eq. NaH and 2 eq. allyl bromide. After overnight stirring, the solution was filtered, concentrated, and precipitated in ice-cold diethylether. The resulting PEG-allyl was dissolved in dichloromethane with 0.5% (w/v) 2,2-dimethoxy-1,2-diphenylethan-1-one and 2 eq. thioacetic acid. The solution was then exposed to ultraviolet light (365 nm, 4 mW cm−2; UVP XX-15S lamp (Upland, CA, USA)) for 1 h and the PEG thioester was precipitated in ice-cold ether. The product was dissolved in ammonium methanol, and 0.5 eq. dithiothreitol were added to avoid disulfide formation. After stirring for 3 h, the product was precipitated in ice-cold ether. A conversion ratio of >95% was confirmed with1H NMR (data not shown).
Protein encapsulation and hydrogel formation
To encapsulate BSA in hydrogels, BSA solution (2% (w/v)) was made by dissolving BSA in phosphate-buffered saline (PBS) with photoinitiator Irgacure 2959 (0.05% (w/v)). To make the chain-growth-polymerized hydrogel precursor solution, PEGDA of different molecular weights (2 kDa, 4 kDa, 5 kDa) and concentrations (10%, 15%, 20% (w/v)) was dissolved in BSA solution (Table S1†). For the step-growth-polymerized hydrogel precursor solution, multi-arm norbornene-terminated PEG and thiol-terminated PEG were dissolved in the BSA solution in a stoichiometrically balanced ratio (Table S1†). To form hydrogels, 50 µL of precursor solution were added to a cylindrical gel mold (5 mm diameter, 3 mm thickness) and exposed to ultraviolet light (365 nm, 4 mW cm−2) for 5 min. Four replicates were constructed for each combination of molecular weight and concentration.
Protein release
Hydrogels with preloaded BSA were incubated in fresh PBS (200 µL). At each time point, the supernatant was collected, and the hydrogels were moved to new wells with 200 µL of fresh PBS to mimic infinite sink conditions until protein release plateaued. The BioRad Protein Assay was used to quantify the initial BSA concentration in the precursor solution and the BSA concentration in the collected supernatant. The BSA loading amount (MLoad) for each hydrogel was calculated by multiplying the measured BSA concentration in the precursor solution by the volume of each gel (50 µL). BSA released into the supernatant at each time point was calculated by multiplying the measured BSA concentration by the supernatant volume (200 µL). Cumulative BSA release (Mt) at time t was obtained by summing the amount of BSA released up to time t. Total cumulative releasable BSA (M∞) was defined as the value of Mt at plateau.
BSA release profiles were examined in terms of releasable BSA (Mt/MLoad) and fractional BSA release (Mt/M∞). To determine the efficiency of BSA release, total releasable BSA (M∞/MLoad) was estimated by accumulating releasable BSA (Mt/MLoad) until the data plateaued. BSA diffusivity (D) was calculated by fitting the fractional BSA release profile (Mt/M∞) to a three-dimensional Fickian diffusion model of a disk-shaped gel with a uniform initial concentration and an equal surface concentration:14
The geometry of the final, swollen hydrogel was taken into account when calculating protein diffusivity (a: gel diameter, l: gel thickness).
Statistical analysis
Data are presented as mean ± standard deviation (n = 4). Unpaired Student’s t-test was used for between-group comparisons. P-Values less than 0.05 were considered statistically significant.
Results and discussion
Effect of varying the PEG MW on BSA release from chain-growth- and step-growth-polymerized hydrogels
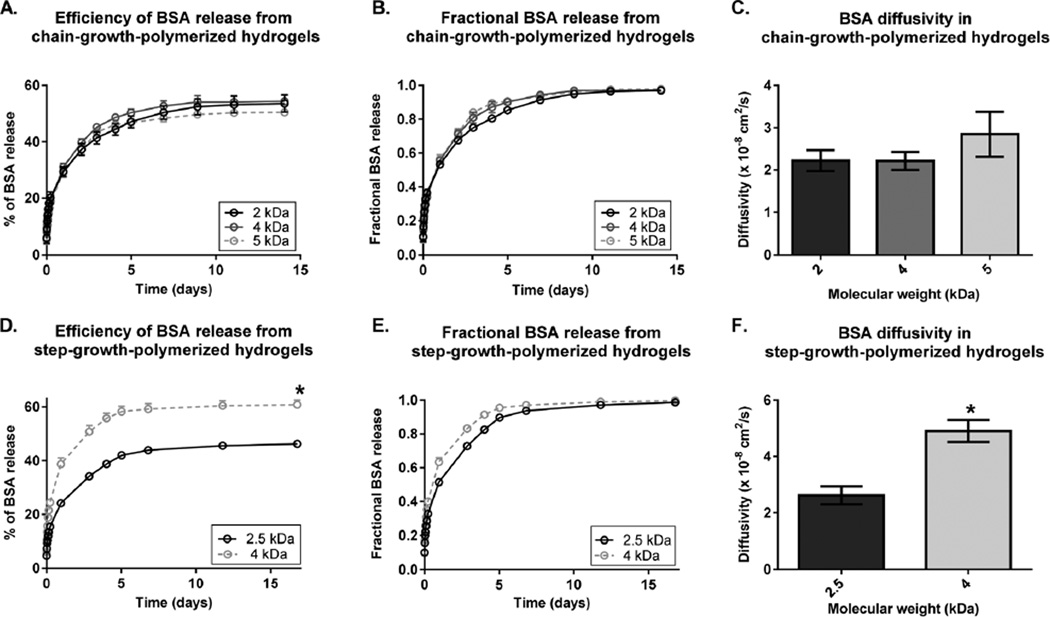
First, we examined the effect of varying the PEG MW on BSA release from chain-growth- and step-growth-polymerized hydrogels (Fig. 2). Surprisingly, varying the PEG MW in chain-growth-polymerized hydrogels did not lead to significant differences in the release efficiency of BSA or in BSA diffusivity. All chain-growth-polymerized hydrogel groups released ~50% of loaded BSA (Fig. 2A). There were no significant differences between fractional BSA release profiles (Fig. 2B), which led to no significant differences in BSA diffusivity (Fig. 2C). On the other hand, varying the PEG MW in step-growth-polymerized hydrogels significantly changed the efficiency of BSA release and BSA diffusivity. Increasing the PEG MW from 2.5 kDa to 4 kDa led to a 1.3-fold increase in the efficiency of BSA release (Fig. 2D). Increasing the PEG MW also led to a faster fractional BSA release, thereby increasing BSA diffusivity by 1.9-fold (Fig. 2E and F).
Fig. 2.
Effect of varying the PEG MW on BSA release from chain-growth-polymerized hydrogels (A–C) and step-growth-polymerized hydrogels (D–F). All chain-growth-polymerized hydrogels are 10% (w/v) PEGDA of MW 2 kDa (black), 4 kDa (dark grey), or 5 kDa (light grey). All step-growth-polymerized hydrogels are 10% (w/v) 8-arm PEG of MW 2.5 kDa (black) or 4 kDa (grey). (A, D) Effect of varying the PEG MW on the BSA release efficiency (Mt/MLoad × 100%). (B, E) Effect of varying the PEG MW on fractional BSA release (Mt/M∞). (C, F) Effect of varying the PEG MWon BSA diffusivity. Data are presented as mean ± standard deviation (n = 4). *p < 0.05 compared to the lowest-MW groups
Effect of varying the PEG concentration on BSA release from chain-growth-and step-growth-polymerized hydrogels
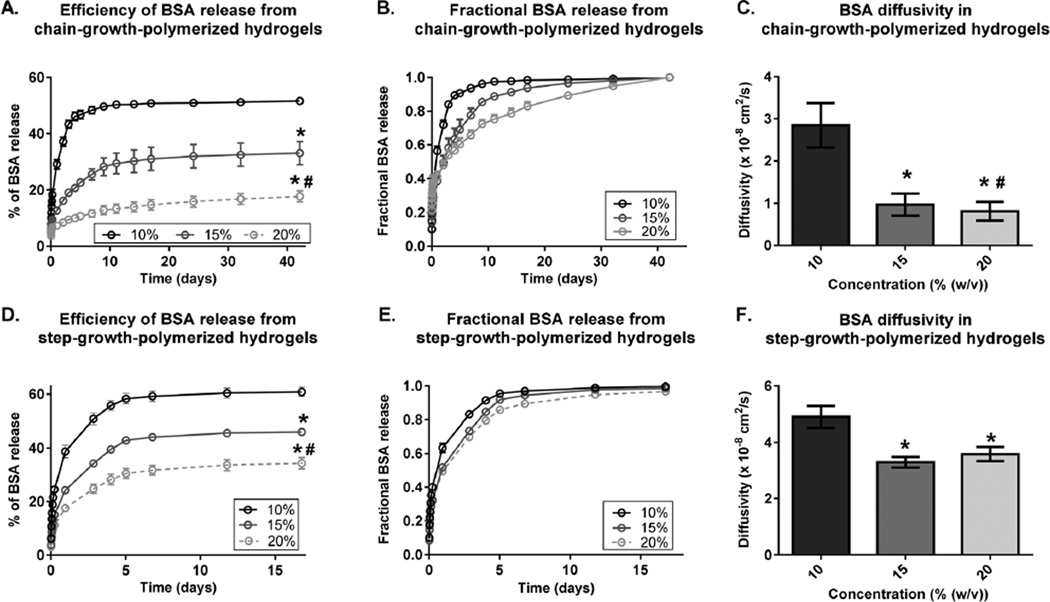
Next, we examined the effect of varying the PEG concentration on BSA release from chain-growth- and step-growth-polymerized hydrogels. In contrast to the effect of varying the PEG MW (Fig. 2), increasing the PEG concentration decreased the efficiency of BSA release and BSA diffusivity for both chain-growth- polymerized and step-growth-polymerized hydrogels (Fig. 3). In chain-growth-polymerized hydrogels, increasing the PEG concentration from 10% to 20% decreased the efficiency of BSA release by 3.6-fold (Fig. 3A) and delayed fractional BSA release, which decreased BSA diffusivity by 3.5-fold (Fig. 3B and C). The same trends were observed in step-growth-polymerized hydrogels (Fig. 3D–F).
Fig. 3.
Effect of varying the PEG concentration on BSA release from chain-growth-polymerized hydrogels (A–C) and step-growth-polymerized hydrogels (D–F). All chain-growth-polymerized hydrogels are MW 5 kDa with PEGDA concentrations of 10% (black), 15% (dark grey), or 20% (light grey) (w/v). All step-growth-polymerized hydrogels are MW 4 kDa with 8-arm PEG concentrations of 10% (black), 15% (dark grey), or 20% (light grey) (w/v). (A, D) Effect of varying the PEG concentration on the BSA release efficiency (Mt/MLoad × 100%). (B, E) Effect of varying the PEG concentration on fractional BSA release (Mt/M∞). (C, F) Effect of varying the PEG concentration on BSA diffusivity. Data are presented as mean ± standard deviation (n = 4). *p < 0.05 compared to the corresponding 10% gels;#p < 0.05 compared to the corresponding 15% gels.
Effect of the PEG hydrogel crosslinking mechanism on BSA release
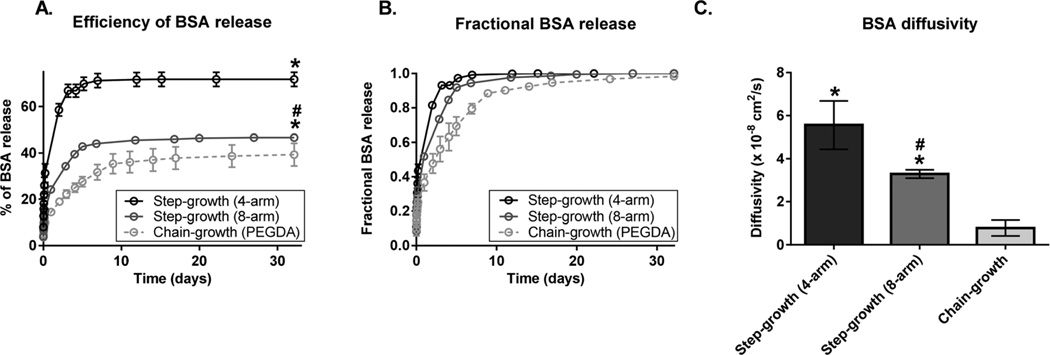
Finally, we investigated the effect of the PEG hydrogel crosslinking mechanism on BSA release (Fig. 4). To evaluate the effect of the PEG hydrogel crosslinking mechanism, the same PEG MW and PEG concentration were used to form chain-growth- and step-growth-polymerized hydrogels, and the calculated mesh sizes of hydrogels were statistically comparable (Fig. S2†). Step-growth-polymerized hydrogels displayed more efficient BSA release and faster fractional BSA release than chain-growth-polymerized hydrogels (Fig. 4A and B). For example, the efficiency of BSA release was 1.8 times greater from step-growth-polymerized hydrogels (4-arm) than from chain-growth-polymerized hydrogels (PEGDA) (Fig. 4A). The same trend was observed for 10% and 20% gels (Fig. S3†). BSA diffusivity was 7.1 times higher from step-growth-polymerized hydrogels (4-arm) than from chain-growth-polymerized hydrogels (PEGDA) (Fig. 4C).
Fig. 4.
Effects of the crosslinking mechanism and crosslink functionality on BSA release. Chain-growth-polymerized hydrogels (light grey) were constructed from 4 kDa PEGDA (15% (w/v)) and step-growth-polymerized hydrogels were constructed from 4 kDa 4-arm (black) or 8-arm (dark grey) PEG (15% (w/v)). (A) Effects of the crosslinking mechanism and crosslink functionality on the BSA release efficiency (Mt/MLoad × 100%). (B) Effects of the crosslinking mechanism and crosslink functionality on fractional BSA release (Mt/M∞). (C) Effects of the crosslinking mechanism and crosslink functionality on BSA diffusivity. Data are presented as mean ± standard deviation (n = 4). *p < 0.05 compared to chain-growth-polymerized hydrogels;#p < 0.05 compared to step-growth-polymerized hydrogels constructed from 4-arm PEG.
Since step growth polymerization allows further control over crosslink functionality, we studied the effect of crosslink functionality on BSA release using 4-arm and 8-arm PEG. As expected, increasing crosslink functionality led to less and slower BSA release (Fig. 4A and B). For example, increasing crosslink functionality from 4-arm PEG to 8-arm PEG (4 kDa, 15%) led to a significant decrease in the efficiency of BSA release (1.5 fold) and in BSA diffusivity (1.7 fold).
Discussion
Since the hydrogel crosslinking mechanism influences the structure of the resulting network, it is crucial to understand how the crosslinking mechanism affects protein release. Here we studied how varying the PEG MW or concentration affected BSA release from chain-growth-polymerized or step-growth-polymerized hydrogels. Although previous studies have shown that increasing the PEG MW or lowering the PEG concentration can increase protein diffusivity,11,17 it has not been studied how the crosslinking mechanism can change the effect of varying the PEG MW and concentration on protein release.
For step-growth-polymerized hydrogels, we found that increasing the PEG MW led to a simultaneous increase in the BSA release efficiency (M∞/MLoad) and BSA diffusivity. In contrast, for chain-growth-polymerized hydrogels, varying the PEG MW had minimal influence on the BSA release efficiency and diffusivity (Fig. 2). Our observations with chain-growth-polymerized gels were in agreement with a previous report, where the PEG MW did not significantly influence protein diffusivity.8 In chain-growth polymerized hydrogels, crosslink functionality is determined by the degree of polymerization (Fig. S1†). On one hand, since increasing the PEG MW decreases the initial molar concentration of acrylate groups, the degree of polymerization decreases according to the Mayo equation. On the other hand, increasing the PEG MW can increase the degree of polymerization by increasing the polymer solution viscosity and chain entanglement. Therefore, these confounding factors contribute to minimizing the effect of varying the PEG MW on BSA release from chain-growth polymerized hydrogels. In contrast, because crosslink functionality of step-growth-polymerized hydrogels is predetermined by the number of arms in the PEG monomer, increasing the PEG MW decreases the molar concentration of active sites (norbornene and thiol groups) as well as crosslinking points, thereby facilitating BSA release from step-growth-polymerized hydrogels.
For both chain-growth- and step-growth-polymerized hydrogels, we found that increasing the PEG concentration decreased the BSA release efficiency (M∞/MLoad) and BSA diffusivity (Fig. 3). Several previous reports have shown that increasing the PEG concentration decreases the hydrogel swelling and mesh size of the hydrogel network, leading to a reduced BSA release from chain-growth-polymerized,9,10,18 or step-growth polymerized hydrogels.11,19 In addition, it has been suggested that an increase in PEG concentrations can lead to an increase in chain entanglement during crosslinking.20 The entangled points can act as a secondary set of crosslinks in the network and further contribute to reduced BSA release.
We investigated the effect of the crosslinking mechanism on BSA release, and found that the BSA release efficiency and diffusivity of chain-growth polymerized hydrogels were significantly lowered than those of step-growth polymerized hydrogels (Fig. 4), despite an insignificant difference in the calculated network mesh sizes between chain- and step-growth-polymerized hydrogels (Fig. S2†). Since the mesh size was calculated from the measured swelling ratio of bulk hydrogel, we think such a difference in BSA release may be due to local inhomogeneity of the hydrogel network. Given that chain-growth-polymerized hydrogels may contain PEG chain loops and have higher crosslink functionalities per crosslinking point, chain-growth-polymerized hydrogels may be a more restricted environment for BSA release than step-growth-polymerized hydrogels. Further characterization of crosslink functionality and network homogeneity using small-angle neutron scattering and static light scattering may help elucidate how the crosslinking mechanism influences protein release.21,22
Lastly, our result shows that the BSA release efficiency is less than 100% for all the hydrogel groups tested by the end of one month release test. BSA has one free thiol that may contribute to its retention in hydrogel via chemical conjugation. However, many previous studies in the literature have reported that proteins including BSA can be highly recovered from degradable chain-growth-polymerized and step-growth-polymerized PEG hydrogels with retained bioactivity.11, 23–25 Given that the PEG hydrogel is not degradable, it is most likely that it will take more than one month for the entrapped BSA to be completely released.
This study demonstrates that protein release depends not only on varying the monomer MW or concentration but also on the crosslinking mechanism. We believe the results of this study will help guide the rational design of more sophisticated hydrogel carriers for protein delivery. For example, in chain-growth-polymerized hydrogels (2 kDa, 20%), BSA diffusivity was shown to be the lowest (0.361 × 10−8 cm2 s−1) with only 13.4% of loaded BSA released over 42 days (Fig. S4†). This formulation can be used to design hydrogels for sustained delivery vehicles to avoid initial burst release. Although non-degradable PEG hydrogel was used as a model hydrogel system to examine the effect of the crosslinking mechanism on BSA release, degradation can be easily introduced to control the release by incorporating hydrolytically degradable units such as lactic acid or enzymatically degradable units such as matrix metalloproteinase (MMP)-cleavable peptides. On the other hand, step-growth-polymerized hydrogels can be beneficial for fine-tuning hydrogel network structures and subsequent protein release by controlling the monomer MW, concentration, or functionality.
PEG has been widely used for a variety of medical applications, and PEG-based materials have already undergone an extensive biocompatibility test, which led to many FDA-approved PEG-based products on the market.26 Furthermore, previous in vivo biodegradation and biocompatibility tests have shown that PEG degradation products resulted in minimal inflammatory and immune response.27,28 Towards broadening the use of PEG hydrogels as protein delivery vehicles, this study helps deepen the understanding of PEG hydrogel network structures for sophisticated design for controlling protein release. We envision that this study further provides valuable insight into the importance of the hydrogel crosslinking mechanism on tailoring other types of hydrogel network structures.
Conclusions
Here, we demonstrated that the crosslinking mechanism of PEG hydrogels (chain growth vs. step growth) differentially impacted protein release. Protein release was controlled by varying the PEG concentration of chain-growth-polymerized hydrogels, while, in step-growth-polymerized hydrogels, varying the PEG concentration, PEG MW, and crosslink functionality mediated protein release. This study further suggests that the PEG hydrogel crosslinking mechanism influences protein release by varying the network structure, which cannot be predicted by the calculated mesh size. These findings may be used to guide the rational design of PEG hydrogels to achieve desired protein release for specific delivery applications.
Acknowledgments
This work was supported by the Stanford Bio-X Interdisciplinary Initiatives grant, the California Institute for Regenerative Medicine (Grant #TR3-05569), and the Basil O’ Connor Starter Scholar Research Award from the March of Dimes Foundation. S. L. would like to thank Stanford Bio-X for fellowship support. The authors would like to thank Michael Keeney, Christine Wang, and Heather Rogan for helpful discussions on the manuscript.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c5bm00256g
Also, the authors declare that there are no conflicts of interest.
References
- 1.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv. Mater. 2006;18:1345–1360. [Google Scholar]
- 2.Drury JL, Mooney DJ. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 3.Mellott MB, Searcy K, Pishko MV. Biomaterials. 2001;22:929–941. doi: 10.1016/s0142-9612(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 4.Peppas NA, Keys KB, Torres-Lugo M, Lowman AM. J. Controlled Release. 1999;62:81–87. doi: 10.1016/s0168-3659(99)00027-9. [DOI] [PubMed] [Google Scholar]
- 5.Lin CC, Anseth KS. Pharm. Res. 2009;26:631–643. doi: 10.1007/s11095-008-9801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermonden T, Censi R, Hennink WE. Chem. Rev. 2012;112:2853–2888. doi: 10.1021/cr200157d. [DOI] [PubMed] [Google Scholar]
- 7.Lin CC, Metters AT. Adv. Drug Delivery Rev. 2006;58:1379–1408. doi: 10.1016/j.addr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Weber LM, Lopez CG, Anseth KS. J. Biomed. Mater. Res., Part A. 2009;90:720–729. doi: 10.1002/jbm.a.32134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruise GM, Scharp DS, Hubbell JA. Biomaterials. 1998;19:1287–1294. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 10.Bal T, Kepsutlu B, Kizilel S. J. Biomed. Mater. Res., Part A. 2014;102:487–495. doi: 10.1002/jbm.a.34701. [DOI] [PubMed] [Google Scholar]
- 11.Zustiak SP, Leach JB. Biotechnol. Bioeng. 2011;108:197–206. doi: 10.1002/bit.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malkoch M, Vestberg R, Gupta N, Mespouille L, Dubois P, Mason AF, Hedrick JL, Liao Q, Frank CW, Kingsbury K, Hawker CJ. Chem. Commun. 2006:2774–2776. doi: 10.1039/b603438a. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Tong X, Yang F. Acta Biomater. 2014;10:4167–4174. doi: 10.1016/j.actbio.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 14.Ritger PL, Peppas NA. J. Controlled Release. 1987;5:23–36. [Google Scholar]
- 15.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv. Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. Biomaterials. 2011;32:3564–3574. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leach JB, Schmidt CE. Biomaterials. 2005;26:125–135. doi: 10.1016/j.biomaterials.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Sokic S, Papavasiliou G. Tissue Eng., Part A. 2012;18:2477–2486. doi: 10.1089/ten.tea.2012.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metters A, Hubbell J. Biomacromolecules. 2005;6:290–301. doi: 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- 20.Beamish JA, Zhu J, Kottke-Marchant K, Marchant RE, Biomed J. Mater. Res., Part A. 2010;92:441–450. doi: 10.1002/jbm.a.32353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsunaga T, Sakai T, Akagi Y, Chung U-i, Shibayama M. Macromolecules. 2009;42:6245–6252. [Google Scholar]
- 22.Matsunaga T, Sakai T, Akagi Y, Chung U-i, Shibayama M. Macromolecules. 2009;42:1344–1351. [Google Scholar]
- 23.Rehmann MS, Garibian AC, Kloxin AM. Macromol. Symp. 2013;329:58–65. doi: 10.1002/masy.201200133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdick JA, Mason MN, Hinman AD, Thorne K, Anseth KS. J. Controlled Release. 2002;83:53–63. doi: 10.1016/s0168-3659(02)00181-5. [DOI] [PubMed] [Google Scholar]
- 25.Tong X, Lee S, Bararpour L, Yang F. Macromol. Biosci. 2015 doi: 10.1002/mabi.201500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alconcel SNS, Baas AS, Maynard HD. Polym. Chem. 2011;2:1442–1448. [Google Scholar]
- 27.Kim J, Dadsetan M, Ameenuddin S, Windebank AJ, Yaszemski MJ, Lu L, Biomed J. Mater. Res., Part A. 2010;95:191–197. doi: 10.1002/jbm.a.32810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjugstad KB, Lampe K, Kern DS, Mahoney M, Biomed J. Mater. Res., Part A. 2010;95:79–91. doi: 10.1002/jbm.a.32809. [DOI] [PubMed] [Google Scholar]