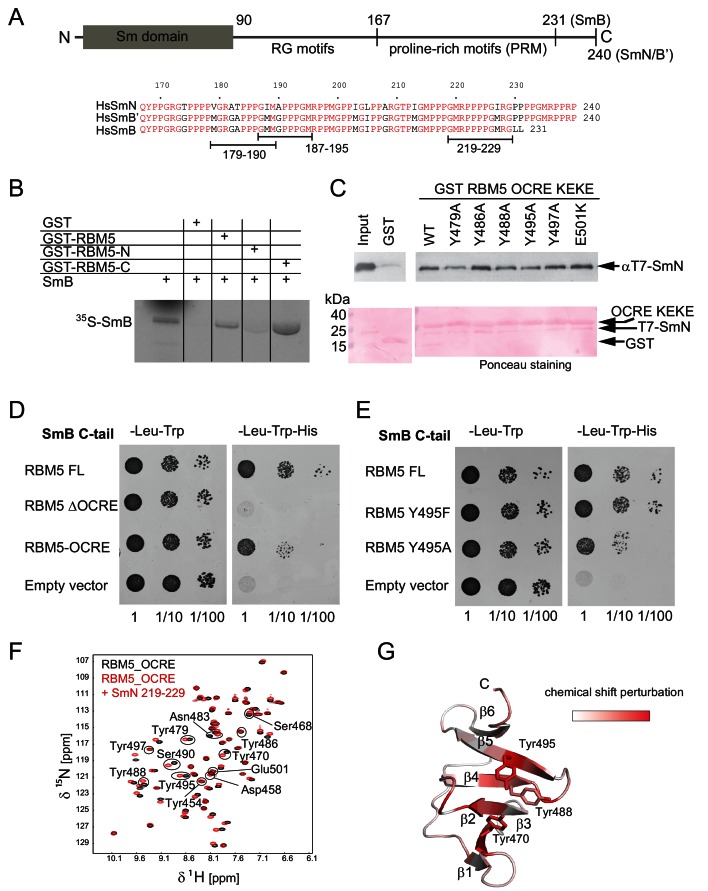
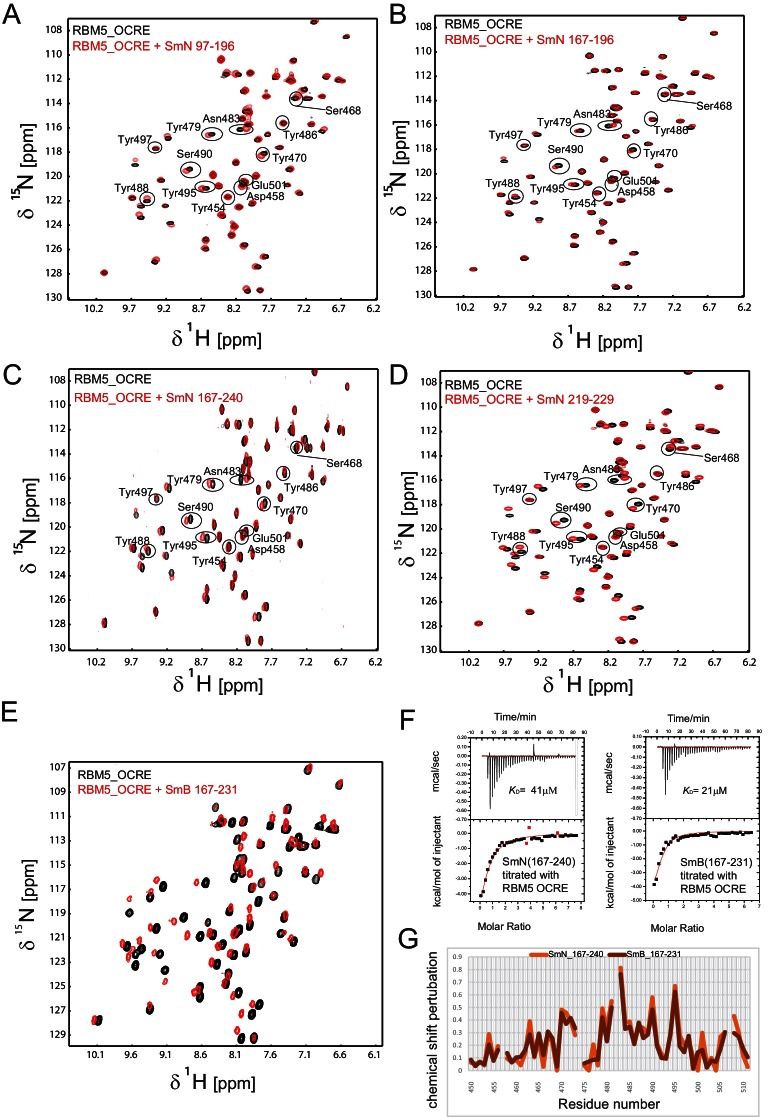
Figure 2. Interaction of RBM5 OCRE with SmN/B/B’ proteins.
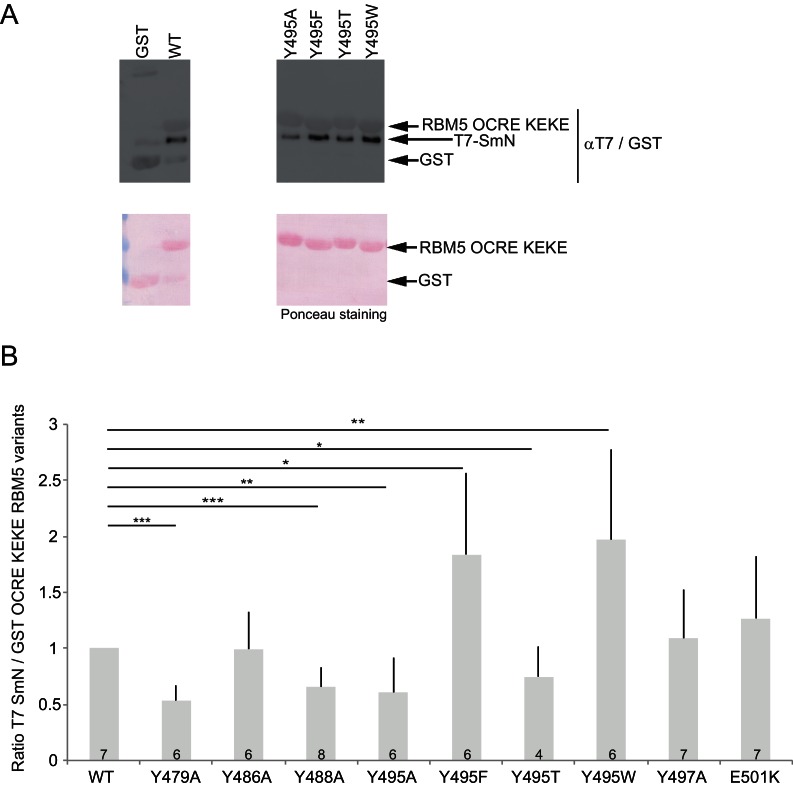
(A) Domain structure and multiple sequence alignment of the C-terminal tails of human SmN/B/B’ proteins. (B) In vitro pull down assays of 35S-labeled SmB with GST-RBM5 full-length, N- and C-terminal halves of human RBM5. First lane represents 20% of the input used in the pull down. (C) Conserved tyrosine residues in the RBM5 OCRE domain are important for the interaction with SmN protein. GST-pull down assays were carried out with different variants of RBM5 comprising the OCRE and KEKE domains harboring the mutations indicated. The detection of the protein was carried out by western blot with T7 epitope antibody and Ponceau staining was performed as a loading control. Quantification is provided in Figure 2—figure supplement 1. (D) The C-terminal domain of human SmB binds to the OCRE domain of RBM5 in vivo. Yeast two hybrid plasmids encoding the C-terminal domain of human SmB (SmB C-tail) and the indicated RBM5 coding regions were transformed into yeast. Serial dilutions of equivalent amounts of exponentially growing yeast were plated on double and triple dropout media. Growth on -Leu -Trp -His is indicative of an interaction between the tested proteins: RBM5-FL: full length RBM5 protein; RBM5 ∆OCRE: deletion of the RBM5 OCRE domain (amino-acids 452–535); RBM5 OCRE: RBM5 OCRE domain (amino-acids 452–535). (E) Surface-exposed tyrosine residues are important for SmB C-tail binding in vivo. RBM5-FL: full length RBM5 protein; RBM5 Y495F: RBM5-FL carrying a tyrosine to phenylalanine substitution at position 495; RBM5-FL Y495A: RBM5 carrying a tyrosine to alanine substitution at position 495. The two-hybrid assay was performed as described in panel d. (F) NMR titrationof 15N-labeled RMB5 OCRE domain (0.2 mM- black) with SmN residues 219–229 (red) at two-fold molar excess. (G) Mapping of NMR chemical shift perturbations upon titration of the OCRE domain with SmN (residues 219–229) onto a surface representation of the RBM5 OCRE domain structure.