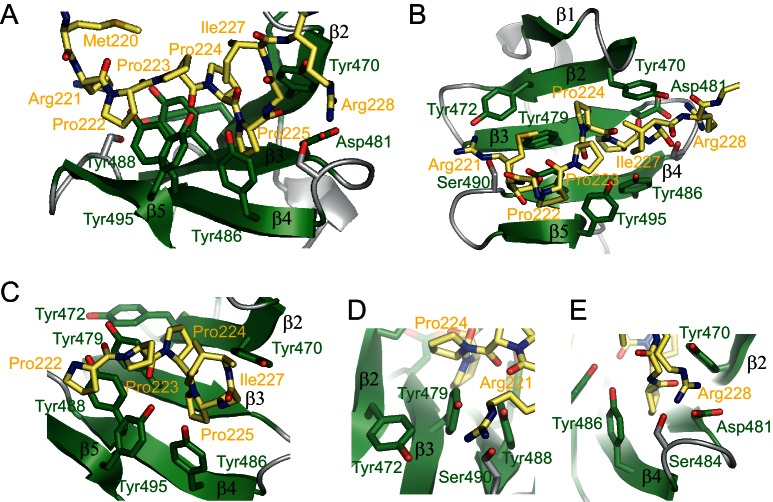
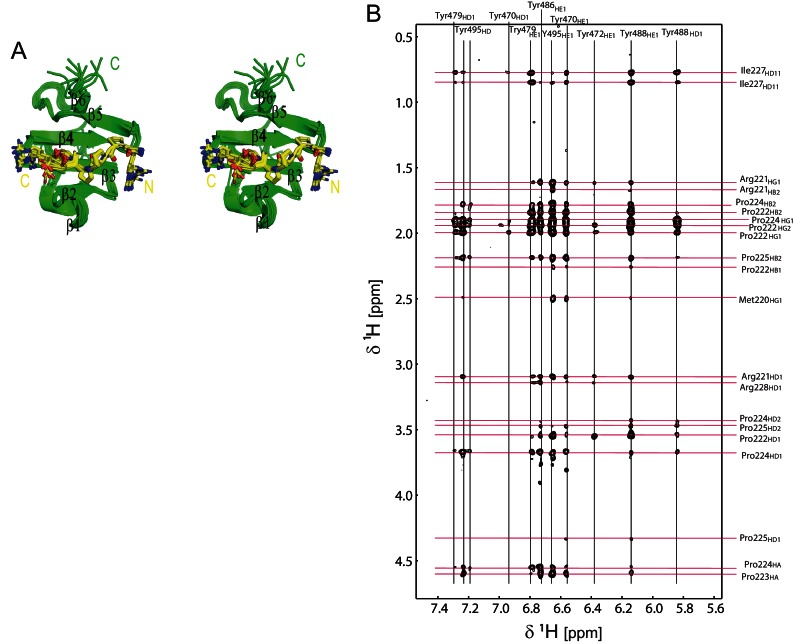
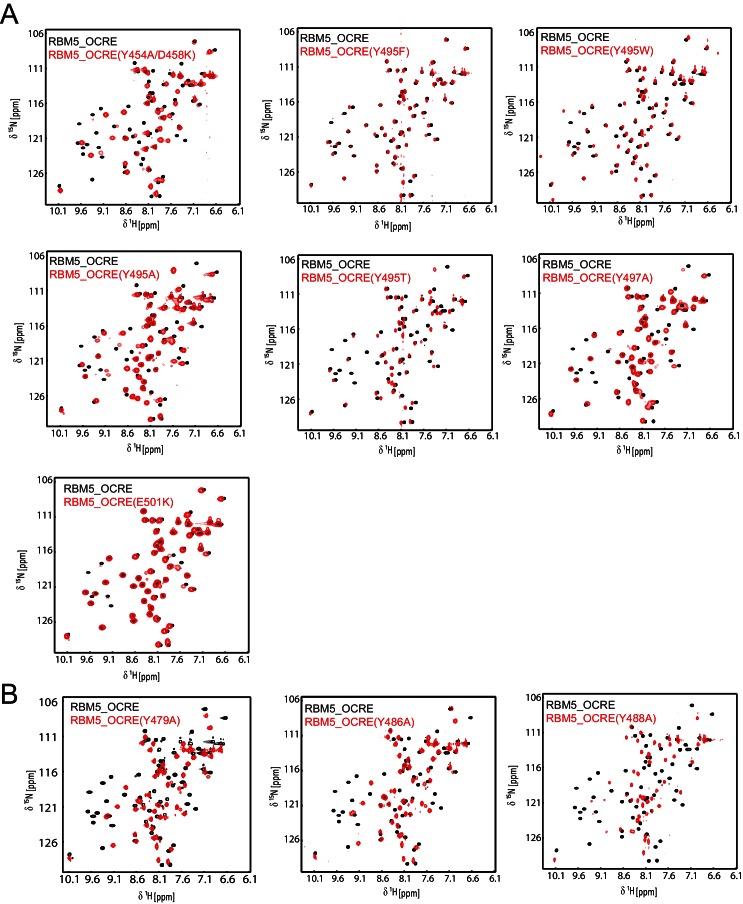
Figure 4. Structure of the RBM5 OCRE/SmN peptide complex.
(A,B) Side and top views in a cartoon presentation of the RBM5 OCRE/SmN peptide complex.The secondary structure and loops in the RBM5 OCRE domain are colored in green and grey, respectively, the proline-rich motif (PRM) peptide corresponding to SmN residues 221–229 is shown in yellow. (C–E) Zoomed views of key interaction sites. (C) The SmN PRM adopts a proline-type II helical conformation and is recognized by stacking with key tyrosine residues from the OCRE domain. The side chain of SmN Ile227 packs against the hydrophobic surface of the PPII helix. (D) Recognition of SmN Arg221 by interactions with hydroxyl groups of Tyr472, Tyr479 and Ser490 (E) Arg228 forms electrostatic contacts with the side chains of Asp481 and Ser484.