Abstract
MHC class I–associated peptides (MAPs) define the immune self for CD8+ T lymphocytes and are key targets of cancer immunosurveillance. Here, the goals of our work were to determine whether the entire set of protein-coding genes could generate MAPs and whether specific features influence the ability of discrete genes to generate MAPs. Using proteogenomics, we have identified 25,270 MAPs isolated from the B lymphocytes of 18 individuals who collectively expressed 27 high-frequency HLA-A,B allotypes. The entire MAP repertoire presented by these 27 allotypes covered only 10% of the exomic sequences expressed in B lymphocytes. Indeed, 41% of expressed protein-coding genes generated no MAPs, while 59% of genes generated up to 64 MAPs, often derived from adjacent regions and presented by different allotypes. We next identified several features of transcripts and proteins associated with efficient MAP production. From these data, we built a logistic regression model that predicts with good accuracy whether a gene generates MAPs. Our results show preferential selection of MAPs from a limited repertoire of proteins with distinctive features. The notion that the MHC class I immunopeptidome presents only a small fraction of the protein-coding genome for monitoring by the immune system has profound implications in autoimmunity and cancer immunology.
Introduction
MHC class I (MHCI) molecules present thousands of peptides at the surface of nucleated somatic cells (1). These MHCI-associated peptides (MAPs), collectively referred to as the immunopeptidome, regulate each step in the development and function of CD8+ T cells (2, 3). Indeed, real-time monitoring of the immunopeptidome is a vital process that allows CD8+ T cells to discriminate between self and nonself and to swiftly reject infected or transformed cells (4–6). Genesis of the immunopeptidome can be broadly divided into 2 events: (a) the processing of MAPs and (b) their binding to MHCI molecules (7, 8). The rules that regulate the second event, binding of MAPs to MHCI, are well defined: MHCI alleles are highly polymorphic, and each allotype has a specific peptide-binding motif that can be accurately predicted by several algorithms (9, 10). However, the first event, processing of MAPs, is a complex multistep process whose overall outcome cannot be predicted (1). Some proteins appear to generate more MAPs than others, but the mechanistic underpinning for these discrepancies remains elusive (11).
Classic biochemical studies have shown that MAP processing is initiated in the cytoplasm by proteasomal protein degradation followed by further trimming by cytosolic peptidases, transport in the ER, and final trimming by ER peptidases (8, 12–15). According to the dominant paradigm, MAPs preferentially originate from defective ribosomal products (DRiPs) which can be created by several mechanisms such as nonsense-mediated decay (NMD), mRNA destabilization, or noncanonical translation in the cytosol or the nucleus (16–20). Large-scale mass spectrometry (MS) offers the sole direct approach to analyzing the global molecular composition of the immunopeptidome. Previous large-scale MS studies of MAPs presented by one or a few MHCI allotypes have shown that thousands of proteins located in all cell compartments can be the source of MAPs (21–24). However, the rules of MAP processing cannot be figured out by studying the immunopeptidome presented by individual HLA allotypes because each allotype can only bind peptides containing a specific motif (25, 26).
The goals of our study were to assess the extent of MAP generation from the entire set of protein-coding genes and to determine whether specific features influence the ability of discrete genes to generate MAPs. We used a well-validated high-throughput proteogenomic approach to identify MAPs presented by 27 HLA-A and HLA-B allotypes on B lymphoblastoid cell lines (B-LCLs) derived from 18 subjects. Overall, we identified 25,270 nonredundant MAPs, which derived from 6,195 out of the 10,575 genes expressed in B-LCLs. Hence, while 59% of genes were the source of 1–64 MAPs per gene, 41% of expressed genes were not represented in the immunopeptidome. Overall, we estimate that the immunopeptidome presented by 27 alleles covered only 10% of exomic sequences expressed in B-LCLs. We then used a series of bioinformatic tools to understand how identifiable features of genes, transcripts, and proteins could influence MAP generation. With these data we built a logistic regression model that was able to predict whether or not a given gene will produce MAPs with a receiver operating characteristic (ROC) AUC of 0.81 ± 0.02 (95% CI). Our results show that the immunopeptidome is forged from a limited repertoire of gene products with distinct features influencing transcription, translation, and proteasomal degradation.
Results
Proteogenomic-based definition of the MAP repertoire presented by 27 HLA allotypes.
To obtain a comprehensive representation of the immunopeptidome presented by HLA-A and HLA-B molecules, we applied a well-validated high-throughput proteogenomic approach that hinges on a combination of next-generation sequencing and high-throughput MS (20, 27, 28). Transcriptome and exome-sequencing data were used to build personalized protein databases for B-LCLs of 18 subjects using the Python package pyGeno (29). These personalized databases were used for peptide identification by MS. MAPs were eluted from the cell surface by mild acid elution, and stringent quality filters were applied to the list of MAPs assigned by MS: (a) a peptide length of 8–14 amino acids, (b) a 1% false discovery rate based on searches against concatenated target/decoy databases (30), (c) assignment to single genetic origin among the 10,575 protein-coding genes expressed and annotated in B-LCLs, and (d) a predicted MHCI IC50 of less than or equal to 1,250 nM according to the NetMHC or NetMHCcons algorithms (31, 32) (Supplemental Figure 1C, see details in Methods; supplemental material available online with this article; doi:10.1172/JCI88590DS1). About 99.8% of individuals of European descent bear at least one of the 27 HLA-A,B allotypes studied (33).
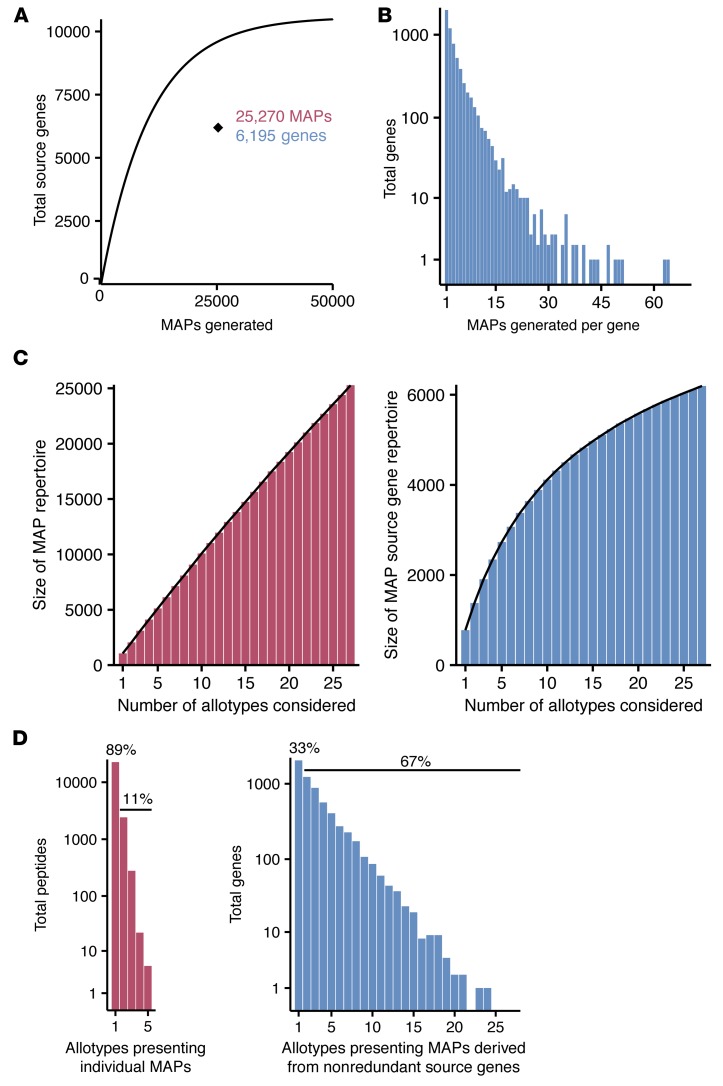
We identified 25,270 nonredundant MAPs derived from 6,195 source genes in, to the best of our knowledge, the largest set of MHCI-associated peptides reported to date (Figure 1A and Supplemental Tables 1 and 2). Strikingly, only 59% expressed and annotated genes in B-LCLs were capable of generating detectable MAPs. MAP source genes produced up to 64 individual MAPs, and 68% of these genes produced more than 1 MAP (Figure 1B). To estimate the diversity of a multiallelic immunopeptidome, we computed the size of the MAP repertoire and MAP source gene repertoire as a function of the number of HLA allotypes considered (Figure 1C). We counted the number of unique identifications when a given number of randomly selected allotypes was considered. For MAPs, the nearly linear nature of this relationship demonstrated little redundancy in the peptides presented by different allotypes (Figure 1C). Conversely, the redundancy of the genes generating MAPs across all 27 HLA allotypes was much greater (Figure 1C). As more allotypes were considered, a diminishing number of additional genes were represented in the immunopeptidome. A simulation examining the size of the peptide and gene repertoires as various numbers of subjects were considered showed similar results (Supplemental Figure 1, A and B). Most MAPs (89%) were presented by a single HLA allotype (Figure 1D). The few promiscuous binders were presented by HLA allotypes with similar peptide-binding motifs (i.e., same “superfamily”), such as A*03:01 and A*11:01 (34). However, the majority of MAP source genes (67%) produced MAPs for multiple allotypes, some for up to 24 of the 27 allotypes studied (Figure 1D).
Figure 1. The immunopeptidome presented by 27 HLA allotypes.
(A) Total number of nonredundant MAPs and their source genes in the immunopeptidome of 18 B-LCLs compared with an expected binomial distribution. The curve depicts the expected number of source genes if all genes had a similar ability to generate MAPs. The black diamond shows the actual number of source genes (n = 6,195) observed for 25,270 MAPs (P < 1 × 10-250, binomial test). (B) Histogram showing the number of MAPs generated per MAP source gene (range = 1–64). (C) The number of unique identifications of MAPs (left panel) and MAP source genes (right panel) was counted for various numbers of randomly selected HLA allotypes. Results show the average of 1,000 simulations. (D) The promiscuity of antigen presentation for MAPs (left panel) and their source genes (right panel). Histograms show the number of allotypes associated with each peptide or gene.
Since MS analyses can be subject to some stochastic variations, would it be possible that no MAPs were assigned to 41% of expressed genes because these MAPs were missed by MS? We reasoned that if our MS analyses randomly missed some MAPs, the proportion of MAP source versus nonsource genes should nevertheless follow a binomial distribution where the number of source genes increases as a function of the number of detected MAPs (Figure 1A). Notably, we found that the 25,270 MAPs that we identified by MS derived from significantly fewer genes (n = 6,195) than predicted by a binomial distribution (Figure 1A, exact binomial test P < 10–250). Hence, random failure to detect some MAPs cannot explain that only 59% of genes were found to generate MAPs. In addition, we used internal standard triggered–parallel reaction monitoring (IS-PRM) in order to compare the detection threshold for 2 sets of stable isotopically labeled synthetic peptides (35). Peptides AEIEQKIKEY, EEINLQRNI, EEIPVSSHY and EESAVPERSW (underlined residues indicate 13C, 15N-labeled amino acids) had the amino acid sequence of MAPs presented by B*44:03. The other synthetic peptides AESQELLTF, EESHLNRHF, HESAEGKEY, and TESSDITEY corresponded to amino acid sequences from nonsource genes (i.e., not detected in our initial shotgun MS analyses) that were randomly chosen among peptides predicted to be strong binders for B*44:03 (IC50 < 50 nM). These synthetic peptides were spiked (100 fmoles each) in mild acid elution extracts from 3 different B-LCLs to correlate the identification of the corresponding endogenous MAPs. IS-PRM analyses showed that the detection threshold was similar for the 2 groups of synthetic peptides (Supplemental Figure 2). Furthermore, none of the selected B*44:03 strong binders coded by nonsource genes were detected in the 3 different B-LCLs using IS-PRM. In contrast, endogenous peptides from source genes presented by B*44:03 were all correlated with their corresponding synthetic peptides (Supplemental Figure 2). These results provide compelling evidence that failure to detect MAPs from nonsource genes cannot be ascribed to MS bias against the product of nonsource genes.
Two major points can be made from these data: (a) a distinct subset of genes produced most MAPs, and (b) our method captured the majority of MAP source genes (Figure 1C). As a corollary, these results suggest a model whereby a common pool of source proteins selectively enters the antigen-processing pathway and can generate MAPs with suitable motifs for most MHCI allotypes.
Discrete protein regions are preferential sources of MAPs.
We next asked whether there might be “hot spots” in MAP source genes, i.e., regions or domains that provide disproportionately high amounts of MAPs. To this end, we analyzed the spatial distribution of MAPs along proteins that generated more than one MAP. We first identified 3,682 pairs of overlapping MAPs formed by 5,046 individual peptides (20% of the entire data set). In a given pair, MAPs differed from each other at their N and/or C terminus (Figure 2A). These pairs may result from differential trimming of a common precursor by various peptidases in the cytosol and ER. Notably, 71% of MAP pairs bound different allotypes; of these, 82% bound allotypes from different superfamilies (Figure 2B). Hence, from the perspective of an MHCI allotype, generation of overlapping MAP pairs is generally not redundant: members of a pair are seldom presented by the same MHCI allotype. At the population level, the net result is that some protein regions are included in the immunopeptidome of many people who do not share the same HLA alleles.
Figure 2. Spatial distribution of MAPs along source proteins.
(A) Distribution of overlap types for 3,682 pairs of overlapping MAPs formed by 5,046 individual peptides: pairs with any overlapping residues and no common ends; pairs with a common C terminus (C term); pairs with a common N terminus; and pairs with 1 peptide contained within the other. (B) Proportion of overlapping MAP pairs presented by the same allotype or different allotypes. For MAP pairs presented by different allotypes, whether the 2 allotypes belong to the same superfamily is indicated (34). (C) Distances between MAP start sites along proteins generating more than 1 MAP compared with a matched, random distribution. Distances are shown up to 30 residues. Distances are significantly shorter in the actual distribution (Wilcoxon rank sum test, P = 7 × 10-52). (D) Exome coverage by the immunopeptidome. A window of 50 or 25 amino acids (left and right panel, respectively) was moved residue by residue along proteins of the transcribed exome of B-LCLs. Histograms show the number of MAPs found in each window; the proportion of windows containing 0 versus at least 1 MAP is indicated.
To further evaluate whether selected protein regions were preferential sources of MAPs, we analyzed the spatial distribution of MAPs along proteins. For each protein, distances between adjacent MAP start sites were calculated. A control distribution was generated by randomly placing the same number of MAPs along the same protein length. We found that MAPs colocalized along proteins more than expected (P = 7 × 10-52, Figure 2C). The fact that no MAPs were assigned to 41% of expressed protein-coding genes, together with the clustering of MAP-coding sequences in source genes, suggests that the immunopeptidome covers a limited portion of the whole exome. To estimate global exome coverage, (a) we moved a walking window of 150 base pairs (50 amino acids) along the exome coding for the 10,575 genes expressed in B-LCLs, and (b) we calculated the number of MAPs seen in each window. We found that 83% of windows generated no MAPs, whereas 17% of windows covered 1–30 MAPs per window (Figure 2D). When we reduced the window size to 75 base pairs, only 10% of windows were a source of MAPs (Figure 2D). From this, we conclude that the immunopeptidome presented by 27 HLA-A,B allotypes covers an unexpectedly small portion of the whole transcribed exome.
Gene expression cannot solely account for differential ability of genes to generate MAPs.
Understanding the genetic origins of the immunopeptidome is of paramount importance fundamentally and in the search for MAPs that could be used as therapeutic targets. What distinguishes the 6,195 genes that were capable of generating MAPs compared with the other 41% of genes from which no MAPs were detected? To answer this question, we applied a variety of analyses and prediction algorithms to study the features of MAP source and nonsource genes, transcripts, and proteins. We first asked whether MAP source proteins simply contained more potential HLA-binding peptides, i.e., peptides with the right binding motif for the 27 HLA allotypes considered here. This was not the case: the density of predicted nonamer MHCI binders was no greater in source genes than nonsource genes (Supplemental Figure 1D). Since the difference between MAP source and nonsource genes is unrelated to the number of potential MHC binders, it must therefore involve discrepancies in the processing of MAP source proteins.
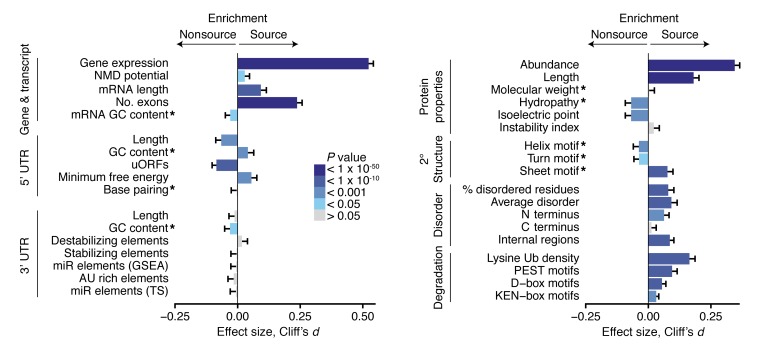
Whether gene expression influences MAP generation is a controversial issue, as shown by previous studies based on smaller data sets. According to some reports, MAPs derive preferentially from highly abundant mRNAs or proteins (19, 25, 36), but other reports cast some doubts on this contention (23, 37). By analyzing RNA-sequencing data of the 18 B-LCLs studied herein, we found that the average gene expression was significantly higher for MAP source genes (Figure 3). However, expression alone provided an incomplete portrait of antigen presentation: some highly expressed genes generated no MAPs, and some lowly expressed genes were capable of generating MAPs. Since the proteome is an imperfect mirror of the transcriptome (38, 39), we also analyzed the relationship between protein abundance in human B cells (40) and MAP generation. MAP source proteins are more abundant than nonsource proteins (Figure 3), yet the fact that some proteins with similar expression belonged to source or nonsource groups suggested that other factors were at play.
Figure 3. Features of MAP source and nonsource genes, transcripts, and proteins.
Error bars represent a 95% CI based on bootstrapping for Cliff’s d value, a nonparametric measurement of effect size. P values derived from 2-sided Wilcoxon tests; 6,195 source and 4,380 nonsource genes and gene products were studied for each comparison. * indicate features that were normalized for the respective transcript, UTR, or protein lengths. See Methods for details of how each feature was calculated. miR, microRNA; TS, TargetScan software; Ub, ubiquitination site.
MAP source transcripts are enriched in features conferring greater translation efficiency.
Ultimately, MAP generation must be regulated at the level of translation and protein degradation (41). To gain further insights into the mechanisms regulating MAP generation, we analyzed the potential role of factors regulating protein metabolism. We first asked whether features enhancing translation efficiency and transcript stability may distinguish source from nonsource transcripts. Coherent with the concept that NMD is a source of MAPs (18), we observed that the proportion of genes with at least one transcript with an NMD biotype (determined with the ENSEMBL regulatory build) was higher in source relative to nonsource genes (Figure 3). Also, consistent with the positive correlation between the number of exons and translation efficiency (42), we found that MAPs derived from transcripts composed of more exons than nonsource transcripts (Figure 3), even when normalized for transcript length (P = 5 × 10-49).
We next examined features of the 5′ UTR for evidence of translational regulation of antigen processing. Upstream open reading frames (uORFs) tend to negatively influence translation by destabilizing transcripts and by acting as physical obstacles that slow ribosomal scanning (43). The 5′ UTRs of MAP source transcripts were significantly shorter and contained fewer predicted uORFs. Similarly, the secondary structure predicted with Vienna RNAfold (44) revealed greater free energy scores in spite of enriched GC content for MAP source 5′ UTRs. No definitive differences between the amount of base pairing in 5′ UTR structures were found (Figure 3). These findings suggest that MAP source 5′ UTRs are structurally fluid and contain fewer obstacles to translation.
The 3′ UTR is a critical site of translational control containing regulatory elements such as adenylate-uridylate–rich (AU-rich) elements and binding sites for microRNAs and RNA-binding proteins (45). Despite this regulatory potential, we initially remarked no difference in the lengths of 3′ UTRs (Figure 3). The density of AU-rich elements was similar; however, our analyses could not take into account the distinction between AU elements involved in rapid decay and finer stability regulation (46). Greater GC content was found in nonsource 3′ UTRs and along the entire mRNA transcript in general; this may reflect a positive association with mRNA levels in a degradation-independent manner (47). Stabilizing and destabilizing regulatory elements were queried in the 3′ UTRs of all transcripts (48) and revealed similar prevalence in source and nonsource transcripts (Figure 3). Moreover, we were unable to confirm previous results that MAPs derive preferentially from transcripts with microRNA-binding sites using 2 different tools: Gene Set Enrichment Analysis (GSEA) and TargetScan (19, 49, 50) (Figure 3). However, our negative findings regarding binding sites for microRNAs and RNA-binding proteins must be considered with some reservations. First, microRNA regulation is highly cell-type specific, while the methods used to predict microRNA involvement operate at an organism-wide level (50). Second, since the effects of 3′ UTR regulatory elements are heavily context dependent (45), the role of 3′ UTR regulation in MAP generation may be obscured by the specific activity of predicted elements in B-LCLs. Nonetheless, in contrast with the 5′ UTR, these findings indicate at least limited regulation of MAP generation by 3′ UTR elements.
Notably, features enriched in MAP source transcripts and UTRs had minimal correlations with protein abundance (absolute Spearman’s rank correlation coefficient r of 0.22 for number of exons and r < 0.14 for others, Supplemental Figure 3). This led us to postulate that gene expression and transcript features may provide nonredundant information for the modeling of MAP generation.
The primary and secondary structure of proteins regulates MAP generation.
Next, we assessed the electrochemical and structural features of MAP-generating proteins. We confirmed previous reports that longer proteins generate more MAPs (25, 36) (Figure 3). This may reflect that, relative to shorter proteins, longer proteins (a) contain more MHCI-binding sequences, (b) have a greater chance of forming DRiPs, and (c) bind more ribosomes (25,42). MAP source proteins had lower hydropathy scores, indicating more polar amino acid composition. Furthermore, the predicted isoelectric point revealed greater acidic composition of source proteins (Figure 3). At the next level of complexity, the predicted secondary structure of MAP source proteins showed distinct contributions of helix, turn, and sheet motifs. In particular, MAP source proteins showed a conspicuous enrichment in sheet motifs (Figure 3).
The ubiquitin proteasome system is a key entry point for proteins into the MHCI-processing pathway (7, 51). We first examined MAP proteins for proteasomal degradation motifs. We found that, compared with nonsource proteins, MAP source proteins contained higher frequencies of (a) KEN-box and D-box motifs targeted by the anaphase-promoting complex ubiquitin ligase (52), (b) PEST motifs, which serve as proteolytic signals for the proteasome and other proteases (53), and (c) canonical lysine ubiquitination sites (54) (Figure 3).
Unstructured protein regions serve as initiation sites for proteasomal degradation (55), and intrinsically disordered segments favor proteasome degradation (56). Therefore, to analyze the potential influence of protein disorder on MAP generation, we computed the disorder status of proteins in our data set with the neural network predictor PONDR VLXT (57). Whether the proportion of disordered residues, the average disorder of all residues, the length of N-terminal disorder, or the presence of internally disordered regions longer than 30 residues was considered, MAP source proteins consistently contained greater disorder compared with nonsource proteins (Figure 3). Similar results were obtained using 2 other disorder predictors: DISOPRED (58) and IUPRED (59) (Supplemental Figure 4B). We conclude that primary and secondary structure of proteins and particularly features linked to proteasomal degradation have a strong influence on MAP generation.
GO terms analysis.
We next compared the enrichment of gene ontology (GO) terms in MAP source and nonsource genes using the topGO algorithm (60) to eliminate redundancies (Figure 4). Our findings here confirm and extend reports based on smaller data sets (19, 22, 25). The source gene population was highly enriched in genes coding for intracellular proteins interacting with DNA, RNA, and other proteins (Figure 4A and Supplemental Table 3). This may have resulted from a relatively greater expression of genes implicated in housekeeping functions, such as poly(A) RNA binding, mitotic cell cycle, and mRNA processing. Non–mutually exclusive hypotheses are that these genes have a preferential access to the MHC-processing machinery, for example, via “immunoribosomes,” or that components of macromolecular complexes have a greater propensity to form DRiPs (17). Nonsource proteins were enriched in membrane components and related signaling processes, demonstrating that proteins traversing the secretory pathway are poorly represented in the MHCI immunopeptidome (Figure 4B and Supplemental Table 4).
Figure 4. GO analysis of source and nonsource genes.
Enrichment in source (A) and nonsource (B) groups was calculated on a background of both groups using the topGO algorithm to eliminate redundancies (60). The top 15 most enriched functions are shown for each group including all 3 ontology categories. For all GO terms significantly enriched in source and nonsource gene categories, see Supplemental Tables 3 and 4. PR, positive regulation; RNP, ribonucleoprotein; CC, cellular component; MF, molecular function; BP, biological process.
Modeling MAP generation.
Having identified features that differentiate MAP source versus nonsource genes, we asked whether it might be possible to build a model for predicting whether a given gene generates MAPs. Taking into account features listed in Supplemental Table 5 (see also Supplemental Figure 3), we trained a logistic regression model on 80% of our data set using 10-fold cross-validation and tested its ability to discriminate MAP source versus nonsource genes on the remaining 20% of our data set. The process was repeated 1,000 times with randomly divided training and test data sets (see Methods for details). Prediction scores, falling between 0 and 1, demonstrated a considerable ability to correctly discriminate between MAP source and nonsource genes (Figure 5A). Although the model was blind to the number of MAPs produced by source genes, we found that the predictions corresponded to the rate of MAP production (Figure 5B).
Figure 5. A logistic regression model to predict whether or not a gene will generate MAPs.
(A) Prediction scores for each gene grouped by experimentally defined source classification. (B) Prediction scores for each gene and the number of MAPs generated. (C) Model performance measured by a ROC plot of sensitivity (the rate of true positives) as a function of specificity (the rate of true negatives); the AUC is 0.81 ± 0.02 (95% CI). (D) Frequency of input variable selection in a logistic regression model using recursive feature elimination; frequencies above 0.05 are shown. (E) The relative weight of all input variables in the 2-class logistic regression model. Variables normalized by the length of the corresponding UTR, transcript, or protein are denoted by * and GO terms denoted by #. EC, extracellular; IC, intracellular; Mem., membrane; MFE, minimum free energy; MM, macromolecular; NR, negative regulation of; PR, positive regulation of; TS, TargetScan software. All metrics are averaged over 1,000 models (see Methods for details).
To assess the overall predictive power of the model, we constructed ROC plots with averaged prediction scores and found an AUC of 0.81 ± 0.02 (95% CI) (Figure 5C). By examining the parameters of the model, we assessed the relative contribution of each feature to learning (Figure 5E). We found that gene expression was the most informative variable, followed by protein length, the presence of sheet motifs, and various GO terms. Features of genes, transcripts, and proteins were included in the group of relatively less important but significant variables, suggesting that a wide range of fine-tuning processes contribute to MAP generation. Since estimates of relative importance can be influenced by related variables, we used a second method to assess feature importance. We assessed the predictive capacity of a logistic regression model, selecting only the top 10 most informative features. Despite this constraint, the model demonstrated comparable predictive power. The frequency with which features were selected in this model (Figure 5D) coincided with the relative weight when all input variables were considered (Figure 5E).
A 2-class distinction of MAP source and nonsource genes does not take into consideration that some source genes generate up to 64 nonredundant MAPs, while other genes produce only one (Figure 1B). To integrate these findings, we produced a nuanced version of the classification model that made predictions for 3 ordered groups: none (no MAPs), low (1–2 MAPs), and high (≥ 3 MAPs). Predictions were most accurate for the high category, which obtained an AUC of 0.86 ± 0.02, while the low and none groups had AUCs of 0.64 ± 0.01 and 0.81 ± 0.02, respectively (Supplemental Figure 5A). Clearly, the model had difficulty distinguishing the low group, for which its predictions reached a maximum probability of 0.43 compared with 0.99 for the high and none categories (Supplemental Figure 5C). Interestingly, when we compared the relative contribution of different input parameters between the 2-class and 3-class models, we found a very similar hierarchy (Figure 5E and Supplemental Figure 5B). We conclude that no particular feature within the model distinguishes genes that generate few versus numerous MAPs.
Model validation with independent data sets and human cancer cell lines.
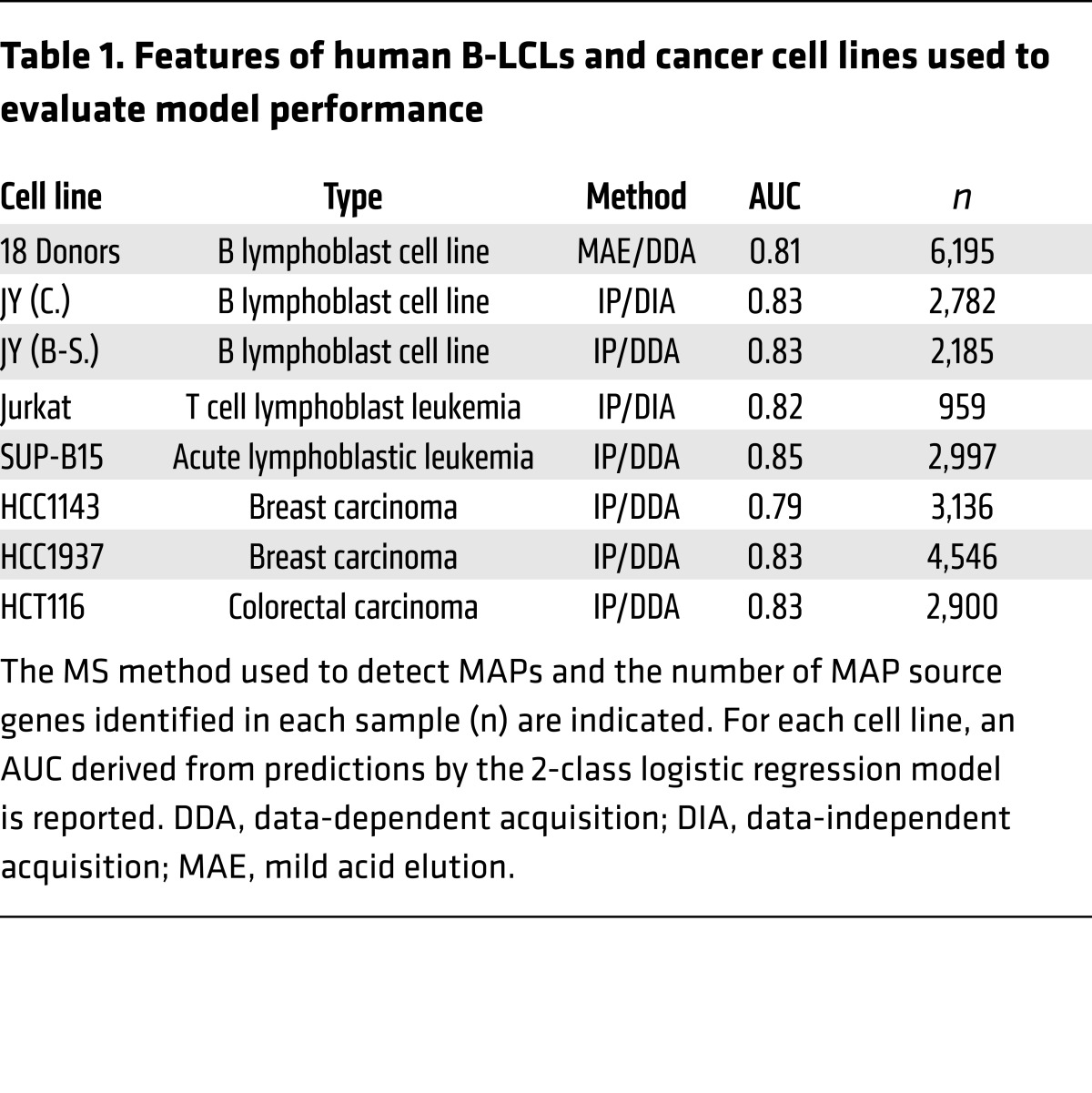
The various strategies used for high-throughput MS analyses of the immunopeptidome present strengths and limitations (61). In the present study, MAPs were isolated from 18 B-LCLs by mild acid elution and analyzed by data-dependent MS. To gauge the robustness of the model, we tested it on MAPs identified by 2 other groups in JY B-LCLs. MAPs in these 2 data sets were isolated by MHCI immunoprecipitation; one study used data-dependent MS (36), and the other used data-independent MS (21). While our data set contained MAPs presented by 27 HLA-A,B allotypes, JY B-LCLs express just 2 of these: HLA-A*02:01 and HLA-B*07:02. Transcriptomic data from JY B-LCLs (62) defined a set of candidate genes on which we performed predictions with the 2-class logistic regression model. Notably, 82% of source genes for the 2 other data sets were included in our own set of source genes (Figure 6A). Moreover, our model effectively predicted MAP origin in these 2 independent data sets (Table 1 and Figure 6, B and C) despite differences in methods of MAP isolation and MS analyses.
Figure 6. Evaluation of model performance with independent data sets on human cancer cell lines.
(A) Overlap in source gene identifications between the present study and 2 independent studies of JY B-LCLs using different MS techniques: JY (C.) and JY (B-S.). (B) Distribution of prediction scores for MAP source genes in B-LCLs and cancer cell lines (details in Table 1); median value is shown with whiskers extending to the extremes of the interquartile range x 1.5; outliers are hidden. (C) Proportion of MAP source genes captured as a function of prediction score threshold.
Table 1. Features of human B-LCLs and cancer cell lines used to evaluate model performance.
We further challenged the model trained on 18 donor B-LCLs to predict MAP generation in 5 human cancer cell lines: 2 leukemias, 2 breast carcinomas, and 1 colorectal carcinoma (Table 1). To evaluate the performance of our model, we used previous analyses of the transcriptome (63) and the immunopeptidome (21, 36) of these cell lines. The models’ ability to predict MAP source genes was very good for the 5 cancer cell lines, with ROC AUC ranging from 0.79 to 0.85, similar to the accuracy observed with B-LCLs (Table 1). The distribution of the prediction scores for MAP source genes was similar in the various cell lines (Figure 6B), though the rate at which source genes were captured at different probabilities of MAP generation revealed slight divergence at the lower prediction scores (Figure 6C). These data suggest that MAP processing follows consistent rules with limited variations between cell types. Overall, we conclude that our prediction model is robust for cells of various lineages and that its accuracy is not biased by the methods used for MAP isolation or identification.
Discussion
To the best of our knowledge, this study reports the largest data set of MAPs to date. Several points can be made from our comprehensive analyses of 25,270 MAPs presented by 27 HLA-A,B allotypes, which illustrate how there can be “strength in numbers” (64). Indeed, while analyses of smaller data sets suggested that individual genes were represented in the immunopeptidome by only a single MAP (25), we found that MAP source genes generated up to 64 nonredundant MAPs. Importantly, we found that MAPs presented by 27 MHCI allotypes together cover an unexpectedly small fraction of the protein-coding exome (10%) because (a) 41% of genes did not generate detectable MAPs, and (b) MAPs derived from the same gene tend to originate from adjacent sequences. At the population level, one implication is that even though HLA allotypes have different peptide-binding motifs, a large fraction of MAPs presented by different subjects (2 to 4 HLA-A,B allotypes/subject) will originate from common genomic regions. Further studies are certainly warranted in order to explore whether, relative to the whole exome, MAP “hot spots” have distinctive features that would make their monitoring by T cells of special importance or whether these regions are simply opportunistically captured. For instance, are these hot spots preferential sites of somatic mutations in cancer cells or do they resemble viral genes? Notably, we observed that some features enriched in MAP source genes are also common in viral genes (e.g., internal disorder and 5′ UTR secondary structures). Indeed, disorder is prevalent in viral genomes (65), and several viral transcripts contain complex 5′ UTR secondary structures that stall ribosomal translation (66).
Our results suggest that at the systems level, MAP generation is regulated by specific features of transcripts and proteins that often affect translation and proteasomal degradation. For example, features of the 5′ UTR, such as shorter length, looser secondary structure, and fewer uORFs, which are easier for ribosomes to navigate, may confer efficient translation and consequently greater MAP generation. The importance of proteasomal processing is underscored by the prevalence of disorder and degradation motifs in MAP source proteins. Additionally, that MAPs originate preferentially from abundant transcripts is consistent with the fact that the immunopeptidome is different from one cell lineage to another and is affected by the metabolic status of cells (5, 51). The relation between transcript abundance and MAP presentation may also be relevant to the establishment of self-tolerance in the thymic medulla. Indeed, central self-tolerance depends on promiscuous gene expression by medullary thymic epithelial cells that collectively express almost all protein-coding genes (67, 68). Remarkably, this promiscuous gene expression follows a mosaic pattern: individual medullary thymic epithelial cells promiscuously express a limited number of genes, but at a high level (67, 69). A mosaic pattern of highly expressed genes may be instrumental in increasing the breadth of the MAP repertoire that can thereby induce central self-tolerance.
By taking into account the various features enriched in MAP source genes, we were able to build a logistic regression model that predicts whether or not a given gene will produce MAPs with a ROC AUC of 0.81 ± 0.02. The robustness of this model was validated by predicting MAP generation in 7 independent data sets. Notably, when the model was applied to predict MAP generation in 5 human cancer cell lines, it performed comparably well, suggesting strong potential for predicting MAP generation in a clinical context. Would it be possible to build an in silico antigen-processing machine that would predict with even greater accuracy sources and sites of MAP generation? We speculate that this may be possible if we trained the model with more quantitative data and more accurate assessment of features. Indeed, there are certain limitations to a rather coarse 2-class output, not the least of which is a lack of precision for the number of MAPs produced and their location along a protein. Recent developments in MS now enable quantification of MAPs in terms of number of copies per cell (61). High-throughput quantitative analyses of immunopeptidomes could thereby pave the way to the development of improved predictive models, and community-based efforts to achieve this goal should be encouraged (21).
Our demonstration that the immunopeptidome covers only a small fraction of the protein-coding exome has special relevance to cancer immunology. There is a general consensus that cancer-specific neo-MAPs derived from somatic mutations represent ideal targets for cancer immunotherapy (70). However, discovery of cancer-specific MAPs is currently fraught with major difficulties. Typically, neo-MAP discovery strategies adopt the following path: exome sequencing, identification of mutations, and selection of mutations located in peptide regions predicted to have a good MHC-binding affinity. However, when putative neo-MAPs are tested experimentally, by MS or immune assays, the hit rate is below 10% (71–73). Our contention is that this low success rate is simply due to the fact that few mutations are strategically located in MAP-generating regions and that most mutations are in exomic sequences that are not covered by the immunopeptidome. We believe that progress in the field of neo-MAP discovery will be greatly facilitated by large-scale analyses of cancer cell immunopeptidomes.
Methods
Cell lines.
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples of 18 volunteers. High-resolution HLA genotyping was performed at the Hôpital Maisonneuve-Rosemont using 500 ng of genomic DNA. B-LCLs were derived from PBMCs as described (27).
Proteogenomic identification of MAPs derived from B-LCLs.
We applied our previously described proteogenomic approach to isolate and sequence MAPs. The methods of cell culture, transcriptome sequencing, mild acid elution, and MS have been described previously (27, 28). RNA-sequencing data were mapped using kallisto version 0.42.5 to ENSEMBL assembly 37.75 (NCBI Bioproject database http://www.ncbi.nlm.nih.gov/bioproject/; accession PRJNA286122) (74, 75). Transcriptome sequencing revealed no genetic polymorphisms in the regions coding for the mature (active) form of PSMB5 and PSMB8, the proteasome subunits that are mainly responsible for MAP processing (data not shown). We defined the B-LCL transcriptome as 10,575 expressed (averaged transcripts per million > 2) and annotated protein-coding genes. To mitigate the risk of false positives, stringent quality filters were applied to the list of identified MAPs: a peptide length of 8–14 amino acids; a 1% false discovery rate; and a predicted IC50 of 1250 nM or less. The binding affinity threshold was chosen to optimize inclusivity and stringency; a less stringent threshold of 5,000 nM included 8.6% more MAPs and 2.3% more source genes (Supplemental Figure 1C). When possible, binding affinities were predicted with NetMHC 3.4 (21 allotypes); otherwise, NetMHCcons 1.1 was applied (6 allotypes). For each individual, peptides were assigned to the allele with the strongest binding affinity. Peptides were mapped to proteins using pyGeno (29, 74). We applied further filtering steps to facilitate bioinformatic analysis; peptides assigned to more than one gene origin, transcripts with incomplete 5′ and 3′ annotation, and proteins with internal stop codons were all excluded. Where multiple isoforms were identified for a gene, MAPs were assigned to the most abundant transcript. Estimates of HLA allele frequency were derived from the European Caucasian population registered in the National Marrow Donor Program (33). The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (PXD004023). In addition, the list of MAP sequences was deposited in the Immune Epitope Database (http://www.iedb.org/; 1000704). For IS-PRM analyses, nonsource peptides were chosen randomly from the lowest predicted quintile of nonsource genes generating a peptide that bound HLA-B*44:03 with an affinity IC50 of less than 50 nM. IS-PRM analyses for 2 sets of stable isotopically labeled synthetic peptides were performed as described (35).
Simulations of the redundancy in MAP and MAP source gene repertoires.
HLA allotypes were randomly ordered, and either peptides or genes were considered. The number of nonredundant identifications was counted, considering the repertoires of each subsequent allotype. The simulation was repeated 1,000 times; average repertoire sizes are shown. The same simulation considering subjects instead of allotypes was also performed (Supplemental Figure 1, A and B). We noted greater redundancy in this simulation due to some subjects sharing the same allotypes.
Spatial localization of MAPs along source proteins.
Every pair of overlapping MAPs was extracted for each protein generating more than 1 MAP. Overlapping MAP pairs were classified as sharing the same beginning “C-terminal extensions,” sharing the same end “N-terminal extensions,” being contained within another peptide, “Internal,” or sharing at least 1 amino acid, “Overlap.” Alleles presenting each peptide pair and their superfamilies were compared (34). Distances between adjacent MAP start sites on the same protein were computed for the actual distribution. For the random distribution, an equivalent number of MAPs was randomly placed within the same protein length and adjacent distances between start sites computed. To estimate exome coverage, a window of 50 amino acids or 25 amino acids was moved residue by residue along each of the 10,575 proteins expressed in our B-LCLs; the number of MAPs seen in each window was counted.
Evaluating features of transcripts and proteins.
To ensure the quality and relevance of our source and nonsource gene sets, we considered all genes expressed on average more than 2 transcripts per million. For each gene, the most expressed protein-generating transcript with complete HAVANA annotation and the corresponding protein were selected. Feature assembly was executed in Python version 2.7.10; pyGeno was used to extract transcript and protein sequences (29). Annotation translation was determined with the ENSEMBL BioMart extension (74). To calculate MAP density, NetMHC was used to predict the binding affinity of overlapping nonamers from each protein for all 27 allotypes expressed by the B-LCLs. NetMHC 3.4 was applied preferentially to predict binding affinities for 21 allotypes; NetMHCcons 1.1 was applied for the remaining 6 allotypes. The fraction of 9mers binding any of the 27 allotypes with an affinity of 1,250 nM or less was calculated for each protein.
B cell protein abundance in average spectral counts per gene evaluated by MS analysis of whole cell extracts was extracted from the Human Proteome Map (40). Genes with at least 1 transcript with an NMD biotype based on Vega annotation in ENSEMBL were considered to have NMD potential, that is, if a coding sequence finished more than 50 bp from a downstream splice site using any exon structure (http://vega.sanger.ac.uk/info/about/gene_and_transcript_types.html). uORFs were defined as nonoverlapping sequences within the 5′ UTR beginning with the cognate start codon AUG and ending with an in-frame stop codon. 5′ UTR secondary structure was predicted using RNAfold within the ViennaRNA Package version 2.1.7 (44). The percentage of AU-rich elements was defined as the number of A and/or U sequence of at least 5 nucleotides in length within the 3′ UTR. Stabilizing and destabilizing elements identified by Zhao et al. were queried in the 3′ UTR (48). TargetScan 7.0 was employed to predict microRNA-binding sites within the 3′ UTR (50). 3′ UTRs were prepared by removing ORFs; the number of nonoverlapping microRNA-binding sites was computed for all families of microRNAs. MicroRNA-binding sites were retrieved from the Molecular Signatures Database of GSEA (http://software.broadinstitute.org/gsea/msigdb/) and queried in all 3′ UTRs. To analyze the structural features of proteins, we used BioPython’s package SeqUtils (specifically the ProtParam tool) to predict the proportion of residues conforming to a helix, turn, or sheet motif as well as the isoelectric point, instability index, and hydropathy for each protein sequence (76).
Protein degradation prediction softwares.
Anaphase-promoting complex target sequences were predicted using GPS-ARM version 1.0 using default thresholds for D-box and KEN-box motif (52). PEST motifs were predicted using the function epestfind within EMBOSS version 6.5.7 (77). Ubiquitination sites were predicted with UbiProber (54) with a stringency of 70%. Three disorder prediction software programs were selected for the complementarity of their approaches: PONDR VLXT is a neural network predictor trained on missing residues in x-ray structures as well as known terminal and long disordered segments; DISOPRED, version 3.16, is a support vector machine and neural network predictor also trained on missing residues in x-ray structures; and IUPRED, version 1.0, is a biophysical model based on local interaction energies (78). Where residues were assigned to be disordered or not, disorder cutoff values were determined to equate the total disorder of the B-LCL proteome for PONDR-VLXT, DISOPRED, and IUPRED at 0.7, 0.3, and 0.5 respectively (Supplemental Figure 4A). Where lengths of N or C terminus disorder or internally disordered regions were computed, stretches up to 3 aa of ordered residues were allowed.
Data visualization.
Graphics were made in R, version 3.2.2, using ggplot2, version 1.0.0 (https://cran.r-project.org/web/packages/ggplot2/index.html).
GO analysis.
We compared either source or nonsource genes on a background of both groups using the R package topGO (60). The Fisher weight algorithm was used to reduce redundancies and compute P values.
Statistics.
To generate Figure 1A, a binomial distribution of the probability of detecting between 0 and 50,000 peptides in a repertoire of 10,575 genes was computed. An exact binomial test was used to compare the expected distribution with the experimental values. A nonparametric effect size measure, Cliff’s d, was used to compare enrichment of features in source and nonsource groups. The 95% CI was calculated based on 100 bootstraps using the orddom package, version 3.1, in R (https://cran.r-project.org/web/packages/orddom/orddom.pdf). Unless otherwise noted, we employed 2-sample Wilcoxon rank sum tests to compare continuous variables for robustness. P values of less than 0.05 were considered significant. All statistical analyses were performed in R, version 3.2.2.
Modeling.
The variables listed in Supplemental Table 5 were used as input variables for logistic regression models run with the R packages caret and MASS (79, 80). The top 50 most enriched GO terms from the source and nonsource groups were included (Supplemental Tables 3 and 4). Near-zero variance parameters were excluded; this excluded the majority of GO terms. To limit the extent of correlation in input variables that can obscure their relative weight, further variables were excluded. Spearman’s rank correlation coefficient r was calculated for each pair of input parameters, and a maximum absolute r of 0.6 was permitted (Supplemental Figure 3). As noted, input variables were also normalized by length of the appropriate UTR, transcript, or protein. The data were divided into training and testing sets containing 80% and 20% of genes, respectively. A logistic regression model with or without recursive feature elimination was built with centered and scaled training data using 10-fold cross-validation. The model then predicted the probability of generating MAPs for each gene in the testing set. Relative variable weight was computed based on the t statistic for all model parameters. An ordered logistic regression model with 3-class outcomes was built using the same protocol; categories were selected to optimize class balance (number of genes: 4,380, none; 3,164, low; 3,031, high). All metrics reported are averages of 1,000 iterations of data division and model building. An R script is included in Supplemental Methods (source code) that trains and applies the 2-class logistic regression model using the data frame in Supplemental Table 6 and that reproduces the panels of Figure 5.
Validation in independent data sets and human cancer cell lines.
Transcriptomic data for JY (62) and 5 other human cancer cell lines (63) were combined with the respective immunopeptidomes described by other groups (21,36). Transcriptomic mapping was performed with kallisto, version 0.42.5, and the most expressed transcript for each gene was selected for analysis (75). Features of each gene and its gene products were annotated. Protein abundance was extracted from the Human Proteome Map (40) defined by the closest matching tissue (B cells for the JY and SUP-B15 cell lines, CD4 cells for Jurkat cells, and adult colon for HCT116) or using cell line–specific data for HCC1143 and HCC1937 (81). The 2-class logistic regression model using all features trained using 10-fold cross-validation on all genes from 18 B-LCL samples was used to predict MAP generation in each cell line.
Study approval.
This study was approved by the Comité d’Éthique de la Recherche de l’Hôpital Maisonneuve-Rosemont (permit number CÉR 14095). Volunteers provided written, informed consent.
Author contributions
HP was responsible for conception and design, acquisition of data, analysis and interpretation of data, and drafting and revising the article. DPG, TD, CD, EB, MC, AR, JPL, CC were responsible for acquisition of data, analysis and interpretation of data, and revising the article. SL, PT, CP were responsible for conception and design, analysis and interpretation of data, and revising the article. SM was responsible for analysis and interpretation of data and revising the article.
Supplementary Material
Acknowledgments
This work was supported by grants from the Quebec Breast Cancer Foundation (to CP and SM) and from the Genome Canada Innovation Network (to PT). We are most grateful to our blood donors. CP and PT hold Canada Research Chairs in Immunobiology, and Proteomics and Bioanalytical Spectrometry, respectively. SM holds the CIBC Breast Cancer Research Chair at Université de Montréal. The CP lab is supported in part by the Katelyn Bedard Bone Marrow Association.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(12):4690–4701. doi:10.1172/JCI88590.
See the related Commentary at I’ve got algorithm: predicting tumor and autoimmune peptide targets for CD8+ T cells.
Contributor Information
Hillary Pearson, Email: hillary.pearson@umontreal.ca.
Tariq Daouda, Email: tariq.daouda@umontreal.ca.
Diana Paola Granados, Email: dp.granados@umontreal.ca.
Chantal Durette, Email: chantal.durette@umontreal.ca.
Mathieu Courcelles, Email: mathieu.courcelles@umontreal.ca.
Anja Rodenbrock, Email: anja_rodenbrock@yahoo.ca.
Jean-Philippe Laverdure, Email: jean-philippe.laverdure@umontreal.ca.
Caroline Côté, Email: caroline.cote.8@umontreal.ca.
Sylvie Mader, Email: sylvie.mader@umontreal.ca.
Sébastien Lemieux, Email: s.lemieux@umontreal.ca.
Pierre Thibault, Email: pierre.thibault@umontreal.ca.
Claude Perreault, Email: c.perreault@videotron.ca.
References
- 1.Granados DP, Laumont CM, Thibault P, Perreault C. The nature of self for T cells-a systems-level perspective. Curr Opin Immunol. 2015;34:1–8. doi: 10.1016/j.coi.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Govern CC, Paczosa MK, Chakraborty AK, Huseby ES. Fast on-rates allow short dwell time ligands to activate T cells. Proc Natl Acad Sci U S A. 2010;107(19):8724–8729. doi: 10.1073/pnas.1000966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15(9):798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler TC, Kardar M, Chakraborty AK. Quorum sensing allows T cells to discriminate between self and nonself. Proc Natl Acad Sci U S A. 2013;110(29):11833–11838. doi: 10.1073/pnas.1222467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron E, et al. The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol Syst Biol. 2011;7:533. doi: 10.1038/msb.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vrisekoop N, Monteiro JP, Mandl JN, Germain RN. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity. 2014;41(2):181–190. doi: 10.1016/j.immuni.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3(12):952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 8.Hammer GE, Kanaseki T, Shastri N. The final touches make perfect the peptide-MHC class I repertoire. Immunity. 2007;26(4):397–406. doi: 10.1016/j.immuni.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Sidney J, Buus S, Sette A, Nielsen M, Peters B. Dataset size and composition impact the reliability of performance benchmarks for peptide-MHC binding predictions. BMC Bioinformatics. 2014;15:241. doi: 10.1186/1471-2105-15-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Verteuil D, Granados DP, Thibault P, Perreault C. Origin and plasticity of MHC I-associated self peptides. Autoimmun Rev. 2012;11(9):627–635. doi: 10.1016/j.autrev.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Eisenlohr LC, Huang L, Golovina TN. Rethinking peptide supply to MHC class I molecules. Nat Rev Immunol. 2007;7(5):403–410. doi: 10.1038/nri2077. [DOI] [PubMed] [Google Scholar]
- 13.Vigneron N, Van den Eynde BJ. Proteasome subtypes and the processing of tumor antigens: increasing antigenic diversity. Curr Opin Immunol. 2012;24(1):84–91. doi: 10.1016/j.coi.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Rock KL, Farfán-Arribas DJ, Colbert JD, Goldberg AL. Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 2014;35(4):144–152. doi: 10.1016/j.it.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodenough E, et al. Cryptic MHC class I-binding peptides are revealed by aminoglycoside-induced stop codon read-through into the 3’ UTR. Proc Natl Acad Sci U S A. 2014;111(15):5670–5675. doi: 10.1073/pnas.1402670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antón LC, Yewdell JW. Translating DRiPs: MHC class I immunosurveillance of pathogens and tumors. J Leukoc Biol. 2014;95(4):551–562. doi: 10.1189/jlb.1113599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apcher S, Millot G, Daskalogianni C, Scherl A, Manoury B, Fåhraeus R. Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci U S A. 2013;110(44):17951–17956. doi: 10.1073/pnas.1309956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granados DP, et al. MHC I-associated peptides preferentially derive from transcripts bearing miRNA response elements. Blood. 2012;119(26):e181–e191. doi: 10.1182/blood-2012-02-412593. [DOI] [PubMed] [Google Scholar]
- 20.Laumont CM, et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun. 2016;7:10238. doi: 10.1038/ncomms10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caron E, et al. An open-source computational and data resource to analyze digital maps of immunopeptidomes. Elife. 2015;4:e07661. doi: 10.7554/eLife.07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickman HD, et al. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172(5):2944–2952. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 23.Mester G, Hoffmann V, Stevanović S. Insights into MHC class I antigen processing gained from large-scale analysis of class I ligands. Cell Mol Life Sci. 2011;68(9):1521–1532. doi: 10.1007/s00018-011-0659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassan C, et al. The human leukocyte antigen-presented ligandome of B lymphocytes. Mol Cell Proteomics. 2013;12(7):1829–1843. doi: 10.1074/mcp.M112.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoof I, van Baarle D, Hildebrand WH, Keşmir C. Proteome sampling by the HLA class I antigen processing pathway. PLoS Comput Biol. 2012;8(5):e1002517. doi: 10.1371/journal.pcbi.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 2013;191(12):5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granados DP, et al. Impact of genomic polymorphisms on the repertoire of human MHC class I-associated peptides. Nat Commun. 2014;5:3600. doi: 10.1038/ncomms4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granados DP, et al. Proteogenomic-based discovery of minor histocompatibility antigens with suitable features for immunotherapy of hematologic cancers. Leukemia. 2016;30(6):1344–1354. doi: 10.1038/leu.2016.22. [DOI] [PubMed] [Google Scholar]
- 29. Daouda T, Perreault C, Lemieux S. pyGeno: A Python package for precision medicine and proteogenomics. F1000Research 2016;5(381). https://f1000research.com/articles/5-381/v2 Accessed October 31, 2016.
- 30.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 31.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36(Web Server issue):W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karosiene E, Lundegaard C, Lund O, Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64(3):177–186. doi: 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
- 33.González-Galarza FF, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43(Database issue):D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallien S, Kim SY, Domon B. Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM) Mol Cell Proteomics. 2015;14(6):1630–1644. doi: 10.1074/mcp.O114.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics. 2015;14(3):658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinzierl AO, et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface. Mol Cell Proteomics. 2007;6(1):102–113. doi: 10.1074/mcp.M600310-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Jovanovic M, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347(6226):1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Aebersold R. The interdependence of transcript and protein abundance: new data--new complexities. Mol Syst Biol. 2016;12(1):856. doi: 10.15252/msb.20156720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Princiotta MF, et al. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18(3):343–354. doi: 10.1016/S1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 42.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016;5:e10921. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106(18):7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szostak E, Gebauer F. Translational control by 3’-UTR-binding proteins. Brief Funct Genomics. 2013;12(1):58–65. doi: 10.1093/bfgp/els056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schott J, Stoecklin G. Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA. 2010;1(3):432–456. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]
- 47.Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6):e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. Massively parallel functional annotation of 3’ untranslated regions. Nat Biotechnol. 2014;32(4):387–391. doi: 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Verteuil D, et al. Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Mol Cell Proteomics. 2010;9(9):2034–2047. doi: 10.1074/mcp.M900566-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z, et al. GPS-ARM: computational analysis of the APC/C recognition motif by predicting D-boxes and KEN-boxes. PLoS One. 2012;7(3):e34370. doi: 10.1371/journal.pone.0034370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21(7):267–271. doi: 10.1016/S0968-0004(96)10031-1. [DOI] [PubMed] [Google Scholar]
- 54.Chen X, Qiu JD, Shi SP, Suo SB, Huang SY, Liang RP. Incorporating key position and amino acid residue features to identify general and species-specific Ubiquitin conjugation sites. Bioinformatics. 2013;29(13):1614–1622. doi: 10.1093/bioinformatics/btt196. [DOI] [PubMed] [Google Scholar]
- 55.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11(9):830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 56.van der Lee R, et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014;8(6):1832–1844. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42(1):38–48. doi: 10.1002/1097-0134(20010101)42:1<38::AID-PROT50>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 58.Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31(6):857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 60. Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.24.0. https://bioconductor.org/packages/release/bioc/html/topGO.html Accessed October 5, 2016.
- 61.Caron E, Kowalewski DJ, Chiek Koh C, Sturm T, Schuster H, Aebersold R. Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Mol Cell Proteomics. 2015;14(12):3105–3117. doi: 10.1074/mcp.M115.052431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Concha M, et al. Identification of new viral genes and transcript isoforms during Epstein-Barr virus reactivation using RNA-Seq. J Virol. 2012;86(3):1458–1467. doi: 10.1128/JVI.06537-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33(3):306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 64.Benoist C, Germain RN, Mathis D. A plaidoyer for ‘systems immunology’. Immunol Rev. 2006;210:229–234. doi: 10.1111/j.0105-2896.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- 65.Longhi S. Structural disorder in viral proteins. Protein Pept Lett. 2010;17(8):930–931. doi: 10.2174/092986610791498975. [DOI] [PubMed] [Google Scholar]
- 66.Murat P, Tellam J. Effects of messenger RNA structure and other translational control mechanisms on major histocompatibility complex-I mediated antigen presentation. Wiley Interdiscip Rev RNA. 2015;6(2):157–171. doi: 10.1002/wrna.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sansom SN, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014;24(12):1918–1931. doi: 10.1101/gr.171645.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential Features of AIRE-Induced and AIRE-Independent Promiscuous Gene Expression in Thymic Epithelial Cells. J Immunol. 2015;195(2):498–506. doi: 10.4049/jimmunol.1500558. [DOI] [PubMed] [Google Scholar]
- 69.Brennecke P, et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol. 2015;16(9):933–941. doi: 10.1038/ni.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 71.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 73.Blankenstein T, Leisegang M, Uckert W, Schreiber H. Targeting cancer-specific mutations by T cell receptor gene therapy. Curr Opin Immunol. 2015;33:112–119. doi: 10.1016/j.coi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 76.Wilkins MR, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 77.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 78.Dosztányi Z, Mészáros B, Simon I. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Brief Bioinformatics. 2010;11(2):225–243. doi: 10.1093/bib/bbp061. [DOI] [PubMed] [Google Scholar]
- 79. Kuhn M, et al. caret: Classification and Regression Training. https://github.com/topepo/caret/ Published August 5, 2016. Accessed October 5, 2016.
- 80. Venables WN, Ripley BD. Modern applied statistics with S. Fourth edition. New York: Springer-Verlag; 2002. [Google Scholar]
- 81.Lawrence RT, et al. The proteomic landscape of triple-negative breast cancer. Cell Rep. 2015;11(4):630–644. doi: 10.1016/j.celrep.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.