Abstract
Mutations and deletions in components of ubiquitin ligase complexes that lead to alterations in protein turnover are important mechanisms in driving tumorigenesis. Here we describe an alternative mechanism involving upregulation of the microRNA miR-424 that leads to impaired ubiquitination and degradation of oncogenic transcription factors in prostate cancers. We found that miR-424 targets the E3 ubiquitin ligase COP1 and identified STAT3 as a key substrate of COP1 in promoting tumorigenic and cancer stem-like properties in prostate epithelial cells. Altered protein turnover due to impaired COP1 function led to accumulation and enhanced basal and cytokine-induced activity of STAT3. We further determined that loss of the ETS factor ESE3/EHF is the initial event that triggers the deregulation of the miR-424/COP1/STAT3 axis. COP1 silencing and STAT3 activation were effectively reverted by blocking of miR-424, suggesting a possible strategy to attack this key node of tumorigenesis in ESE3/EHF–deficient tumors. These results establish miR-424 as an oncogenic effector linked to noncanonical activation of STAT3 and as a potential therapeutic target.
Introduction
Cancer of the prostate is a leading cause of cancer death in developed countries (1). Clinical behavior of prostate cancer ranges from indolent disease to very aggressive tumors that rapidly progress to castration resistance (2). The molecular mechanisms contributing to this heterogeneity are still poorly understood (3). Cancer stem-like cells within the primary tumors may contribute to metastasis, treatment failure, and aggressive clinical behavior (4). Thus, the identification of pathways leading to the expansion of stem-like cancer cells could provide innovative strategies for targeting this cell subpopulation and prevent disease progression and recurrence.
Chromosomal translocations involving oncogenic ETS transcription factors (TFs), like ERG, are found in about 50% of prostate cancers (5–7). Deregulation of endogenous ETS TFs, like ESE3/EHF, has also a relevant role in prostate tumorigenesis (8–11). We reported that in ETS translocation–negative primary prostate tumors, reduced expression of ESE3/EHF was associated with increased expression of epithelial-to-mesenchymal transition (EMT) and stem cell–like genes and clinical features of aggressive disease (8). Defining the mechanisms leading to dedifferentiation and acquisition of stem-like properties in this context could provide relevant insights for the management of prostate cancer.
MicroRNAs (miRNAs) are naturally occurring small noncoding RNAs that regulate gene expression by inhibiting mRNA translation into proteins (12). Deregulated expression of miRNAs is very frequent in human cancers, including prostate cancer (13–15). Mutations and deletions of genes encoding components of the ubiquitin ligase complexes resulting in altered protein ubiquitination and turnover are also emerging as important mechanisms driving prostate tumorigenesis (16–19).
In this study we defined a novel oncogenic axis linking miRNA deregulation and altered protein ubiquitination. This axis involved microRNA-424–induced posttranscriptional silencing of the E3 ubiquitin ligase COP1 leading to reduced degradation and activation of STAT3, a critical TF involved in tumor progression (20, 21). This pathway was initiated by loss of ESE3/EHF and led to acquisition of tumor-initiating and metastatic properties in prostate epithelial cells. Thus, this study establishes miR-424 as a key oncogenic effector in prostate cancer and uncovers a novel mechanism controlling STAT3 protein level and activity. Furthermore, we identify an important pathway leading to aggressive prostate cancer, opening new perspectives for the treatment of this disease.
Results
miR-424 is upregulated in prostate tumors and associated with aggressive features.
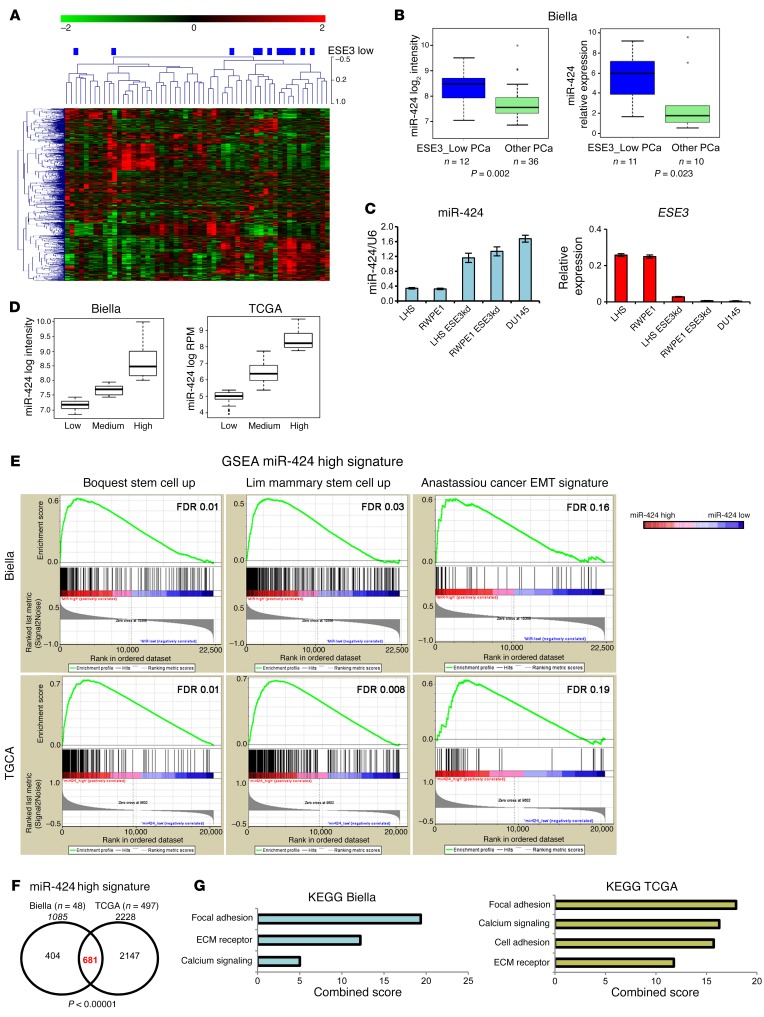
To understand the mechanisms leading to dedifferentiation in prostate tumors and the role of ETS factors in this context, we profiled miRNA expression in human normal prostate (n = 21) and prostate tumor (n = 48) samples that we previously analyzed for the global gene expression and ETS gene status (8, 10). We observed a substantial heterogeneity of the miRNA profiles across tumor samples, in line with the notion of subtype-specific miRNA deregulation. Interestingly, most of the ESE3lo tumors displayed a similar miRNA profile and clustered together in unsupervised hierarchical clustering analysis (Figure 1A). We reported previously that these tumors represented a distinct subgroup characterized by low ESE3/EHF expression and the absence of ETS gene rearrangements. ESE3lo tumors showed distinctive biological features, including increased expression of EMT and stem cell genes (8). Low ESE3/EHF expression was also associated with more aggressive features and poor clinical outcome (8). To identify miRNAs whose altered expression was causally linked to ESE3/EHF downregulation, we profiled miRNAs in normal prostate epithelial cells with and without stable knockdown of ESE3/EHF (ESE3kd cells). Stable ESE3kd cells recapitulated the EMT and cancer stem cell–like features observed in ESE3lo tumors and provided an adequate model to study the transition from normal to transformed prostate epithelial cells (8). Notably, we found miR-424 as the top miRNA upregulated in both ESE3kd cells and ESE3lo tumors. miR-424 was significantly overexpressed in primary tumors compared with normal prostate (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI86505DS1) and was preferentially upregulated in the ESE3lo tumor subgroup as indicated by both microarray and quantitative real-time RT-PCR (qRT-PCR) analysis (Figure 1B). miR-424 level was also inversely correlated to ESE3/EHF expression in a panel of prostate cell lines (Figure 1C). Furthermore, miR-424 expression was particularly low in androgen receptor–positive (AR-positive) LNCaP and VCaP cells compared with AR-negative DU145 cells, consistent with the higher expression of ESE3/EHF in the first 2 cell lines (Supplemental Figure 1C). Also, treatment of LNCaP cells with dihydrotestosterone did not lead to a substantial increase of miR-424 level, indicating that this miRNA was not androgen regulated (data not shown).
Figure 1. miR-424 is elevated in prostate tumors and is associated with EMT and stem cell–like features.
(A) Unsupervised hierarchical clustering (UHC) of primary prostate tumors from the Biella data set according to their miRNA profile. ESE3lo tumors (blue rectangles) cluster together and have similar miRNA profile. (B) miR-424 log2 intensity levels evaluated by microarray (left) and relative expression levels evaluated by qRT-PCR (right) in prostate tumors. (C) miR-424 level evaluated by qRT-PCR in indicated prostate cell lines. The results were normalized to RNU6 internal control. (D) Prostate samples divided according to miR-424 expression in 2 independent primary prostate tumor data sets, The Cancer Genome Atlas (TCGA) project and the in-house Biella data set (see Methods for details). (E) Gene set enrichment analysis (GSEA) comparing miR-424hi with miR-424lo prostate tumors in the 2 indicated microarray data sets (total = 545), using the indicated stem cell–like and EMT gene signatures. (F) Venn diagram crossing genes upregulated in miR-424hi signatures extracted from Biella and TCGA data sets and showing a significantly high convergence (P < 0.00001). (G) Functional analysis of miR-424hi signatures in Biella and TCGA data sets using ENCODE pathway analysis tools. Signatures were extracted by comparison of miR-424hi versus miR-424lo samples. See Methods for experimental details.
To further support these findings we examined the expression of miR-424 in the large TCGA (The Cancer Genome Atlas) primary prostate tumor data set (n = 419). In both the TCGA and the Biella data set the expression of miR-424 was highly heterogeneous with tumors exhibiting a low, intermediate, or high level of the miRNA (Figure 1D). Interestingly, in both data sets high–miR-424–expressing tumors were prevalently ERG negative (Supplemental Figure 1B). To understand the potential impact of miR-424 upregulation on the tumor phenotype, we applied differential gene expression analysis and compared tumors with high and low miR-424 level in the available data sets to identify molecular and biological differences between the 2 subgroups. Gene set enrichment analysis (GSEA) of the extracted miR-424 signatures revealed that miR-424–high tumors compared with miR-424–low tumors were significantly enriched of genes involved in mesenchymal transition (Anastassiou_cancer_mesenchymal_transition_signature) and stem cell–like features (Boquest_stem_cell_up and Lim_mammary_stem_cell_up) (Figure 1E). In contrast, GSEA with androgen-related gene lists (wang_response_to_androgen_up; nelson_response_to_androgen_up; nelson_response_to_androgen_dn) revealed no significant enrichment in both data sets (data not shown). Furthermore, we did not find any significant association between miR-424 level and the recently described AR score in the TCGA data set (22) (Supplemental Figure 1D). Strikingly, the gene signatures of miR-424–high tumors in distinct data sets were significantly convergent (n = 681, P < 0.000001), supporting miR-424 as a key driver of these features (Figure 1F). Functional annotation analysis of the miR-424 gene signatures also revealed a significant overlap between the 2 data sets with enrichment of common pathways, including focal adhesion, extracellular membrane receptor, and calcium signaling (Figure 1G). Notably, analysis of miRNA expression data sets from additional tumor types revealed that miR-424 was significantly upregulated also in other tumors, including lung, gastric, and breast cancer (Supplemental Figure 1D). Collectively, these findings indicated that miR-424 could act as an oncogenic miRNA deregulated in prostate cancer and potentially in multiple other epithelial tumors.
miR-424 is repressed by ESE3/EHF in normal prostate epithelial cells.
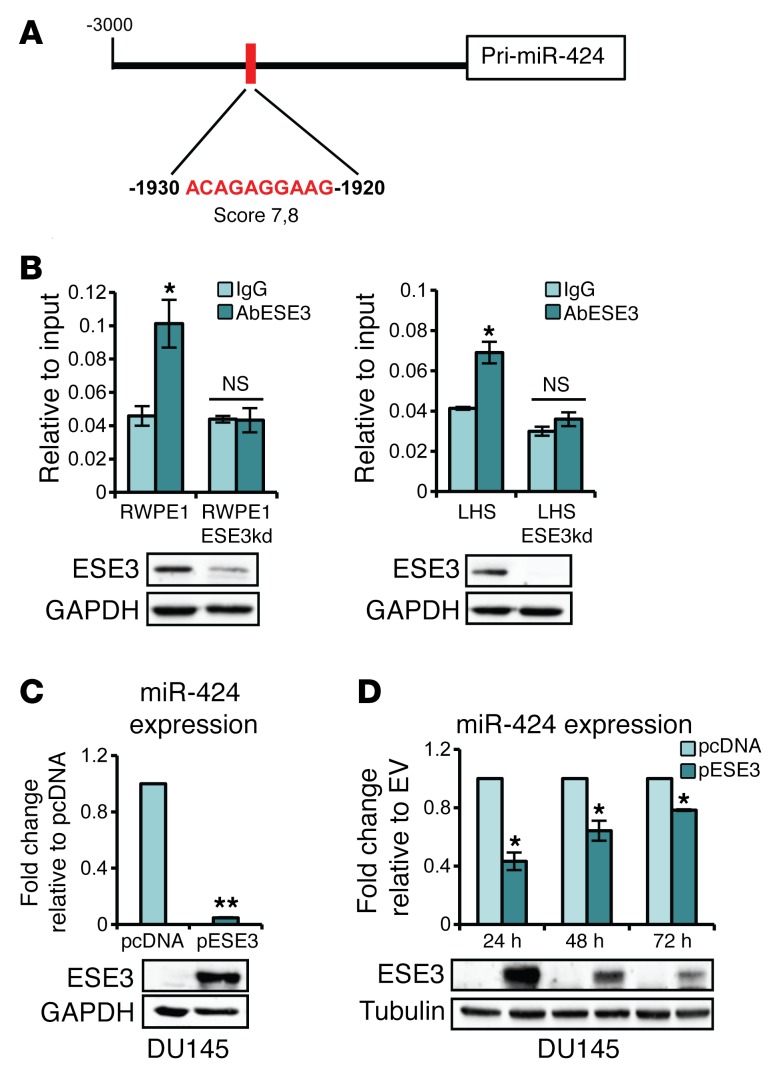
Our data in prostate tumors and cell lines indicate an inverse correlation between ESE3/EHF and miR-424 and suggested that ESE3/EHF could negatively regulate miR-424. We found multiple ETS binding sites with high score in the MIR424 promoter (Supplemental Figure 2A). ChIP showed binding of ESE3/EHF to the highest-score ETS binding site, in the MIR424 promoter in RWPE1 and LHS cells and not in the derivative ESE3kd lines (Figure 2, A–D, and Supplemental Figure 2B). Consistent with a repressive role of ESE3/EHF, we found significant enrichment of repressive histone marks on the MIR424 promoter in parental ESE3/EHF–expressing RWPE1 and LHS cells (Supplemental Figure 2C). In contrast, enrichment of active chromatin marks was found on the MIR424 promoter in ESE3kd cells. Consistently with a repressive role, stable (Figure 2C) and transient expression (Figure 1D) of ESE3/EHF in DU145 cells reduced miR-424 level. Furthermore, transient overexpression of ESE3/EHF reduced miR-424 level in a time-dependent manner in RWPE1-ESE3kd cells (Supplemental Figure 2D). Thus, collectively these data demonstrate that ESE3/EHF prevented the expression of miR-424 in normal prostate epithelial cells and loss of ESE3/EHF triggered miR-424 upregulation.
Figure 2. ESE3/EHF occupies MIR424 promoter region and represses miR-424 transcription.
(A) Schema of MIR424 promoter and position of the evaluated ETS binding site relative to the 5′ end of pri-miR-424 (RefSeq NR_029946). (B) ESE3/EHF occupancy on MIR424 promoter evaluated by ChIP and quantitative PCR in indicated cell lines. The results are represented relative to input. Bottom panels: Immunoblots of ESE3 in indicated cell lines. (C) miR-424 level in DU145 with stable ESE3/EHF (pESE3) expression compared with control cells (pcDNA). The results were normalized to RNU6 and are represented as miR-424 expression relative to pcDNA. Bottom panel: Immunoblot of ESE3. (D) miR-424 level evaluated by qRT-PCR in DU145 following transfection with pESE3 or pcDNA at the indicated time points (top), and immunoblot of ESE3/EHF (bottom). *P < 0.05; **P < 0.01.
miR-424 promotes oncogenic properties in prostate epithelial cells.
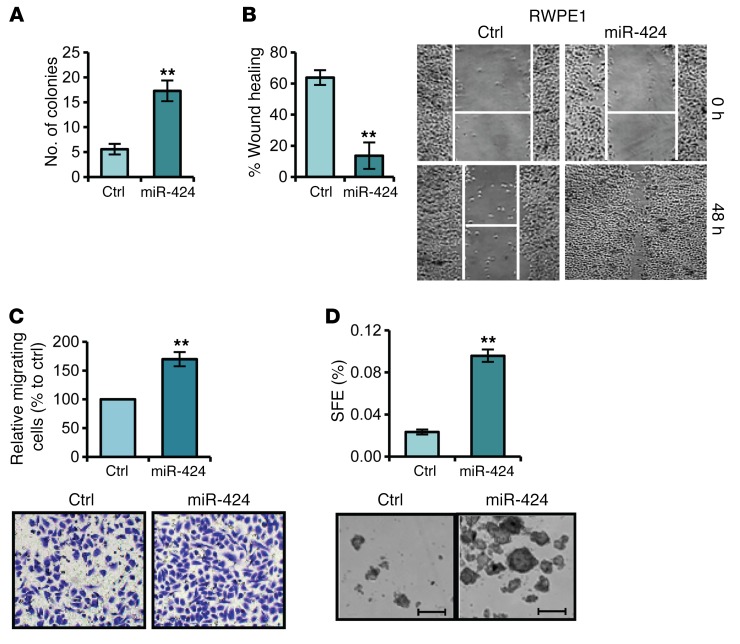
Our data identified miR-424 as a potential oncogenic miRNA in prostate cancer. Consistently, we found that overexpression of miR-424 in RWPE1 cells promoted anchorage-independent growth as assessed by the soft agar assay (Figure 3A). miR-424 promoted cell migration and invasion as shown by wound healing and Boyden chamber assays (Figure 3, B and C). miR-424–transfected RWPE1 cells exhibited also a dramatic change in morphology and expression of EMT markers consistent with reprogramming toward a more mesenchymal phenotype (Supplemental Figure 3A). Furthermore, formation of prostatospheres (PSs) and expression of stem cell–like markers, features associated with a cancer stem cell–like phenotype (8, 23), were increased by miR-424 in RWPE1 cells (Figure 3D and Supplemental Figure 3B). Notably, stable expression of miR-424 in the low-expressing LNCaP cells induced similar consequences, including increased cell migration and PS formation (Supplemental Figure 3, C–G). Interestingly, both stable and transient expression of miR-424 in LNCaP cells induced a comparable increase of anchorage-independent growth in the soft agar assay (Supplemental Figure 3H).
Figure 3. miR-424 promotes transforming phenotypes in prostate epithelial cells.
miR-424 precursor (miR-424) or negative control (Ctrl) was transiently transfected in RWPE1 cells. (A) Colony formation in soft agar. (B and C) Cell migration by wound healing (WH) assay (B) and Boyden chamber assay (C). (D) Sphere-forming efficiency (SFE) (top) and representative images of spheroids (bottom). **P < 0.01. Scale bar: 200 µm.
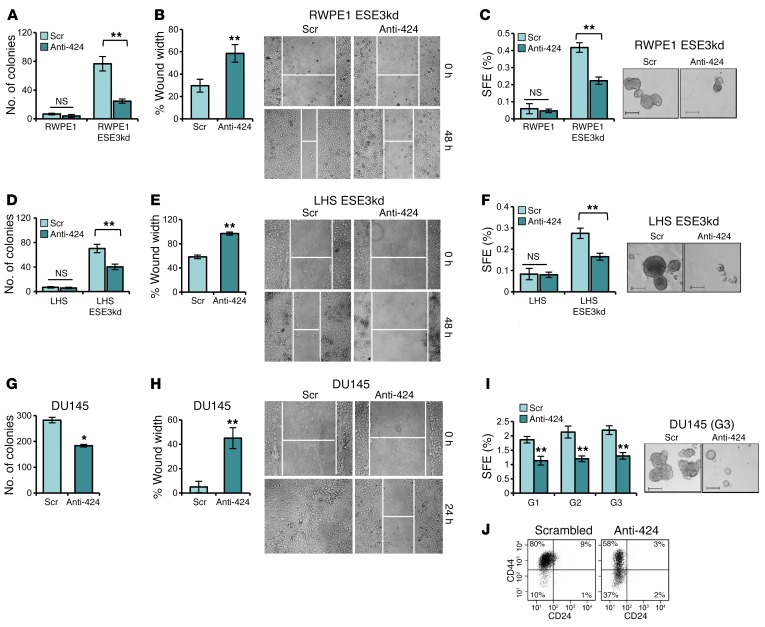
Conversely, a miR-424 antagonist (anti–miR-424), which effectively reduced miR-424 level (Supplemental Figure 4, A and B), decreased anchorage-independent growth, migration, and PS formation in the tumorigenic RWPE1-ESE3kd (Figure 4, A–C) and LHS-ESE3kd (Figure 4, D–F), sustaining the role of miR-424 as a key oncogenic driver in the ESE3lo context. Furthermore, in metastatic DU145 cells expressing high miR-424 and low ESE3/EHF, anti–miR-424 decreased miR-424 (Supplemental Figure 4C), anchorage-independent growth, and cell migration (Figure 4, G–H, and Supplemental Figure 4E). PS-forming ability of DU145 cells was also reduced by anti–miR-424, and the effect remained in the following PS generations, indicating a persistent impairment of the stem-like properties (Figure 4I). Consistently, the fraction of CD44hiCD24lo DU145 cells (Figure 4J), marking the stem-like cancer cell subpopulation (23), and the expression of stem cell–like markers (Supplemental Figure 4D) were also significantly reduced by anti–miR-424. Notably, modulating miR-424 level did not affect proliferation and cell cycle in RWPE1, LNCaP, and DU145 cells (Supplemental Figure 5, A–C), indicating that miR-424 impacted prominently on specific properties of the malignant cells such as the anchorage-independent growth and stem cell–like phenotype.
Figure 4. miR-424 inhibition reverts transforming phenotypes in transformed cell models.
(A–F) Colony formation in soft agar (A and D), WH (B and E), and SFE and representative images of spheroids (C and F) in ESE3kd cell line models following inhibition of miR-424 by anti–miR-424 (Anti-424) or scrambled control (Scr). (G–J) Colony formation in soft agar (G), WH assay (H), SFE and self-renewal potential (I), and representative images of spheroids (I, right). (J) Evaluation of CD44hiCD24lo fraction by FACS following inhibition of miR-424 in DU145. Data show mean ± SD (n = 3) of 1 representative experiment. *P ≤ 0.05, **P ≤ 0.01 by 2-tailed Student’s t test. Scale bars: 200 μm.
miR-424 targets the E3 ubiquitin ligase COP1.
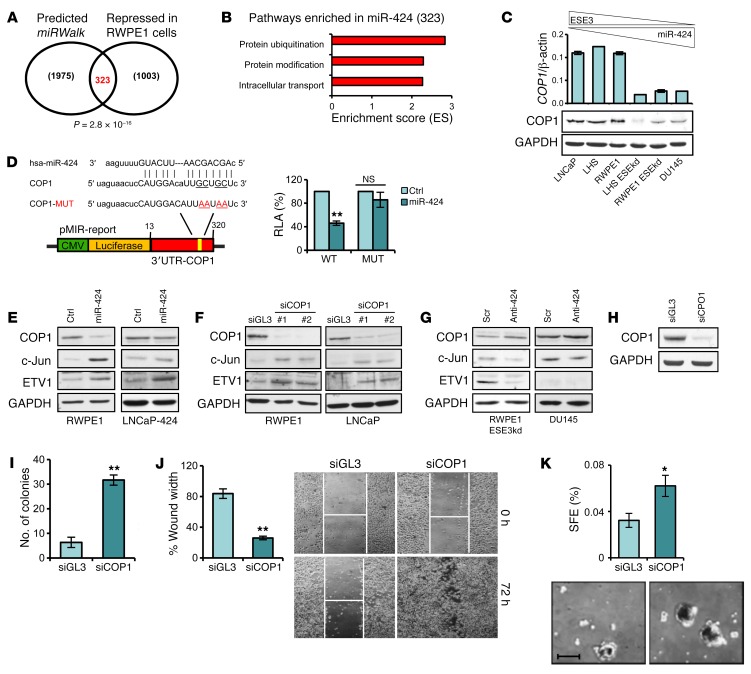
To identify the relevant targets of miR-424 in prostate epithelial cells, we performed gene expression analysis in RWPE1 cells transfected with or without miR-424. The list of miR-424–repressed genes was then merged with a list of genes with predicted miR-424 targets in their 3′-UTR nominated using multiple prediction algorithms to enrich for putative direct targets. Predicted targets converged significantly with miR-424–repressed genes (P = 2.8 × 10–16), yielding a total of 323 candidate miR-424 targets (Figure 5A and Supplemental Table 1). Intriguingly, functional annotation analysis revealed among the 323 putative targets a striking prevalence of genes involved in protein ubiquitination and specifically encoding components of the E3 ubiquitin ligase complexes (n = 29) (Figure 5B and Supplemental Table 2). Proteins of the E3 ubiquitin ligase complex are emerging as important oncogenic or tumor suppressor factors deregulated as a consequence of genetic events in various cancers, including prostate cancer (16, 18, 19). Notably, among the top miR-424 targets we found the E3 ubiquitin ligase COP1 (also known as RFWD2), which is a known tumor suppressor deleted in 3%–8% of prostate cancers and other types of tumors (16, 19). In keeping with the hypothesis of a link between miR-424 and COP1, we found a positive correlation between COP1 and ESE3/EHF in multiple prostate cancer data sets, with the most significant correlation (P < 0.00001) observed in the 2 larger data sets examined (Supplemental Figure 6, A and B). COP1 correlated positively with ESE3/EHF also in other cancers, including kidney, breast, and bladder cancer (Supplemental Figure 6A). An inverse correlation was also observed between MIR424 and COP1 (n = 419; P < 0.0009) in the TCGA prostate cancer data set (Supplemental Figure 6C). Furthermore, COP1 expression was directly correlated with ESE3/EHF and inversely correlated with miR-424 levels in prostate cell lines (Figure 5C).
Figure 5. miR-424 targets the E3 ubiquitin ligase COP1, leading to multiple oncogenic TF activation.
(A) Venn diagram of predicted targets (miRWalk) and genes repressed by miR-424 in RWPE1 cells. (B) Functional annotation analysis by DAVID of the putative miR-424 target genes. (C) Level of COP1 by qRT-PCR (top) and immunoblotting (bottom) in prostate cell lines. (D) Diagram of 3′-UTR COP1 reporter construct with WT and mutated (MUT) sequence of the miR-424 binding site (left) and relative luciferase activity (RLA) following transfection of miR-424 in LNCaP cells (right). (E) COP1, c-Jun, and ETV1 expression in RWPE1 cells following transient transfection of miR-424 and LNCaP cells stably overexpressing miR-424. Replicate samples were run on separate gels for blotting with the different antibodies. (F) COP1, c-Jun, and ETV1 expression following transfection of siRNAs targeting COP1 (siCOP1#1 and #2) or control siRNA (siGL3) in RWPE1 and LNCaP cells. (G) COP1, c-Jun, and ETV1 expression following transfection of Anti-424 or Scr in RWPE-ESE3kd and DU145 cells. (H–K) Immunoblot of COP1 (H), colony formation in soft agar (I), cell migration by WH (J), and SFE and representative images of spheroids (K), following knockdown of COP1 by siRNA (siCOP1#1) and control (siGL3) in RWPE1 cells. Data show mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01. Scale bar: 200 µm.
These findings were in line with the hypothesis that ESE3/EHF and miR-424 deregulation resulted in downregulation of COP1. In support of this hypothesis, miR-424 inhibited the luciferase reporter containing the highly conserved miR-424 target sequence in the COP1 3′-UTR, while it was ineffective on the mutated 3′-UTR reporter, indicating that miR-424 acted directly on the 3′-UTR of the COP1 mRNA to block protein translation (Figure 5D). Consistently, transfection of miR-424 decreased COP1 protein level and increased concomitantly the level of c-Jun and ETV1, known COP1 targets (16, 19), in both RWPE1 and LNCaP cells (Figure 5E). Importantly, knockdown of COP1 had the same effect on c-Jun and ETV1 expression in these cells (Figure 5F). Furthermore, COP1 knockdown with and without miR-424 overexpression had similar effects on ETV1 and c-Jun, indicating that miR-424 was acting mainly by repressing COP1 (Supplemental Figure 7). On the other hand, anti–miR-424 increased COP1 and decreased ETV1 or c-Jun in the miR-424–expressing RWPE1-ESE3kd and DU145 cells (Figure 5G). Notably, COP1 downregulation was important for the phenotypic effects induced by miR-424 upregulation. Indeed, knockdown of COP1 by siRNAs reproduced the effect of miR-424 on anchorage-independent growth, migration, and PS formation in RWPE1 cells (Figure 5, H–K). Conversely, expression of COP1 in DU145 cells reduced anchorage-independent growth, migration, and PS formation (Supplemental Figure 8, A–D). Notably, coexpression of COP1 in RWPE1 cells reversed the induction of anchorage-independent growth by miR-424 (Supplemental Figure 8, E and F). Thus, modulating COP1 level reproduced the major effects on target proteins and cell phenotypes associated with deregulation of miR-424, and COP1 prevented the transforming effects of miR-424, indicating that it was a key target of this oncogenic axis.
miR-424 increases STAT3 protein level by targeting COP1.
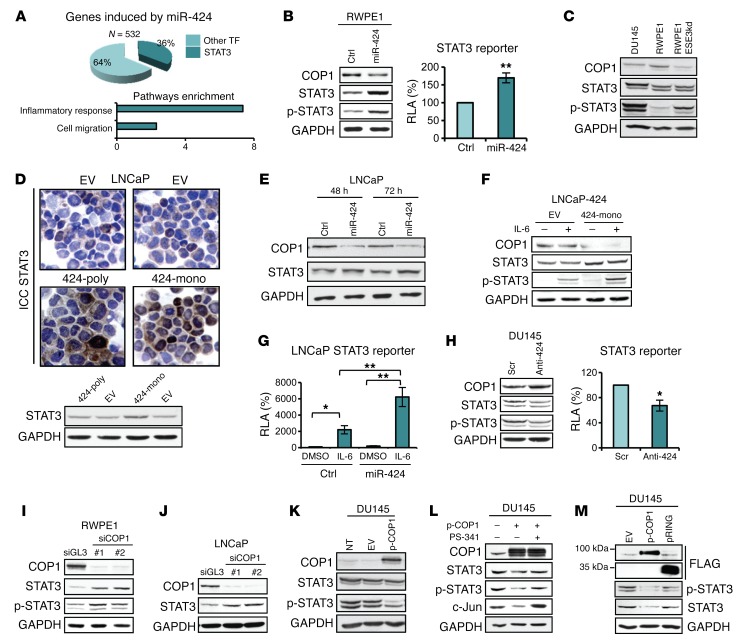
miR-424 upregulation was associated with broad transcriptional reprogramming in both prostate epithelial cells and human tumors. To further investigate this aspect, we applied the ENCODE transcription factor target tool to identify the TFs putatively controlling the transcription of miR-424–activated genes in RWPE1 cells. We found STAT3 among the top-ranking enriched TFs (P = 7.06–20). Interestingly, the genes induced by miR-424 were significantly enriched of STAT3 targets, which represented 36% of the total induced genes (Figure 6A, top). Abnormal activation of STAT3 signaling is implicated in many cancers, including prostate cancer (21, 24–26). Functional annotation analysis of miR-424–activated genes in RWPE1 cells showed significant enrichment of genes in inflammatory response and cell migration (Figure 6A, bottom), 2 features associated with activation of STAT3 signaling (20). These findings suggested a strong link between STAT3 and the oncogenic activity of miR-424 in prostate cancer. In support of this, transfection of miR-424 in RWPE1 cells increased STAT3, phosphorylated STAT3 (p-STAT3), and STAT3-dependent reporter activity (Figure 6B). Notably, STAT3 and p-STAT3 were also higher in RWPE1-ESE3kd and DU145 cells than in RWPE1 control cells, in line with the high expression of miR-424 in the former cell lines (Figure 6C). An increase of total STAT3 was also observed in LNCaP cells with stable expression of miR-424 (Figure 6D) and following transient transfection of miR-424 (Figure 6E). Notably, the effect of miR-424 on total STAT3 protein in LNCaP cells was seen in both the presence and the absence of IL-6–induced phosphorylation (Figure 6F). Furthermore, the induction of p-STAT3 by IL-6 was significantly higher in the cells expressing miR-424 (Figure 6F) and resulted in hyperinduction of STAT3 reporter activity in the presence of both IL-6 and miR-424 (Figure 6G), suggesting that miR-424 enhanced the cell response to IL-6 by increasing the STAT3 protein level. Importantly, treatment with anti–miR-424 reduced total STAT3 and p-STAT3 along with STAT3 reporter activity in DU145 cells (Figure 6H and Supplemental Figure 9).
Figure 6. miR-424 induces STAT3 stability and activity by targeting COP1.
(A) Pie chart showing the number of STAT3 targets among the genes induced by miR-424 extracted with Enrich (A, top) and functional annotation analysis by DAVID (A, bottom) of genes induced by miR-424 in RWPE1 cells. (B) COP1, STAT3, and p-STAT3 level (left) and relative luciferase activity (RLA) of STAT3 reporter (right) following transfection of miR-424 or control (Ctrl) in RWPE1 cells. (C) Immunoblot (IB) of COP1, STAT3, and p-STAT3 in indicated cell lines. (D) STAT3 level in LNCaP-424 stable cells by IB (bottom) and immunocytochemistry (ICC; top). (E) IB of COP1 and STAT3 in LNCaP cells after transient transfection of miR-424 or control (Ctrl) at the indicated time points. (F) IB of COP1, STAT3, and p-STAT3 in control (EV) and LNCaP-424 monoclonal cells with or without IL-6 (10 ng/ml) stimulation. (G) RLA of STAT3 reporter in cells transfected with negative control (Ctrl) or miR-424 with or without IL-6 stimulation. (H) IB of COP1, STAT3, and p-STAT3 (left) and RLA of STAT3 reporter (right) in DU145 cells following transfection with Anti-424 or Scr. (I and J) IB of COP1, STAT3, and p-STAT3 in RWPE1 (I) and LNCaP (J) cells after COP1 silencing. (K) IB of COP1, STAT3, and p-STAT3 in DU145 cells after transfection with full-length FLAG-COP1 (p-COP1) or empty vector (EV). (L) IB of DU145 cells transfected with p-COP1 with or without treatment with proteasome inhibitor PS-341 (10 μM) for 5 hours. c-Jun used as control. (M) IB of STAT3 and p-STAT3 in DU145 transfected with EV, p-COP1, or a deletion mutant (pRING). *P ≤ 0.05, **P ≤ 0.01 by 2-tailed Student’s t test. The experiments were performed in triplicate, and data are shown as mean ± SD.
These results showed that miR-424 induced changes in STAT3 protein level and activity. These effects were strikingly concomitant with the downregulation of COP1 by miR-424. Thus, we hypothesized that these effects could depend on reduced COP1 activity. In support of a direct link between COP1 and STAT3, knockdown of COP1 by siRNAs increased STAT3 level in RWPE1 (Figure 6I) and LNCaP cells (Figure 6J). Conversely, expression of COP1 reduced STAT3 in DU145 cells as seen after anti–miR-424 treatment (Figure 6K and Supplemental Figure 9). This suggested that STAT3 is a substrate of the E3 ligase COP1, leading to its ubiquitination and degradation by the ubiquitin-proteasome system. Consistently, proteasome inhibition by PS-341 prevented STAT3 downregulation induced by COP1 in DU145 cells (Figure 6L and Supplemental Figure 9). Furthermore, expression of a deletion mutant of COP1 (RING) unable to bind and ubiquitinate substrate proteins (16) did not affect STAT3 level (Figure 6M and Supplemental Figure 9). COP1 knockdown significantly delayed turnover of STAT3 in RWPE1 cells, indicating that COP1 was an important regulator of STAT3 degradation in prostate epithelial cells (Supplemental Figure 10).
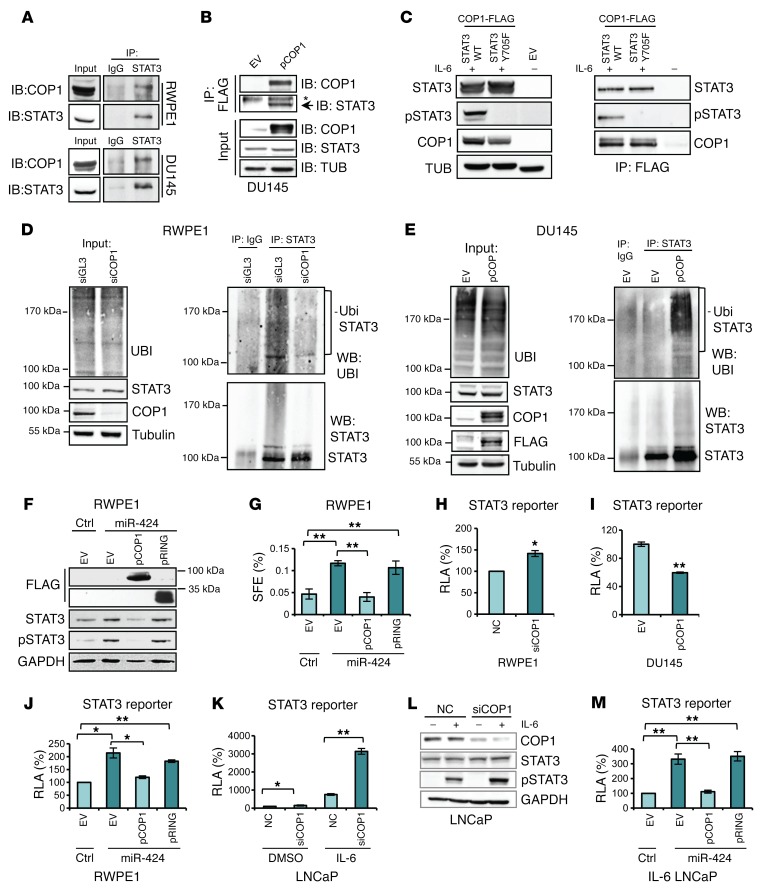
Consistent with a direct effect on STAT3, COP1 and STAT3 interacted in RWPE1 and DU145 cells in coimmunoprecipitation experiments (Figure 7, A and B). Interestingly, coimmunoprecipitation experiments with WT STAT3 and a phosphorylation-defective Y705F mutant showed that the interaction with COP1 did not depend on the STAT3 phosphorylation status (Figure 7C).
Figure 7. COP1 interacts with STAT3 and mediates STAT3 ubiquitylation and degradation.
(A) Immunoprecipitation of STAT3 and IB of COP1 and STAT3 (B) Immunoprecipitation of COP1 (p-COP1) with anti-FLAG and IB with anti-STAT3 in cells treated with PS-341 (10 μM for 6 hours). Asterisk: aspecific band. TUB, tubulin. (C) IB of STAT3 and COP1 in HEK293T cells transfected with indicated constructs. IP of COP1 with anti-FLAG and IB using indicated antibodies. Cells were treated with IL-6 (10 ng/ml for 45 minutes) and with 10 μM PS-341 for 3 hours. (D) STAT3 ubiquitination in RWPE1 cells transfected with siCOP1#1 or siGL3 for 48 hours. Lysates were immunoprecipitated with anti-STAT3 or IgG followed by IB with indicated antibodies. UBI,ubiquitin. (E) STAT3 ubiquitination in DU145 cells transfected for 48 hours and treated with PS-341 (10 μM for 3 hours). Lysates were immunoprecipitated with anti-STAT3 or IgG followed by IB with indicated antibodies. (F) IB of STAT3 and p-STAT3 following sequential transfection of miR-424 or control and p-COP1, the mutant construct (pRING), or empty vector (EV). (G) SFE in cells transfected as described in F. (H and I) RLA of STAT3 reporter in RWPE1 cells following transfection of siCOP1#1 or negative control (NC) (H) and in DU145 cells transfected with p-COP1 or EV (I). (J) STAT3 reporter activity following sequential transfection of miR-424 or control and p-COP1, mutant construct (pRING), or EV. (K) STAT3 reporter activity following transfection with siCOP1#1 or NC with or without IL-6. (L) IB of indicated proteins following IL-6 with and without COP1 knockdown. (M) STAT3 reporter activity following transfection of miR-424 or control and pCOP1, pRING, or EV in presence of IL-6. *P ≤ 0.05, **P ≤ 0.01 by 2-tailed Student’s t test. The experiments were performed in triplicate, and data are shown as mean ± SD.
In RWPE1 cells the level of ubiquitinated STAT3 was reduced after COP1 knockdown (Figure 7D). Conversely, ubiquitinated STAT3 increased in DU145 cells after expression of COP1 (Figure 7E). Collectively, these data provided evidence of COP1-dependent ubiquitination and proteasome-mediated degradation of STAT3 in prostate epithelial cells. Significantly, COP1 overexpression in RWPE1 cells reduced STAT3 and p-STAT3 (Figure 7F) and concomitantly blocked PS formation promoted by miR-424 (Figure 7G). The COP1 deletion (RING) mutant was ineffective in these assays, underlining the important role of the ubiquitin ligase activity of COP1 in mediating miR-424 effects (Figure 7, F and G). Thus, COP1, by preventing the increase in STAT3 protein level, blocked the miR-424–induced expansion of the cancer stem-like cell compartment in the PS-forming assay. These data support the functional relevance of the COP1 effects on STAT3 ubiquitination and degradation.
Altered STAT3 protein turnover increases basal and cytokine-induced STAT3 activity.
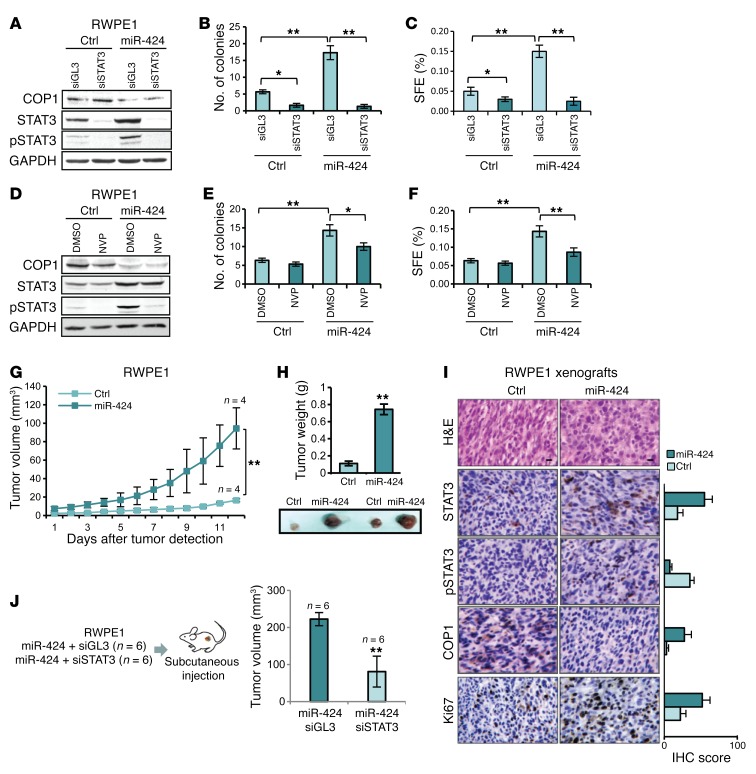
Next, we assessed whether the increase in STAT3 protein level dependent on loss of COP1 resulted in increased STAT3 activity. To assess STAT3 transcriptional activity in an unbiased manner, we used a STAT3-responsive luciferase reporter. This assay had the advantage of being independent of the expression of specific targets that could be highly cell type dependent. Knockdown of COP1 by siRNA increased STAT3 reporter activity in RWPE1 cells (Figure 7H), while COP1 expression in DU145 cells decreased it (Figure 7I). Moreover, in RWPE1 cells miR-424–induced STAT3 activity was blocked by the concomitant expression of WT COP1 and not the RING mutant (Figure 7J), indicating that STAT3 activation by miR-424 in this context was a direct consequence of COP1 repression and impaired STAT3 degradation. We similarly assessed the role of COP1 in response to IL-6 stimulation in LNCaP cells. Knockdown of COP1 slightly increased STAT3 activity in unstimulated LNCaP cells (Figure 7K). However, addition of IL-6 to COP1-silenced cells resulted in hyperinduction of the reporter and hyperinduction of p-STAT3 (Figure 7, K and L, and Supplemental Figure 9), mimicking the effect of miR-424 on STAT3. Furthermore, expression of WT COP1, but not the RING mutant, prevented STAT3 activation by IL-6 in miR-424–expressing LNCaP cells (Figure 7M). Thus, controlling STAT3 protein turnover, COP1 affected the level of both basal and cytokine-induced STAT3 activity. These data suggest that increased total STAT3 level primes the cells to respond more strongly to IL-6 stimulation. Consistent with a key role of STAT3 in mediating the effects of miR-424, knockdown of STAT3 by 2 distinct siRNAs blocked the induction of anchorage-independent growth and PS formation by miR-424 in RWPE1 cells (Figure 8, A–C, and Supplemental Figure 11, A–C). Interestingly, blocking STAT3 phosphorylation with the JAK inhibitor NVP-BSK805 (ref. 27 and Figure 8D) decreased the effects of miR-424 on growth in soft agar and PS formation (Figure 8, E and F), although less effectively than STAT3 knockdown. Thus, depletion of total STAT3 could be more effective than indirect inhibition of p-STAT3 in tumors with deregulated miR-424/COP1/STAT3 axis.
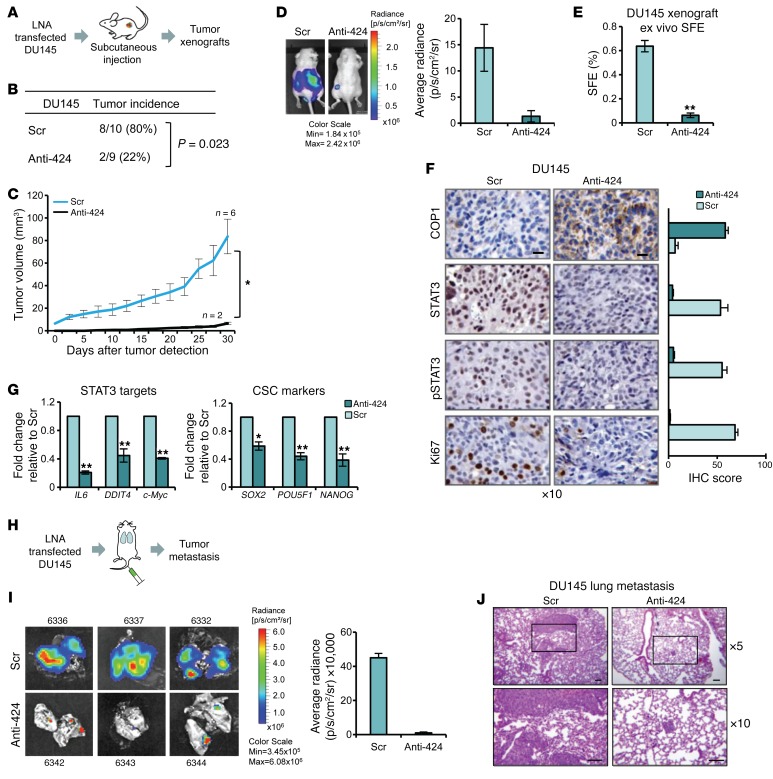
Figure 8. miR-424 promotes tumor initiation recapitulating in vivo miR-424/COP1/STAT3 axis.
(A–C) IB of COP1, STAT3, and p-STAT3 (A), colony formation in soft agar (B), and SFE (C) in RWPE1 cells following cotransfection with miR-424 or Ctr and siRNA targeting STAT3 (siSTAT3) or control siRNA (siGL3). (D–F) IB of COP1, STAT3, and p-STAT3 (D), colony formation in soft agar (E), and SFE (F) in RWPE1 cells 48 hours after transfection with miR-424 or Ctr and treatment with 5 μM NVP or DMSO for 16 hours. (G and H) Tumor growth (G), tumor weight (H), and representative images of tumors (bottom) of subcutaneous xenografts in nude mice, from RWPE1 cells transfected with miR-424 (n = 4) or Ctr (n = 4). (I) H&E and IHC stain and scores (percentage of positive cells) of STAT3, p-STAT3, COP1, and Ki67 stain of control and miR-424 induced xenografts described above. Scale bars: 10 μm. (J) Schematic of the experimental plan (left) and tumor volume (right). *P < 0.05; **P < 0.01.
miR-424 promotes tumor initiation, and its inhibition disrupts the tumorigenic and metastatic properties of prostate cancer cells in vivo.
miR-424 through deregulation of COP1 and STAT3 induced profound changes in the prostate epithelial cell phenotype in vitro, promoting tumorigenic, invasive, and stem-like properties. Next, we sought to recapitulate in vivo the activation of this oncogenic axis induced by transient overexpression of miR-424. To this end, RWPE1 cells transfected with miR-424 were engrafted subcutaneously in mice. miR-424–transfected RWPE1 cells, unlike control-transfected cells, acquired the ability to form subcutaneous tumors that grew in nude mice, indicating that miR-424 was a potent inducer of tumor-initiating properties in prostate epithelial cells (Figure 8, G and H). Notably, tumor xenografts derived from miR-424–transfected cells exhibited increased levels of STAT3 and p-STAT3 and a reduced level of COP1 compared with control-derived xenografts (Figure 8I). Thus, elevation of miR-424 led to persistent activation of the miR-424/COP1/STAT3 axis in vivo, sustaining transformation and tumorigenicity. To further assess the role of STAT3 in the tumorigenic phenotype induced by miR-424, we cotransfected RPWE1 cells with miR-424 and either a siRNA targeting STAT3 (siSTAT3) or control siRNA (siGL3). Relevantly, STAT3 knockdown significantly reduced the growth of miR-424–induced tumors (Figure 8J and Supplemental Figure 11, D and E), demonstrating that STAT3 was a key mediator of the oncogenic effects of miR-424 in vivo.
We hypothesized that targeting miR-424 using a miRNA antagonist could be an effective therapeutic strategy to restore COP1-mediated degradation of STAT3 and counteract its oncogenic effects. To test this, we assessed the tumor-initiating capability of DU145 cells transfected with anti–miR-424 or a scrambled control and implanted in nude mice (Figure 9A). Anti–miR-424–transfected cells formed significantly fewer tumors than control cells (Fisher exact test 2-tailed P = 0.023) (Figure 9B). Only 2 tumors formed from anti–miR-424–transfected cells, and their in vivo growth was considerably impaired in comparison with control cells (P < 0.001) (Figure 9C). This was also confirmed using IVIS Spectrum (Figure 9D). Tumor cells harvested from anti–miR-424–treated tumors exhibited ex vivo considerably reduced PS-forming ability compared with cells from control tumors (Figure 9E), indicating a persistent loss of stem cell–like properties as a consequence of miR-424 inhibition and disruption of the miR-424/COP1/STAT3 axis. IHC staining and quantitative scores of xenograft sections showed the reactivation of COP1 expression and reduced levels of STAT3, p-STAT3, and the proliferation marker Ki67 in the anti–miR-424–treated xenografts compared with control (Figure 9F), confirming stable reversion of the activation of the miR-424, COP1, and STAT3 axis in vivo. Furthermore, selected STAT3 targets and cancer stem cell markers were also significantly downregulated in anti–miR-424–treated xenografts (Figure 9G).
Figure 9. miR-424 inhibition disrupts the miR-424/COP1/STAT3 axis in vivo.
(A) Schematic of the experimental plan. (B) Tumor incidence in control-treated (Scr n = 10) and anti–miR-424–treated (Anti-424 n = 9) DU145 cells. Significance was calculated by Fisher exact test. Two-tailed P = 0.023. (C and D) Tumor growth monitored by tumor size (C) and in vivo bioluminescence (D) of subcutaneous xenografts (mean ± SEM, Scr n = 6 and Anti-424 n = 2) of DU145-Luc cells pretreated in vitro with anti–miR-424 or Scr. (E) Ex vivo SFE from Scr or Anti-424 dissociated tumor xenografts. (F) IHC staining for STAT3, p-STAT3, COP1, and Ki67 in tumor xenografts of DU145 cells treated with Anti-424 or Scr. Scale bars: 10 μm. (G) qRT-PCR evaluation of selected STAT3 targets (top) and selected cancer stem cell markers (bottom) in tumor xenografts from control (Scr) and miR-424–ablated (Anti-424) xenografts. (H) Schematic of the experimental plan. (I) Lung metastasis from DU145-Luc cells pretreated in vitro with Anti-424 or Scr and injected into the tail vein of nude mice. Representative bioluminescent images (left); quantification after necropsy of lung metastatic foci (right). (J) Representative images of H&E, immunostaining, and IHC scores (percentage of positive cells). Areas within the boxes in top panels are magnified in the lower samples. Scale bars: 100 μm. *P < 0.05; **P < 0.01.
Next, we assessed the metastatic capability of DU145 cells transfected with anti–miR-424 or a scrambled control and injected into the tail vein in mice. Importantly, treatment with anti–miR-424 significantly compromised the ability of DU145 cells to metastasize to the lung (Figure 9H). Formation of lung metastasis by anti–miR-424–treated cells was almost completely prevented as shown by bioluminescence imaging and histopathology evaluation (Figure 9, I and J). Taken together, these results implicated that miR-424 had a relevant impact on in vivo tumor growth and tumor-initiating and metastatic properties. Furthermore, its ablation might be a novel context-dependent therapeutic strategy for prostate tumors characterized by elevation of miR-424 expression.
COP1/STAT3 deregulation is associated with adverse prognosis in prostate cancer.
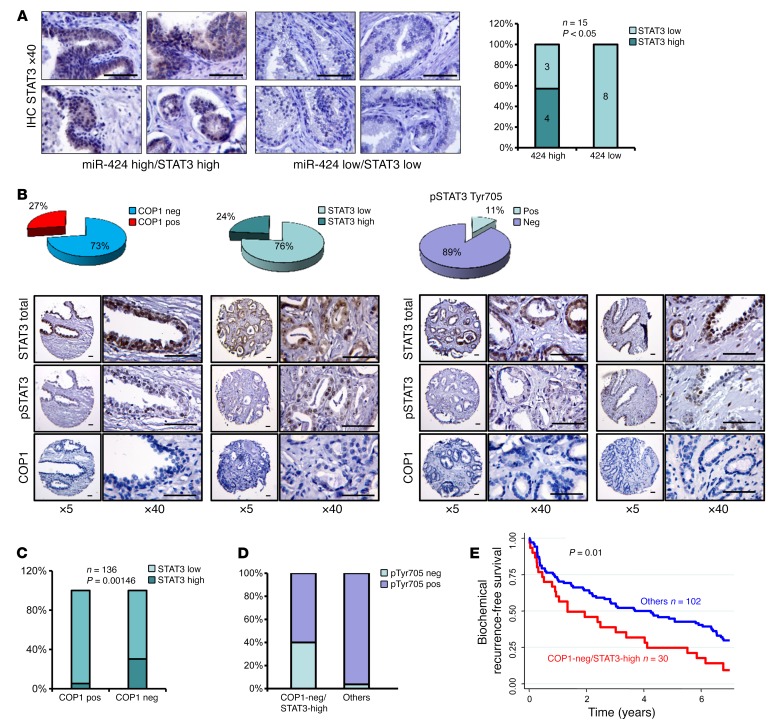
To asses the clinical relevance of our findings, we examined the relation between miR-424, COP1, and STAT3 in human prostate tumors. In support of the link between miR-424 and STAT3 observed in experimental models, we found a positive correlation (Fisher test P = 0.025) between high miR-424 expression determined by qRT-PCR and STAT3 level assessed by IHC in a set of prostate tumors (n = 15) (Figure 10A). Next, we examined the level of COP1, STAT3, and p-STAT3 by IHC in primary prostate tumors (n = 136) from patients with long-term clinical follow-up (Figure 10B). Low COP1 expression was significantly associated with high STAT3 level (Fisher test P < 0.001) (Figure 10C). In line with our in vitro data, p-STAT3 was prevalently observed in the group of COP1-negative/STAT3-high tumors indicative of STAT3 activation (Figure 10D). Moreover, the combination of low COP1 and high STAT3 expression identified patients with a significantly higher risk of biochemical relapse after prostatectomy (P ≤ 0.01) (Figure 10E). Thus, loss of COP1 was associated with increased STAT3 level, and their association was clinically relevant. Collectively, these data support the notion that miR-424 has key oncogenic functions in prostate tumors largely determined by its ability to silence COP1 and activate STAT3, promoting clinically aggressive tumors (Figure 11).
Figure 10. Prognostic impact of COP1 loss and STAT3 activation in prostate tumors.
(A) Representative IHC images of total STAT3 in prostate tumors (left) and STAT3/miR-424 high/low association in prostate tumors (right). The t test P value is shown. Scale bars: 100 μm. (B) IHC score of COP1 and STAT3 and p-STAT3 Tyr705 in TMA of primary prostate tumors (n = 136) (top) and representative IHC images (bottom). Scale bars: 100 μm. (C) STAT3/COP1 and COP1-negative/STAT3/p-STAT3 association analysis in the TMA. The t test P value is shown. (D) Association between p-STAT3 Tyr705–negative and –positive tumors and COP1 and STAT3 staining (t test P < 0.00001). (E) Survival plots showing significant increase in biochemical recurrence in COP1-negative/STAT3-high tumors compared with the other tumors in the TMA cohort analyzed above. Log-rank test P value is shown.
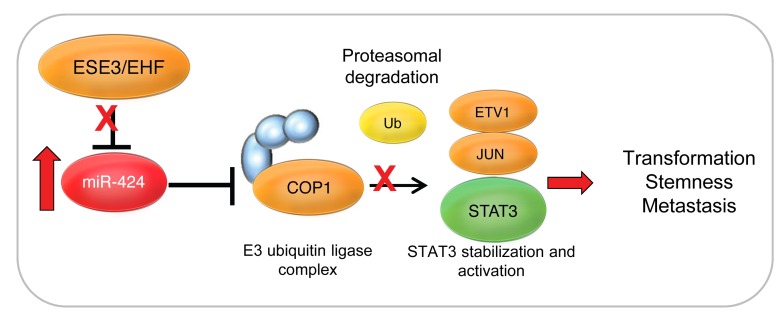
Figure 11. Proposed model for induction of miR-424 and establishment of the miR-424/COP1/STAT3 oncogenic axis.
Discussion
In this study we uncovered a novel oncogenic pathway of broad biological and clinical relevance involving deregulation of miR-424 and leading to altered ubiquitination and stability of key oncogenic TFs in prostate tumors. Activation of this oncogenic axis promotes tumor-initiating and metastatic properties and provides opportunities for targeted therapeutic intervention. This study highlights the oncogenic role of miR-424 in prostate cancer and shows that miR-424–mediated silencing is a relevant mechanism of inactivation of the E3 ubiquitin ligase COP1. Furthermore, we provide evidence that this represents an alternative, noncanonical pathway of activation of STAT3 through decreased ubiquitination and degradation by the ubiquitin-proteasome system.
miR-424 belongs to the miR-15/107 family of miRNAs (28). In line with our data, miR-424 was recently reported to be expressed in highly metastatic subclones of DU145 cells and promote the EMT and metastatic phenotype (29). Furthermore, miR-424 was shown to be overexpressed and to exert oncogenic functions in breast cancer (30). In this study, for the first time to our knowledge, we identified COP1 as a biologically relevant target of miR-424 in prostate epithelial cells. COP1 acts as a tumor suppressor in prostate cancer (16, 19), and our study reveals that posttranscriptional silencing by miR-424 is a frequent mechanism of attenuation of COP1 expression, in addition to the genetic deletions identified in prostate and other tumors. Interestingly, we found that miR-424 upregulation and COP1 silencing increased ETV1 protein level in both ETV1 translocation–positive LNCaP and translocation-negative RWPE1 cells. ETV1 is translocated in 2%–6% of prostate cancer (7). Thus, these data suggest an alternative pathway of ETV1 upregulation, independent of ETV1 translocation and COP1 deletion linked to miR-424 posttranscriptional silencing. Furthermore, the miR-424/COP1 oncogenic axis was preferentially activated in prostate tumors with reduced expression of ESE3/EHF. Indeed, we found that ESE3/EHF repressed transcription of miR-424 in normal prostate epithelial cells and loss of ESE3/EHF resulted in increased expression of miR-424 with consequent acquisition of tumorigenic and stem-like properties. Interestingly, the contribution of ESE3/EHF to miR-424 deregulation might not be limited to prostate cancer, and this pathway might be relevant in other cancer types, in which we found frequent overexpression of miR-424.
A major finding of this study is the demonstration that STAT3 is a substrate of COP1. Activation of STAT3 signaling is frequent in human tumors and is associated with poor clinical outcome (20, 21). STAT3 promotes cancer cell proliferation and survival, metastasis, and tumor progression (20, 21). In prostate cancer STAT3 contributes to the expansion and maintenance of stem-like cancer cells and progression to castration resistance (24–26). In line with these data, we found that miR-424 overexpression was associated with transcriptional signatures of stem-like cells and EMT compatible with aggressive features in human prostate tumors. Furthermore, both COP1 and STAT3 deregulation was associated with increased risk of biochemical relapse in prostate cancer patients. STAT3 activation is ascribed to the activity of receptor-associated and non–receptor-associated tyrosine kinases that phosphorylate STAT3 at Tyr705 (20, 21). However, alternative pathways of activation that are independent or cooperate with this canonical pathway are emerging (21, 31, 32). In light of these recent findings, our evidence of a direct impact of miR-424 and COP1 deregulation on STAT3 protein turnover is highly relevant. Indeed, we show here that altered turnover and accumulation of STAT3 due to miR-424 overexpression and silencing of COP1 increased basal activity and enhanced responsiveness to cytokine stimulation. These results establish that the activity of this key oncogenic TF is also regulated at the level of protein turnover and not exclusively by posttranslational phosphorylation at Tyr705.
Consequently, tumors with high miR-424 expression might be more prone to aberrant activation of STAT3 in response to various stimuli, like inflammatory cytokines, which could lead to the establishment of feed-forward loops promoting stem-like behavior and tumor progression. Our data do not rule out the possibility that, particularly in an aggressive cell context, STAT3 level is maintained aberrantly elevated by additional or concomitant mechanisms including alterations of other ubiquitin ligases beside COP1. Altogether, these findings suggest the necessity of identifying these tumors and devising strategies to prevent their progression.
Our results, therefore, also have important therapeutic implications, as compounds that inactivate or deplete STAT3 might be highly effective in tumors exhibiting miR-424/COP1/STAT3 deregulation. To this end, we showed that a miR-424 antagonist restored COP1 expression and reduced STAT3 protein level and activity both in vitro and in tumor xenografts. Indeed, the ability of the miR-424 antagonist to restore COP1 function and promote STAT3 represents a new approach to selectively block STAT3 signaling effectively and selectively in miR-424–expressing cancers. Furthermore, anti–miR-424 treatment was highly effective in reversing the malignant phenotype of miR-424–expressing cancer cells with a prominent and persistent effect on the stem-like cancer cell subpopulation both in vitro and in vivo. Anti-miRNA strategies are an area of active investigation and innovative therapeutic opportunities (33, 34). Our present data strongly support the efficacy of miR-424 targeting in the context of prostate cancers with deregulation of the miR-424/COP1/STAT3 axis.
Collectively, our data identify a key oncogenic axis initiated by loss of ESE3/EHF and a novel mechanism leading to activation of STAT3 and other oncogenic TFs through altered ubiquitination and degradation. Monitoring the state of this miR-424/COP1/STAT3 axis could identify patients at risk of progression and disease recurrence and provide insights for the development of specific therapeutic strategies.
Methods
Cell cultures.
LNCaP and DU145 cells were obtained from ATCC and maintained in RPMI-1640 (Gibco) supplemented with 10% FBS. Immortalized normal prostate epithelial LHS cells were maintained in PrEC growth medium (PrEGM; Cambrex, Lonza) and RWPE1 cells in Keratinocyte Serum-Free Growth Medium (KSF; Gibco) with specific supplements (Epidermal Growth Factor and Bovine Pituitary Extract; Gibco) (8). Cell lines with stable ESE3 knockdown (ESE3kd) were established and maintained as previously described (8). LNCaP cells with stable expression of miR-424 (LNCaP-424) and empty vector as control (LNCaP-EV) were established by infection with the pCDH lentiviral vector containing both GFP and a puromycin-resistant gene (vector provided by M. Soleimani, Tehran, Iran) (35). HEK293T cells were used for lentiviral amplification, and viruses were harvested at 48 and 72 hours after transfection, filtered, and used for infection of cells in the presence of 5 μg/ml polybrene (Sigma-Aldrich). After drug selection with puromycin (0.5 μg/ml), resistant LNCaP-424 and control cells were subsequently infected with the pMMP-LucNeo retrovirus and selected with 2 mg/ml of G418 (Gibco). Monoclonal cell lines (LNCaP-424m) were obtained by serial dilution in a 96-well plate. Clones were selected based on highest GFP expression measured by FACS and miR-424 expression levels by qRT-PCR. For in vivo experiments, DU145-Luc cells were established by infection of cells with a constitutively expressing luciferase vector (pMMP-LucNeo retrovirus). LucNeo-expressing cells were selected with 2 mg/ml G418 (Gibco). Where indicated, LNCaP cells were stimulated with 10 ng/ml IL-6 for 1 hour. Cells were regularly checked for mycoplasma contamination using MycoAlert Mycoplasma detection kit (Lonza).
Cellular proliferation, clonogenicity, migration, and invasion assays.
Growth in anchorage-independent condition was assessed by colony formation in soft agar as previously described (8). Cell migration was assessed using scratch/wound-healing assay (8). Cells were grown to confluence and serum-starved overnight before introduction of the scratch in the cell monolayer. Pictures of cell migration were taken at different time points until the closure of the wound. Cell motility was further evaluated in Boyden chamber assays using 6.5-mm Transwell chambers with 8.0-μm pore polycarbonate membrane (3422; Corning Inc.) coated with rat tail Collagen Type I Solution (60 μg/ml; Sigma-Aldrich). Briefly, following transfection with the miR-424 precursor or miR-424 inhibitor and relative controls, cells were incubated for 24 hours in RPMI without serum (DU145) or in KSF without supplements (RWPE1). Then cells were plated (5 × 105 cells per well in 24-well plates) on the top chamber of Transwells in the medium described above with the addition of 0.1% BSA. RPMI with 10% FBS or KSF with supplements was added to the lower chamber, and cells were incubated at 37°C. After 20 hours (DU145) or 24 hours (RWPE1) of incubation, nonmigrated cells were carefully removed from the top of each insert with a cotton swab. Migrated cells were fixed in 4% formaldehyde at room temperature for 15 minutes and stained with 0.1% crystal violet. The number of cells in 5 random fields (×200 magnification) was counted for each chamber. The stained cells were dissolved in 10% acetic acid, and absorbance (proportional to the number of cells) was measured at 590 nm. The migrated cells were represented as percentage of absorbance value relative to the control. Representative images of invading cells stained with crystal violet were taken. The assay was done in triplicate and repeated in 2 independent experiments. For cell proliferation assay, cells were seeded in 12-well plates, collected, and counted at the indicated time points using an automated cell counter (Beckman Coulter Counter).
In vitro and ex vivo sphere formation assay.
For sphere-forming assay (SFA), single-cell suspensions were plated in poly-HEMA–treated dishes in serum-free Mammary Epithelial Basal Medium (MEBM; Cambrex) supplemented with specific supplements for the selection of cancer stem-like cells (8). Prostatospheres at least 50 μm in diameter were counted after 7 days, and the sphere-forming efficiency was evaluated as previously described (8). For ex vivo SFA, xenograft tissues were chopped, incubated with Collagenase-Dispase mix solution (1:2) in Hank’s buffer for 3 hours at 37°C, and passed through a 40-μm filter, and single cells were counted by hemocytometer. Single-cell suspensions were used to perform ex vivo SFA (8 × 103 cells/ml).
RNA extraction and qRT-PCR.
RNA was extracted by Direct-zol RNA Mini-prep kit (Zymo Research). Quantitative real-time RT-PCR (qRT-PCR) was carried out using 20 ng of RNA as template and SYBR Green Fast One Step kit (Qiagen). Primer sets used for qRT-PCR (designed using Primer3, http://frodo.wi.mit.edu/primer3/) are reported in Supplemental Table 3. The expression of each gene was normalized to β-actin.
For miRNA expression analysis, 400 ng of purified RNA was retro-transcribed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) with specific primers (TaqMan MicroRNA Assays ID: 4427975-000604; Applied Biosystems), and the cDNA was subjected to TaqMan Probe–based real-time PCR (TaqMan Universal PCR Master Mix; Applied Biosystems). The expression was normalized to RNU6 (Control miRNA assay, ID: 4427975-001093; Applied Biosystems).
miRNA precursor, antagomiR, and siRNAs.
For miR-424 overexpression, RWPE1 and LNCaP cells were transiently transfected with 40 nM miR-424 precursor (PremiR-424, AM17100-PM10306; Ambion) or Negative Control #1 (AM17110; Ambion). For miRNA inhibition, DU145 and ESE3kd cells were transiently transfected with 40 nM of a specific LNA antagomiR (Mercury LNA Power Inhibitor; Exiqon) or a scrambled control (Negative Control A; Exiqon). For COP1 knockdown, cells were transfected with 50 nM of 2 distinct COP1-directed siRNAs (siCOP1#1 and siCOP1#2; Ambion) and siRNA directed to luciferase gene (siGL3; Ambion) or Negative Control #1 siRNA (4611; Ambion) as controls. For STAT3 knockdown, cells were transfected with 50 nM of STAT3-directed siRNA (siSTAT3, ID s743; Ambion). All transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. In vitro functional assays were performed 24 hours after the indicated transfection. Unless otherwise specified, cell lysates were prepared 72 hours after miRNA precursor or antagomiR transfection and 48 hours after siRNA transfection.
miRNA target prediction.
miR-424 target prediction was performed using miRWalk (http://mirwalk.uni-hd.de/) with a comparative analysis of different prediction programs. We selected targets predicted by at least 5 of the 8 prediction tools included in miRWalk. miR-424 binding site on COP1 3′-UTR was identified by 6 of the 8 programs (miRWalk, miRanda, Targetscan, Diana, Pictar4, PITA) with an 8-bp perfect match between the seed region and the mRNA target sequence. The miR-424/COP1 binding was further confirmed by RNAhybrid (36).
Expression vectors, reporter constructs, and luciferase assays.
FLAG-tagged COP1 WT and RING-overexpressing plasmid (provided by J.C. Marine, VIB Laboratory for Molecular Cancer Biology, Leuven, Belgium) (16) was transfected using jetPRIME (Polyplus) according to the manufacturer’s protocol. Overexpression of ESE3/EHF was performed using the pRc/CMV-ESE3b-FLAG vector as previously described (8). The COP1 3′-UTR reporter construct was generated by cloning of the 3′-UTR region of the gene downstream from the luciferase reporter in the pMIR-REPORT vector (Life Technologies). The specific region was amplified by PCR from human genomic DNA extracted from LNCaP (PCR primers: COP1-SacI forward, 5′-GAGCTCAGTCAAATTGTACTTGATCCTGCTG-3′; COP1-HindIII reverse, 5′-AAGCTTCAAGCTCAACTGTGGCTCAA-3′), cloned in pGEM-T Easy vector, and then subcloned in pMIR-REPORT into the SacI and HindIII sites. Point mutations were introduced in the putative binding site of COP1 reporter using GENEART Site Directed Mutagenesis System (Invitrogen) to generate COP1-MUT (primers: COP1 forward, 5′-TGTAACTCCATGGACATTAATAATCTTGGTGGTGTTATC-3′; COP1 reverse, 5′-GATAACACCACCAAGATTATTAATGTCCATGGAGTTACA-3′). For STAT3 activity we used the STAT3-responsive luciferase reporter (pGL4.32/luc2P/STAT3-RE/Hygro from Promega; ID CS154201). Luciferase reporter assays were performed using the Dual-Glo Luciferase kit (Promega) as previously described (8). Results were expressed as relative luciferase activity corrected for Renilla luciferase activity and normalized to the control.
Full-length WT EGFP-STAT3 was provided by Nadya I. Tarasova (Georgetown University, Washington, DC, USA). The EGFP-STAT3 Y705F mutant construct was generated by site-directed mutagenesis using GENEART Site-Directed mutagenesis (Life Technologies).
Flow cytometry.
All steps for flow cytometry were performed in PBS supplemented with 0.5% BSA, and 2 mM EDTA. Single cells were stained with anti–human CD44 (1:100; BD Pharmingen; 555477) and anti–human CD24 (1:20; BD Pharmingen; 555427). Cell cycle analysis was performed by staining with 7-AAD (25 mg/ml; Sigma-Aldrich). Samples were analyzed with a FACS Fortessa (BD Biosciences) and FlowJo software as previously described (23).
Immunoblotting and immunoprecipitation.
Cell lysates were prepared using RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% Na-deoxycholate, 1 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 0.1% SDS) with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (PhosStop; Roche) as previously described (8). Total cell extracts were separated by SDS-PAGE and transferred to nitrocellulose membranes (Protran). The following antibodies were used for immunoblot analysis: anti-COP1 (ab56400; Abcam), anti-STAT3 (124H6, 9139; Cell Signaling), anti–p-STAT3 Tyr705 (D3A7, 9145; Cell Signaling), anti-ETV1 (ab81086; Abcam), anti–c-Jun (Calbiochem), anti-GAPDH (Millipore), anti-ubiquitin (Enzo Life Sciences), anti-FLAG (F3165; Sigma-Aldrich), anti-ESE3 (ESE-3A H-205, sc-367574; Santa Cruz Biotechnology). Immunoprecipitation was performed as previously described (37). Cell lysates were incubated with antibody against STAT3 (sc-482; Santa Cruz Biotechnology) or FLAG tag (F3165; Sigma-Aldrich), and immunoblotting was performed using antibody against COP1 and STAT3. To detect ubiquitylated STAT3, cells were lysed in denaturing buffer (1% SDS, 20 nM HEPES pH 7.2, 2 mM EGTA, 20 nM NaF, 1 mM Na3VO4) supplemented with 20 mM NEM (N-Ethylmaleimide; Sigma-Aldrich) and protease inhibitor and phosphatase inhibitor cocktails. After incubation for 10 minutes at 65°C, lysates were sonicated and diluted 10 times in 1% Triton buffer (1% Triton, 20 nM HEPES pH 7.2, 2 mM EGTA, 20 nM NaF, 1 mM Na3VO4) and subjected to immunoprecipitation with anti-STAT3 antibody (sc-482; Santa Cruz Biotechnology). Where indicated, PS-341 treatment was done at 10 μM concentration for the indicated time.
Chromatin immunoprecipitation.
A computational search for ETS binding sites on the MIR424 promoter was performed using MotifViz (https://biowulf.bu.edu/MotifViz/). ChIP was performed as previously described (8, 10). Chromatin was immunoprecipitated with antibodies anti-ESE3 (Clone 5A5.5; Lab Vision), anti-H3K27me3 (Millipore), anti-H3Ac (Millipore), and anti-H3K9me3 (Millipore), and IgG antibody (Millipore) as control. Samples were analyzed by quantitative PCR performed with SYBR Green PCR Master Mix (Applied Biosystems) using primers for the MIR424 promoter: forward, 5′-GGACGAAGGCATATGCAAAA-3′; reverse, 5′-TCTGAACTGCAATGTGTTTGG-3′. Analysis of the TWIST1 promoter including the ETS binding site (forward, 5′-GAATTTCTGCCGCATTCTCT-3′; reverse, 5′-GCCGCTGCCTTACTGTAAAA-3′) and of the GAPDH promoter region (forward, 5′-TCCTCCTGTTTCATCCAAGC-3′; reverse, 5′-TAGTAGCCGGGCCCTACTTT-3′) was performed as positive and negative control, respectively.
Animals and tumor xenografts.
Athymic nude mice (BALB/c nu/nu, 4–6 weeks old; Harlan Laboratories) were used for in vivo experiments. 2 × 106 RWPE1 cells were subcutaneously injected (n = 4 per group) with Matrigel (1:1) and 2 × 106 UGSM2 cells 24 hours after in vitro transfection of 40 nM PremiR-424 or negative control. For the in vivo rescue experiment with siSTAT3, 4 × 106 RWPE1 cells were injected subcutaneously in mice (n = 6 per group) with Matrigel (1:1) and 2 × 106 UGSM2 cells 24 hours after in vitro transfection of 40 nM PremiR-424 with RNA targeting STAT3 (siSTAT3n_1) or control siRNA (siGL3). Tumor growth was monitored every 2 days with a caliper, and final tumor weight was measured. DU145-Luc cells (1 × 106) were subcutaneously injected in mice (n = 6 per group) after in vitro transfection of 40 nM LNA anti-424 or scrambled control. Tumor growth was monitored every 2 days with a caliper and IVIS Spectrum (PerkinElmer). Results were analyzed using Living Image software 4.2 (PerkinElmer). To evaluate lung metastatic capacity, in vitro–transfected DU145-Luc cells (1.5 × 106) were injected into the tail vein of nude mice (n = 7 per group). Animals were sacrificed after 5 weeks. Formation of lung metastasis was monitored by in vivo and postnecropsy bioluminescence imaging with the IVIS Spectrum.
Patient samples and data sets.
The in-house patient cohort (Biella cohort) was used for miRNA expression profiling and included snap-frozen tissue samples collected from normal prostate biopsies (n = 21) and primary prostate tumors (n = 48). The clinical parameters of the 48 prostate cancer samples are described in Supplemental Table 4. Gene expression profile data from these patients were previously described (10). The TCGA (The Cancer Genome Atlas) data set was downloaded from http://gdac.broadinstitute.org/, and included 419 primary prostate tumors. Data for other tumors were downloaded from data sets publicly available on the GEO database (GSE60371). See above for further details.
miRNA expression profiling.
miRNA expression profiling was carried out in normal (n = 21) and primary prostate tumors (n = 48) and in ESE3kd and control cells using the 1-color labeling method and following the manufacturer’s protocols (Agilent Technologies). Briefly, 100 ng of total RNA was dephosphorylated and denatured; then a ligation and labeling step with Cy3 was performed. Samples were hybridized to oligonucleotide glass arrays (Agilent Human miRNA Microarray 8x15K,V3) with sequences representing probes for 866 human and 89 human viral miRNAs from the Sanger database version 12.0 (Human miRNA Microarray V3, Agilent Technologies). Slides were washed and scanned using an Agilent B dual-laser microarray scanner. Images obtained were analyzed using Feature Extraction software. Raw data were processed using R; briefly, background subtraction was not applied, and log2 intensities were normalized using the “invariants” methodology (38). Differentially expressed miRNAs were obtained by selection of probes with absolute log2 fold change greater than 0.37 and adjusted P value less than 0.01. Data are compliant with Minimum Information About a Microarray Experiment (MIAME) and have been deposited in GEO (GSE60371).
Hierarchical clustering.
Hierarchical clustering with Pearson correlation as similarity metric and “complete” as linkage method was applied to the log intensity matrix of miRNAs × samples, with selection of miRNAs based only on their coefficient of variation along samples (>0.016, i.e., greater than the median of all coefficients of variation). This selection yielded to 1,367 probes (corresponding to 698 miRNAs), including those for hsa-miR-424-5p.
Gene set enrichment analysis.
Gene set enrichment analysis (GSEA) was conducted as previously described (8, 11). To identify gene signatures associated with increased miR-424 expression, we used the GSEA algorithm and the Molecular Signatures Database (MSigDB) C2 collection (version 4) of chemical and genetic perturbations (n = 3,402 gene sets) (http://software.broadinstitute.org/gsea/msigdb). The metric used for gene ranking was the correlation of the expression values of a gene with the miR-424 expression values across the human prostate samples. The metric was calculated for all genes within the microarray, after collapsing of the probes to the one with maximum absolute value. GSEA results are shown using the normalized enrichment score, which accounts for differences in gene set size and correlations between gene sets in different data sets. The androgen receptor (AR) score available in TCGA was applied to tumors with high, intermediate, and low levels of miR-424 in the same data set.
Gene expression profiling.
RNA was collected from RWPE1 cells 48 hours after transfection with 30 nM pre–miR-424 or Negative Control #1. We performed 8x60k Sure Print G3 Human GE Arrays. RNA was amplified, labeled, and hybridized according to the 2-color microarray-based gene expression analysis protocol (Agilent Technologies). Slides were scanned with the dual-laser scanner Agilent G2505B and analyzed as previously described (8). Differentially expressed genes were obtained by selection of probes with absolute log2 fold change greater than 0.37 and adjusted P value less than 0.05. Data are MIAME compliant and have been deposited in GEO (GSE60371).
Comparative transcriptome analysis of miR-424.
For comparative transcriptome analysis of miR-424-5p in several data sets (lung, gastric, breast, and prostate tumors vs. normal matched controls), gene expression data sets publicly available in the GEO database were examined (GSE60371). Moderated t test P values were obtained using linear models for microarray analysis (LIMMA). In case of multiple probes for miR-424-5p, the one with the lowest P value was used. In addition we retrieved RNA sequencing (RNA-Seq) data from the TCGA project (https://gdcportal.nci.nih.gov/projects/TCGA-PRAD). Levels of miR-424-5p were expressed as reads per million.
Extraction of miR-424 signatures in primary prostate tumors.
RNA-Seq data were retrieved from TCGA as described above. Samples were subdivided according to the level of miR-424-5p. Two-class comparison was performed on prostate tumors with high (n = 50) versus low (n = 50) mir-424-5p levels, using the LIMMA package available within Bioconductor. For the Biella data set, 2-class comparison was performed on prostate tumors with high (n = 17) versus low (n = 17) miR-424-5p level as described above.
Functional annotation and TF enrichment analysis.
For functional annotation we used the ENCODE project database to search for the most enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in selected gene lists. In addition we used the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/summary.jsp) to search for the most enriched biological processes of the Gene Ontology Biological Process (GoBP) terms. Enrichment scores were used to indicate the enrichment level of genes assigned to the indicated GoBP in each subgroup. To select for putative TFs involved in the regulation of selected gene lists (e.g., genes induced by miRNA-424), we used Enrichr, a database within the ENCODE project that includes a collection of genes whose promoters are bound by specific TFs as determined by ChIP sequencing. In addition, we used ChIP Enrichment Analysis (ChEA) software, which searches for TF enrichment in the ChIP-X database containing a collection of ChIP-chip, ChIP-seq, ChIP-PET, and DamID experiments.
ESE3/EHF–COP1 mRNA expression correlation analysis.
Correlation analysis was performed in indicated human prostate cancer data sets and various carcinoma data sets within TCGA. Pearson test was used, which estimates a correlation value “r” and a significance P value (r > 0 < 1, direct correlation; r < 0 > –1, inverse correlation).
Immunohistochemistry and immunocytochemistry.
Immunohistochemistry (IHC) was performed as previously described (11) using anti-STAT3 (124H6; Cell Signaling; catalog 9139); anti–p-STAT3 Tyr705 (D3A7; Cell Signaling; catalog 9145); anti-COP1 (B-12; Santa Cruz Biotechnology; catalog sc-166799); anti-Ki67 (Lab Vision Corp.; ready-to-use RT-9106-R7); anti–cleaved caspase-3 (Cell Signaling; 9664); and anti-CD31 (PECAM-1; Cell Signaling; 3528). The specificity of all antibodies was previously confirmed by Western blot analysis. Cell nuclei were counterstained with hematoxylin solution. Slides were evaluated by at least 2 investigators in a blinded manner. Immunocytochemistry was performed on cells (4 × 106 to 5 × 106 cells/ml in PBS) fixed in acetone/methanol (1:1) using anti-STAT3 antibody. Cell nuclei were counterstained by hematoxylin solution.
Tissue samples for IHC.
Single tissue sections from primary prostate tumors (n = 15) were obtained from the Cancer Genomics laboratory (Biella, Italy). Tissue microarrays (TMA) were constructed from formalin-fixed paraffin-embedded tissue specimens as previously described (39).
Survival analysis.
Differential survival between patient subgroups was plotted and calculated using Kaplan-Meier curves. P values were calculated using log-rank test. For TMA staining, patient samples were stratified based on percentage of positive cells in primary prostate tumors. The “high STAT3” group is defined as samples being within the 75th to 100th percentiles of positive cells. The “low COP1” group is defined as samples being between 0 and the 25th percentile of positive cells. Patients included in the Kaplan-Meier analysis had been treated with radical prostatectomy and had complete clinical record and follow-up data and evaluable IHC staining. Biochemical recurrence was defined as a 0.2-ng/ml increase in prostate-specific antigen (PSA) with a second confirmatory PSA measurement greater than 0.2 ng/ml or recurrence of disease after prostatectomy, such as development of metastatic cancer, if biochemical recurrence information was not available. For biochemical recurrence-free survival we included all the deaths that occurred within 10 years after radical retropubic prostatectomy.
Statistics.
The significance of differences between groups was measured with 2-tailed Student’s t test, and P values of less than 0.05 were considered statistically significant. Significance in overlapping gene lists was calculated using Fisher’s exact test. Statistical analysis was done using Stata 12.1 and SPSS software. For correlation analysis the Pearson correlation was calculated in Stata 13 software. The t test is used to establish if the correlation coefficient is significantly different from 0, and hence that there is evidence of an association between the 2 variables.
Study approval.
The protocol for tissue collection was approved by the Ethics Committee of Regione Piemonte (Italy) and by patients’ written informed consent. Tissue samples for TMA were collected with the approval of the Institutional Ethics Committees at Insespital (Bern, Switzerland) and patients’ written informed consent. Histopathological and clinical data relative to the TMA specimens were collected and continuously updated (Insespital, Bern). All animal experiments were approved by the Swiss Cantonal Veterinary Committee and were in accordance with the National Policy on Humane Care and Use of Laboratory Animals.
Author contributions
CD, DA, and G. Civenni conducted experiments, and acquired and analyzed data. PO and MMG acquired and analyzed data. JM conducted experiments, and acquired and analyzed data. GD, FS, and GNT provided samples and analyzed data. SR, ML, and RGE acquired and analyzed data. AZ provided samples and analyzed data. G. Chiorino acquired and analyzed data. CVC and GMC designed studies, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank A. Tizzani, P. Gontero, P. Pierini, V.A. Accardo, and I. Gregnanin for patient samples. We thank S. Pinton for technical support. This work was supported by grants from the Swiss Cancer League (KLS 3243-08-2010 and KLS 3243-08-2013), the Swiss National Science Foundation (FNS 310030-146560), and the Ticino Foundation for Cancer Research (to GMC); and from the Virginia Boeger Foundation and the Fidinam Foundation (to CVC). DA was supported by a Novartis Fellowship. MMG and G. Chiorino were supported by Compagnia di San Paolo, Torino, Italy.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(12):4585–4602. doi:10.1172/JCI86505.
Contributor Information
Cecilia Dallavalle, Email: cecilia.dallavalle@ior.iosi.ch.
Domenico Albino, Email: domenico.albino@ior.iosi.ch.
Gianluca Civenni, Email: gianluca.civenni@ior.iosi.ch.
Jessica Merulla, Email: jessica.merulla@ior.iosi.ch.
Paola Ostano, Email: paola.ostano@fondoedotempia.it.
Maurizia Mello-Grand, Email: maurizia.mellogrand@gmail.com.
Simona Rossi, Email: simona.rossi@ior.iosi.ch.
Marco Losa, Email: marco.losa@ior.iosi.ch.
Fausto Sessa, Email: Fausto.Sessa@uninsubria.it.
Ramon Garcia-Escudero, Email: ramon.garcia@ciemat.es.
Andrea Zitella, Email: azitella@cittadellasalute.to.it.
Giovanna Chiorino, Email: giovanna.chiorino@gmail.com.
Carlo V. Catapano, Email: carlo.catapano@ior.iosi.ch.
Giuseppina M. Carbone, Email: pina.carbone@ior.iosi.ch.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349(4):366–381. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 3.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24(18):1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39(1):41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 8.Albino D, et al. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res. 2012;72(11):2889–2900. doi: 10.1158/0008-5472.CAN-12-0212. [DOI] [PubMed] [Google Scholar]
- 9.Cangemi R, et al. Reduced expression and tumor suppressor function of the ETS transcription factor ESE-3 in prostate cancer. Oncogene. 2008;27(20):2877–2885. doi: 10.1038/sj.onc.1210953. [DOI] [PubMed] [Google Scholar]
- 10.Kunderfranco P, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One. 2010;5(5):e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longoni N, et al. ETS transcription factor ESE1/ELF3 orchestrates a positive feedback loop that constitutively activates NF-κB and drives prostate cancer progression. Cancer Res. 2013;73(14):4533–4547. doi: 10.1158/0008-5472.CAN-12-4537. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catto JW, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59(5):671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 15.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33(2):135–147. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 16.Migliorini D, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest. 2011;121(4):1329–1343. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013;31(9):1231–1238. doi: 10.1200/JCO.2012.44.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theurillat JP, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346(6205):85–89. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitari AC, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474(7351):403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Civenni G, et al. RNAi-mediated silencing of Myc transcription inhibits stem-like cell maintenance and tumorigenicity in prostate cancer. Cancer Res. 2013;73(22):6816–6827. doi: 10.1158/0008-5472.CAN-13-0615. [DOI] [PubMed] [Google Scholar]
- 24.Mora LB, et al. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62(22):6659–6666. [PubMed] [Google Scholar]
- 25.Schroeder A, et al. Loss of androgen receptor expression promotes a stem-like cell phenotype in prostate cancer through STAT3 signaling. Cancer Res. 2014;74(4):1227–1237. doi: 10.1158/0008-5472.CAN-13-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroon P, et al. JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res. 2013;73(16):5288–5298. doi: 10.1158/0008-5472.CAN-13-0874. [DOI] [PubMed] [Google Scholar]
- 27.Baffert F, et al. Potent and selective inhibition of polycythemia by the quinoxaline JAK2 inhibitor NVP-BSK805. Mol Cancer Ther. 2010;9(7):1945–1955. doi: 10.1158/1535-7163.MCT-10-0053. [DOI] [PubMed] [Google Scholar]
- 28.Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banyard J, et al. Regulation of epithelial plasticity by miR-424 and miR-200 in a new prostate cancer metastasis model. Sci Rep. 2013;3:3151. doi: 10.1038/srep03151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin HR, et al. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68(19):7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sellier H, Rébillard A, Guette C, Barré B, Coqueret O. How should we define STAT3 as an oncogene and as a potential target for therapy? JAKSTAT. 2013;2(3):e24716. doi: 10.4161/jkst.24716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng CJ, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15(12):2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 35.Rahimian A, et al. Bypassing the maturation arrest in myeloid cell line U937 by over-expression of microRNA-424. Hematology. 2011;16(5):298–302. doi: 10.1179/102453311X13085644680140. [DOI] [PubMed] [Google Scholar]
- 36.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(Web Server issue):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longoni N, et al. Aberrant expression of the neuronal-specific protein DCDC2 promotes malignant phenotypes and is associated with prostate cancer progression. Oncogene. 2013;32(18):2315–2324.e1. doi: 10.1038/onc.2012.245. [DOI] [PubMed] [Google Scholar]
- 38.Pradervand S, et al. Impact of normalization on miRNA microarray expression profiling. RNA. 2009;15(3):493–501. doi: 10.1261/rna.1295509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prtilo A, et al. Tissue microarray analysis of hMSH2 expression predicts outcome in men with prostate cancer. J Urol. 2005;174(5):1814–1818. doi: 10.1097/01.ju.0000176796.47988.64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.