Abstract
The rising success of cancer immunotherapy has produced immense interest in defining the clinical contexts that may benefit from this therapeutic approach. To this end, there is a need to ascertain how the therapeutic modulation of intrinsic cancer cell programs influences the anticancer immune response. For example, the role of autophagy as a tumor cell survival and metabolic fitness pathway is being therapeutically targeted in ongoing clinical trials that combine cancer therapies with antimalarial drugs for the treatment of a broad spectrum of cancers, many of which will likely benefit from immunotherapy. However, our current understanding of the interplay between autophagy and the immune response remains incomplete. Here, we have evaluated how autophagy inhibition impacts the antitumor immune response in immune-competent mouse models of melanoma and mammary cancer. We observed equivalent levels of T cell infiltration and function within autophagy-competent and -deficient tumors, even upon treatment with the anthracycline chemotherapeutic doxorubicin. Similarly, we found equivalent T cell responses upon systemic treatment of tumor-bearing mice with antimalarial drugs. Our findings demonstrate that antitumor adaptive immunity is not adversely impaired by autophagy inhibition in these models, allowing for the future possibility of combining autophagy inhibitors with immunotherapy in certain clinical contexts.
Introduction
Autophagy is a tightly regulated cellular program that results in the engulfment and sequestration of cytoplasmic protein and organelle cargo into double-membrane structures termed autophagosomes, which are subsequently delivered to the lysosome for degradation (1). Autophagy occurs both at basal conditions to maintain cellular homeostasis, and in response to environmental stresses such as nutrient starvation or hypoxia. Most studies investigating autophagy in cancer have focused on its cell-intrinsic effects, including aiding cancer cell survival during extrinsic stress (2, 3) as well as promoting drug resistance (4). Importantly, several clinical trials have combined traditional chemotherapy or targeted therapy with antimalarial lysosomotropic agents, such as hydroxychloroquine, which block the late stages of autophagic proteolysis (5–12). Despite interest in inhibiting autophagy in the clinical oncology setting in combination with chemotherapies or other targeted therapies, emerging evidence has raised questions with regard to the efficacy of such approaches to cancer treatment. It is now appreciated that certain chemotherapies, particularly anthracycline agents, can kill tumors through combined cytotoxic and immunogenic mechanisms, via the process of immunogenic cell death (ICD), in which dying cells release damage-associated molecular patterns (DAMPs) to elicit an immune response (13, 14). ICD is believed to be an important effector of both chemotherapy and radiation therapy, but the precise contributions of ICD to treatment-mediated tumor killing are varied and context-dependent (15).
Importantly, autophagy has been shown to promote certain hallmarks of ICD in vitro, including the secretion of ATP and high-motility group protein B1 (HMGB1), both of which act as DAMPs (16). In support of the notion that autophagy facilitates antitumor immunity in vivo are the observations that loss of either autophagy or adaptive immunity impairs the regression of some mouse tumors during anthracycline therapy, and autophagy-deficient mouse colon tumors exhibit decreases in recruitment and activation of T cells (16, 17).
Overall, these potentially adverse effects on the adaptive immune system argue against the use of autophagy inhibitors in anticancer therapy. Nonetheless, autophagy inhibition may be a very useful anticancer therapy not only in undermining tumor cell growth but also in preventing the survival of quiescent cells during chemotherapy (18). These discrepancies become especially important considerations in light of the recent success of cancer immunotherapies. Notably, immune checkpoint blockade therapies, which leverage monoclonal antibodies targeting cytotoxic T lymphocyte–associated protein 4 (CTLA4) (19, 20) and programmed cell death protein 1 (PD-1) (21, 22) to reinvigorate existing T cell responses within tumors, have produced long-term remission in certain patients (23, 24). Currently, the best indication of a powerful response to these therapies is a high number of cancer-expressed neoantigens; thus, melanomas and smoking-associated lung carcinomas, which carry a high mutational load and express high levels of neoantigens, exhibit the highest response rates to immune checkpoint blockade (25). Even so, only a portion of patients with these cancers responds well to immunotherapy. With the promise of the impressive durable responses achieved with these therapeutic modalities comes a pressing need to better define the clinical contexts in which immunotherapies will be effective (26). Furthermore, leveraging the durable responses in immunotherapy in combination with short-term responses in other strategies is an alluring approach to further improve cancer treatment outcomes (27).
In this study, we sought to more fully delineate the effects of autophagy on the tumor-associated T cell response in order to better ascertain whether autophagy inhibition can be effectively combined with chemotherapy in the clinic. Using established immunogenic models of mouse melanoma and mammary cancer, we assessed the effects of autophagy inhibition on the functional status of tumor-infiltrating T cells, both at baseline and following doxorubicin (Dox) chemotherapy. We interrogated the functional status of tumor-associated CD4+ T helper cells and CD8+ cytotoxic T cells by flow cytometry for a variety of markers of T cell activation (28, 29) as well as immune checkpoint modulators. Our results demonstrate that genetic autophagy inhibition in tumor cells in vivo does not significantly impact T cell infiltration, activation, or immune checkpoint regulation. Similarly, we observe no changes in the T cell response upon systemic treatment of tumor-bearing mice with antimalarials that inhibit autophagy. We further confirm that autophagy inhibition does not impact the functional activation of adoptively transferred ovalbumin-specific transgenic T cell receptor (OT-I) CD8+ T cells (30) in ovalbumin-expressing tumors. Hence, autophagy does not modulate the antitumor T lymphocyte response in multiple immune-competent mouse cancer models. Based on these results, we propose that certain clinical contexts exist in which autophagy inhibition may be utilized as a therapeutic strategy against cancer without adversely affecting the antitumor immune response.
Results
Genetic inhibition of autophagy in tumor cells does not alter primary tumor growth.
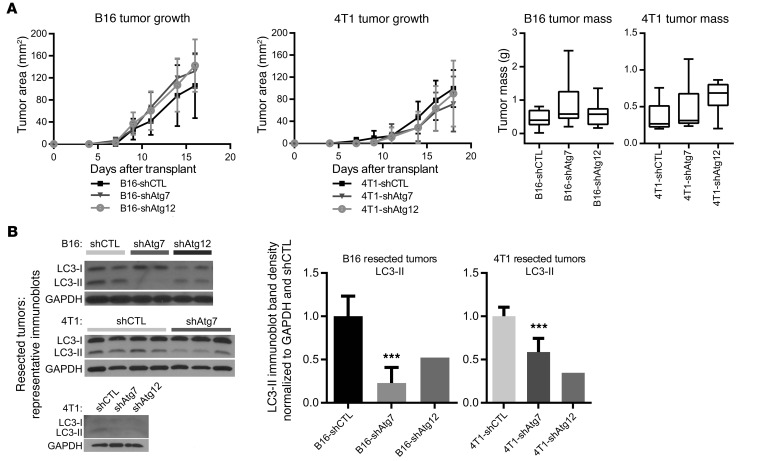
To dissect the effect of tumor cell–intrinsic autophagy on the extrinsic antitumor immune response, we generated B16 murine melanoma cells stably expressing shRNAs against the essential autophagy-related genes Atg7 and Atg12. Cells expressing nontargeting shRNA (shCTL) served as autophagy-competent controls. Stable ATG7 or ATG12 knockdown (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI85705DS1) led to reduced autophagic flux, evidenced by reduced LC3-II formation and lysosomal turnover (Supplemental Figure 1B). Control and autophagy-deficient B16 tumors were introduced s.c. into C57BL/6 mice, and palpable tumors formed 7–10 days after transplantation. Primary tumor growth was unchanged between autophagy-competent and -deficient tumors, based on caliper measurements of tumor area over time as well as resected tumor mass at experimental endpoint (Figure 1A). Similarly, autophagy inhibition in 4T1 mammary cancer cells, achieved via stable ATG7 or ATG12 knockdown (Supplemental Figure 1A), significantly impaired autophagic flux (Supplemental Figure 1C) but did not impact primary tumor growth following orthotopic transplantation into the mammary fat pad of 6- to 7-week-old female BALB/c mice (Figure 1A). In both models, we confirmed that robust autophagy inhibition was maintained over the duration of the experiment, evidenced by the loss of LC3-II in lysates generated from resected tumors harvested at 2–3 weeks after transplant (Figure 1B).
Figure 1. Genetic models of autophagy deficiency in mouse melanoma and mammary cancer.
B16 mouse melanoma or 4T1 mouse mammary cancer cells bearing nontargeting shRNA (shCTL) or shRNA directed against autophagy-related genes (ATGs) were transplanted into immune-competent host mice. (A) Top: Primary tumor growth of autophagy-competent and -deficient subcutaneous B16 tumors (shCTL: n = 6; shATG7: n = 7; shATG12: n = 6) and orthotopic 4T1 tumors (shCTL: n = 5; shATG7: n = 5; shATG12: n = 8) in syngeneic host mice, as assessed by caliper measurements of tumor area. Error bars represent SD. Bottom: Tumor mass at experimental endpoint (day 16–18). For B16 tumors, shCTL: n = 14; shATG7: n = 15; shATG12: n = 9; and for 4T1 tumors, shCTL: n = 13; shATG7: n = 13; shATG12: n = 8. Box and whisker plots indicate minimum, median, and maximum values. Two-way ANOVA not significant. (B) Lysates from resected tumors subject to α-LC3 immunoblotting; band densities for LC3-II were normalized to GAPDH and to the control average within each experiment. For B16 tumors, shCTL: n = 5; shATG7: n = 6; shATG12: n = 2; and for 4T1 tumors, shCTL: n = 6; shATG7: n = 5; shATG12: n = 2. Error bars represent SD. ***P < 0.001 using unpaired t test.
Autophagy-competent and -deficient tumors elicit equivalent T cell responses.
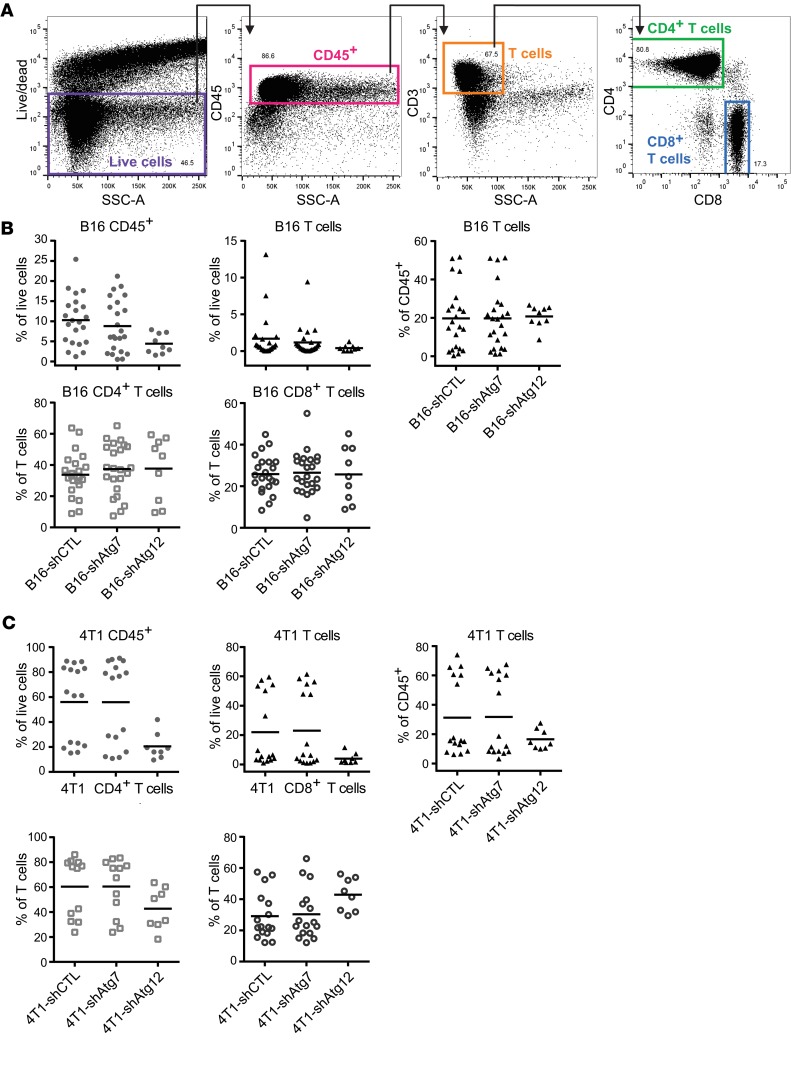
Having developed 2 models of tumor cell autophagy deficiency in distinct mouse genetic backgrounds, we evaluated the immune response to tumors arising from these cells. To quantify and interrogate the function of tumor-associated T cells, we prepared single-cell suspensions from resected tumors and subsequently stained a variety of surface and intracellular markers for flow cytometric analysis. To accurately measure intracellular cytokines in T cells, we injected mice with brefeldin A (BFA) before tumor resection to prevent cytokine secretion. T cells were identified as the CD45+CD3+SSC-Alo fraction of total live cells defined by a live/dead marker; from these, CD4+ and CD8+ T cell subpopulations were identified (Figure 2A). Autophagy-deficient B16 and 4T1 tumors did not exhibit significant differences in the infiltration of CD45+ cells, total T cells, or CD4+ or CD8+ subsets, when compared with autophagy-competent controls (Figure 2, B and C). While biological variation occurred across the littermates used for the experimental repeats, autophagy-competent and -deficient tumors generated in littermates exhibited equivalent levels of immune infiltration; hence, statistical analyses of both individual and batched data sets were not significant.
Figure 2. T cell infiltration is unchanged in autophagy-deficient mouse tumors.
Subcutaneous B16 and orthotopic 4T1 tumors were allowed to form for 2–3 weeks. Tumors were resected and digested enzymatically, and T cell infiltration was measured by flow cytometry. (A) Representative flow cytometry gating strategy to define T cell populations. A live/dead marker was used to define live cells as a subset of singlets. CD45+ cells were defined from live cells, and T cells were defined as the CD3+SSC-Alo fraction of CD45+ cells. CD4+ and CD8+ single-positive T cell populations were subdivided from total T cells. (B) Infiltration of CD45+ cells and T cell populations into primary mouse tumors in autophagy-competent and -deficient B16 melanomas. Each data point represents a distinct tumor from an individual host mouse. Bars represent mean values with 2-way ANOVA not significant. (C) Infiltration of CD45+ cells and T cell populations into primary mouse tumors in autophagy-competent and -deficient 4T1 mammary tumors. Each data point represents a distinct tumor from an individual host mouse. Bars represent mean values with 2-way ANOVA not significant.
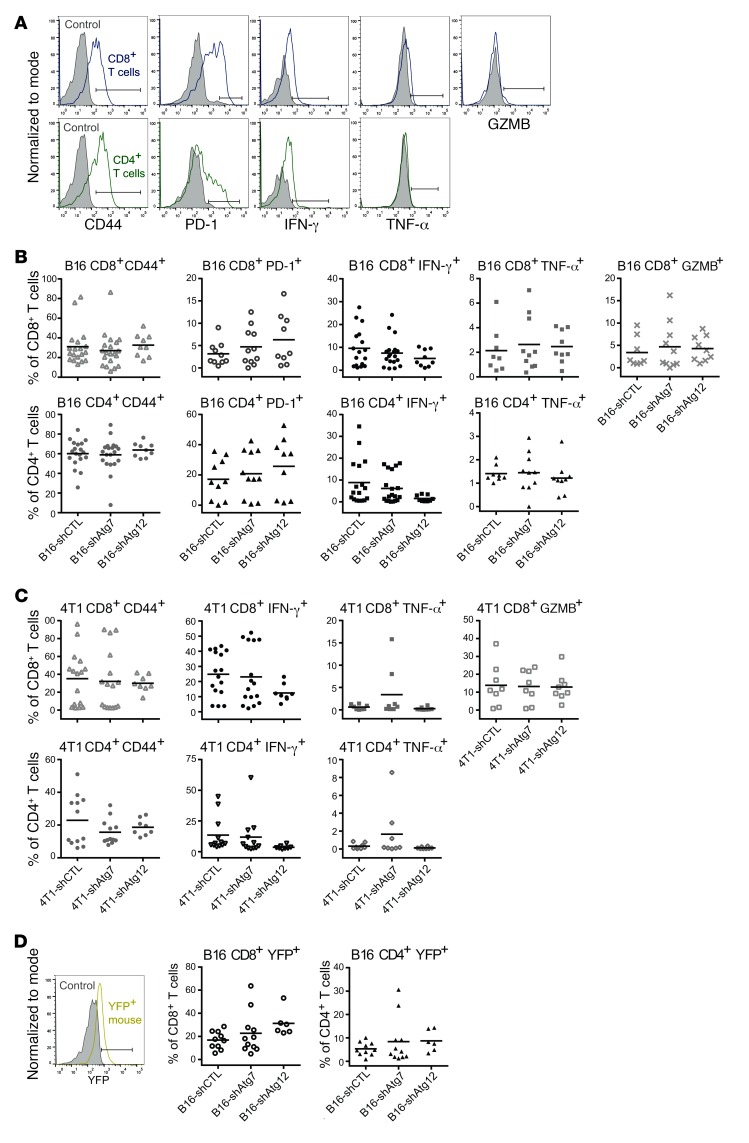
We next assessed the functional activation status of CD8+ and CD4+ T cells that had infiltrated B16 tumors by analyzing cell surface expression of the activation and memory marker CD44 and intracellular expression of the inflammatory cytokines IFN-γ and TNF-α. By these measures, we observed no differences in the T cell activation phenotype between control and autophagy-deficient B16 tumors (Figure 3, A and B, and Supplemental Figure 2A). Similarly, we found no differences between cohorts in the activation of the CD8+ T cell cytotoxic program based on intracellular expression of the serine protease granzyme B (GZMB) (Figure 3, A and B, and Supplemental Figure 2A). We also measured the surface expression of the immune checkpoint regulator programmed cell death protein 1 (PD-1) and found no differences in either CD4+ or CD8+ T cells between autophagy-competent and -deficient tumors (Figure 3, A and B, and Supplemental Figure 2A). In agreement with these results from B16 tumors, T cell functional status and activation phenotype were unchanged between autophagy-competent and -deficient 4T1 tumors (Figure 3C). As before, statistical analyses of both individual and batched data sets were not significant. Together, these findings indicate that reduced tumor cell autophagy in both mouse melanomas and mammary tumors does not influence the ability of the host adaptive immune system to infiltrate and become functionally active within these tumors.
Figure 3. T cell functional status is unchanged in autophagy-deficient mouse tumors.
Expression of T cell activation markers (CD44, IFN-γ, TNF-α, GZMB) and immune checkpoint marker PD-1 was measured by flow cytometry in CD8+ and CD4+ T cell populations. (A) Representative histograms of functional marker staining of CD8+ and CD4+ T cell populations. Solid gray plots represent unstained controls (CD44, CD4, CD8) and isotype controls (PD-1, IFN-γ, TNF-α, GZMB). The same unstained control was used for the CD4+ and CD8+ T cell panels. Positive staining is indicated by gate and defined as that above the unstained or isotype control. (B) Functional status of T cells isolated from autophagy-competent (shCTL) and -deficient (shAtg7 or shAtg12) B16 tumors. Each data point represents a distinct tumor from an individual host mouse. Bars represent mean values with 2-way ANOVA not significant. (C) Functional status of T cells isolated from autophagy-competent (shCTL) and -deficient (shAtg7 or shAtg12) 4T1 tumors. Each data point represents a distinct tumor from an individual host mouse. Bars represent mean values with 2-way ANOVA not significant. (D) B16 tumors were grown in GREAT reporter mice bearing the IFN-γ-IRES-eYFP reporter cassette. Representative histogram shows YFP-negative control (solid gray plot) and cells isolated from a GREAT reporter mouse (yellow plot). Positive signal was defined as that above the control and is indicated by a gate. Endogenous eYFP expression was measured in tumor-infiltrating T cell populations by flow cytometry without BFA injection. Each data point represents a distinct tumor from an individual host mouse. Bars represent mean values with 2-way ANOVA not significant.
To further corroborate the observation that T cell activation was independent of autophagy status, we generated autophagy-competent and -deficient B16 tumors in mice carrying an IFN-γ-IRES-eYFP reporter cassette (also known as the “IFN-γ reporter with endogenous polyA transcript,” or GREAT allele), in which expression of IFN-γ and expression of enhanced yellow fluorescent protein (eYFP) are separated by an internal ribosome entry site (IRES) and dually controlled by the endogenous Ifng promoter/enhancer region (31). While IFN-γ is a secreted protein, eYFP accumulates intracellularly upon IFN-γ transcription; thus, T cell activation in resected tumors is measured using endogenous eYFP fluorescence, obviating the need for BFA treatment or intracellular staining. No differences in eYFP expression in CD4+ or CD8+ T cells were observed by this method, providing further evidence for an autophagy-independent T cell response (Figure 3D). Remarkably, based on eYFP reporter levels, the IFN-γ response was comparable to that obtained with intracellular IFN-γ staining.
To address whether tumor cell autophagy status modulates immunosuppression, we evaluated the quantity and function of regulatory T cells (Tregs) associated with autophagy-competent and -deficient B16 tumors (Supplemental Figure 3A). Once again, neither the number of Tregs (CD45+CD3+CD4+Foxp3+) nor their activation (CD44+) was changed upon genetic autophagy inhibition, further supporting that T cell responses were unaffected by autophagy status in these tumors.
Finally, to assess tumor-associated T cell activation potential, we isolated CD8+ T cells from 4T1 tumors by FACS or negative bead selection and cultured them with CD3 and CD28 antibodies to incite robust ex vivo antigen-independent restimulation. We measured IFN-γ secretion in conditioned medium by ELISA and observed equivalent levels of secretion from T cells derived from either autophagy-competent or -deficient 4T1 tumors (Supplemental Figure 3B). Thus, tumor-associated T cells possess equivalent activation in situ as well as equivalent activation potential, regardless of the autophagy status of tumor cells.
Autophagy-competent and -deficient OVA-expressing tumors stimulate comparable transgenic T cell responses.
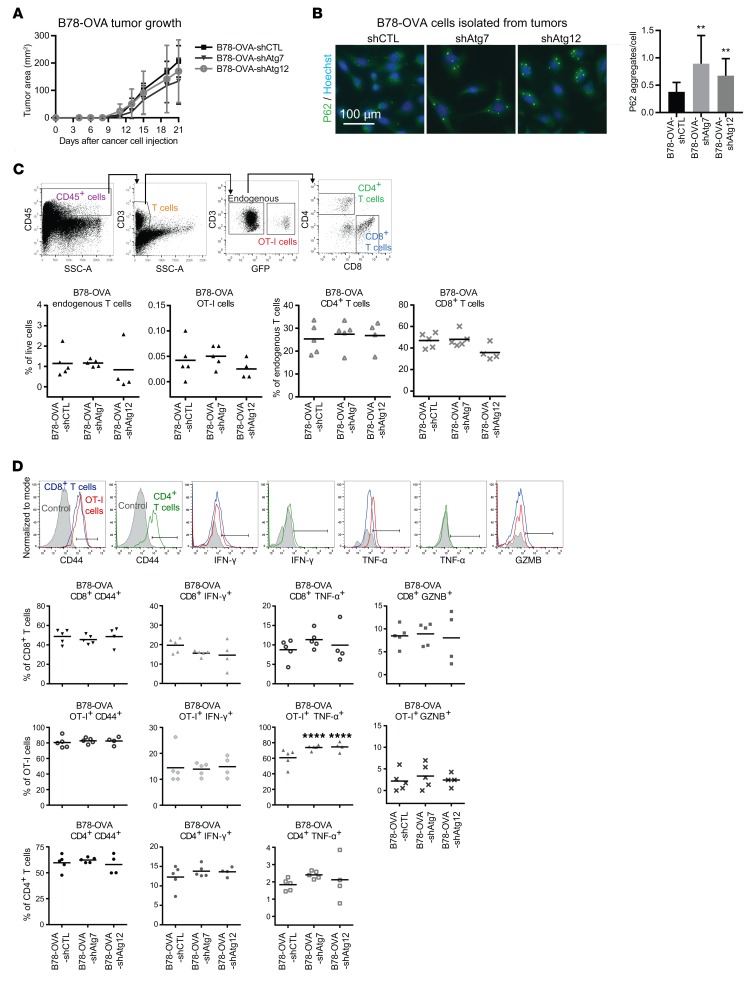
Studies using T cells expressing a transgenic T cell receptor matched to a specific antigen (e.g., ovalbumin) have been valuable in defining a tumor-specific adaptive immune response. To evaluate cancer cell recognition by T cells, we generated autophagy-competent (shCTL) and autophagy-deficient (Atg7 or Atg12 shRNA) B78 melanoma cells (a variant of B16) expressing ovalbumin (OVA). We confirmed stable suppression of ATG7 and ATG12 (Supplemental Figure 4A) and reduction of autophagic flux by reduced LC3-II and increased P62 (also known as SQSTM1), an autophagic cargo receptor that accumulates upon autophagy inhibition, in cell culture lysates (Supplemental Figure 4B). B78-OVA cells were injected s.c. into WT C57BL/6 mice and allowed to form palpable primary tumors. Freshly isolated OVA-specific OT-I CD8+ T cells expressing green fluorescent protein (GFP) were then adoptively transferred by retro-orbital injection into tumor-bearing mice, and tumors were resected 1 week later. Tumor growth kinetics was unchanged between autophagy-competent and -deficient groups (Figure 4A). Autophagy inhibition was confirmed by the accumulation of P62 aggregates in B78-OVA cells isolated from digested tumors (Figure 4B).
Figure 4. Transgenic OT-I cell activation is unchanged in autophagy-deficient OVA-expressing tumors.
Freshly isolated transgenic OT-I CD8+ T cells expressing GFP were adoptively transferred by retro-orbital injection into mice bearing 2-week subcutaneous autophagy-competent and -deficient B78 melanomas expressing OVA. (A) Primary tumor growth of autophagy-competent and -deficient subcutaneous B78-OVA tumors in syngeneic host mice as assessed by caliper measurements of tumor area (n = 5 per cohort). (B) Autophagy deficiency was confirmed in B78-OVA cells isolated from digested tumors by immunofluorescence for P62. Accumulation of P62 aggregates was quantified per cell nucleus. Error bars represent SD; **P < 0.01 using unpaired t test. (C) Total T cells were defined as the CD45+CD3+SSC-Alo fraction of live cells and were further subdivided into OT-I (GFP+) and endogenous (GFP–) populations; the latter was analyzed for CD4 and CD8 surface expression. Endogenous and OT-I T cell populations were equivalent between autophagy-competent (shCTL) and -deficient (shAtg12) B78-OVA tumors. Error bars represent SD with 2-way ANOVA not significant. (D) Activation status was measured by surface CD44 and intracellular IFN-γ, TNF-α, and GZMB expression in endogenous and OT-I T cell populations from autophagy-competent and -deficient B78-OVA tumors. Representative histograms of functional marker staining of endogenous and OT-I T cell populations: solid gray plots represent unstained controls (for surface staining of CD44) and isotype controls (for intracellular stains of IFN-γ, TNF-α, GZMB). Positive staining is defined as that above the unstained or isotype control and is indicated by gates. In graphs, each data point represents a distinct tumor from an individual host mouse, and bars represent mean values. ****P < 0.0001 using 2-way ANOVA multiplicity-adjusted P values (Dunnett’s correction).
Endogenous and adoptively transferred tumor-associated T cells were defined by flow cytometric analysis based on expression of CD45, CD3, CD4, CD8, and GFP; no differences were observed in the tumor infiltration of total T cells, endogenous CD4+ or CD8+ T cells, or adoptively transferred OT-I CD8+ T cells (Figure 4C). We next measured T cell activation by expression of surface CD44 and intracellular IFN-γ, TNF-α, and GZMB. We observed no significant differences in expression of activation markers in either OT-I or endogenous populations of T cells between autophagy-competent and -deficient B78-OVA tumors, with the exception of an increase in TNF-α–expressing OT-I cells associated with autophagy-deficient tumors (Figure 4D and Supplemental Figure 2B). Importantly, autophagy deficiency did not blunt the T cell response. As expected, the activation of OT-I T cells was higher than that of endogenous CD8+ T cells, as evidenced by higher percentages of CD44+ and TNF-α+ cells. Overall, the T cell response remained intact in both the endogenous and the antigen-matched setting upon genetic autophagy inhibition.
Dox-treated autophagy-competent and -deficient tumors stimulate equivalent T cell responses.
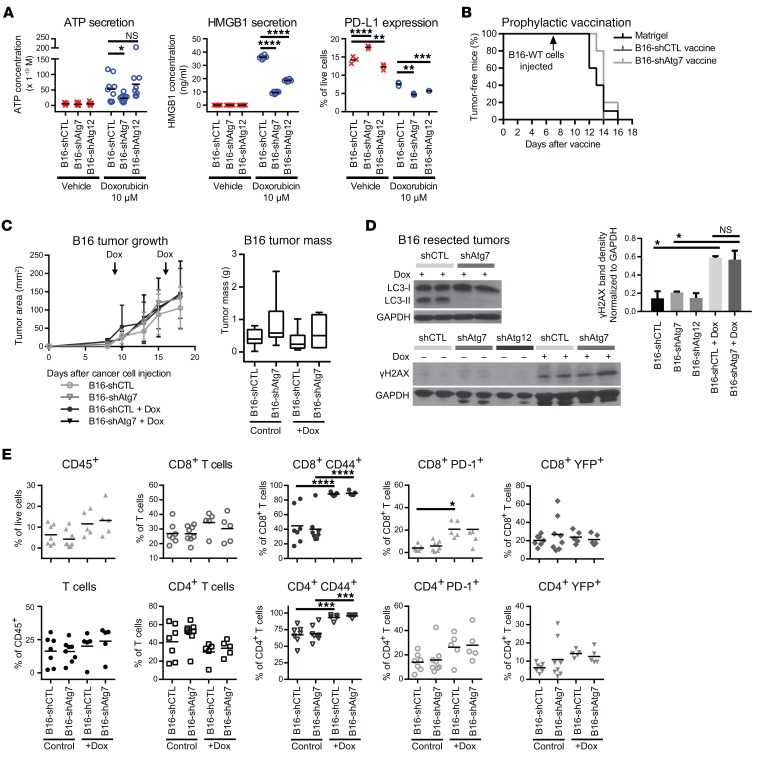
Because our results indicated that tumor cell–intrinsic autophagy is dispensable for stimulating a T cell response, we next tested whether autophagy was required for the immune response in tumor-bearing mice following acute treatment with chemotherapy. Previous work with murine colon cancer suggests that autophagy is necessary for the immunogenic cell death (ICD) associated with the efficacy of anthracycline chemotherapy (16). Thus, genetic inhibition of autophagy may hinder the chemotherapeutic drug response by blunting immune responses. We treated autophagy-competent and -deficient B16 melanoma cells with the anthracycline Dox in vitro, and measured the secreted levels of ATP and HMGB1, 2 immunomodulatory factors implicated in ICD (Figure 5A). Consistent with existing literature, we observed that Dox treatment induced ATP and HMGB1 secretion, which was attenuated in autophagy-deficient cells as compared with autophagy-competent controls. Interestingly, we noted that Dox treatment also led to a modest, albeit significant, reduction in the surface expression of the immunosuppressive protein programmed death-ligand 1 (PD-L1) in autophagy-deficient tumor cells (Figure 5A). Notably, B16-shAtg7 cells exhibited higher levels of death upon Dox treatment relative to B16-shCTL cells (Supplemental Figure 5A), indicating increased sensitivity to autophagy impairment; similarly, autophagy-deficient 4T1 cells showed reduced cellular viability following 24 hours of Dox treatment (Supplemental Figure 5B).
Figure 5. Dox-treated autophagy-deficient tumors elicit equivalent T cell responses despite altered secretion of immunomodulatory factors.
(A) Autophagy-competent (shCTL) and -deficient (shAtg7 or shAtg12) B16 cells treated with 10 μM Dox or vehicle control. ATP and HMGB1 secretion measured in conditioned medium and surface PD-L1 measured by flow cytometry. Data points represent biological replicates; bars represent mean values. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; not significant (NS) using 2-way ANOVA multiplicity-adjusted P values (Dunnett’s correction). (B) Autophagy-competent (shCTL) and -deficient (shAtg7) B16 cells were treated for 24 hours with 8.8 μM and 7.5 μM Dox, respectively, and subsequently injected into WT C57BL/6 mice (n = 5). Control mice were injected with Matrigel (n = 10). Mice were rechallenged with WT B16 cells 1 week later; tumor incidence was monitored daily. (C) Autophagy-competent and -deficient B16 cells were injected into GREAT reporter mice; mice were then treated with 5 mg/kg Dox weekly. Tumor growth curves: arrows indicate Dox treatment; error bars represent SD (B16-shCTL: n = 6; B16-shATG7: n = 7; B16-shCTL + Dox: n = 8; B16-shATG7 + Dox: n = 8). Resected tumor mass: box and whisker plots indicate minimum, median, and maximum values. Untreated tumors are also included in Figure 1A. (D) Resected and digested tumors were immunoblotted for LC3 (autophagy deficiency) and γH2AX (DNA damage). Expression quantified normalized to GAPDH. Error bars represent SD. *P < 0.05; NS, not significant, using unpaired t test. (E) T cell infiltration and functional status (CD44, PD-1, eYFP) in control and Dox-treated autophagy-competent and -deficient B16 tumors from 2 experimental cohorts. Each data point represents a distinct tumor from an individual host mouse. Bars represent mean values. *P < 0.05; ***P < 0.001; ****P < 0.0001, using 2-way ANOVA multiplicity-adjusted P values (Tukey’s correction). Measurements of untreated tumors are also included in Figures 2B and 3B.
Because these in vitro findings suggested that autophagy status in B16 melanoma cells influenced their expression of several markers of ICD, we evaluated the effects of genetic autophagy inhibition on ICD in vivo. To assess whether autophagy-competent and -deficient B16 melanomas underwent equivalent levels of ICD in vivo, we used the prophylactic vaccination experimental design described previously by Michaud et al. (16). Based on dose-response curves to determine appropriate treatment regimens (Supplemental Figure 5A), we pretreated B16-shCTL and B16-shAtg7 with Dox in vitro and injected cocktails s.c. into mice in which 70% of cells were apoptotic; Matrigel was injected as a control “vaccination.” One week later, healthy WT B16 cells were injected s.c. into contralateral flanks, and tumor incidence was measured by daily palpation. Tumor-free survival was unchanged between mice vaccinated with Matrigel, B16-shCTL, or B16-shAtg7 cells (Figure 5B). Thus, in spite of expressing classic ICD markers in vitro, Dox-treated B16 cells were unable to vaccinate mice against rechallenge irrespective of autophagy status. These results supported that ICD is a highly dynamic, context-dependent process (15), and further broached that autophagy inhibition does not universally perturb immunogenic responses following chemotherapy.
Based on these results, we sought to directly evaluate the effects of genetic autophagy inhibition on the immune response during in vivo Dox treatment. We generated autophagy-competent and -deficient B16 melanoma tumors in GREAT reporter mice; following the development of palpable tumors, mice were treated with Dox. There were no significant differences in tumor growth between cohorts, either by caliper measurements or tumor mass at resection (Figure 5C). Phosphorylated H2a histone family member X (γH2AX) expression, a marker of DNA damage, was significantly elevated in Dox-treated tumors compared with untreated controls, but was unchanged between autophagy-competent and -deficient Dox-treated cohorts (Figure 5D). Thus, this dose regimen instigated DNA damage in B16 tumors, a prerequisite for cell death and immunogenic potential. We also confirmed autophagy deficiency in resected tumors by immunoblotting for LC3-II (Figure 5D).
Upon assessment of immune infiltration, we found that percentages of CD45+ cells, total T cells, and CD4+ and CD8+ subsets were equivalent between untreated and Dox-treated cohorts (Figure 5E). T cell activation phenotype was measured by flow cytometry using surface CD44 expression and endogenous eYFP fluorescence as a reporter for IFN-γ levels, while T cell immune checkpoint regulation was measured by surface PD-1 expression. However, no differences were observed in the levels of any of these functional markers between control and ATG7 knockdown Dox-treated tumors (Figure 5E).
Importantly, we observed evidence of a chemotherapy-induced immune response in Dox-treated tumors compared with untreated controls. Most strikingly, CD44 expression was significantly elevated in both CD8+ and CD4+ T cell populations, in both Dox-treated B16-shCTL and B16-shAtg7 tumors, as compared with untreated counterparts (Figure 5E). Thus, Dox treatment of B16 tumor–bearing mice elicited immune responses within the tumor site, as evidenced by direct measurement of tumor-associated T cell activation. Because autophagy inhibition did not blunt this enhanced immune response during Dox treatment, our data suggest that autophagy inhibition and anthracycline chemotherapy can be safely combined in certain tumor types.
Antimalarial treatment of tumor-bearing mice does not alter the antitumor T cell response.
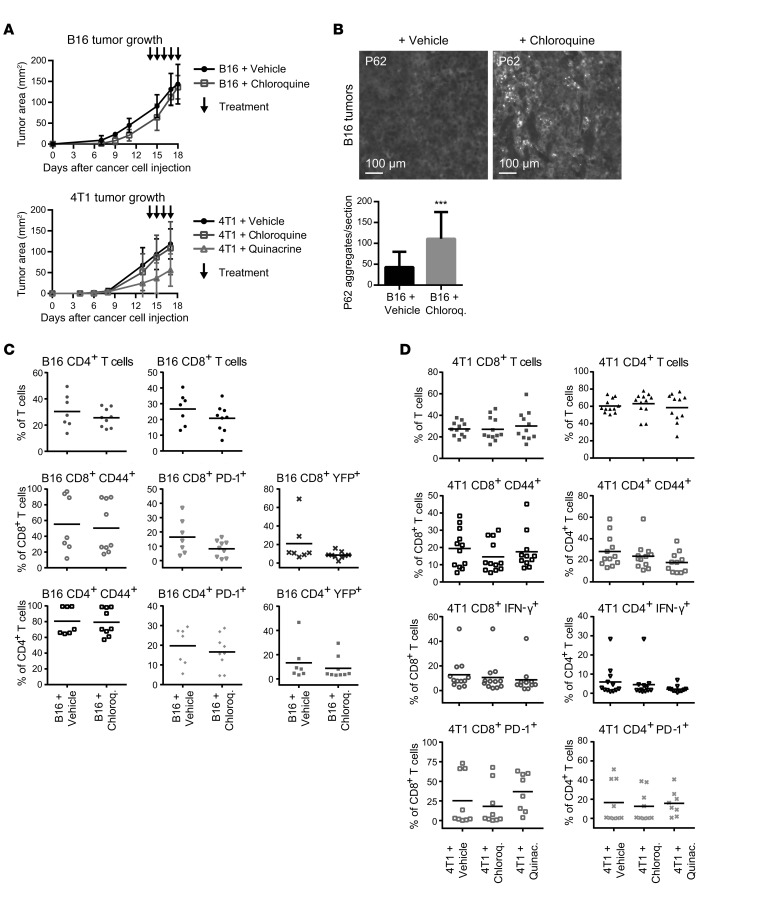
Autophagy inhibition is currently accomplished in the clinical setting by systemic treatment with antimalarial drugs such as chloroquine and hydroxychloroquine (32). These drugs inhibit acidification of intracellular vesicular compartments such as the lysosome, thereby blocking the terminal stages of autophagic proteolysis. To ascertain the effects of systemic antimalarial treatment on the antitumor immune response, we evaluated the effects of chloroquine treatment on subcutaneous B16 melanomas in GREAT reporter mice. Mice bearing palpable tumors were treated with daily i.p. injections of 60 mg/kg chloroquine or vehicle control for 4–5 days before tumor resection. As with genetic autophagy inhibition, tumor growth kinetics was unchanged upon pharmacological autophagy inhibition (Figure 6A), which was confirmed by the accumulation of P62 aggregates in resected tumors (Figure 6B). T cell infiltration and activation, measured by CD44 and eYFP reporter activity, were unchanged in chloroquine-treated B16 melanomas compared with vehicle-treated controls (Figure 6C). In addition, chloroquine treatment did not change the levels of immune checkpoint regulator PD-1 (Figure 6C). To extend these results, we generated orthotopic 4T1 mammary tumors in BALB/c mice and evaluated the effects of chloroquine as well as quinacrine, another FDA-approved antimalarial demonstrated to inhibit autophagy in preclinical models (18, 33). Mice bearing palpable tumors were treated with daily i.p. injections of either 60 mg/kg chloroquine, 50 mg/kg quinacrine, or vehicle control. Tumor growth was not significantly different between the 3 cohorts (Figure 6A). Consistent with our results in B16 melanomas, T cell infiltration and activation were unchanged in both chloroquine- and quinacrine-treated 4T1 mammary tumors compared with vehicle-treated controls (Figure 6D). Overall, these data demonstrate that systemic pharmacological inhibition of autophagy using antimalarials does not adversely impact the antitumor T cell response.
Figure 6. Antimalarial-treated tumors exhibit equivalent T cell responses.
Subcutaneous B16 melanomas were raised in GREAT reporter mice and orthotopic 4T1 mammary tumors in WT BALB/c mice. Primary tumors were allowed to form for 7–10 days and were subsequently treated daily with chloroquine, quinacrine, or vehicle control by i.p. injection. (A) Primary tumor growth of B16 (n = 5 per cohort) and 4T1 tumors (Vehicle, n = 9; Chloroquine, n = 9; Quinacrine, n = 8) as assessed by caliper measurements of tumor area. Error bars represent SD, and arrows indicate treatment days. (B) Autophagy deficiency was confirmed in resected B16 tumors by immunofluorescence for P62, and accumulation of P62 aggregates was quantified. Error bars represent SD; ***P < 0.001 using unpaired t test. (C) Infiltration and functional phenotype of CD4+ and CD8+ T cell populations were measured by flow cytometry in vehicle- and chloroquine-treated B16 tumors. Data points represent distinct tumors from individual mice; bars represent mean values with 2-way ANOVA not significant. (D) Infiltration and activation of CD4+ and CD8+ T cell populations were measured by flow cytometry in vehicle-, chloroquine-, and quinacrine-treated 4T1 tumors. Data points represent distinct tumors from individual mice, and bars represent mean values with 2-way ANOVA not significant.
Discussion
In this study, we demonstrate that the T cell immune response in preclinical models of melanoma and breast cancer is not dependent on autophagy activity of the tumor cell. This suggests the ability to safely combine autophagy inhibition with chemotherapy during cancer treatment, and opens the possibility of future combination with immunotherapy. While ongoing clinical trials are testing the efficacy of combining autophagy inhibition with chemotherapy in multiple cancer types, some recent studies have argued against such combinations because of autophagic regulation of immunogenic mechanisms associated with chemotherapeutic efficacy (14, 16, 17). Because of the limited scope of those studies with regard to interrogating autophagy and T cell functional status, as well as the clinical importance of delineating the consequences of autophagy inhibition, we further addressed this critical issue in additional mouse models.
Our study design primarily included 2 tumor types: B16 melanoma in C57BL/6 mice and 4T1 mammary cancer in BALB/c mice (34, 35). These models have been extensively used in studies of the tumor microenvironment and immune response and provided us with the opportunity to interrogate the effects of autophagy inhibition in 2 distinct immune-competent genetic backgrounds. We found that the levels of autophagy within tumor cells did not affect either the quantities of infiltrating T cell populations or their functional status. As a further validation of our measurements of the functional analysis of T cells via antibody staining, we raised B16 melanomas in GREAT reporter mice and measured eYFP reporter activity. This model allowed us to use an entirely parallel technical approach to assessing T cell activity, and eliminated any technical caveats that could be introduced by the reliance on antibody reagents. Once again, we found equivalent tumor-associated T cell responses as measured by eYFP reporter expression between autophagy-competent and -deficient tumors. We also demonstrated that autophagy deficiency did not alter the quantity or quality of Tregs associated with B16 tumors, and did not alter the activation potential of restimulated CD8+ T cells isolated from 4T1 tumors.
Because of the ongoing efforts to repurpose antimalarials as autophagy inhibitors to treat cancer, we interrogated the effects of these agents on the antitumor T cell response. Similar to genetic autophagy inhibition in tumor cells, systemic treatment with the autophagy-inhibiting antimalarial agents chloroquine and quinacrine produced equivalent numbers of tumor-infiltrating T cells, which were functionally equivalent. Remarkably, antimalarials are used clinically in the treatment of autoimmune disorders, and it has been proposed these agents can act as immune suppressors by interfering with immune cell function (36, 37). However, in daily treatments of tumor-bearing mice for a short duration, we observed no evidence of a blunted T cell response. Thus, our results point to a therapeutic window in which antimalarials may be effectively combined with immunotherapies without antagonistic effects. Future preclinical and clinical studies will be necessary to define such combinatorial approaches.
In addition to assessing the endogenous, heterogeneous T cell response in B16 and 4T1 tumors, we used a more targeted approach to address whether autophagy-deficient cancer cells were less inherently immunogenic. It has been speculated that impaired autophagy leads to changes in antigen availability, processing, or presentation (38, 39), or to changes in immune-modulatory secreted proteins; such effects would potentially dampen cancer cell immunogenicity and subsequent T cell responses. Hence, we used the antigen-matched OT-I system (30), in which OVA-specific OT-I CD8+ T cells expressing a traceable fluorescent marker were adoptively transferred into mice bearing OVA-expressing B78 melanoma tumors, a derivative cell line of the B16 model. While the OT-I cells mounted a stronger antitumor response than endogenous T cells, the overall T cell response was comparable between autophagy-competent and -deficient B78-OVA tumors. The majority of activation markers measured, most notably IFN-γ, were unchanged between tumor types, indicating that autophagy inhibition did not impair T cell function in this model of a highly active tumor antigen–specific T cell response. Taken together, the comparable antitumor responses of both endogenous and antigen-matched T cells indicate the autophagy independence of these programs.
Our results differ from previous work demonstrating that tumor cell autophagy promotes T cell infiltration and activation in response to anthracycline chemotherapy. These phenotypic differences may partly be due to the aggressive nature of the models used in our studies, which may diminish the immune modulatory effects of tumor cell autophagy following therapy. Importantly, autophagy promotes the in vitro secretion of key factors associated with ICD, including ATP and HMGB1 (15); consistent with those previous findings, we observe that ATP and HMGB1 secretion is impaired in Dox-treated autophagy-deficient B16 cells in vitro. However, in contrast to tumor models used previously to study ICD, the introduction of Dox-treated apoptotic B16 cells, either autophagy-competent or -deficient, is unable to vaccinate mice against rechallenge with healthy WT B16 cells, which is considered the benchmark assay for ICD (15). Accordingly, our results demonstrate that autophagy inhibition does not adversely impact immune cell function in the absence of a robust ICD response. Our results highlight the highly context-specific nature of ICD and suggest that more precise corroboration of this death pathway in individual patients may be needed to predict how autophagy inhibitors influence anticancer immunity in the clinical setting (15). Nevertheless, despite the absence of significant ICD, B16 tumor–bearing mice treated with Dox still exhibited significantly elevated immune activation compared with untreated tumor-bearing mice, which once again is unaffected by autophagy status. This raises the likely possibility that specific clinical contexts and therapeutic windows exist in which autophagy-dependent immune modulation will not compromise chemotherapeutic efficacy. We propose that autophagy inhibition can be safely combined with chemotherapy and still stimulate a productive antitumor T cell response in certain tumor types. Future studies analyzing patient-derived T cells from clinical trials of autophagy inhibitors will be crucial to determine whether this is truly the case. Finally, our studies raise the possibility that anticancer immunotherapies can be combined with autophagy inhibition; such a combination would be a dual-pronged treatment strategy that would unleash the power of the immune system upon tumor cells weakened by autophagy inhibition.
Methods
Antibodies.
Commercial antibodies included the following: eFluor 450, Alexa 700, and APC-Cy7 anti-CD45 (clone 30-F11, 1:400); PE and APC anti-CD3ε (clone 145-2C11, 1:400); PE-Cy7 anti-CD4 (clone RM4-5, 1:400); Percp-Cy5.5 anti-CD8α (clone 53-6.7, 1:400); FITC and eFluor 450 anti–PD-1 (clone RMP1-30, 1:400); eFluor 660 anti–IFN-γ (clone XMG1.2, 1:400); FITC and eFluor 450 anti–TNF-α (clone MP6-XT22, 1:400); and eFluor 450 anti-Foxp3 (clone FJK-16s, 1:400) were obtained from eBioscience. APC-Cy7 anti-CD45 (clone 30-F11, 1:400); FITC anti-CD8α (clone 53-6.7, 1:400); Brilliant Violet 785 anti-CD44 (clone IM7, 1:800); Pacific Blue anti–granzyme B (clone GB11, 1:400); and Brilliant Violet 421 anti–PD-L1 (clone 10F.9G2, 1:800) were obtained from BioLegend. Anti-ATG7 (2631, 1:500) was obtained from Cell Signaling Technology. Anti-ATG5 (NB110-53818, 1:2,000) was obtained from Novus Biologicals. Anti-P62 (GP62-C, 1:100 for immunofluorescence and 1:1,000 for immunoblotting) was obtained from Progen. Anti–phospho-histone H2A.X (clone JBW301, 1:1,000) and anti-GAPDH (AB2302, 1:5,000) were obtained from Millipore. A rabbit polyclonal anti-LC3 antibody was created using a conserved N-terminal peptide between human, rat, and mouse (40), which is also commercially available from Millipore (ABC232, 1:1,000).
Cell culture.
Lewis Lanier (UCSF) provided B16 murine melanoma cells, 4T1 mammary carcinoma cells were purchased from the American Type Culture Collection, and Matthew Krummel (UCSF) provided B78-OVA murine melanoma cells, which express OVA via the mCherry-p2A-OVA sequence (41). All cells were cultured in D10 culture medium (DMEM, 10% FBS, penicillin/streptomycin), verified to be free of Mycoplasma, and authenticated via transplantation in the appropriate syngeneic host. For stable RNA interference, pLKO.1-puro (puromycin) lentiviral plasmids containing nontargeting shRNA (shCTL) or shRNA against mouse Atg7 (NM_028835) and mouse Atg12 (NM_026217) were purchased from Sigma-Aldrich. The target sequence for shRNA directed against mouse Atg7 (TRCN0000092163) is CCAGCTCTGAACTCAATAATA, and directed against Atg12 (TRCN0000257708) is TGGTAAACTGGTCCTGCATTA. Viral particles were produced using a third-generation lentiviral packaging system in HEK293T cells, and used to infect B16 murine melanoma, 4T1 mammary carcinoma, and B78-OVA murine melanoma cells. After infection and drug selection, early-passage stable pools of ATG knockdown cells were used for both in vitro and in vivo assays. To confirm autophagy deficiency following ATG knockdown, cells were cultured in full growth medium or starvation medium (HBSS), with or without bafilomycin A. Cell lysates were then assessed for autophagic flux by LC3-II turnover via immunoblot.
For analysis of secreted factors associated with ICD, cells were treated with 10 μM Dox (Sigma-Aldrich, 44583) for 24 hours; conditioned medium was collected and analyzed for HMGB1 secretion by ELISA (GENTAUR, ST51011) or ATP secretion by Enliten ATP assay (Promega, FF2000). Viability of Dox-treated cells was assessed by crystal violet staining. Cultured cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes at room temperature, incubated with 0.3% crystal violet for 20 minutes at room temperature, washed with deionized water, air-dried overnight, and resolubilized with 45% methanol in deionized water for 30 minutes at room temperature before absorbance reading.
For T cell restimulation, tissue culture plates were incubated overnight with 1 μg/ml purified CD3 antibody in PBS. Excess antibody was aspirated and plates were blocked with R10 medium (RPMI, 10% calf serum, penicillin/streptomycin) at 4°C. Tumor-associated CD8+ T cells were isolated by FACS and cultured overnight on CD3-coated plates with R10 medium and 0.5 μg/ml purified CD28 antibody. Conditioned medium was collected and analyzed for IFN-γ secretion by ELISA (R&D Systems, MIF00). Experiments were also repeated in which untouched tumor-associated CD8+ T cells were isolated by negative bead selection kit (Miltenyi Biotec, 130-104-075).
Animals.
Richard Locksley (UCSF, Howard Hughes Medical Institute, San Francisco, California, USA) provided the IFN-γ reporter (GREAT) mouse strain [Ifngtm3(EYFP)Lky], which carries a bicistronic IFN-γ-IRES-eYFP reporter allele under the control of the endogenous Ifng promoter/enhancer region. Murine cells expressing IFN-γ also express cytoplasmic eYFP, which can be detected by flow cytometry. Matthew Krummel (UCSF) provided OT-I mice specific for the OVA peptide SIINFEKL (SL8) in the context of H-2Kb. WT C57BL/6 and BALB/c mice were purchased from Jackson Laboratory.
For the generation of tumor-bearing mice, B16 and B78-OVA cells were injected s.c. (150,000 cells per injection in 50% growth factor–reduced Matrigel in PBS) into the back flanks of 6- to 7-week-old male and female WT C57BL/6 mice or GREAT reporter mice on the C57BL/6 background. For studies of intracellular cytokine staining, mice were injected by tail vein with 10 μg/g body weight of brefeldin A (BFA; Sigma-Aldrich, B6542) at 6 hours before tumor resection. Tumors were resected, minced, and subjected to enzymatic digest with Collagenase IV (Sigma-Aldrich, C5138; 500 U/ml), Collagenase A (Worthington Biochemical, LS004197; 100 U/ml), and DNase (200 μg/ml) in RPMI medium for 30 minutes at 37°C with shaking followed by passage through a 70-μm cell strainer. BFA was also added to reagents to prevent ex vivo secretion during tissue processing.
Mice bearing 2-week B78-OVA tumors received an adoptive transfer of 2 × 106 freshly isolated OT-I T cells expressing CD2-GFP by retro-orbital injection. 4T1 cells were injected orthotopically into mammary fat pads of 6- to 7-week-old female WT BALB/c mice (100,000 cells per injection in 50% growth factor–reduced Matrigel in PBS). To confirm autophagy deficiency in resected B78-OVA tumors at endpoint, cancer cells were isolated by ex vivo culture from tumor digests in puromycin, plated on coverslips, and subjected to immunocytochemistry as described below to measure P62 accumulation.
For prophylactic vaccination, mice were injected s.c. with Matrigel or 106 B16 cells pretreated with Dox to achieve 70% dead and dying cells, as assessed by annexin V and DAPI staining. Interpolation from a dose-response curve indicated that 24 hours of treatment with 8.8 μM or 7.5 μM Dox yielded 70% dead and dying cells for B16-shCTL and -shAtg7 cells, respectively. One week after vaccination, mice were injected s.c. on the contralateral flank with 100,000 healthy WT B16 cells. Tumor incidence was assessed by daily palpation.
For chemotherapy, mice bearing palpable tumors were injected by tail vein with 5 mg/kg body weight of Dox (Sigma-Aldrich, 44583) once per 7 days for 2 weeks before tumor resection. For antimalarial treatment, mice bearing palpable tumors were injected i.p. with 60 mg/kg chloroquine, 50 mg/kg quinacrine, or vehicle control, daily for 4–6 days before tumor resection. B16-bearing mice were treated for either 4 or 5 consecutive days; 4T1-bearing mice were treated for 3 days on and 2 days off for 6 total injections. Only chloroquine treatment was used for the studies of B16 melanoma in GREAT mice because quinacrine is autofluorescent in a similar range to eYFP.
Flow cytometry and FACS.
Red blood cell lysis was performed with 175 mM ammonium chloride on ice. Zombie NIR Fixable Viability kit (BioLegend, 423105) was applied to cells for 30 minutes on ice. Subsequent steps were performed in flow buffer (PBS, 2% calf serum, penicillin, streptomycin, glutamine, 2 mM EDTA, 0.01% sodium azide). Cells were incubated in flow buffer with FcR blocking reagent (Miltenyi Biotec, 130-092-575), 2% FBS, 2% Armenian hamster serum (Innovative Research, IGHMA-SER), and antibodies against surface markers for 30 minutes on ice. Cells were then fixed with 2% PFA in flow buffer for 15 minutes at 25°C, permeabilized with 0.02% saponin in flow buffer for 10 minutes at 25°C, and incubated with 0.02% saponin and antibodies against intracellular markers in flow buffer for 30 minutes at 25°C. Fixation and permeabilization of cells for intracellular Foxp3 staining were performed using the Foxp3 transcription factor staining buffer set (eBioscience, 00-5523-00). All flow cytometry was performed on a BD LSR II flow cytometer. Flow cytometry data analysis was done with FlowJo software (version 10.1). Fluorescence-activated cell sorting (FACS) of tumor-associated CD8+ T cells was performed using a Beckman Coulter MoFlo XDP.
Immunoblotting.
Cells were lysed in RIPA buffer with 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO4, 10 nM calyculin A, 0.1 mM E64-D, 10 μg/ml pepstatin A, and 20 nM bafilomycin A. Lysates were cleared by centrifugation for 15 minutes at 4°C, boiled in sample buffer, resolved by SDS-PAGE, and transferred to PVDF membrane. Membranes were blocked in 5% milk in PBS with 0.1% Tween-20 (PBS-T), incubated with primary antibodies overnight at 4°C in blocking buffer, incubated with HRP-conjugated secondary antibodies, and analyzed by chemiluminescence. Image analysis was performed on raw images, and image brightness was adjusted for publication. See complete unedited blots in the supplemental material.
Immunohistochemistry and immunofluorescence.
For immunohistochemistry, tumors were resected and fixed in formalin overnight at 4°C, incubated in 30% sucrose for cryoprotection for 24 hours at 4°C, embedded in OCT, and stored at –80°C before and after tissue sectioning. Thawed tissue slides were incubated in 4% PFA for 5 minutes, washed with PBS-T, and incubated in 1X target retrieval solution (Dako, S1699) at 96°C for 20 minutes in a plastic Coplin jar submerged in a beaker of boiling water. Slides were cooled for 20 minutes at 25°C, washed with PBS-T, blocked with 10% goat serum in PBS-T, and incubated with primary antibody in blocking serum overnight at 4°C. Slides were washed with PBS-T and incubated with fluorescent secondary antibody for 1 hour at 25°C, washed with PBS-T, incubated with 10 mg/ml Hoechst 33342 nuclear stain (Thermo Fisher Scientific, H1399) for 5 minutes at 25°C, and washed with PBS-T and distilled water. Coverslips were mounted with Prolong Gold antifade mountant (Thermo Fisher Scientific, P36934) and sealed with nail polish.
For immunofluorescence, cells were cultured on fibronectin-coated coverslips for 24 hours. Cells were fixed with 4% PFA for 10 minutes at 25°C, incubated with 0.1 M glycine in PBS for 5 minutes at 25°C to quench PFA autofluorescence, permeabilized with 0.5% Triton X-100 for 5 minutes at 25°C, and blocked for 1 hour at 25°C with 10% goat serum in PBS. Primary antibody incubation was performed overnight at 4°C, and secondary antibody incubation for 1 hour at 25°C, and nuclei were stained with Hoechst 33342 nuclear stain (Thermo Fisher Scientific, H1399) for 10 minutes at 25°C. Coverslips were mounted with Prolong Gold antifade mountant (Thermo Fisher Scientific, P36934) and sealed with nail polish. Washes were performed between each incubation with 1X immunofluorescence wash buffer (PBS, 0.1% BSA, 0.2% Triton X-100, 0.04% Tween-20).
Fluorescence imaging was performed using a Zeiss Axiovert 200 microscope equipped with a SPOT RT camera (Diagnostic Instruments) and mercury lamp; images were acquired and prepared using SPOT and ImageJ (NIH) software. Image analysis was performed on raw images, and brightness was adjusted for publication.
Statistics.
Where representative images are shown, experiments were performed at least 3 times and no data were excluded from analysis. Statistical analysis was done using GraphPad Prism software (version 7.01). Error bars represent SD from at least triplicate experimental conditions. P values were determined by unpaired 2-tailed t test for comparisons of 2 groups or by 2-way ANOVA with Dunnett’s or Tukey’s correction for multiple comparisons as indicated. P values for all tests are indicated on graphs as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and not significant (NS).
Study approval.
All animal studies were performed in accordance with a protocol (AN107285) approved by the UCSF Institutional Animal Care and Use Committee.
Author contributions
HS and JD conceived the study and designed the experiments. HS performed the experiments and analyzed data. JY performed cell culture, tissue harvests, and tissue processing for experimental repeats. MB and KB performed B78-OVA experiments and assisted with flow cytometry. JG assisted with tissue harvests and performed cell culture assays for experimental repeats. TM and FR assisted with mouse experiments. MK assisted with design of experiments and provided materials. HS and JD wrote the manuscript, with input from the other authors. JD supervised the study.
Supplementary Material
Acknowledgments
We thank Zena Werb, Gabriele Bergers, and Dean Sheppard, as well as members of the Debnath laboratory and Timur Starobinets, for critical discussions. Grant support to JD includes the NIH (CA188404) and the Department of Defense Breast Cancer Research Program (W81XWH-11-1-013). HS was supported by a National Science Foundation Graduate Research Fellowship (DGE-1144247) and a UCSF Discovery Fellowship. Flow cytometry and FACS were performed in the UCSF Flow Cytometry Core Facility. Tissue sectioning was performed by Jennifer Bolen in the UCSF Mouse Pathology Core Facility.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(12):4417–4429. doi:10.1172/JCI85705.
Contributor Information
Hanna Starobinets, Email: hannastarobin@gmail.com.
Jordan Ye, Email: jordan.ye@ucsf.edu.
Miranda Broz, Email: Miranda.broz.1988@gmail.com.
Kevin Barry, Email: Kevin.Barry@ucsf.edu.
Juliet Goldsmith, Email: juliet.goldsmith@ucsf.edu.
Timothy Marsh, Email: Tim.marsh@ucsf.edu.
Fanya Rostker, Email: fanya.rostker@ucsf.edu.
Matthew Krummel, Email: matthew.krummel@ucsf.edu.
Jayanta Debnath, Email: jayanta.debnath@ucsf.edu.
References
- 1.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 2.Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015;25(1):37–45. doi: 10.1016/j.tcb.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rebecca VW, Amaravadi RK. Emerging strategies to effectively target autophagy in cancer. Oncogene. 2016;35(1):1–11. doi: 10.1038/onc.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10(9):1533–1541. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10(8):1415–1425. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahalingam D, et al. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10(8):1403–1414. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rangwala R, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangwala R, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenfeld MR, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogl DT, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10(8):1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolpin BM, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. Oncologist. 2014;19(6):637–638. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity. 2013;39(1):74–88. doi: 10.1016/j.immuni.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Sistigu A, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20(11):1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 15.Garg AD, Martin S, Golab J, Agostinis P. Danger signalling during cancer cell death: origins, plasticity and regulation. Cell Death Differ. 2014;21(1):26–38. doi: 10.1038/cdd.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaud M, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334(6062):1573–1577. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- 17.Michaud M, Xie X, Bravo-San Pedro JM, Zitvogel L, White E, Kroemer G. An autophagy-dependent anticancer immune response determines the efficacy of melanoma chemotherapy. Oncoimmunology. 2014;3(7):e944047. doi: 10.4161/21624011.2014.944047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta A, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST) Proc Natl Acad Sci U S A. 2010;107(32):14333–14338. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 20.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brahmer JR, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 26.Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12(4):307–313. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12(4):237–251. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity. 2010;32(1):104–115. doi: 10.1016/j.immuni.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 31.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10(4):385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L. Chloroquine and hydroxychloroquine for cancer therapy. Mol Cell Oncol. 2014;1(1):e29911. doi: 10.4161/mco.29911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salas E, Roy S, Marsh T, Rubin B, Debnath J. Oxidative pentose phosphate pathway inhibition is a key determinant of antimalarial induced cancer cell death. Oncogene. 2016;35(22):2913–2922. doi: 10.1038/onc.2015.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overwijk WW, Restifo NP. B16 as a mouse model for human melanoma. Curr Protoc Immunol. 2001;Chapter 20:Unit 20.1. doi: 10.1002/0471142735.im2001s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr Protoc Immunol. 2001;chapter 20:unit 20.2. doi: 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 36.Salmeron G, Lipsky PE. Immunosuppressive potential of antimalarials. Am J Med. 1983;75(1A):19–24. doi: 10.1016/0002-9343(83)91266-4. [DOI] [PubMed] [Google Scholar]
- 37.Fox RI. Mechanism of action of hydroxychloroquine as an antirheumatic drug. Semin Arthritis Rheum. 1993;23(2 suppl 1):82–91. doi: 10.1016/s0049-0172(10)80012-5. [DOI] [PubMed] [Google Scholar]
- 38.Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8(8):607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demachi-Okamura A, Torikai H, Akatsuka Y, Miyoshi H, Yoshimori T, Kuzushima K. Autophagy creates a CTL epitope that mimics tumor-associated antigens. PLoS One. 2012;7(10):e47126. doi: 10.1371/journal.pone.0047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19(3):797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelhardt JJ, et al. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 2012;21(3):402–417. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.