Abstract
Small nucleolar RNAs (snoRNAs) are non-coding RNAs that form ribonucleoproteins to guide covalent modifications of ribosomal and small nuclear RNAs in the nucleus. Recent studies have also uncovered additional non-canonical roles for snoRNAs. However, the physiological contributions of these small RNAs are largely unknown. Here, we selectively deleted four snoRNAs encoded within the introns of the ribosomal protein L13a (Rpl13a) locus in a mouse model. Loss of Rpl13a snoRNAs altered mitochondrial metabolism and lowered reactive oxygen species tone, leading to increased glucose-stimulated insulin secretion from pancreatic islets and enhanced systemic glucose tolerance. Islets from mice lacking Rpl13a snoRNAs demonstrated blunted oxidative stress responses. Furthermore, these mice were protected against diabetogenic stimuli that cause oxidative stress damage to islets. Our study illuminates a previously unrecognized role for snoRNAs in metabolic regulation.
Introduction
Box C/D snoRNAs are short noncoding RNAs containing conserved C and D box consensus motifs that form ribonucleoproteins with NOP56, NOP58, 15.5 kDa, and the methyltransferase fibrillarin (1). These ribonucleoproteins localize to nucleoli, where their canonical function is to serve as guides to target specific sites on ribosomal RNAs (rRNAs) or small nuclear RNAs (snRNAs) for 2′-O-methylation through a short stretch of antisense complementarity (10–21 nucleotides). In vertebrates, this function has been demonstrated experimentally for some box C/D snoRNAs and is predicted for others. The observation that animals homozygous for knockout of fibrillarin are inviable indicates that functions of box C/D snoRNAs as a class are essential for normal development (2).
Genetic studies have suggested an expanding functional repertoire for snoRNAs beyond their well-established roles in modification and processing of rRNAs and snRNAs (3). Dysregulated snoRNA expression has been associated with altered splicing in the developmental syndromes, Prader-Willi and tetralogy of Fallot (4, 5), and snoRNAs U50, ACA11, and ACA42 have been shown to modulate tumor proliferation in vivo (6–9). Nonetheless, the physiological consequences of perturbation of expression of most mammalian snoRNAs remain unexplored, and no animal models exist with selective and complete loss of function for any snoRNAs.
Our laboratory’s studies have demonstrated that box C/D snoRNAs encoded by introns of the Rpl13a locus function as critical mediators of cell death in response to metabolic and oxidative stress in cultured cells (10–12). The observations that this role can be dissociated from changes in the 2′-O-methylation status of predicted ribosomal RNA targets and that the Rpl13a snoRNAs accumulate in the cytosol during oxidative stress suggest that the Rpl13a snoRNAs may function through noncanonical mechanisms. The goal of this study was to determine the physiological role of these noncoding RNAs.
Results
Generation of Rpl13a-snoless mice.
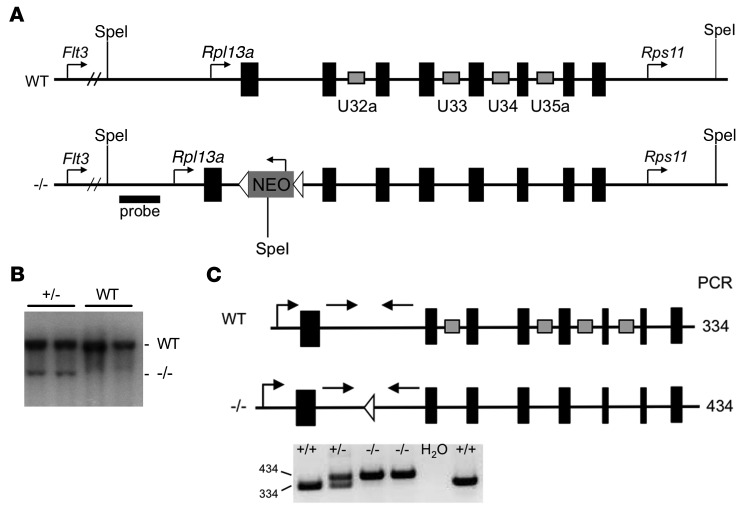
Our previous work demonstrated a critical role for box C/D snoRNAs U32a, U33, U34, and U35a, embedded within 4 introns of the Rpl13a locus (Figure 1A), in the cellular response to lipotoxic and oxidative stress (10). The observation that loss of function of the individual snoRNAs in cultured cells is not sufficient to confer resistance to metabolic stress suggests that these 4 snoRNAs function in concert in stress response pathways. Therefore, to probe the long-term physiological consequences of loss of function of Rpl13a snoRNAs, we generated a model with simultaneous loss of all 4 box C/D sno-RNAs encoded in this locus. Since deletion of the RPL13a protein in Drosophila is embryonic lethal (13), our goal was to selectively modify 4 snoRNA-hosting introns without perturbing expression of the exon-encoded ribosomal protein. This was accomplished using a single recombination event to replace the locus with an allele from which the snoRNAs were selectively deleted (Figure 1, A–C, and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI88069DS1). No known regulatory regions overlap with these 4 snoRNAs, and these intronic sequences are relatively depleted of H3K27 histone marks that are often found near regulatory regions.
Figure 1. Generation of mice lacking Rpl13a snoRNAs.
(A) WT locus and targeting construct (–/–) with exons in black and intronic snoRNAs in gray rectangles. (B) Representative Southern blot from F1 (+/–) and WT offspring (n = 5 experiments) using probe indicated in A. (C) Primers (straight arrows) and predicted size (nucleotides) of genotyping PCR products, above. Representative agarose gel of PCR products from tail DNA is shown below (n > 10 experiments).
Homozygous Rpl13a snoRNA loss-of-function mice (referred to hereafter in text as Rpl13a-snoless and in figures as -/-) were live-born at normal Mendelian ratios, were fertile, and displayed no overt pathophysiological phenotype under standard housing, feeding, and breeding conditions. Body weight and body composition were indistinguishable between Rpl13a-snoless and WT mice (Supplemental Figure 2, A and B). The Rpl13a-snoless mice were deficient in expression of the 4 intronic snoRNAs from this locus, but Rpl13a mRNA and protein levels were indistinguishable from those of WT controls (Figure 2, A and B), and expression of other snoRNAs was unchanged (Supplemental Figure 2C). Fibroblasts from Rpl13a-snoless embryos (MEFs) had significantly lower levels of cellular superoxide and hydrogen peroxide under normal growth conditions (Figure 2, C and D). Furthermore, following brief exposure to exogenous hydrogen peroxide, Rpl13a-snoless MEFs had diminished ability to amplify ROS. Lower basal ROS tone and resistance to ROS stimulation are consistent with the phenotype of the previously reported mutant Chinese hamster ovary cell line, in which decreased expression of the Rpl13a snoRNAs caused diminished oxidative stress responses (10).
Figure 2. Rpl13a-snoless mice demonstrate lower ROS tone.
(A) RT-qPCR-quantified expression of Rpl13a snoRNAs and mRNA (relative to Rplp0) in WT and Rpl13a-snoless (–/–) fibroblasts. Mean + SEM for n = 3 independent experiments. (B) Representative Western blot of RPL13a protein in embryos (n = 4 mice/genotype). (C) MitoSOX staining of embryonic fibroblasts. Mean fluorescence of 104 cells/sample (+SEM) for n = 4 independent experiments. (D) DCF staining of embryonic fibroblasts under basal growth conditions and following exposure to 100 μM H2O2 for 10 minutes. n = 3 independent experiments. *P < 0.005 for –/– vs. WT determined by unpaired t test. rel, relative.
Altered glucose homeostasis in Rpl13a-snoless mice.
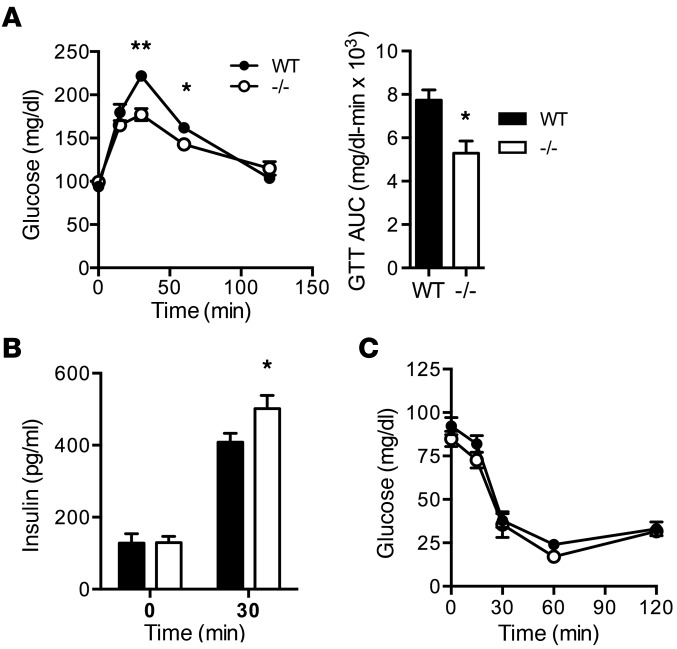
ROS participate in physiologic signaling cascades that regulate insulin secretion. Therefore, we assessed systemic glucose metabolism in Rpl13a-snoless mice. Fasting plasma glucose and insulin levels were indistinguishable between WT and Rpl13a-snoless mice; however, glucose tolerance test (GTT) revealed enhanced glucose tolerance with lower peak glucose excursion and decreased AUC following an intraperitoneal glucose bolus (Figure 3A). To understand the cause of these changes in glucose metabolism, we used a sensitive Singulex single molecule tracking method to quantify fasting serum insulin and insulin levels at the 30-minute time point of the GTT, the point at which the greatest difference in serum glucose was observed between the genotypes. Serum insulin was significantly increased in Rpl13a-snoless mice at the 30-minute time point (Figure 3B), implicating enhanced release of insulin into the circulation in response to a glucose challenge as the cause of improved glucose tolerance. By contrast, glucose levels during insulin tolerance test (ITT) were similar between the genotypes (Figure 3C), further suggesting that the lower peak glucose excursion resulted not from alterations in peripheral insulin sensitivity, but rather from a difference in insulin secretion.
Figure 3. Rpl13a-snoless mice have improved glucose tolerance and insulin secretion.
(A) Mean (+SEM) blood glucose and AUC during GTT in 8-week-old female WT and –/– mice. n = minimum of 9 mice/genotype. (B) Mean serum insulin (+SEM) at 0 and 30 minutes of GTT. n = minimum of 8 mice/genotype. (C) Mean blood glucose (+SEM) during ITT. n = minimum of 7 mice/genotype. **P < 0.0005; *P < 0.05 for –/– vs. WT determined by unpaired t test.
Rpl13a-snoless islets have enhanced insulin secretion.
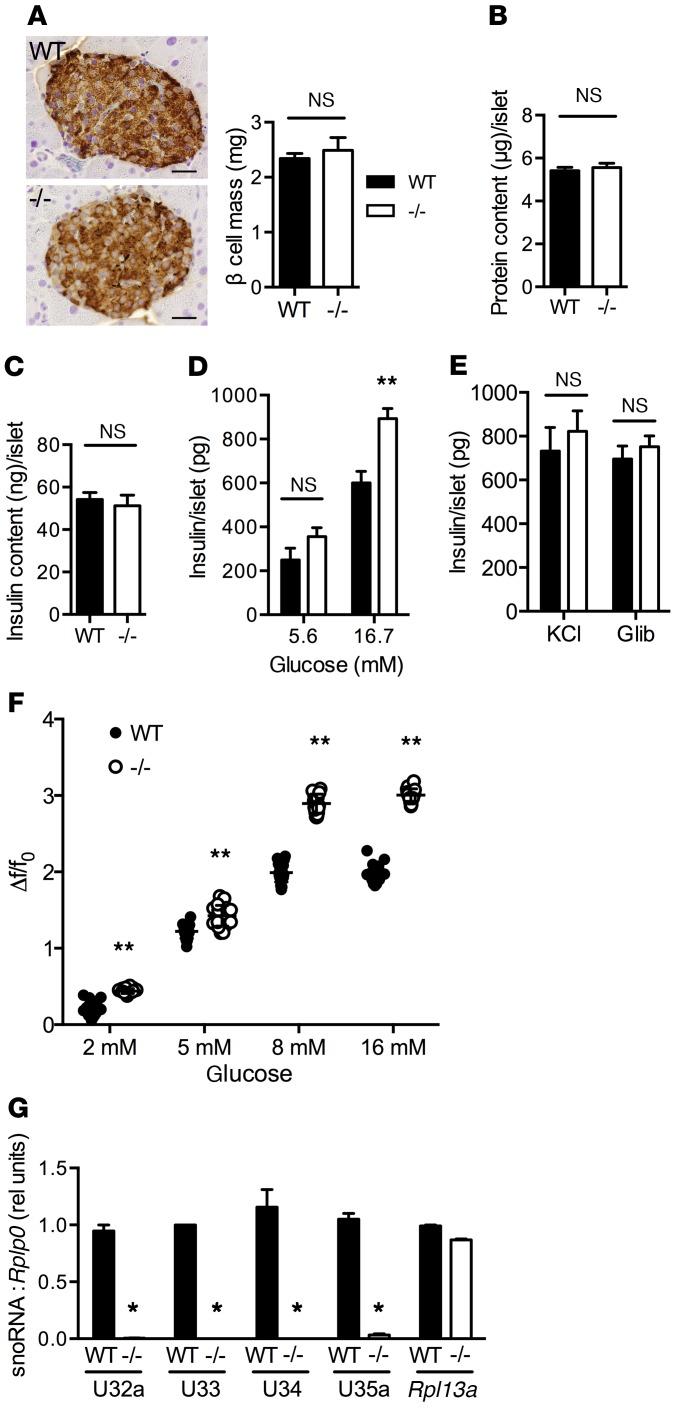
To investigate the physiological basis of increased insulin levels following a glucose challenge, we examined pancreatic tissue of 8-week-old WT and Rpl13a-snoless animals. β Cell mass as determined by morphometric analysis was indistinguishable between the genotypes (Figure 4A). Protein and insulin content of isolated islets were also not different between WT and Rpl13a-snoless mice (Figure 4, B and C). Nonetheless, consistent with enhanced insulin secretion and glucose tolerance in vivo, Rpl13a-snoless islets had a 51% increase in glucose-stimulated insulin secretion (GSIS; Figure 4D). Similar insulin release from islets of the two genotypes in response to potassium chloride and glibenclamide indicated that the secretory machinery downstream of the KATP channel is intact in Rpl13a-snoless islets (Figure 4E). Consistent with increased insulin secretion at stimulatory glucose concentrations, Rpl13a-snoless islets demonstrated increased calcium oscillations, and these differences were greatest at 8 mM and 16 mM glucose (Figure 4F and Supplemental Figure 3). We confirmed that Rpl13a snoRNAs are expressed in WT, but not Rpl13a-snoless, islets (Figure 4G). Together, these findings suggest that enhanced glucose tolerance in Rpl13a-snoless mice results from greater islet insulin secretion following glucose challenge.
Figure 4. Rpl13a-snoless islets demonstrate enhanced glucose-stimulated insulin secretion.
(A) Representative insulin-stained pancreas sections with morphometric quantification of β cell mass for WT and Rpl13a-snoless (–/–) mice (n = 4 mice/genotype). Mean + SEM; n = 4 mice/genotype; scale bars: 25 μm. (B) Mean islet protein content (+SEM) quantified in 8 mice/genotype (10 islets/mouse) obtained from 3 independent experiments. (C) Mean islet insulin content (+SEM) quantified in 12 mice/genotype (10 islets/mouse) obtained from 3 independent experiments. (D) Mean islet insulin secretion (+SEM) in response to low and high glucose quantified in a minimum of 9 mice/genotype over 3 independent experiments. (E) Mean islet insulin secretion (+SEM) in response to 30 mM KCl and 1 μM glibenclamide quantified in 3–4 mice/genotype over 3 independent experiments. (F) Changes in Fluo-4–quantified calcium activity of islets relative to basal, expressed as Δf/f0 for differing glucose concentrations. Data from 20 independent cells from 4 mice/genotype. (G) RT-qPCR-quantified expression of Rpl13a snoRNAs and mRNA in islets (relative to Rplp0). n = 3 independent islet preparations/genotype. **P < 0.001; *P < 0.05; for –/– vs. WT determined by unpaired t test.
Increased mitochondrial leak and lower ROS in Rpl13a-snoless islets.
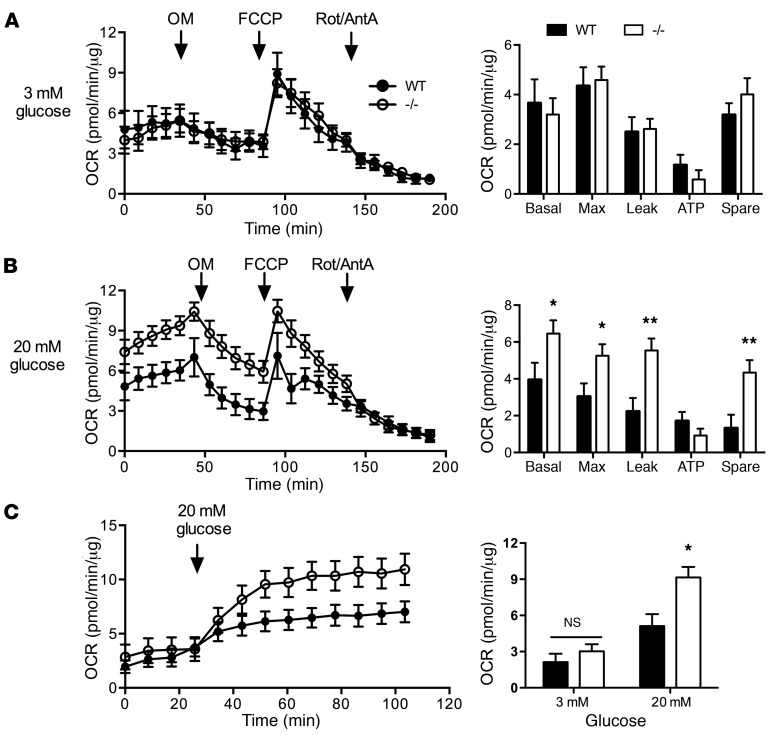
Glucose stimulates insulin secretion through its metabolism to produce “metabolic coupling factors” that regulate the cellular machinery for insulin secretion (14). To assess islet glucose metabolism and mitochondrial function, we quantified oxygen consumption rates (OCR) in isolated WT and Rpl13a-snoless islets at baseline and following serial addition of modulators that target specific components of the electron transport chain. At 3 mM glucose, basal and maximal respiration, ATP production, and spare respiratory capacity were comparable between the genotypes, with a large proton leak relative to ATP production in both genotypes (Figure 5A), similar to results previously described in WT islets (15). At 20 mM glucose, basal and maximal respiration were substantially increased in Rpl13a-snoless islets relative to WT, which was due to a large increase in proton leak, but oxygen consumption for ATP production was not significantly different (Figure 5B). Dynamic OCR measurements — for which WT and Rpl13a-snoless islets were initially incubated in 3 mM glucose, followed by addition of glucose to 20 mM in the same wells — demonstrated similar OCR under basal conditions, with divergence in response only under stimulatory conditions (Figure 5C). These results indicate that Rpl13a-snoless islets have altered mitochondrial efficiency specifically during metabolic stress conditions. Nonetheless, morphology of β cell mitochondria and quantity of mitochondrial DNA per cell were indistinguishable between WT and Rpl13a-snoless mice (Supplemental Figure 4, A and B).
Figure 5. Increased mitochondrial leak in Rpl13a-snoless islets.
(A) OCR of WT and –/– islets at 3 mM glucose with injection of oligomycin (OM), FCCP, and rotenone and antimycin A (Rot/AntA). Data points in line graphs (left) represent mean values (±SEM) from 3 independent experiments in which islets pooled by genotype were used to generate a total of 9 (WT) or 8 (–/–) technical replicates from n = 11 WT and n = 12 –/– mice. Bar graphs (right) report calculated mean (+SEM) oxygen consumption for basal and maximal (Max) respiration, proton leak, ATP production and spare respiratory capacity. (B) OCR of WT and –/– islets at 20 mM glucose, as in A. Data points from 3 independent experiments in which islets pooled by genotype were used to generate a total of 9 (WT) or 11 (–/–) technical replicates from n = 13 WT and n = 14 –/– animals. Graphs as in A. (C) OCR of WT and –/– islets initially maintained at 3 mM glucose; this was changed at the time indicated to 20 mM glucose. Data points represent mean values (±SEM) from 3 independent experiments in which islets pooled by genotype generated 12 (WT) or 8 (–/–) technical replicates from n = 15 WT and n = 10 –/– animals. *P < 0.05 for –/– vs. WT; **P < 0.01 for –/– vs. WT determined by unpaired t test.
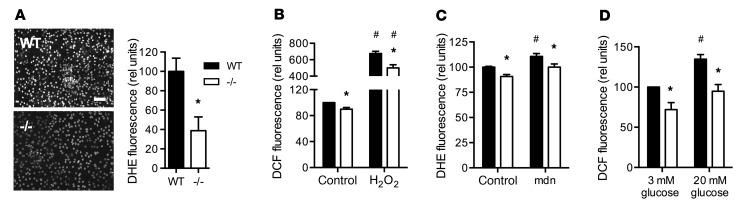
Mitochondria are a major source of ROS in all cell types during substrate oxidation and oxidative phosphorylation, with production of superoxide or hydrogen peroxide at 11 distinct mitochondrial sites (16). Given that uncoupled respiration decreases mitochondrial hydrogen peroxide production during metabolic challenge (17, 18), we hypothesized that increased proton leak in Rpl13a-snoless islets could lead to lower islet ROS. Similar to our observations in MEFs, isolated, intact islets from Rpl13a-snoless mice demonstrated a 60% decrease in basal superoxide content compared with WT islets, as assessed by staining with dihydroethidium (DHE) (Figure 6A). Dissociated islets demonstrated significantly lower intracellular hydrogen peroxide (DCF) and superoxide (DHE) not only under basal conditions, but also following exposure to exogenous hydrogen peroxide (Figure 6B) or menadione, a rapid inducer of superoxide (Figure 6C). Consistent with the known ability of high glucose to induce islet ROS (19), dispersed WT islets had higher hydrogen peroxide content under stimulatory (20 mM) compared with basal (3 mM) glucose conditions (Figure 6D). Dispersed Rpl13a-snoless islets demonstrated significantly lower ROS under both conditions. General antioxidant capacity measured in lysates of WT and Rpl13a-snoless islets was indistinguishable as determined using the Trolox equivalent antioxidant capacity assay (Supplemental Figure 4C), suggesting that differences in ROS related to differences in ROS production. Given that ROS can function to inhibit GSIS (20, 21), our findings suggest a model in which enhanced mitochondrial proton leak and the resulting lower ROS in Rpl13a-snoless islets leads to greater insulin secretion.
Figure 6. Rpl13a-snoless islets are ROS resistant.
(A) Representative images of WT and –/– islets (n = 3 mice/genotype) stained for superoxide with DHE. Scale bar: 50 μm. Graph shows mean fluorescence (+SEM) of WT and –/– islets for n = 6 random fields/genotype in each of 2 independent experiments. (B and C) DCF and DHE staining of dispersed islets at baseline and following treatment for 2 hours with 100 μM H2O2 (B) or 25 μM menadione (mdn; C). Mean (+SEM) of data from a minimum of 3 independent experiments. (D) DCF staining (mean + SEM) of dispersed islets incubated for 2 hours with 3 mM or 20 mM glucose. Data from 3 independent experiments. *P < 0.05 for –/– vs. WT; #P < 0.05 for treated vs. untreated (within a genotype) determined by unpaired t test.
To gain further mechanistic insight into these observations, we carried out RNAseq analysis of Rpl13a-snoless and WT islets. We observed 2-fold or greater differences in the expression of only 6 genes, none of which encodes a mitochondrial protein or is known to have direct effects on the response to ROS (Supplemental Tables 1 and 2). None is predicted bioinformatically to have strong binding sites for U32a, U33, U24, or U35a (22). Our findings suggest these 6 genes are unlikely to be direct targets of the Rpl13a snoRNAs, and effects of Rpl13a snoRNAs on gene expression may be post-transcriptional.
Rpl13a-snoless mice are resistant to diabetogenic stimuli.
ROS have also been implicated in the pathogenesis of diabetes in animal models and in humans (23–27). The enhanced ROS tolerance of Rpl13a-snoless islets suggested that Rpl13a-snoless mice would have improved responses to diabetogenic stimuli. We therefore examined the susceptibility of Rpl13a-snoless mice to treatment with low dose streptozotocin, an antibiotic known to cause β-cell oxidative stress and cell death (28). After administration of streptozotocin or sodium citrate as control, mice were followed for an additional 8 weeks. Hyperglycemia was less severe in streptozotocin-treated Rpl13a-snoless mice compared with WT mice (Figure 7A). GTT performed 8 weeks after streptozotocin treatment revealed lower blood glucose in Rpl13a-snoless mice at each time point (Figure 7B). In sodium citrate–treated mice, fasting blood glucose was indistinguishable between the genotypes, with persistence of improved glucose tolerance in Rpl13a-snoless mice at 16 weeks of age (Supplemental Figure 5). To gain insight into the mechanism of this protection, we harvested pancreas tissue from an independent cohort of animals 5 days after streptozotocin treatment. While markers of inflammation and β cell replication were indistinguishable between the genotypes (Supplemental Figure 5), streptozotocin-induced accumulation of the oxidatively modified lipids 4-hydroxynonenal and 3β,5α,6β-cholestanetriol (triol) was prevented in Rpl13a-snoless mice (Figure 7, C and D). Furthermore, TUNEL staining of insulin-positive β cells was decreased by 50% in Rpl13a-snoless animals compared with WT controls (Figure 7E). This indicates that Rpl13a snoRNAs function in propagation of oxidative stress in the pancreas and in β cell apoptosis in response to streptozotocin.
Figure 7. Rpl13a-snoless mice are resistant to diabetogenic stimuli.
(A–E) streptozotocin- (STZ-) or vehicle-treated (control) 8-week-old WT and –/– mice. (A) Mean fasting blood glucose over time (+SEM) for n = 9 mice per genotype/treatment. (B) Mean blood glucose (+SEM) during GTT at 16 weeks for n = 9 mice per genotype/treatment. (C) Representative pancreas sections stained for 4-hydroxynonenal 5 days after STZ. Scale bar: 25 μm (n = 4 mice per genotype/treatment). (D) Pancreas tissue triol 5 days after STZ for n = 7 mice per genotype/treatment. (E) Representative pancreas sections stained for TUNEL 5 days after STZ. Scale bar: 25 μm. Quantification for n = 4 mice per genotype/treatment. Magnification ×2. (F) Ins2C96Y Akita allele bred into Rpl13a-snoless model. Mean (+SEM) random blood glucose at indicated time points for a minimum of n = 7 per group. (G) Rpl13a-snoless alleles were bred into the NOD model. Diabetes-free survival over 24 weeks for NOD (n = 26) vs. NOD Rpl13a-snoless (n = 16) female mice. *P < 0.05 for (–/–, STZ) vs. (WT, STZ); **P < 0.05 for (WT, STZ) vs. (WT); #P < 0.001 for (WT, C96Y) vs. (WT); †P < 0.0005 for (–/–, C96Y) vs. (WT, C96Y); determined by unpaired t test. §P < 0.04 by log-rank testing.
We further evaluated the effect of loss of Rpl13a snoRNAs in other murine models of diabetes. In the Akita model of diabetogenesis, the Ins2C96Y allele causes endoplasmic reticulum and oxidative stress, leading to β cell apoptosis and diabetes (29, 30). Loss of function of the Rpl13a snoRNAs decreased the degree of hyperglycemia in both male and female mice carrying the Ins2C96Y allele (Figure 7F). In the non-obese diabetic (NOD) model, oxidative damage in pancreatic β cells, β cell destruction, and the development of diabetes are mitigated by treatment with inhibitors of ROS-producing enzymes, treatment with chemical ROS scavengers, or β cell–specific overexpression of antioxidants, independent of immune cell infiltration into islets (24, 31, 32). Cross of the Rpl13a-snoless alleles into the NOD strain revealed that loss of the snoRNAs was also protective against the development of hyperglycemia in the NOD background (Figure 7G). Taken together, our findings provide evidence that the Rpl13a snoRNAs contribute to hyperglycemia in 3 independent murine models of diabetogenesis.
Discussion
Here, we identified noncanonical metabolic roles for box C/D snoRNAs in the physiological function and pathophysiological responses of β cells. Although broad loss of box C/D snoRNA–directed 2′-O-methyl modifications of ribosomal RNAs that results from fibrillarin knockout is embryonic lethal (2), we demonstrate that selective loss of 4 snoRNAs in the Rpl13a-snoless model is compatible with normal murine development. Unexpectedly, this model revealed a role for Rpl13a-snoRNAs in regulation of islet responses to metabolic stress that impacts whole body glucose homeostasis. Furthermore, the snoRNAs also modulate the response to stimuli that damage islets through oxidative stress, thereby impacting diabetes susceptibility. Together, these results establish that box C/D snoRNAs from the Rpl13a locus have noncanonical functions in pancreatic β cells beyond a housekeeping role in the biosynthesis of ribosomal RNAs. Our findings provide new insights into islet biology, systemic metabolic homeostasis, and the pathophysiology of diabetes.
Loss of the Rpl13a-snoRNAs in this model led to enhanced GSIS in isolated pancreatic islets. Loss of the snoRNAs was also associated with increased mitochondrial proton leak, lower ROS tone, and diminished amplification of cellular ROS in response to high glucose, as well as chemicals that more directly induce ROS. Uncoupled respiration has been shown to decrease mitochondrial hydrogen peroxide production during metabolic challenge (17, 18), providing a precedent for our observations of increased leak and decreased ROS in the islets. Furthermore, high levels of ROS and oxidative stress are known to inhibit β cell release of insulin (19–21, 33). Our findings suggest that under stimulatory glucose conditions, Rpl13a-snoless islets escape this inhibition, because they maintain lower oxidative tone. Together, our results suggest that in the Rpl13a-snoless model, mitochondrial proton leak leads to lower islet ROS and greater insulin secretion in the face of metabolic challenge, with resulting enhanced efficiency of systemic glucose metabolism.
The relationship between ROS and insulin secretion from pancreatic β cells is complex. These specialized cells are characterized by low-level expression of many antioxidant enzymes yet support high-level synthesis of a secreted protein that must undergo proper folding and disulfide bond formation in the endoplasmic reticulum, a process that is exquisitely sensitive to changes in cellular redox state (34–36) Furthermore, ROS function in several different ways to regulate insulin secretion. Findings that stimulatory glucose levels induce mitochondrial ROS, and antioxidants that blunt this ROS inhibit GSIS, support a role for ROS as a stimulus required for GSIS (20, 37). On the other hand, high levels of ROS that induce oxidative stress impair GSIS, and loss of function of ROS-producing enzymes or scavenging of ROS potentiates GSIS (20, 21, 33). Specifically, high levels of ROS have been proposed to inhibit insulin granule exocytosis and to interfere with stimulus-secretion coupling in β cells (38, 39). Our observations of lowered provoked ROS and improved GSIS in Rpl13a-snoless islets, without a change in antioxidant potential, suggest that the major impact of loss of these noncoding RNAs is on production of ROS species that serve to inhibit insulin secretion.
Rpl13a-snoless mice have germline disruption of Rpl13a snoRNAs. Thus, although ITT responses were comparable between the genotypes, effects on systemic glucose homeostasis from tissues other than islets cannot be formally excluded. Given our goal of precisely removing 4 distinct intronic sequences (ranging from 66 to 89 nucleotides in length) within a genomic space of 2.8 kb, without disturbing expression of the host protein-coding sequences, we chose to knock in a “snoless” allele. Neither Cre-lox technology nor CRISPR approaches would have been feasible for generation of the 4 simultaneous conditional mutations, as this would have required generation and interbreeding of 4 distinct knockout lines to achieve tissue specificity. In our knockin model, however, the observations that the snoRNAs are normally expressed in islets and that their disruption alters GSIS in isolated islets indicate an islet-autonomous role for these noncoding RNAs in GSIS. These effects at the level of the islet are consistent with the physiological observation of enhanced insulin secretion after glucose challenge in Rpl13a-snoless mice.
Destruction of pancreatic islets during the development of type 1 diabetes has been modeled in mice by chemical, genetic, and immunological perturbations. Although each class of models invokes different pathways leading to β cell destruction and eventual loss of insulin secretion and hyperglycemia, a common feature of these pathways is oxidative stress–mediated tissue damage (24–26, 29, 40). Moreover, improved outcomes following genetic interventions to decrease β cell ROS provide evidence that oxidative damage from excessive ROS contributes to destruction of pancreatic islets. Observations in humans of elevated plasma and red blood cell markers of ROS during the first weeks following the onset of symptoms of type 1 diabetes also implicate oxidative stress in the pathogenesis of diabetes (27). Our findings in Rpl13a-snoless mice of lower oxidative stress, less β cell death, and improved glucose tolerance following chemical provocation of diabetes and decreased hyperglycemia in genetic models of diabetogenesis raise the possibility that these noncoding RNAs function in the progressive β cell damage that leads to diabetes.
Based on the well-characterized biology of canonical snoRNAs, the Rpl13a snoRNAs likely form ribonucleoprotein particles that target cellular RNAs for modifications. Although our study links the snoRNAs to altered mitochondrial function and ROS production, the precise molecular targets of these snoRNAs responsible for the observed physiological phenotypes remain to be determined. Beyond the predicted 28S and 18S ribosomal RNA targets for these snoRNAs (41), the Rpl13a snoRNAs may also function non-canonically to target mRNAs, given recent studies demonstrating that some snoRNAs target mRNAs (42, 43) and our previous observation that the Rpl13a snoRNAs accumulate in the cytoplasm under metabolic stress conditions (12). Our RNAseq analysis suggests that rather than affecting the abundance of potential mRNA targets, the Rpl13a snoRNAs may impact their translation potential. Ongoing efforts are focused on high-throughput biochemical analyses of snoRNAs engaged with their relevant targets in WT tissues, studies for which the Rpl13a-snoless model will serve as a rigorous control. However, these approaches will be addressed in future studies, as they require substantially greater material than can be obtained from isolated islets (44). Furthermore, the Rpl13a snoRNAs may also contribute to metabolic stress–induced alterations in β cell growth and survival by binding to and activating protein kinase RNA-activated (PKR) to affect downstream protein translation and signaling pathways (45–48).
This study provides insights into the disease relevance and underlying cellular functions of snoRNAs in vivo. Our findings challenge the paradigm that box C/D snoRNAs serve solely to catalyze nucleotide modifications and processing of structural RNAs. The striking effects of loss of function of the Rpl13a snoRNAs raise the possibility that genetic variation affecting the production or function of these and other snoRNAs may contribute to metabolic disease phenotypes in mammals. Furthermore, given that snoRNAs can be targeted for antisense knockdown, our findings suggest that molecular therapies could be designed to target these noncoding RNAs.
Methods
Mice.
To generate mice lacking all 4 Rpl13a snoRNAs, we created a targeting vector containing the Rpl13a sequence between exons 1 and 8 with a loxP-flanked neomycin phosphotransferase (NEO) cassette in intron 1, and lacking the U32a, U33, U34, and U35a snoRNAs in introns 2, 4, 5, and 6, respectively. The construct was electroporated into 129S5/SvEv ES cells. ES cells with the correct homologous recombination event were selected by neomycin resistance, PCR, and Southern blot analysis. ES clones with normal karyotype were microinjected into B6/FBN blastocysts that were implanted into pseudopregnant females. F1 progeny from the resulting chimeras were tested for the presence of the NEO cassette in the Rpl13a locus by Southern blot analysis. The NEO cassette was removed by breeding these mice with mice expressing CRE under the adenovirus EIIa promoter. Heterozygote progeny were backcrossed into the FVB/N strain more than 10 generations. See Supplemental Table 3 for genotyping primers. Mice generated from heterozygote-heterozygote breeding pairs were used for experiments. The Rpl13a-snoless mice were crossed with Akita mice bearing the Ins2C96Y allele, which had previously been backcrossed onto the FVB background for more than 9 generations (FVB.B6-Ins2Akita/MlnJ; The Jackson Laboratory). NOD (NOD/ShiLtJ) mice from the Jackson Laboratory were bred in the Immunology of Type 1 Diabetes Core of the Washington University Diabetes Research Center. Speed Congenics (49) was used to move the Rpl13a-snoless allele onto the NOD background strain, using a marker-assisted breeding strategy. At each generation, multiple offspring were analyzed for genomic single nucleotide polymorphisms to select subsequent breeders with the highest percentage of NOD. After 6 generations, animals were >98.2% NOD. NOD/Rpl13a+/– breeders were used to generate female progeny for studies. Diabetic mice were identified based on two consecutive random blood glucose measures of ≥250 mg/dl.
RT-qPCR.
Total RNA was isolated using TRIzol reagent. For mRNA analyses, cDNA synthesis used the SuperScript First-Strand Synthesis System with oligo(dT) priming (Invitrogen). For snoRNA analyses, stem-loop primers were used for reverse transcription, as previously described (10, 50). Quantitative RT-PCR (RT-qPCR) was performed using SYBR Green PCR Master Mix (Applied Biosystems) and an ABI Prism 7500 Fast Real-Time PCR System. Relative quantification of gene expression was performed using the comparative threshold method. See Supplemental Table 3 for RT-qPCR primers.
Immunoblot analyses.
Tissues were homogenized in RIPA buffer (150 mM NaCl, 50 mM HEPES pH 7.4, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 1× Protease Complete [Roche], 1 mM phenylmethylsulfonyl fluoride). Proteins were separated by SDS-PAGE, transferred to nitrocellulose using the semi-dry method, blocked with dry milk, and immunoblotted with polyclonal rabbit α–hamster RPL13a antibody (1:2,000) (10) and monoclonal α-actin (Sigma-Aldrich). Detection used HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and Western Lightning Plus-ECL (PerkinElmer Life Sciences) reagent. Images were digitally captured using a Bio-Rad ChemiDoc MP and analyzed with Bio-Rad Image Lab software.
Islet isolation.
Pancreases were inflated with ice-cold collagenase (Sigma-Aldrich) buffer (0.45 mg/ml collagenase in HBSS without Ca2+) and digested at 37°C. Islets were collected after sequential washes and incubated overnight in RPMI media (with 10% FBS and 2 mM l-glutamine) until use (51). Islets of equal size were hand picked to generate 3–5 technical replicates (measures, wells) for all experiments. Very large and very small islets were excluded. Results are reported for a minimum of 3 independent experiments.
Insulin secretion assay.
Hand-picked islets were distributed into 0.4-μm Transwells of a 24-well plate containing Krebs-Ringer buffer (KRB) without glucose. After washing twice in 2.8 mM glucose, islets were sequentially incubated in 5.6 mM and 16.7 mM glucose for 1 hour each (52). For KCl and glibenclamide experiments, islets were incubated first in 5.6 mM glucose and then 30 mM KCl or 1 μM glibenclamide (Sigma-Aldrich). A Rat/Mouse Insulin ELISA kit (Millipore) was used to measure the insulin levels in media.
Calcium imaging of islets.
Hand-picked islets were loaded with 4 μM Fluo-4 (Invitrogen) in KRB with 20 mM HEPES and 0.1% BSA, pH 7.4, for 40 minutes and then imaged on a Zeiss LSM 880 confocal microscope (Plan-Apochromat 20× M27, NA0.8 objective) in 2, 5, 8, and 16 mM glucose, with analyses as previously described (53). Following a 6-minute incubation at each concentration, a 5-minute time series was acquired, and 20 mM KCl was injected at the end of acquisition to prime maximum calcium release. Data were analyzed using ImageJ (NIH) and averaged over 20 cells in 6 different islets from each of 4 different mice per genotype. Data are expressed as fluorescence intensity under each condition relative to basal (nontreated) conditions.
ROS detection from whole islets or dispersed islet cells.
For whole islets, equivalent numbers of hand-picked islets of each genotype were incubated in media containing 10 μM DHE (Molecular Probes) for 1 hour, washed with PBS, and mounted. Epifluorescence images were obtained using a Zeiss Axioskop, and DHE fluorescence was quantified in 6 random fields per sample using ImageJ. For dispersed islets, equivalent numbers of cells of each genotype (pooled from multiple animals) were plated in RPMI media in 96-well plates overnight, incubated with 10 μM DHE or 10 μM H2DCFDA (DCF; Molecular Probes) for 1 hour, and then treated with 100 μM H2O2 (Sigma-Aldrich) for 2 hours. Fluorescence was quantified in each well using a TECAN plate reader. To detect ROS production in low- (3 mM) or high-glucose (20 mM) conditions, dispersed islets were plated on poly-d-lysine–coated (Sigma-Aldrich) glass-bottom culture dishes (MatTek Corp.). After overnight culture, islets were incubated in KRB containing 3 mM or 20 mM glucose with 10 μM DCF for 1 hour at 37°C. Immediately after washing, fluorescence images were obtained using a Leica DMIRB microscope. DCF fluorescence of individual islet cells from 8–10 random fields per condition was quantified using Leica Application Suite AF (19).
Trolox equivalent antioxidant capacity.
Antioxidant capacity was quantified in lysates from hand-picked islets (n = 4 mice/genotype) as described previously, and values are reported per microgram islet protein (54).
RNAseq analysis of islets.
High-quality total RNA (RNA integrity number [RIN] ≥8.5) was prepared from hand-picked islets (n = 4 mice/genotype) using TRIzol reagent, treated with TURBO DNAse (Thermo Fisher), and used to prepare SeqPlex RNAseq libraries (Sigma-Aldrich). Sequencing was performed by the Washington University Genome Technology Access Center using two lanes of Illumina HiSeq 2500. Reads were demultiplexed and trimmed, and STAR alignment and quantification analysis were carried out using the Partek Flow platform. Uniquely aligned reads were quantified to identify genes with at least a 2-fold change between genotypes, with P < 0.05 and an FDR step-up of 0.05. Genes identified were further analyzed for potential U32a, U33, U34, and U35a snoRNA target sites using the PLEXY program and a cut-off of minimum free energy for predicted duplexes of less than –20.4 kcal/mol (22). Sequence data are available in the NCBI’s Gene Expression Omnibus (GEO GSE87354).
Streptozotocin treatment.
Eight-week-old female mice were injected with streptozotocin intraperitoneally (Sigma-Aldrich; 50 mg/kg in 0.1 M Na citrate, pH 4.5) for 5 consecutive days as previously described (55). Control animals received buffer only. Serial fasting blood glucose and body weight were quantified every 2 weeks; GTT was performed at 8 weeks after the streptozotocin injection, and pancreases were harvested for histological analysis.
Intraperitoneal GTT and ITT.
In vivo dynamic testing was carried out following a 4-hour fast using standard procedures of the NIH-sponsored National Mouse Metabolic Phenotyping Centers (http://www.mmpc.org/shared/protocols.aspx). Mice were injected intraperitoneally with 1 g/kg glucose (GTT) or 1 U/kg insulin (ITT). Blood glucose was measured at 0, 15, 30, 60, and 120 minutes after the injection using a glucometer (Contour). Serum was analyzed for insulin at 0 and 30 minutes of GTT at the Washington University Diabetes Research Center Immunoassay Core by a sensitive Singulex single molecule tracking method that has a lower limit of quantification of 19 pg/ml.
Histological and immunofluorescence analyses.
For histological analyses, 5-μm sections of formalin-fixed, paraffin-embedded pancreas tissues were stained with H&E. For TUNEL, sections were labeled with TUNEL reagent (Cell Death Detection Kit TMR, Roche), followed by overnight incubation with α-insulin antibodies (polyclonal guinea pig antibody, 1:200, Dako IR002). TUNEL-positive signals were quantified from >75 islets per condition (n = 4 mice [>5,000 nuclei]/genotype/condition). Proliferation was analyzed with α–Ki-67 antibodies (anti-rabbit monoclonal IgG, 1:200, Cell Signaling Technology 9129), quantified from >75 islets per condition (n = 4 mice [>3,500 nuclei]/genotype/condition). Secondary antibodies were Alexa Flour 488–goat anti-guinea pig and Alexa Fluor 594–goat anti-rabbit (Molecular Probes A-11073 and A-11037). Stained sections were examined using an Olympus FSX100 microscope.
Morphometry.
Morphometric analysis of pancreases from 8-week-old mice was performed as previously reported (56). For measurement of β cell mass, every 40th pancreatic section was immunostained with guinea pig anti-insulin antibody (1:100, Invitrogen) and counterstained with hematoxylin. The β cell mass for each mouse was quantified using Image Pro Plus software (Media Cybernetics) by obtaining the fraction of the cross-sectional area of pancreatic tissue (exocrine and endocrine) positive for insulin staining and then multiplying by the pancreatic weight.
Isolation of MEFs and ROS detection.
MEFs were harvested from embryos at E13.5 and cultured in DMEM with 10% FBS, 2 mM l-glutamine, 1× MEM non-essential amino acids, and 150 μM monothioglycerol. ROS was quantified in low-passage (≤5) MEFs by staining with 10 μM DCF or 5 μM MitoSOX Red (Life Technologies) for 5 minutes in PBS with Ca2+/Mg2+. Fluorescence intensity was quantified by flow cytometry (BD FACSCalibur), measuring mean signal intensity for 104 events in each sample, and normalized to WT cells.
Oxidative tissue damage.
Formalin-fixed pancreatic sections were costained for insulin (polyclonal guinea pig antibody, 1:200, Dako) and hydroxynonenal (rabbit polyclonal, 1:100, Abcam) using appropriate Alexa Fluor–coupled secondary antibodies (Molecular Probes). Triol was quantified per mg pancreas tissue and is reported per mg protein (57).
Mitochondrial DNA copy number.
Relative mitochondrial DNA content was measured from isolated islets using RT-qPCR and nuclear DNA content as an internal control, as described previously (58).
OCR.
Oxygen consumption of islets was measured using a Seahorse XF24 analyzer (Seahorse Bioscience) (15). Fifty islets were hand picked and washed once in assay media containing 1% FBS and 1 mM l-glutamine, and 3 mM or 20 mM glucose, and then transferred into the depressed well of an XF24 Islet Capture Microplate. OCR was measured at basal glucose levels, as well as with drugs acting on the respiratory chain: oligomycin (ATP synthase inhibitor; 5 μM), FCCP (uncoupler; 7.5 μM), rotenone (complex I inhibitor; 0.1 μM), and antimycin A (complex III inhibitor; 1 μM) — all from Sigma-Aldrich. After the assay, islets were washed once with PBS, and protein was quantified by microBCA assay (Pierce).
Electron microscopy.
Islets were fixed in a modified Karnovsky’s fixative of 3% glutaraldehyde, 1% paraformaldehyde in 0.1 M sodium cacodylate buffer; post-fixed in 2% osmium tetroxide in 0.1 M sodium cacodylate buffer; stained with 2% aqueous uranyl acetate for 30 minutes; dehydrated in graded acetone; and embedded in Poly/Bed 812 (Polysciences Inc.). 90-nm sections were examined by a JEOL model 1200EX electron microscope. Digital images were acquired using the AMT Advantage HR (Advanced Microscopy Technology) high-definition CCD, 1.3 megapixel TEM camera.
Statistics.
Results are presented as mean ± SEM for a minimum of 3 independent experiments. Statistical significance was assessed by 2-tailed Student’s t test or log-rank test. P < 0.05 was considered significant. Serial glucose and GTT for streptozotocin-treated WT and knockout mice (Figure 7) were compared using multiple unpaired t tests and corrected for multiple comparisons using the Šidák-Bonferroni method, with α of 0.05.
Study approval.
Animal studies were approved by the Institutional Animal Care and Use Committee of Washington University in St. Louis.
Author contributions
JL and JES designed the study in consultation with FU, MSR, DWP, and DSO. Experiments were performed by JL, ANH, CLH, JM, KDP, ZL, DES, HF, RS, JZ, and SCCH. The manuscript was written by JL and JES. All authors contributed to editing the manuscript and support the conclusions.
Supplementary Material
Acknowledgments
This work was supported by NIH 2 R01 DK064989, Fondation Leducq (12CVD04), the Juvenile Diabetes Research Foundation (1-INO-2014-179-A-V), and the American Diabetes Association (7-09-CVD-07) (to JES); by NIH R01 DK098584 (to MSR); and by the Washington University Diabetes Research Center (Transgenic and ES Cell, Morphology and Metabolic Analysis, Immunoassay, Diabetes Models Phenotyping, Mass Spectrometry, and Immunology of Type 1 Diabetes Cores; P30 DK020579). Calcium imaging was performed at the Washington University Center for Cellular Imaging. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with RNAseq analyses. The Center is partially supported by an NCI Cancer Center Support grant to the Siteman Cancer Center (NIH P30 CA91842) and by the Washington University Institute of Clinical and Translational Sciences/Clinical & Translational Science Award (ICTS/CTSA) (NIH UL1 TR000448).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(12):4616–4625. doi:10.1172/JCI88069.
Contributor Information
Jiyeon Lee, Email: nousful@hotmail.com.
Jana Mahadevan, Email: Jmahadev@dom.wustl.edu.
Zeno Lavagnino, Email: zeno.lavagnino@wustl.edu.
Hideji Fujiwara, Email: hideji.fujiwara@gmail.com.
Rohini Sidhu, Email: RSIDHU@DOM.wustl.edu.
Jessie Zhang, Email: jrzhang@wustl.edu.
Fumihiko Urano, Email: urano@dom.wustl.edu.
Jean E. Schaffer, Email: jschaff@wustl.edu.
References
- 1.Kiss T, Fayet E, Jády BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 2.Newton K, Petfalski E, Tollervey D, Caceres JF. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol Cell Biol. 2003;23(23):8519–8527. doi: 10.1128/MCB.23.23.8519-8527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84(8):775–790. doi: 10.1016/S0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien JE. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet. 2012;5(3):279–286. doi: 10.1161/CIRCGENETICS.111.961474. [DOI] [PubMed] [Google Scholar]
- 5.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311(5758):230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 6.Dong XY, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14. Hum Mol Genet. 2008;17(7):1031–1042. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu L, et al. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J Clin Invest. 2012;122(8):2793–2806. doi: 10.1172/JCI63051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mei YP, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31(22):2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siprashvili Z, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48(1):53–58. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel CI, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14(1):33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scruggs BS, Michel CI, Ory DS, Schaffer JE. SmD3 regulates intronic noncoding RNA biogenesis. Mol Cell Biol. 2012;32(20):4092–4103. doi: 10.1128/MCB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holley CL, Li MW, Scruggs BS, Matkovich SJ, Ory DS, Schaffer JE. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J Biol Chem. 2015;290(18):11741–11748. doi: 10.1074/jbc.M115.637413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neely GG, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141(1):142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prentki M, Matschinsky FM, Madiraju SR. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013;18(2):162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Wikstrom JD, et al. A novel high-throughput assay for islet respiration reveals uncoupling of rodent and human islets. PLoS One. 2012;7(5):e33023. doi: 10.1371/journal.pone.0033023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brand MD. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med. doi: 10.1016/j.freeradbiomed.2016.04.001.. [published online ahead of print April 13, 2016]. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Anderson EJ, Yamazaki H, Neufer PD. Induction of endogenous uncoupling protein 3 suppresses mitochondrial oxidant emission during fatty acid-supported respiration. J Biol Chem. 2007;282(43):31257–31266. doi: 10.1074/jbc.M706129200. [DOI] [PubMed] [Google Scholar]
- 18.Boudina S, et al. UCP3 regulates cardiac efficiency and mitochondrial coupling in high fat-fed mice but not in leptin-deficient mice. Diabetes. 2012;61(12):3260–3269. doi: 10.2337/db12-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llanos P, Contreras-Ferrat A, Barrientos G, Valencia M, Mears D, Hidalgo C. Glucose-dependent insulin secretion in pancreatic β-cell islets from male rats requires Ca2+ release via ROS-stimulated ryanodine receptors. PLoS One. 2015;10(6):e0129238. doi: 10.1371/journal.pone.0129238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pi J, et al. Reactive oxygen species as a signal in glucose-stimulated insulin secretion. Diabetes. 2007;56(7):1783–1791. doi: 10.2337/db06-1601. [DOI] [PubMed] [Google Scholar]
- 21.Tang C, et al. Evidence for a role of superoxide generation in glucose-induced β-cell dysfunction in vivo. Diabetes. 2007;56(11):2722–2731. doi: 10.2337/db07-0279. [DOI] [PubMed] [Google Scholar]
- 22.Kehr S, Bartschat S, Stadler PF, Tafer H. PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics. 2011;27(2):279–280. doi: 10.1093/bioinformatics/btq642. [DOI] [PubMed] [Google Scholar]
- 23.Kuo T, Kim-Muller JY, McGraw TE, Accili D. Altered plasma profile of antioxidant proteins as an early correlate of pancreatic β cell dysfunction. J Biol Chem. 2016;291(18):9648–9656. doi: 10.1074/jbc.M115.702183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotta M, et al. Pancreatic β cell-specific expression of thioredoxin, an antioxidative and antiapoptotic protein, prevents autoimmune and streptozotocin-induced diabetes. J Exp Med. 1998;188(8):1445–1451. doi: 10.1084/jem.188.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Li X, Epstein PN. MnSOD and catalase transgenes demonstrate that protection of islets from oxidative stress does not alter cytokine toxicity. Diabetes. 2005;54(5):1437–1446. doi: 10.2337/diabetes.54.5.1437. [DOI] [PubMed] [Google Scholar]
- 26.Xiang FL, Lu X, Strutt B, Hill DJ, Feng Q. NOX2 deficiency protects against streptozotocin-induced β-cell destruction and development of diabetes in mice. Diabetes. 2010;59(10):2603–2611. doi: 10.2337/db09-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domínguez C, Ruiz E, Gussinye M, Carrascosa A. Oxidative stress at onset and in early stages of type 1 diabetes in children and adolescents. Diabetes Care. 1998;21(10):1736–1742. doi: 10.2337/diacare.21.10.1736. [DOI] [PubMed] [Google Scholar]
- 28.Like AA, Rossini AA. Streptozotocin-induced pancreatic insulitis: new model of diabetes mellitus. Science. 1976;193(4251):415–417. doi: 10.1126/science.180605. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell T, et al. Dysfunctional mitochondrial bioenergetics and oxidative stress in Akita(+/Ins2)-derived β-cells. Am J Physiol Endocrinol Metab. 2013;305(5):E585–E599. doi: 10.1152/ajpendo.00093.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oyadomari S, et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109(4):525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suarez-Pinzon WL, Mabley JG, Strynadka K, Power RF, Szabó C, Rabinovitch A. An inhibitor of inducible nitric oxide synthase and scavenger of peroxynitrite prevents diabetes development in NOD mice. J Autoimmun. 2001;16(4):449–455. doi: 10.1006/jaut.2001.0507. [DOI] [PubMed] [Google Scholar]
- 32.Suarez-Pinzon WL, Szabó C, Rabinovitch A. Development of autoimmune diabetes in NOD mice is associated with the formation of peroxynitrite in pancreatic islet β-cells. Diabetes. 1997;46(5):907–911. doi: 10.2337/diab.46.5.907. [DOI] [PubMed] [Google Scholar]
- 33.Li N, et al. NADPH oxidase NOX2 defines a new antagonistic role for reactive oxygen species and cAMP/PKA in the regulation of insulin secretion. Diabetes. 2012;61(11):2842–2850. doi: 10.2337/db12-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic Biol Med. 1996;20(3):463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- 35.Tiedge M, Lortz S, Drinkgern J, Lenzen S. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997;46(11):1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
- 36.Mehmeti I, Lortz S, Elsner M, Lenzen S. Peroxiredoxin 4 improves insulin biosynthesis and glucose-induced insulin secretion in insulin-secreting INS-1E cells. J Biol Chem. 2014;289(39):26904–26913. doi: 10.1074/jbc.M114.568329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leloup C, et al. Mitochondrial reactive oxygen species are obligatory signals for glucose-induced insulin secretion. Diabetes. 2009;58(3):673–681. doi: 10.2337/db07-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiseman DA, Kalwat MA, Thurmond DC. Stimulus-induced S-nitrosylation of Syntaxin 4 impacts insulin granule exocytosis. J Biol Chem. 2011;286(18):16344–16354. doi: 10.1074/jbc.M110.214031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krippeit-Drews P, Kramer C, Welker S, Lang F, Ammon HP, Drews G. Interference of H2O2 with stimulus-secretion coupling in mouse pancreatic β-cells. J Physiol. 1999;514(pt 2):471–481. doi: 10.1111/j.1469-7793.1999.471ae.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thayer TC, et al. Superoxide production by macrophages and T cells is critical for the induction of autoreactivity and type 1 diabetes. Diabetes. 2011;60(8):2144–2151. doi: 10.2337/db10-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicoloso M, Qu LH, Michot B, Bachellerie JP. Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260(2):178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 42.Falaleeva M, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci U S A. 2016;113(12):E1625–E1634. doi: 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engreitz JM, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159(1):188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Youssef OA, Safran SA, Nakamura T, Nix DA, Hotamisligil GS, Bass BL. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci U S A. 2015;112(16):5023–5028. doi: 10.1073/pnas.1424044112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen SS, et al. Activation of double-stranded RNA-dependent protein kinase inhibits proliferation of pancreatic β-cells. Biochem Biophys Res Commun. 2014;443(3):814–820. doi: 10.1016/j.bbrc.2013.12.051. [DOI] [PubMed] [Google Scholar]
- 47.Scarim AL, et al. Mechanisms of β-cell death in response to double-stranded (ds) RNA and interferon-γ: dsRNA-dependent protein kinase apoptosis and nitric oxide-dependent necrosis. Am J Pathol. 2001;159(1):273–283. doi: 10.1016/S0002-9440(10)61693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y, et al. Activated PKR inhibits pancreatic β-cell proliferation through sumoylation-dependent stabilization of P53. Mol Immunol. 2015;68(2 pt A):341–349. doi: 10.1016/j.molimm.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 49.Clements MK. Against time-limited certification. J Am Acad Dermatol. 1989;20(3):515–517. doi: 10.1016/S0190-9622(89)80097-0. [DOI] [PubMed] [Google Scholar]
- 50.Feng J, Wang K, Liu X, Chen S, Chen J. The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene. 2009;437(1–2):14–21. doi: 10.1016/j.gene.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 51.Calderon B, Suri A, Miller MJ, Unanue ER. Dendritic cells in islets of Langerhans constitutively present beta cell-derived peptides bound to their class II MHC molecules. Proc Natl Acad Sci U S A. 2008;105(16):6121–6126. doi: 10.1073/pnas.0801973105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buteau J, Shlien A, Foisy S, Accili D. Metabolic diapause in pancreatic beta-cells expressing a gain-of-function mutant of the forkhead protein Foxo1. J Biol Chem. 2007;282(1):287–293. doi: 10.1074/jbc.M606118200. [DOI] [PubMed] [Google Scholar]
- 53.Rocheleau JV, et al. Critical role of gap junction coupled KATP channel activity for regulated insulin secretion. PLoS Biol. 2006;4(2):e26. doi: 10.1371/journal.pbio.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26(9-10):1231–1237. doi: 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- 55. National Institute of Health. National Mouse Metabolic Phenotyping Centers. MMPC Web site. http://www.mmpc.org/shared/protocols.aspx Accessed October 3, 2016.
- 56.Mahadevan J, et al. Ebselen treatment prevents islet apoptosis, maintains intranuclear Pdx-1 and MafA levels, and preserves β-cell mass and function in ZDF rats. Diabetes. 2013;62(10):3582–3588. doi: 10.2337/db13-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porter FD, et al. Cholesterol oxidation products are sensitive and specific blood-based biomarkers for Niemann-Pick C1 disease. Sci Transl Med. 2010;2(56):56ra81. doi: 10.1126/scitranslmed.3001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9(4):261–265. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.