Abstract
A microRNA (miRNA) is a 21–24 nucleotide RNA product of a non-protein-coding gene. Plants, like animals, have a large number of miRNA-encoding genes in their genomes. The biogenesis of miRNAs in Arabidopsis is similar to that in animals in that miRNAs are processed from primary precursors by at least two steps mediated by RNAse III-like enzymes and that the miRNAs are incorporated into a protein complex named RISC. However, the biogenesis of plant miRNAs consists of an additional step, i.e., the miRNAs are methylated on the ribose of the last nucleotide by the miRNA methyltransferase HEN1. The high degree of sequence complementarity between plant miRNAs and their target mRNAs has facilitated the bioinformatic prediction of miRNA targets, many of which have been subsequently validated. Plant miRNAs have been predicted or confirmed to regulate a variety of processes, such as development, metabolism, and stress responses. A large category of miRNA targets consists of genes encoding transcription factors that play important roles in patterning the plant form.
Keywords: microRNA, Small interfering RNA, DCL1, HEN1, Auxin, Flower development, Leaf development, Developmental transitions
1. What is a microRNA?
A microRNA (miRNA) is a 21–24 nucleotide (nt) small RNA that is the final product of a non-coding RNA gene. miRNA genes resemble protein coding genes in that they may contain introns and that they are transcribed by RNA polymerase II. Like other pol II transcripts, the transcripts from miRNA genes are capped, spliced and polyadenylated (reviewed in [1]). The mature miRNA is located in a hairpin structure within the primary transcript (pri-miRNA) and is processed from the pri-miRNA through at least two RNAse III-mediated steps (reviewed in [1,2]). The miRNA is loaded into a ribonucleoprotein complex named RISC, where it guides the cleavage or translational repression of its target mRNAs by base-pairing with the targets (reviewed in [2]).
Plants are rich in another type of small RNAs (known as siRNAs) that is similar in structure, biogenesis and function to miRNAs. siRNAs originate from transcripts from transgenes [3], endogenous repeat sequences or transposons [4,5]. One key distinction between miRNAs and siRNAs from transgenes and repeat sequences/transposons is that miRNAs target genes other than the ones that give rise to the miRNAs while siRNAs target the very sequences that generate them (reviewed in [2]). Recently, a new class of siRNAs that, like miRNAs, targets mRNAs from other loci was discovered in Arabidopsis and named trans-acting siRNAs (ta-siRNAs) [6,7]. The ta-siRNAs originate from loci that give rise to non-coding transcripts that are themselves targets of miRNAs. The miRNA-mediated cleavage of the transcripts recruits an RNA-dependent RNA polymerse (RdRP) to use the cleaved transcripts as templates to generate long double-stranded RNAs (dsRNAs), which then serve as the source of multiple ta-siRNAs. The miRNA-mediated cleavage of the precursor RNA is crucial for the biogenesis of the ta-siRNAs and sets the register for the cleavage events that generate the ta-siRNAs [8]. Since ta-siRNAs target mRNAs from other genes, the cis/trans relationship between smallRNAs and their targets is no longer a distinction between miRNAs and siRNAs. Now it seems that the only feature that distinguishes miRNAs and siRNAs is the nature of the precursor transcripts. While a miRNA comes from a hairpin pre-miRNA, siRNAs come from a perfect, long dsRNA generated by an RdRP or through the transcription of a hairpin transgene. Usually only one miRNA is generated from the pre-miRNA but several or many siRNAs are generated from long dsRNAs. However, there is a case in which more than one small RNAs come from a single pre-miRNA [9].
2. miRNA biogenesis
2.1. miRNA maturation
In animals, a miRNA is processed from the pri-miRNA by two RNAse III enzymes, Drosha and Dicer. Drosha crops the pri-miRNA into the pre-miRNA, the hairpin structure in the pri-miRNA [10]. Drosha is found in a protein complex named the microprocessor that also contains the DiGeorge Syndrome Critical Region Gene 8 (DGCR8) protein in humans (Pasha in Drosophila) [11–14]. Drosha cleavage leaves in the stem of the hairpin pre-miRNA a 2 nt 3′ overhang [10], a feature that is preferred by Dicer [15], the enzyme that processes the pre-miRNA to produce a duplex of 21 nt small RNAs with 2 nt overhangs at the 3′ end of each strand [16–19]. The strand in which the 5′ end is less stable is selectively incorporated into a protein complex named RISC [20], where the miRNA serves as the effector of gene regulation. The miRNA maturation process is partitioned between the nucleus and the cytoplasm [21]. Drosha processes the pri-miRNA in the nucleus to the pre-miRNA [10], which is then exported to the cytoplasm by exportin 5 [22–24]. In the cytoplasm, Dicer releases the miRNA from the pre-miRNA.
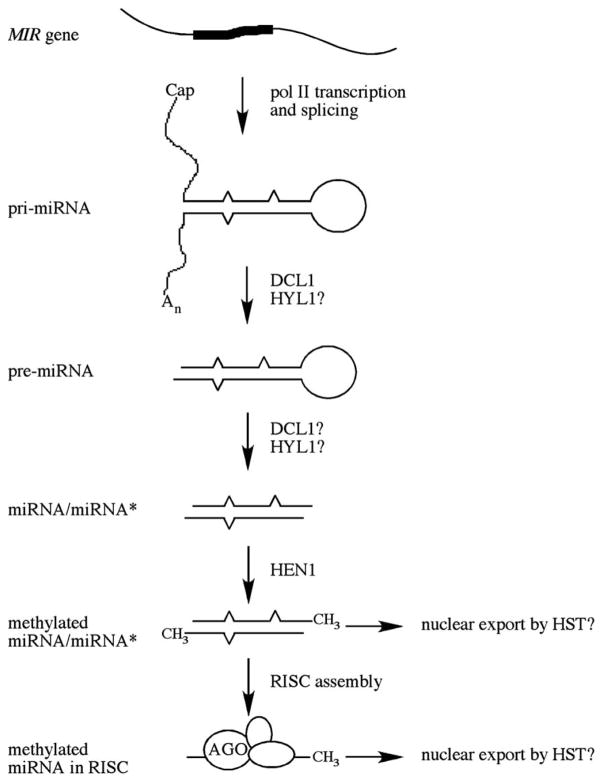
In Arabidopsis, the maturation of the miRNA from the primiRNA is also a stepwise process that involves a Dicer-like protein, DCL1 (Fig. 1). The Arabidopsis genome, however, does not contain homologs of Drosha or DGCR8. In fact, it is found that DCL1 processing pri-miRNA163 to pre-miRNA163 [9]. Although it has not been demonstrated that DCL1 processes pre-miRNA to the mature miRNA, DCL1 is likely the key enzyme that performs this function for the following reasons. First, among the four DCL genes in the Arabidopsis genome, DCL1 is the only essential gene. Null alleles in DCL1 result in embryo lethality while weak dcl1 mutants exhibit pleiotropic developmental defects [25–27]. Mutations in other DCL genes do not result in such severe developmental defects ([5,108–110]). Since miRNAs play key roles in plant development (see below), the phenotypes of the dcl mutants are consistent with DCL1 being the main miRNA-generating Dicer. Second, most miRNAs examined are reduced in abundance in the weak dcl1 mutants, such as dcl1-7 and dcl1-9, while mutations in DCL2 and DCL3 do not affect the abundance of the miRNAs examined [28–30]. However, it should be noted that the accumulation of some miRNAs is not affected in the weak dcl1-9 mutant [31]. Either another DCL protein is responsible for the maturation of these miRNAs or DCL1 is the processing enzyme for these miRNAs but the weak dcl1-9 mutant protein still retains the ability to process the precursors of this set of miRNAs. Like in animal miRNA maturation, the processing of the pre-miRNA yields the miRNA/ miRNA* duplex, in which the miRNA is selectively loaded into RISC. The asymmetric loading of the two strands into RISC probably follows the same rule as for animal miRNAs [20,32]. The presence of the miRNA/miRNA* duplex is supported by the isolation of certain miRNA* sequences from small RNA cloning efforts [30] and the detection of several miRNAs* by filter hybridization [33,34]. Several viral RNA silencing suppressors that affect miRNA biogenesis also provide evidence for the transient presence of the miRNA/ miRNA* duplex. When expressed in Arabidopsis, the Beet yellows virus p21 and the Tomato bushy stunt virus p19, which bind the duplex but not single-stranded miRNAs [35–38], lead to increased abundance of miRNA* and reduced miRNA-mediated cleavage of target mRNAs [35,39]. It is thought that p19 and p21 bind to and stabilize the duplex, therefore preventing RISC assembly.
Fig. 1.
A diagram of miRNA biogenesis in Arabidopsis. A miRNA gene is transcribed by RNA polymerase II. The pri-miRNA is capped and polyadenylated. The pri-miRNA is processed by DCL1, perhaps in two or more steps, to the miRNA/miRNA* duplex, which is then methylated by HEN1. The miRNA strand is incorporated into RISC. The nuclear export of miRNAs may occur before or after RISC assembly.
The subcellular compartmentalization of miRNA biogenesis in Arabidopsis may be different from that in animals. A partial DCL1 protein fused to the green fluorescent protein was localized in the nuclei of union epidermal cells upon transient expression, indicating that DCL1 has a nuclear localization sequence [40]. Although the location of the full-length DCL1 protein in the cell remains to be determined, it is likely that miRNA biogenesis occurs in the nucleus since nuclear localized p19 leads to reduced abundance of miRNAs [40]. While exportin 5 exports pre-miRNAs to the cytoplasm in animals, the Arabidopsis homolog of exportin 5, HASTY (HST; [41]), is proposed to export the miRNA/miRNA* duplex to the cytoplasm based on the assumption that the duplex is produced by DCL1 in the nucleus [2]. hst mutants show reduced accumulation of many but not all miRNAs and are compromised in the cleavage of certain miRNA target genes, consistent with a role of HST in miRNA biogenesis [33]. miRNAs are found in presumably single-stranded forms in both the nuclear and the cytoplasmic compartments (miRNAs* do not accumulate in either compartment) [33]. This implies that miRNAs are present in both compartments as molecules present in RISCs. One possibility is that RISC assembly occurs in the nucleus followed by the export of RISCs (by HST) to the cytoplasm (Fig. 1). Alternatively, miRNA/miRNA* is exported to the cytoplasm (Fig. 1), where RISC assembly occurs, and then some RISCs are imported back to the nucleus. It is also possible that RISC assembly occurs separately in both compartments.
2.2. Methylation of miRNA/miRNA*
The biogenesis of miRNAs in Arabidopsis requires two other proteins, HYL1 and HEN1 [29,42,43] (Fig. 1). HYL1 contains two dsRNA-binding motifs and is homologous to RDE-4 from C. elegans and R2D2 from Drosophila, proteins that play a role in RISC loading in association with Dicer [44,45]. Interestingly, while RDE4 and R2D2 act in siRNA but not miRNA metabolism, HYL1 is required for miRNA but not siRNA biogenesis. HYL1 is a nuclear protein present in a protein complex [42] but its biochemical function in miRNA biogenesis in unknown.
HEN1 has a putative dsRNA-binding motif and a C-terminal methyltransferase domain. The hen1-1, hen1-2, and hen1-4 mutations that compromise miRNA metabolism are all in the methyltransferase region [46,47]. Purified HEN1 protein is able to methylate the miRNA/miRNA* duplex in vitro [48]. The methylation occurs on the ribose of the last nucleotide on each strand of the duplex. HEN1 is highly selective of its substrate: single-stranded miRNA, single-stranded miRNA*, pre-miRNA, dsDNA identical in sequence and structure to miRNA/miRNA* cannot serve as substrates of HEN1. miRNA/miRNA* of different primary sequences can serve as substrates, suggesting that HEN1 recognizes the structure (rather than the sequence) of the duplex produced by Dicer processing of pre-miRNA. The in vitro biochemical activity of HEN1 and the in vivo requirement for HEN1 for miRNA biogenesis suggest that Arabidopsis miRNAs are methylated on their last nucleotides. Indeed, Arabidopsis miRNAs are resistant to chemical reactions that depend on the presence of both hydroxyl groups on the last nucleotide [48]. Mass spectrometry analysis of miR173 purified from Arabidopsis indicates that miR173 possesses one methyl group [48]. For any miRNA species, no unmethylated molecules can be detected in vivo by filter hybridization, indicating that the great majority of any miRNA species is methylated in vivo.
What is the function of miRNA methylation? It is conceivable that the methyl group on the ribose of the last nucleotide, whether at the 2′ or 3′ position, impedes the activities of enzymes that target either hydroxyl group of the last nucleotide, such as ligases, terminal nucleotydyl transferases, or polymerases. One group of enzymes relevant to small RNA metabolism is RdRP. In both C. elegans and in plants, post-transcriptional gene silencing (or RNAi) can be transitive, i.e., silencing can occur in regions outside of the initial region targeted for silencing in an RNA [49,50]. In C. elegans the transitivity only extends to regions 5′ to the initial target region, whereas in plants the transitivity happens in both directions [49,50]. Transitive RNAi in C. elegans requires an RdRP and it has been proposed that primary siRNAs from the silencing trigger serve as primers for the RdRP, which uses the target RNA as the template to synthesize long double-stranded RNAs that in turn are processed to secondary siRNAs [49]. Plant miRNAs are highly complementary to their target mRNAs and many have been shown to guide the cleavage of their target transcripts [28]. It is conceivable that plant RdRPs may use a miRNA as the primer and the target mRNA as the template to synthesize long dsRNA, which in turn may be processed to siRNAs. This may be used as a mechanism to coordinately regulate multiple, highly conserved members of a gene family in which only one or some members have a miRNA target site. This may also be a scenario that the plant should avoid to prevent the unintended silencing of genes sharing high degree of sequence similarity with the real miRNA target gene in regions 5′ to the miRNA target site. The presence of the methyl group on the last nucleotide of plant miRNAs may discourage RdRPs from using them as primers. However, the effect of the methyl group on the ability of the miRNAs to serve as primers for RdRPs needs to be determined biochemically.
One enzymatic activity the methylation appears to protect plant miRNAs against is that of a polymerase or terminal transferase that adds additional nucleotides, primarily U’s to the 3′ end of miRNAs [51]. This finding stemmed from the initial observation that miRNAs become heterogenous in size in addition to being reduced in abundance in hen1 mutants [29,42]. We examined the source of the size heterogeneity in the miRNAs in hen1-1 and found that the 5′ end of the heterogenous miRNAs is the same as the corresponding miRNA species in wild type, suggesting that the size heterogeneity arises from the miRNAs having different 3′ ends. Cloning of specific miRNAs from wild type and hen1 mutants showed that miRNAs in hen1 have additional nucleotides, primarily U’s at their 3′ ends. Perhaps the U tailing of unmethylated miRNAs in hen1 mutants leads to their degradation, which would explain their reduced abundance. The enzyme that adds the additional nucleotides is currently unknown. Intriguingly, it was found that U residues are added to the 5′ fragments of target mRNA after miRNA-mediated cleavage [52].
3. Role of miRNAs in growth and development
Plant miRNAs have a high degree of sequence complementarity to their target mRNAs. This has facilitated the bioinformatic prediction of miRNA target genes [8,29–31,34,53–57], some of which have been subsequently validated experimentally to be true targets of the miRNAs (see below). Plant miRNAs have been predicted or confirmed to regulate genes encoding various types of proteins. A major category of miRNA target genes consists of transcription factors or other regulatory proteins that function in plant development or signal transduction. Below, I summarize the roles of characterized miRNAs and the predicted roles of some miRNAs in plants.
3.1. Auxin signaling
The small molecule auxin is critical for the plant form and for plants’ response to the environment. Recent years have witnessed a tremendous leap in our understanding of the role of auxin in pattern formation (reviewed in [58–62]) and in the mechanisms of auxin signaling [63–66]. The local concentration of auxin, as established by polar auxin transport, appears to pattern the axis of the embryo, establish root stem cells, and control primordia outgrowth from meristems, lateral root formation, and gravitropic responses. Auxin leads to the degradation of a class of transcription repressor proteins known as Aux/IAA proteins through the ubiquitin proteasome pathway. The Aux/IAA proteins heterodimerize with members of the auxin response factor (ARF) family of transcription activators and repressors and inhibit the activities of the activating ARFs. Auxin is bound by TIR1, an F-box protein in the ubiquitin protein ligase SCFTIR1. Auxin promotes the interaction between SCFTIR1 and its substrates, Aux/IAA proteins, to lead to their proteolytic degradation.
Intriguingly, a number of genes in auxin signaling are confirmed or predicted as targets of miRNAs. The TIR1 auxin receptor is a predicted target of miR393 ([34,53–55]). Several ARFs contain potential miRNA binding sites, such as ARF10, ARF16 and ARF17 with miR160 sites [56] and ARF6 and ARF8 with miR167 sites [67]. Products of miR160-guided cleavage of ARF10, ARF16 and 17mRNAs and miR167-guided cleavage of ARF8 mRNA were detected in vivo [28,68]. Expression of a miR160-resistant version of ARF17 (5mARF17) leads to pleiotropic developmental abnormalities, such as leaf serration, leaf curling, early flowering, altered floral morphology, and reduced fertility [68]. This indicates that miR160-mediated regulation of ARF17 is critical for various aspects of plant development. Since the expression of a number of early auxin responsive genes is altered in 5mARF17 plants [68], miR160 likely plays a role in auxin signaling. In addition, ARF3 and ARF4 contain the binding sites of ta-siRNAs from the TAS3 locus [8]. The TAS3 ta-siRNA-guided cleavage of ARF3 and ARF4 mRNAs is observed in vivo [8]. Although the biological function of the ta-siRNA-mediated regulation of ARF3 and ARF4 remains to be determined, the importance of this regulation is reflected by the nucleotide conservation of the ta-siRNA binding sites in ARF3/4 sequences from 16 gymnosperm and angiosperm species [8]. NAC1, which encodes a transcription factor acting downstream of TIR1 to promote lateral root formation, is a target of miR164. miR164 guides the cleavage of NAC1 mRNA in vivo [69]. T-DNA insertions in two of the three members of the miR164 family (mir164a, mir164b) lead to 1/ 4-1/3 wild type levels of total miR164 and cause an increase in NAC1 mRNA levels and a corresponding increase in lateral root number [69].
3.2. Boundary formation/organ separation
Three members of the NAC gene family, CUP SHAPED COTYLEDON (CUC)1, 2, and 3, have partially overlapping functions in organ boundary formation and shoot apical meristem (SAM) initiation. All three genes are expressed in the boundary of the two cotyledons in the embryo and later in the boundaries of floral organs [70–72]. Mutations in two of the three genes together lead to a high frequency of cotyledon fusion on both sides of the cotyledons and doubly mutant plants containing cuc2 and one of the other cuc alleles lack the SAM [70,72]. Although cuc1 cuc2 double mutant seedlings lack the SAM and therefore fail to develop organs postembryonically, shoots can be induced from calli derived from hypocotyls of cuc1 cuc2 seedlings. While the leaves and stems of the shoots appear normal, the flowers have fused sepals and stamens [70], suggesting that these two genes prevent sepal and stamen fusion in addition to their early role during embryogenesis.
CUC1 and CUC2 but not CUC3 are targeted by miR164 [28,55]. Overexpression of miR164 from the 35S promoter in wild type plants leads to floral organ fusion [73,74] and, at a lower frequency, cotyledon fusion [73], phenotypes similar to those of cuc1 cuc2 plants. Expression of miR164-resistant CUC2 can restore sepal separation to miR164 overexpressing lines [73]. Expression of a miR164-resistant form of CUC1 in wild type plants results in reduced sepal number, increased petal number, and broadened leaves [74]. Using an ethanol inducible expression system, the expression of a miR164-resistant form of CUC2 was temporally controlled and the effect on sepal boundaries was monitored following the induction of its expression. It was found that expression of miR164-resistant CUC2 led to an increase in the width of the boundary domain between sepals [73]. An increase in sepal boundary domain width was also observed in miRNA biogenesis mutants such as dcl1, hen1 and hyl1 [73]. The expansion of the sepal boundary domain may explain the narrow sepals in dcl, hen1 and hyl1 mutants and the reduced sepal number in lines expressing miR164-resistant forms of CUC genes.
miR164 is potentially encoded by a three-member gene family. Incredibly, despite potential genetic redundancy, MIR164c (one of the three members of the MIR164 family) was identified in a forward genetic screen as a gene regulating petal number in flowers [75]. The mir164c mutant has extra petals in the early arising flowers. Among the six potential targets of miR164, only CUC1 and CUC2 mRNA levels were increased in the mir164c mutant [75]. Furthermore, cuc1 cuc2 mir164 flowers are indistinguishable from cuc1 cuc2 flowers, suggesting that the increased petal number in mir164c is due to the overexpression of CUC1 or CUC2 [75]. How mis-regulation of CUC1 and CUC2 leads to an increase in petal number (either in the mir164c mutant or in transgenic lines expressing miR164-resistant CUC1 or CUC2) is unknown. It is possible that the increase in petal number is caused by a change in the size of the boundary domain between the first whorl organs (which signals to the second whorl) or in the size of the boundary domain between the first whorl and the third whorl, which is formed before the second whorl is formed.
3.3. Organ polarity
Lateral organs such as leaves and floral organs are initiated as primordia on the flanks of the SAM or floral meristems. The lateral organs are polar structures in that the side of the organ facing the meristem in the primordium (called the adaxial side) differs from the side that faces away from the meristem (called the abaxial side). The difference is reflected by the morphology of the epidermis and the underlying mesophyll on the two sides. The vasculature in the stem and in lateral organs also displays polarity in that the xylem is found on the adaxial side while the phloem is found on the abxial side. Polarity of lateral organs is established through the antagonistic interactions between two major groups of genes, the class III homeodomain leucine zipper (HD-zip) family of genes, PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) and the KANADI family (KAN1, 2, and 3) of genes [76–80]. The HD-zip genes are expressed in the meristem and in the adaxial domain of lateral organs while the KAN genes are expressed in the abaxial domain of lateral organs. Dominant, gain-of-function mutations in PHB, PHV and REV genes result in adaxialized leaves and floral organs, phenotypes similar to those of kan loss-of-function mutants. The triple loss-of-function phb phv rev mutant has abaxialized cotyledons and lacks the SAM, phenotypes similar to those of the transgenic lines overexpressing KAN1 from the 35S promoter. In addition to leaf polarity, these genes also pattern the polarity of the vasculature in a similar manner.
The dominant alleles in PHB, PHV and REV genes contain mutations that affect the amino acid sequences of the proteins, which led to the hypothesis that the mutant proteins have activities that are different from the wild type ones. However, when miR165/166 was cloned it became clear that the mutations in these genes are in the binding sites of miRNA165/ 166 such that they may affect the regulation of these genes by the miRNA rather than affecting the activity of the proteins [30,56]. Indeed, when silent mutations that affect miRNA binding but maintain the protein coding potential were introduced into the genes, similar gain-of-function phenotypes were observed [76,81]. In the gain-of-function phb-d allele, the expression domain of the gene expands into the abaxial region [79], suggesting that the miRNA-mediated regulation contributes to the restriction of PHB expression to the adaxial domain. However, how this occurs is currently unknown. miR165/166 guides the cleavage of PHB, PHV, and REV mRNAs in vivo [81], so an obvious mechanism would be that the cleavage of the HD-zip mRNAs by miR165/166 in the abaxial domain clears the mRNAs from this domain. However, it is also found that miR165/166 causes DNA methylation of the PHB and PHV genes [82]. Therefore, an alternative mechanism is that the miRNA represses the transcription of the genes in the abaxial domain.
miR165/166-mediated regulation of HD-zip genes appears to be evolutionarily conserved. The miR165/166-binding site is highly conserved among angiosperms, gymnosperms, ferns, lycopods, mosses, liverworts, and hornworts [83]. In maize, the semi-dominant mutant Rolled leaf1-Original (Rld1-O) has adaxialized leaves. rld1 encodes a homolog of the Arabidopsis REV and the Rld-O mutation is in the miR165/166-binding site [84]. In Nicotiana sylvestris, a semidominant phv allele isolated from ethyl methanesulfonate mutagenesis is phenotypically similar to the Arabidopsis HD-zip gain-of-function mutants. This allele carries a mutation in the miR165/166-binding site but does not affect the coding potential of the gene [85].
3.4. Floral organ identity and reproductive development
Floral organs are initiated in rings (whorls) from the floral meristem. The identities of the floral organ primordia are specified by the combinatorial activities of three major classes of floral homeotic genes known as the A, B, and C genes [86]. The A and C genes specify the identities of the perianth and reproductive organs, respectively. Loss-of-function mutations in the class C gene AGAMOUS (AG) leads to the replacement of reproductive organs by perianth organs while loss-of-function mutations in the class A gene APETALA2 (AP2) lead to the opposite effect, suggesting that A and C genes act antagonistically to restrict each other to their domains of activity within the floral meristem. In fact, the mRNA of AG is found only in the inner two whorls of the floral meristem that will later become the reproductive organs while the mRNA of APETALA1, a class A gene, is only found in the outer two whorls of the meristem that will later become the perianth organs. However, the transcript of the class A gene AP2 is present throughout the floral meristem.
AP2 contains a binding site for miR172 and is indeed regulated by miR172 in vivo. Overexpression of miR172 from the 35S promoter causes a reduction in the levels of AP2 protein and floral homeotic phenotypes similar to those in ap2 loss-of-function mutants [87,88]. Overexpression of a miR172-resistant form of AP2 cDNA but not wild type AP2 cDNA leads to the replacement of reproductive organs by perianth organs [88]. Expression of the miR172-resistant form of AP2 from the AP2 promoter also results in severe floral patterning defects in the inner two whorls (Zhao L. and Chen X., unpublished results). These observations highlight the importance of miR172 in repressing AP2 in the inner two whorls in floral patterning. Intriguingly, the A/C/ miR172 genetic system in the control of floral patterning is very similar to the HD-zip/KAN/miR165/166 system in the specification of organ polarity. In both cases, two genetically antagonistic functions specify the identities of adjacent domains and a miRNA serves as a negative regulator of one of the two functions.
Although miR172 leads to the cleavage of AP2 mRNA in vivo as deduced from the presence of cleavage products [28,87], RNA cleavage cannot solely explain the regulation of AP2 by miR172. Overexpression of miR172 leads to reduced levels of AP2 protein but not mRNA levels [87–89], suggesting that miR172 resembles animal miRNAs in its mode of action, which has been dubbed translational inhibition [90–92]. miR172 may lead to reduced levels of AP2 protein in the inner two whorls despite the uniform accumulation of AP2 RNA throughout the floral meristem.
Another miRNA, miR159, plays a role in reproductive development by regulating two MYB domain transcription factor genes, MYB33 and MYB65. These two genes act redundantly to prevent the hypertrophy of the tapetum during anther development [93]. miR159 is crucial in restricting the expression of MYB33 and MYB65 to anthers [93]. Transgenic plants containing a miR159-resistant version of MYB33 under its own promoter are arrested for growth at various stages, indicating that the restriction of MTB33 expression by miR159 is critical for plant development [93].
3.5. Developmental transitions
The SAM continues to put out primordia on its flanks during the life cycle of the plant but the types of primordia produced differ at different developmental stages. The SAM generates leaves during the vegetative phase and flowers during the reproductive phase. Even during the vegetative phase, the types of leaves that are put out earlier (juvenile leaves) may differ from the ones made later (adult leaves). Therefore, the state of the SAM, as reflected by the identities of the lateral primordia it produces, changes temporally. Recent findings suggest that small RNAs regulate the transitions between the developmental states of the SAM.
In addition to AP2, miR172 regulates several AP2-like genes, TOE1, TOE2, TOE3, SMZ and SNZ. The loss-of-function toe1-1 mutation leads to slightly early flowering and the toe2-1 mutation does not affect flowering time. The toe1-1 toe2-1 double mutant flowers much earlier than wild type, suggesting that TOE1 and TOE2 are redundant repressors of the vegetative-to-reproductive transition [87]. Consistently, overexpression of TOE1 (toe1-1D) leads to delayed flowering [87]. Overexpression of SMZ or SNZ also causes late flowering, but the role of the two genes in flowering needs to be evaluated from loss-of-function mutant phenotypes [94]. Overexpression of miR172 from the 35S promoter, on the other hand, leads to early flowering [87,88] and overcomes the late flowering phenotpye of toe1-1D [87]. This, together with the fact that miR172-guided cleavage products of TOE1 and TOE2 mRNA can be detected in vivo [28,87], suggests that miR172 regulates the vegetative-to-reproductive transition through the TOE genes. However, overexpression of miR172 does not lead to a decrease in TOE1 mRNA level, suggesting that translational inhibition is the likely mode of regulation of miR172-mediated regulation of TOE1 [87,94]. Overexpression of miR172 does lead to a decrease in TOE2 mRNA level, suggesting that this miRNA regulates different targets with different mechanisms [94].
miR156 is another miRNA that when overexpressed affects flowering time. 35S::MIR156 plants are late flowering [89]. miR156 probably targets a group of transcription factor genes known as squamosa promoter binding protein-like (SPL) [56] but the role of SPL genes in floral transition awaits further investigation. miR319/Jaw overexpression also leads to delayed flowering [95]. miR319/Jaw targets a group of TCP transcription factors for which a role in flowering is currently unknown [95]. Overexpression of miR159 leads to delayed flowering under short day but not long day conditions [96].
Vegetative phase change is pronounced in maize. The juvenile leaves differ from adult leaves in many epidermal characteristics (reviewed in [97]). An AP2-like gene, glossy15, promotes juvenile leaf identity and its RNA is only found in juvenile leaves (reviewed in [97]). glossy15 contains a miR172-binding site and miR172-guided cleavage products can be detected in vivo, suggesting that glossy15 is a target of miR172 [98]. The onset of miR172 expression in maize appears to correlate with the specification of adult leaf characteristics. Since the expression of glossy15 at the RNA level mirrors that of miR172, it is likely that miR172 clears glossy15 mRNA in adult leaves to promote the vegetative phase change [98].
Although not as pronounced, vegetative phase change does occur in Arabidopsis. The Poethig lab used a forward genetic approach to identify genes playing a role in vegetative phase change in Arabidopsis. Intriguingly, among the genes identified to play a role in vegetative phase change, many play a role in small RNA metabolism. These include HST, (which may export miRNAs to the cytoplasm), AGO7, (which may be a RISC component), and RDR6 and SGS3, genes previously known to be required for transgene silencing [6,41,99–101]. RDR6 and SGS3 are also required for the biogenesis of trans-acting siRNAs [6], whose production is triggered by miRNAs [8]. These studies suggest that miRNAs and ta-siRNAs play a role in phase transitions.
3.6. Leaf growth
The snapdragon CINCINNATA (CIN) gene is required for the differential regulation of cell division during leaf morphogenesis to make a flat leaf [102]. cin mutants have crinkly leaves. CIN belongs to the TCP family of transcription factors. In Arabidopsis, overexpression of miR319/Jaw leads to crinkly leaves and reduced levels of five TCP genes containing miR319/Jaw-binding sites [95]. Overexpression of a miR319/ Jaw-resistant form of TCP2 fully restores the crinkly leaf phenotype of miR319/Jaw overexpression [95].
3.7. Small RNA metabolism
At least three miRNAs are known to target genes involved in small RNA metabolism or function. DCL1 contains a binding site for miR162 and miR162-guided cleavage products of DCL1 mRNA are detected in vivo [103]. Consistent with a role of miR162 in reducing DCL1 mRNA levels, DCL1 mRNA abundance is elevated in mutants defective in miRNA biogenesis (such as dcl1 or hen1) and in transgenic lines expressing the viral RNA silencing suppressor HC-Pro, which appears to inhibit RISC assembly [35,103]. The AGO1 gene that codes for a RISC component is targeted by miR168. Overexpression of a miR168-resistant version of AGO1 appears to affect miRNA function because miRNA targets overaccumulate and the plants show phenotypes similar to those of miRNA biogenesis mutants such as dcl1, hen1 and hyl1 [104]. AGO2, another argonaute gene, contains a binding site for miR403 in its 3′ UTR and in vivo cleavage products have been detected [8]. However, the role of AGO2 in small RNA biology is currently unknown. The regulation of genes involved in small RNA metabolism or function by miRNAs is perhaps a feedback mechanism to ensure a certain level of activity of small RNA pathways.
3.8. Predicted roles of other miRNAs
While the transcription factor-encoding miRNA targets are the best characterized so far, miRNA targets encoding proteins with various other cellular functions, such as protein degradation, metabolism and stress responses, have been predicted and some have been validated using the criteria that in vivo cleavage products be detectable [8,31,34,53–55,57]. It is therefore expected that miRNAs regulate a variety of cellular processes.
4. Regulation of the miRNA genes
The tissue- or cell-type-specific functions of the miRNAs discussed above suggest that miRNA gene expression is precisely regulated in the plant. It is conceivable that the abundance of a certain miRNA in cells can be regulated at multiple levels, such as the transcription of the gene, the processing of the primiRNA by DCL1, the methylation of the miRNA by HEN1, the loading of the miRNA into RISC, and the export of the miRNA into the cytoplasm, and even the potential transport of the miRNA in or out of cells or tissues [105]. Although how miRNAs themselves are regulated is currently unknown, available data show that miRNA accumulation exhibits tissue or cell type specificity and responses to stimuli. By in situ hybridization, it was shown that miR172 is present throughout the early floral primordia but not in the SAM from which the floral primordia arise [88] and miR165/166 is restricted to the abaxial side of leaves in Arabidopsis and maize [84,106]. By RNA filter hybridization, it was shown that miR159 levels are induced by GA [96] and miR164 levels respond to auxin [69]. The promoter of miR164c endows dynamic expression to the GUS reporter gene during plant development [75], suggesting that the transcription of miRNA genes are regulated. A study in which the promoter activity of MIR171 and the activity of miR171 were both monitored showed that the promoter is highly cell- or tissue-specific and that transcription of MIR171 is largely responsible for the cell- or tissue-specific activities of the miRNA [107]. One future challenge of miRNA biology is to understand how miRNAs themselves are regulated.
References
- 1.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 12.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 17.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 18.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 21.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 24.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen SE, Running M, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 26.Ray A, Lang JD, Golden T, Ray S. SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development. 1996;122:2631–2638. doi: 10.1242/dev.122.9.2631. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz BW, Yeung EC, Meinke DW. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development. 1994;120:3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- 28.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 29.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 33.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Gen Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 38.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Papp I, et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–1390. doi: 10.1104/pp.103.021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- 42.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis doublestranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 44.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 45.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 46.Boutet S, et al. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 50.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′ end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 53.Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V, Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Gen Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 56.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 57.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleming AJ. Formation of primordia and phyllotaxy. Curr Opin Plant Biol. 2005;8:53–58. doi: 10.1016/j.pbi.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Friml J. Auxin transport – shaping the plant. Curr Opin Plant Biol. 2003;6:7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- 60.Kepinski S, Leyser O. Plant development: auxin in loops. Curr Biol. 2005;15:R208–R210. doi: 10.1016/j.cub.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 61.Leyser O. Regulation of shoot branching by auxin. Trends Plant Sci. 2003;8:541–545. doi: 10.1016/j.tplants.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 62.Weijers D, Jurgens G. Auxin and embryo axis formation: the ends in sight. Curr Opin Plant Biol. 2005;8:32–37. doi: 10.1016/j.pbi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 64.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 65.Leyser O. Molecular genetics of auxin signaling. Annu Rev Plant Biol. 2002;53:377–398. doi: 10.1146/annurev.arplant.53.100301.135227. [DOI] [PubMed] [Google Scholar]
- 66.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot (Lond) 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bartel B, Bartel DP. MicroRNAs: at the root of plant development. Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo HS, Xie Q, Fei JF, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- 72.Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 74.Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 75.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 76.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 77.Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- 78.Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 79.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 80.Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- 81.Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bao N, Lye KW, Barton MK. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell. 2004;7:653–662. doi: 10.1016/j.devcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 83.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 84.Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 85.McHale NA, Koning RE. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell. 2004;16:1730–1740. doi: 10.1105/tpc.021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jack T. Molecular and genetic mechanisms of floral control. Plant Cell. 2004;16(Suppl):S1–S17. doi: 10.1105/tpc.017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 90.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 91.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 92.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 93.Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 95.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 96.Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- 97.Kerstetter RA, Poethig RS. The specification of leaf identity during shoot development. Annu Rev Cell Dev Biol. 1998;14:373–398. doi: 10.1146/annurev.cellbio.14.1.373. [DOI] [PubMed] [Google Scholar]
- 98.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA. 2005 doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 100.Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 101.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 102.Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 103.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by micro-RNA-guided mRNA degradation. Curr Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 104.Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- 107.Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 109.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. PNAS. 2005 doi: 10.1073/pnas.0506426102. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005 doi: 10.1101/gad.1352605. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]