Abstract
Long-range projections from the frontal cortex are known to modulate sensory processing in multiple modalities. Although the mouse has become an increasingly important animal model for studying the circuit basis of behavior, the functional organization of its frontal cortical long-range connectivity remains poorly characterized. Here we used virus-assisted circuit mapping to identify the brain networks for top-down modulation of visual, somatosensory, and auditory processing. The visual cortex is reciprocally connected to the anterior cingulate area, whereas the somatosensory and auditory cortices are connected to the primary and secondary motor cortices. Anterograde and retrograde tracing identified the cortical and subcortical structures belonging to each network. Furthermore, using novel viral techniques to target subpopulations of frontal neurons projecting to the visual cortex versus the superior colliculus, we identified two distinct subnetworks within the visual network. These findings provide an anatomical foundation for understanding the brain mechanisms underlying top-down control of behavior.
Long-range projections from the frontal cortex to sensory areas can powerfully modulate sensory processing, which may underlie sensorimotor integration and top-down attentional modulation. In mouse somatosensory cortex, active touch of an object by whiskers evokes large calcium signals in layer 5 pyramidal neuron dendrites, which depend on inputs from the vibrissa motor cortex1. The projection from the vibrissa motor cortex also disinhibits pyramidal neurons by activating vasoactive intestinal peptide-positive (VIP+) interneurons in the somatosensory cortex2. In the auditory cortex, inputs from the motor cortex suppress the auditory responses, through feedforward inhibition mediated by parvalbumin-positive interneurons3,4. In the visual cortex, activating the projection from the cingulate cortex can strongly enhance visual responses, which also depends on disinhibition mediated by local VIP+ neurons5. Thus, top-down modulation of sensory responses by projections from frontal cortical areas is widespread across sensory modalities.
To understand how top-down modulation is implemented during behavior, an important step is to delineate the brain networks organized by long-range axonal projections to and from the frontal cortex. Although frontal projections to individual sensory cortical areas have been studied extensively, the brain networks encompassing both cortical and subcortical structures have not been mapped systematically and compared across modalities. Furthermore, for each modality, the corresponding frontal area projects to multiple targets in addition to the sensory cortex3,6–13. While in some cases similar signals are broadcast to multiple downstream targets through axon collaterals, in other cases different signals are transmitted by subgroups of neurons projecting to distinct postsynaptic targets. Examining the relationship between different output pathways is crucial for understanding how the frontal cortex coordinates activity of multiple brain areas to optimize behavior. In addition to divergent outputs, each frontal region also receives convergent inputs from various sources6,7,10,13–15. Mapping these inputs is essential for understanding the neural mechanisms that regulate the frontal cortical activity and shape the top-down signals.
In this study, we used several virus-assisted circuit mapping techniques to characterize the long-range inputs and outputs of the frontal cortical regions connected to the visual, somatosensory, and auditory cortices of the mouse. We found largely separate brain networks for visual vs. somatosensory/auditory modalities. Furthermore, within the visual network, we delineated two distinct subnetworks for top-down control by mapping the inputs and outputs of two subsets of frontal neurons that project to the visual cortex vs. the superior colliculus.
RESULTS
To label the long-range inputs and outputs of each cortical area with fluorescent proteins, we injected various viral vectors into the mouse brain, as detailed below. After histological sectioning and fluorescence imaging, each brain sample was aligned to the Allen Mouse Brain Atlas to facilitate 3D whole-brain visualization and quantitative analyses (Fig. 1, also see Methods). The labeled neurons and axons were detected, and their locations were registered in the reference atlas (Table 1 and Supplementary Table 1). To facilitate data visualization at multiple levels of detail, we also used interactive sunburst diagrams (adapted from Allen Mouse Brain Atlas, http://www.brain-map.org/api/examples/examples/sunburst/) to represent the distribution of labeled inputs and outputs in all brain structures (http://top-down-network.org/). The brain structures are arranged hierarchically from inner to outer circles in the diagram, and the size of each sector represents the percentage of labeling in the corresponding structure. The numerical values can be read out by pointing the cursor, and each region of interest can be expanded with a mouse click.
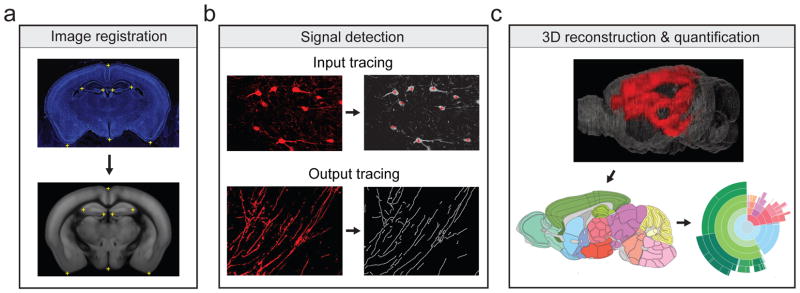
Figure 1.
Steps for data processing. (a) Mapping of raw image (upper panel) onto corresponding coronal section of Allen Mouse Brain Atlas (lower panel). The registration module applied several geometric transformations (translation, rotation and scaling) of the raw image to optimize the match of reference points (yellow crosses) between raw image and atlas. (b) Fluorescence signals (right) detected from raw image (left). Retrogradely labeled neurons were identified manually (red crosses). Anterogradely labeled axons were detected using a ridge detection method (white lines, see Methods). (c) Detected signals were projected to Allen Mouse Brain Atlas and quantified as both the percentage and density of labeling in each brain region (Supplementary table 1).
Table 1.
Abbreviations of Anatomical Structures
| Abbreviation | Definition |
|---|---|
| Cortical areas | |
| ACA | anterior cingulate area |
| AI | agranular insular area |
| AUD | auditory areas |
| ECT | ectorhinal area |
| ILA | infralimbic area |
| MO | somatomotor areas (MOs + MOp) |
| MOp | primary motor area |
| MOs | secondary motor area |
| ORB | orbital area |
| PL | prelimbic area |
| PTLp | posterior parietal association areas |
| RSP | retrosplenial area |
| SS | somatosensory areas |
| TEa | temporal association area |
| VIS | visual areas |
| Thalamic nuclei | |
| AD | anterodorsal nucleus |
| AM | anteromedial nucleus |
| AV | anteroventral nucleus of thalamus |
| LD | lateral dorsal nucleus of thalamus |
| LGd | dorsal part of the lateral geniculate complex |
| LGv | ventral part of the lateral geniculate complex |
| LP | lateral posterior nucleus of the thalamus |
| MD | mediodorsal nucleus of thalamus |
| MG | medial geniculate complex |
| RT | reticular nucleus of the thalamus |
| PF | parafascicular nucleus |
| PO | posterior complex of the thalamus |
| VAL | ventral anterior-lateral complex of the thalamus |
| VM | ventral medial nucleus of the thalamus |
| VP | ventral posterior complex of the thalamus |
| Other structures | |
| PAL | pallidum |
| SC | superior colliculus |
| STR | striatum |
Identification of frontal regions for each sensory modality
To identify the frontal regions directly innervating each sensory cortical area, we used rabies virus (RV)-mediated transsynaptic retrograde tracing, which labels monosynaptic inputs to selected starter cells with high specificity16,17. Avian-specific retroviral receptor (TVA), enhanced green fluorescent protein (EGFP), and rabies glycoprotein (RG) were expressed by injecting two Cre-inducible adeno-associated virus (AAV) vectors (AAV2-EF1α-DIO-TVA-EGFP and AAV2-CAG-DIO-RG) into the primary visual (VIS), somatosensory (SS), or auditory (AUD) cortex of CaMKIIα-Cre mice (Fig. 2a). Two weeks later, we injected a modified RV expressing tdTomato (RV-ΔG-tdTomato+EnvA), which only infects cells expressing TVA and requires RG to spread retrogradely to presynaptic cells.
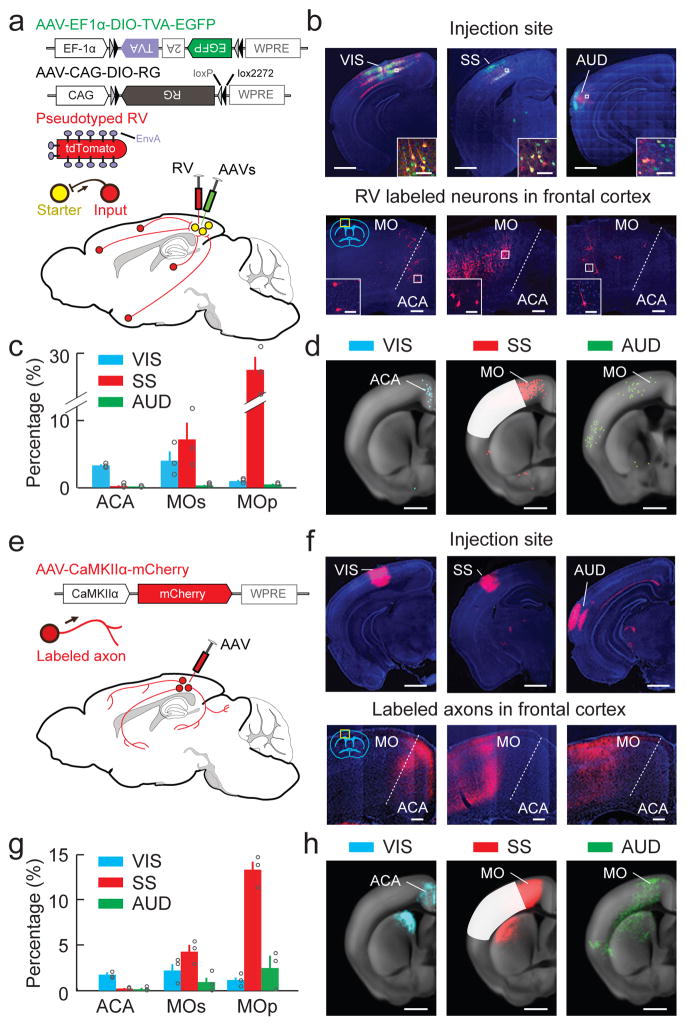
Figure 2.
Mapping connections between sensory and frontal cortices. (a) Viral vectors and injection procedure for RV-mediated transsynaptic retrograde tracing from sensory cortices. (b) Upper panel, injection sites in VIS, SS and AUD (scale bar, 1 mm). Inset, enlarged view of region in white box; Starter cells, yellow; scale bar, 40 μm. Lower panel, fluorescence images of ACA and MO (yellow box in coronal diagram) showing RV-labeled input neurons (red) to each sensory area (scale bar, 200 μm). Inset, enlarged view of region in white box (scale bar, 40 μm). Green, EGFP; red, tdTomato; blue, DAPI. (c) Percentages of input neurons in ACA, MOs, MOp retrogradely labeled from VIS (blue, n = 3 mice), SS (red, n = 3), AUD (green, n = 3). Each circle represents one mouse. Error bar, ±s.e.m. (d) Summary of RV-labeled neurons in all samples of each group (scale bar, 1 mm). Dots, detected neurons. White masks, injection sites excluded from analysis. (e) Viral vector and injection procedure for tracing the axonal projections from each sensory area. (f) Upper panel, injection sites in VIS, SS, AUD (scale bar, 1 mm). Lower panel, fluorescence images of ACA and MO (yellow box in coronal diagram) showing axons from each sensory area (scale bar, 200 μm). Red, mCherry; blue, DAPI. (g) Percentages of labeled axons in ACA, MOs, MOp from VIS (n = 3), SS (n = 3), AUD (n = 3). Each circle represents one mouse. Error bar, ±s.e.m. (h) Summary of axons detected in all samples of each group (scale bar, 1 mm). White masks, injection sites excluded from analysis.
Figure 2b (upper panel) shows examples of starter cells (expressing both tdTomato and EGFP) in each injected sensory area. Across brain samples, the starter cells were distributed over large portions of VIS, SS and AUD (Supplementary Fig. 1), with similar laminar distributions among these areas (Supplementary Fig. 2). Transsynaptically labeled presynaptic neurons (expressing tdTomato only) were found in multiple cortical and subcortical regions (Supplementary Fig. 3, Supplementary Movie 1, Supplementary Table 1, and interactive sunburst diagram at http://top-down-network.org/). Within the frontal cortex, neurons transsynaptically labeled from VIS were found primarily in the anterior cingulate area (ACA) and the medial portion of secondary motor cortex (MOs), whereas those presynaptic to SS and AUD were mainly located in the primary motor cortex (MOp) and the lateral portion of MOs (Fig. 2b, lower panel, c,d and Supplementary Fig. 4). In all three modalities, inputs from the frontal cortex arose primarily from layers 2/3 and 5, consistent with previous studies3,5,8.
We next mapped the brain regions receiving direct axonal projections from each sensory area by injecting AAV expressing mCherry (AAV2-CaMKIIα-mCherry) into VIS, SS, or AUD of wild-type mice (Fig. 2e–h, Supplementary Fig. 5, Supplementary Movie 2, Supplementary Table 1 and sunburst diagram at http://top-down-network.org/). Within the frontal cortex, labeled axons from VIS were found mostly in ACA and medial MOs18, and those from SS or AUD were distributed primarily in MOp and lateral MOs (Fig. 2f, lower panel, g,h). The spatial correspondence between the axonal projections of each sensory area (Fig. 2g,h) and its retrogradely labeled presynaptic neurons (Fig. 2c,d) indicates strong reciprocity of long-range corticocortical connections10. These anterograde and retrograde tracing experiments also indicate a clear segregation between the frontal areas connected to the visual (ACA and medial MOs) and the somatosensory and auditory cortices (MOp and lateral MOs).
Other long-range connections of ACA and MO
In addition to sensory cortices, the frontal areas also project to multiple other brain structures. To label these projections, we injected AAV expressing mCherry (AAV2-CaMKIIα-mCherry) into ACA or MO (Fig. 3a–d, Supplementary Fig. 6, Supplementary Movie 3 and Supplementary Table 1). Among the sensory cortical areas, ACA projects extensively to VIS and sparsely to SS and AUD, whereas MO projects extensively to SS, moderately to AUD and VIS, consistent with retrograde tracing from these sensory areas (Fig. 2b–d). Among other cortical areas, ACA projects extensively, and MO moderately, to the posterior parietal cortex (PTLp) and the retrosplenial area (RSP) (Fig. 3c1–c3). While PTLp makes reciprocal connections with VIS, SS, and AUD, RSP is densely connected only with VIS (Supplementary Fig. 3,5), suggesting that it belongs mainly to the visual network13. Within the frontal cortex, the prelimbic/infralimbic area (PL/ILA) receives more input from ACA, and the orbital area (ORB) receives more input from MO (Fig. 3c6).
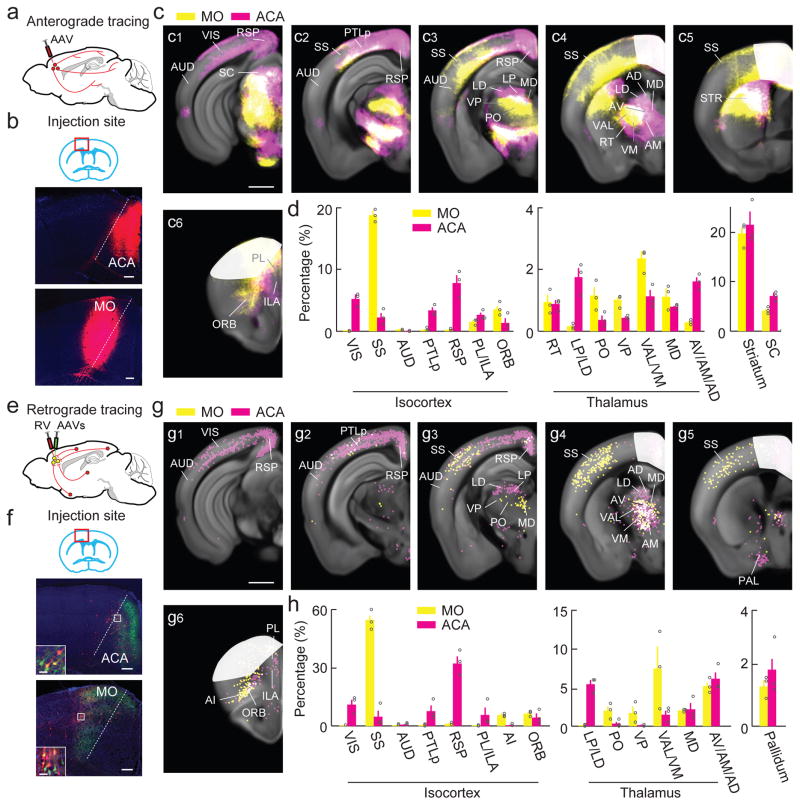
Figure 3.
Whole-brain distributions of axonal projections and input neurons of ACA and MO. (a) Injection procedure for tracing projections. (b) Fluorescence images of ACA and MO (red box in coronal diagram) at injection sites (scale, 200 μm). Red, mCherry; blue, DAPI. (c) Axons detected in all samples of each group (MO, yellow; ACA, magenta; Overlap, white. Scale, 1 mm). White masks, injection sites excluded from analysis. (d) Percentages of labeled axons in cortical and subcortical structures (MO, n = 3; ACA, n = 3). Included are cortical areas with >1% labeling and thalamic structures with >0.8% labeling. Error bar, ±s.e.m. (e) Injection procedure for RV-mediated retrograde tracing. (f) Fluorescence images of ACA and MO (red box in coronal diagram) at injection sites (scale, 200 μm). Inset, enlarged view of region in white box showing starter cells (yellow; scale, 10 μm). Green, EGFP; red, tdTomato; blue, DAPI. (g) RV-labeled neurons detected in all samples of each group (MO, yellow; ACA, magenta. Scale, 1 mm). White masks, injection sites excluded from analysis. (h) Percentages of retrogradely labeled neurons in selected cortical and subcortical regions (MO, n = 3; ACA, n = 3). Included are areas with >2% (cortical) or >1% (thalamic) labeling. Error bar, ±s.e.m.
In the thalamus, the projections from ACA and MO were segregated mainly along the dorsal-ventral axis. For example, ACA projects more to the lateral posterior/lateral dorsal (LP/LD) thalamic nuclei, whereas MO projects more to posterior (PO), ventral posterior (VP), and ventral anterior-lateral/ventral medial (VAL/VM) complex10. Both ACA and MO project to the mediodorsal (MD) nucleus, which is densely connected to the prefrontal cortex19. Other major subcortical targets of the frontal regions include the striatum (STR) and superior colliculus (SC, Fig. 3d), with partially overlapping projections from ACA and MO (Fig. 3c1,c5).
To map the inputs to these frontal areas, we injected the AAV and RV vectors for transsynaptic retrograde tracing (same as in Fig. 2a) into ACA or MO of CaMKIIα-Cre mice (Fig. 3e–h, Supplementary Fig. 6, Supplementary Movie 4 and Supplementary Table 1). Among the sensory cortices, MO injection led to dense labeling in SS but little labeling in VIS and AUD, whereas ACA injection caused the densest labeling in VIS (Fig. 3h), consistent with anterograde tracing (Fig. 2f–g). Among other cortical areas, we found extensive inputs from PTLp and RSP to ACA but only sparse inputs to MO (Fig. 3g,h), further attesting to their membership in the visual network (Fig. 4). Within the frontal cortex, PL/ILA provides more input to ACA than to MO, and ORB provides similar inputs to both regions. The striking similarity between the cortical distributions of inputs to (Fig. 3g,h) and projections from (Fig. 3c,d) each frontal region again demonstrates the reciprocity of corticocortical connections10.
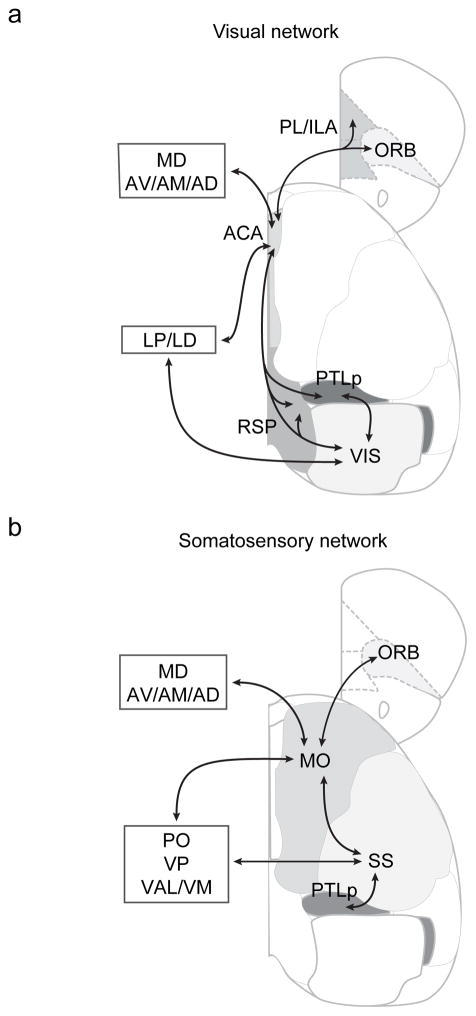
Figure 4.
Schematic diagram of visual and somatosensory networks. Shown are major connections in each network.
In the thalamus, the distributions of inputs to ACA and MO also largely mirrored the distributions of their axonal projections (Fig. 3c,d). ACA receives more inputs from LP/LD, whereas MO receives more inputs from PO, VP, and VAL/VM (Fig. 3g,h). On the other hand, MD projects to both ACA and MO, as expected for these prefrontal cortical areas19. Since LP/LD is reciprocally connected to both ACA and VIS (Fig. 3c,d,g,h and Supplementary Fig. 3,5), it forms an integral part of the visual network (Fig. 4a); PO, VP, and VAL/VM are connected to both MO and SS, suggesting that they are part of the somatosensory network (Fig. 4b). Among other subcortical regions, the pallidum projects to both ACA and MO (Fig. 3g5,h), consistent with the known cholinergic and non-cholinergic projections from the basal forebrain to the entire cortex10,20.
Besides excitatory neurons, several subtypes of inhibitory interneurons have been implicated in long-range corticocortical interactions2–5. To determine whether the long-range axonal projections directly innervate these interneuron subtypes in each cortical area, we injected the AAV and RV vectors for transsynaptic tracing into VIS, RSP, PTLp and ACA of PV-, SOM- and VIP-Cre mice21,22 (Supplementary Table 1 and sunburst diagram at http://top-down-network.org/). Similar to excitatory neurons, all three subtypes of inhibitory neurons in VIS, PTLp and RSP receive monosynaptic inputs from ACA (Supplementary Fig. 7 and Fig. 2b–d), and all of them in ACA receive monosynaptic inputs from VIS, PTLp and RSP (Supplementary Fig. 8 and Fig. 3f–h). Thus, both top-down and bottom-up corticocortical projections directly recruit inhibitory interneurons in their target areas.
Outputs of VIS- and SC-projecting ACA neurons
Different projections from each cortical area in some cases originate from different subsets of neurons23,24, but in other cases may reflect axon collaterals of the same neurons. Distinguishing these possibilities is crucial for understanding how the frontal cortex coordinates the activity of different brain areas for top-down executive control, but this issue has not been addressed systematically in previous efforts mapping mesoscale mouse brain connectivity. In particular, we wondered what other brain regions are also innervated by the frontal cortical neurons projecting to the sensory cortex.
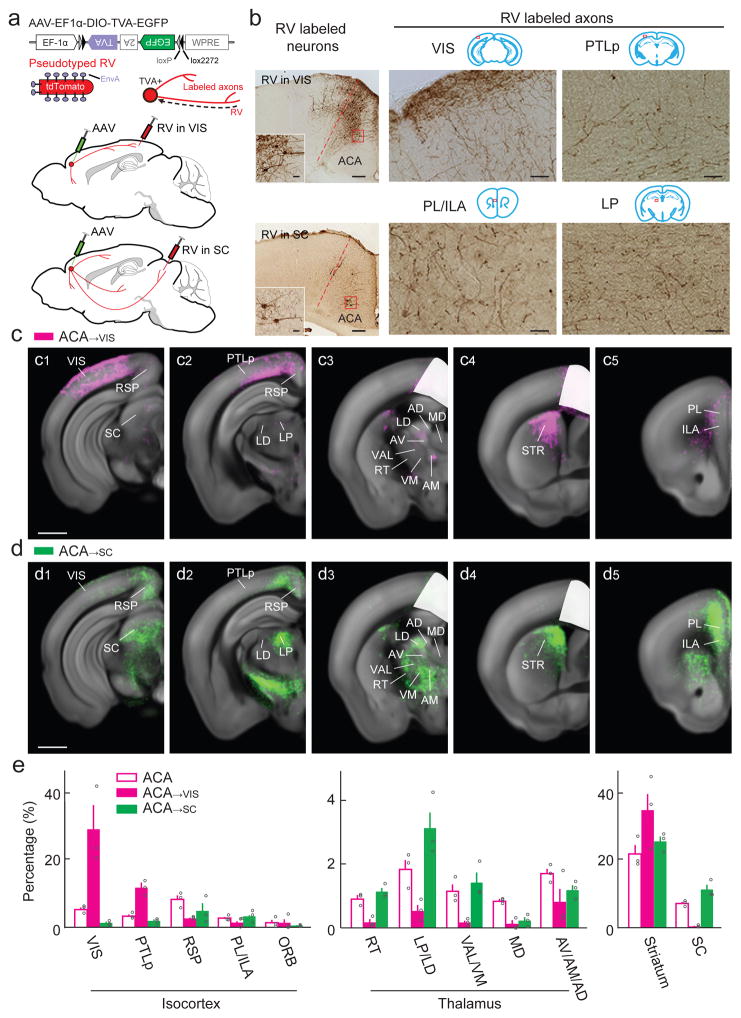
We focused this analysis on the ACA neurons projecting to VIS (ACA→VIS neurons). To label these neurons and their axons, we injected the AAV vector expressing TVA (AAV2-EF1α-DIO-TVA-EGFP) into ACA of CaMKIIα-Cre mice, but the RV vector (RV-ΔG-tdTomato+EnvA) into VIS two weeks after AAV injection (Fig. 5a, upper panel). This allowed RV to enter the TVA-expressing ACA axon terminals in VIS, be transported retrogradely to the ACA neurons, and label all their axon collaterals with tdTomato (Fig. 5b, upper row). To enhance the visibility of labeled thin axons, we performed immunostaining for tdTomato to convert the fluorescence signal into nickel-enhanced diaminobenzidine (DAB) signal.
Figure 5.
Whole-brain distributions of axonal projections from ACA→VIS and ACA→SC neurons. (a) Viral vectors and injection procedure. (b) Upper panel: left, bright field image of ACA and MO showing RV-labeled neurons from VIS (scale, 200 μm). Inset, enlarged view of region in red box (scale, 20 μm). Immunostaining for tdTomato was performed to convert fluorescence signal (tdTomato expressed by RV) into nickel-enhanced DAB signal; right, images of VIS and PTLp (red box in coronal diagram), showing RV-labeled axons of ACA→VIS neurons (scale, 100 μm). Lower panel, similar to upper panel, but the ACA neurons were labeled by RV injection into SC (left), and axons from ACA→SC neurons are concentrated in PL/ILA and LP (right). (c) Axons detected in all ACA→VIS samples (scale, 1 mm). White masks, injection sites excluded from analysis. (d) Similar to (c), for axons of ACA→SC neurons (scale, 1 mm). (e) Percentages of labeled axons in selected cortical and subcortical structures (ACA, n = 3; ACA→VIS, n = 3; ACA→SC, n = 3). Error bar, ±s.e.m. Data for ACA axons are the same as in Fig. 2d, shown here for comparison.
In addition to the dense projection to VIS, we found that the ACA→VIS neurons also project extensively to PTLp and moderately to RSP (Fig. 5c1,c2,e). This suggests that similar modulatory signals are broadcast to VIS, PTLp and RSP, pointing to a tightly coordinated ACA – PTLp – RSP – VIS network for visual processing25 (Fig. 7, Supplementary Movie 5 and Supplementary Table 1). In contrast, the projection from the ACA→VIS neurons to PL/ILA was much sparser than that from the entire ACA population (Fig. 5c5). The thalamus also receives very little inputs from ACA→VIS neurons (Fig. 5c2,c3,e), suggesting that it is not strongly and directly influenced by the modulatory signals sent to VIS. Furthermore, while the SC receives a sizable projection from ACA (Fig. 3d), we found few labeled axons from the ACA→VIS neurons, suggesting that the SC projection originates from a separate ACA neuron population. This is consistent with the finding based on retrograde tracing from VIS and SC in primates26. Targeting the MO→SS neurons using the same technique revealed a similar level of selective axonal projections (Supplementary Fig. 9).
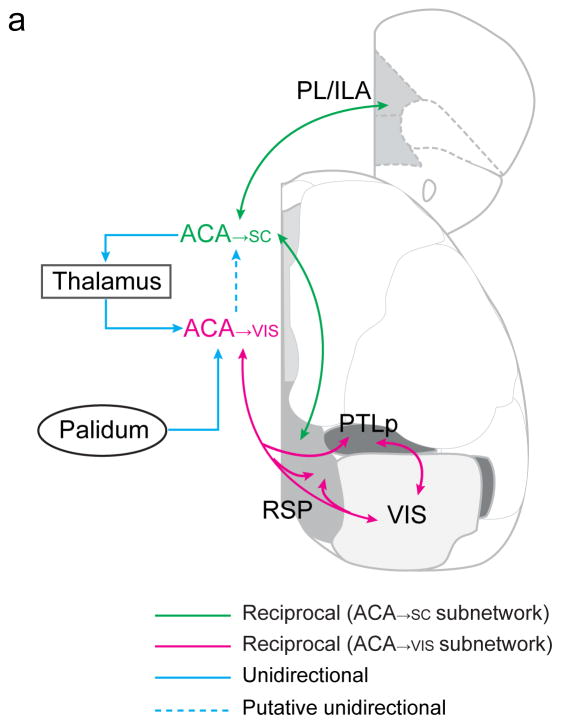
Figure 7.
Schematic diagram of visual subnetworks. Shown are major inputs and outputs of ACA→VIS and ACA→SC neurons. The putative unidirectional connection from ACA→VIS to ACA→SC neurons (dashed blue line) was based on previous literature.
The SC is also known to be important in top-down attentional modulation27–31. We thus examined the outputs of the SC-projecting ACA (ACA→SC) neurons by injecting the AAV vector expressing TVA into ACA and RV vector into SC (Fig. 5a, lower panel). Unlike the ACA→VIS neurons, which were distributed in both layers 2/3 and 5 (Fig. 5b, upper panel), the ACA→SC neurons were found primarily in layer 5 (Fig. 5b, lower panel)5. These spatial distributions are similar to those of intratelencephalic and pyramidal tract neurons in the motor cortex, which form non-overlapping populations of projection neurons with distinct roles in motor control23,24,32. We found little projection from the ACA→SC neurons to VIS (Fig. 5d1,e). In addition, the ACA→VIS but not ACA→SC neurons project to the contralateral ACA through the corpus callosum (Supplementary Movie 5), further supporting the correspondence between ACA→VIS/ACA→SC and intratelencephalic/pyramidal tract neurons. Compared to the ACA→VIS neurons, the ACA→SC neurons project much less to PTLp but more to PL/ILA (Figures 5d2,d5,e). Thus, the two subpopulations of ACA neurons show distinct cortical projection patterns.
For the thalamic nuclei innervated by ACA, the inputs from ACA→SC neurons were much more extensive than those from ACA→VIS neurons (Fig. 5c–e). In contrast, the striatum receives extensive projections from both ACA→VIS and ACA→SC neurons, and their spatial distributions largely overlap. This is reminiscent of the striatal projections from both intratelencephalic and pyramidal tract neurons in the ipsilateral cortex23,24,32.
To assess whether ACA→VIS and ACA→SC neurons form synapses in the identified areas, we expressed membrane-bound GFP (mGFP, for labeling axons) and synaptophysin-mRuby (SYP-mRuby, for labeling putative presynaptic sites) in these neurons33,34. CAV-FLExloxP-Flp was injected into VIS or SC and AAV-hSyn1-FLExFRT-mGFP-2A-synaptophysin-mRuby was injected into ACA of CaMK2α-Cre mice for Flp-dependent expression of mGFP and SYP-mRuby (Supplementary fig. 10). We found synaptophysin-mRuby labeling in all the major cortical and subcortical areas identified above (Fig. 5e), indicating synaptic innervation of those areas.
Inputs to VIS- and SC-Projecting ACA Neurons
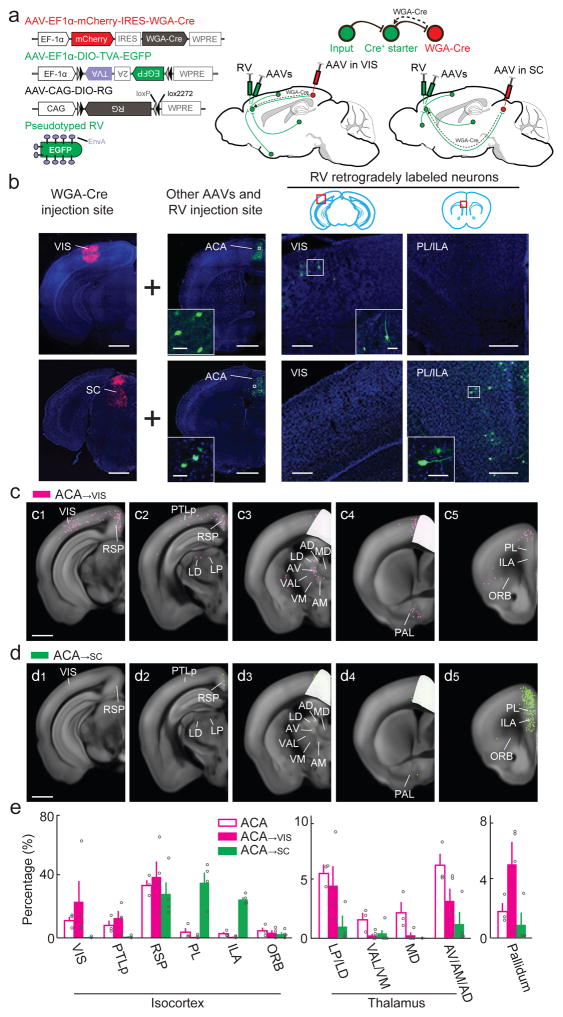
The top-down signals from ACA→VIS and ACA→SC neurons to their distinct postsynaptic targets are determined by their respective inputs. To map the monosynaptic inputs to each subpopulation, we injected AAV expressing the trans-cellular tracer protein wheat germ agglutinin (WGA) fused with Cre recombinase (AAV2-EF1α-mCherry-IRES-WGA-Cre) into VIS or SC of wild-type mice, and AAV vectors with Cre-inducible expression of TVA and RG (same as in Fig. 2a) into their ACA. Four weeks after these AAV injections, RV expressing EGFP was injected into ACA (Fig. 6a). This viral strategy ensured that TVA and RG were expressed only in the ACA→VIS or ACA→SC neurons retrogradely labeled with Cre recombinase35, thus restricting RV labeling to their presynaptic inputs (Supplementary Fig. 11–14).
Figure 6.
Whole-brain distributions of inputs to ACA→VIS and ACA→SC neurons. (a) Viral vectors and injection procedure. (b) Left, fluorescence images of AAV-mCherry-IRES-WGA-Cre injection site in VIS (upper) or SC (lower) (scale, 1 mm). Middle, injection site of other AAVs and RV in ACA (scale, 1 mm). Inset, enlarged view of region in white box showing AAV/RV infected neurons (green; scale, 20 μm). Right, retrogradely labeled neurons (green) in VIS and PL/ILA (scale, 200 μm). Inset, enlarged view of region in white box (scale, 20 μm). Green, EGFP; red, mCherry; blue, DAPI. (c) RV-labeled neurons detected in all ACA→VIS samples (scale, 1 mm). White masks, injection sites excluded from analysis. (d) Similar to (c), for inputs to ACA→SC neurons (scale, 1 mm). (e) Percentages of retrogradely labeled neurons in selected cortical and subcortical brain structures (ACA, n = 3 mice; ACA→VIS, n = 4; ACA→SC, n = 4). Error bar, ±s.e.m. Data for ACA inputs are the same as in Fig. 2h.
We found that ACA→VIS but not ACA→SC neurons receive extensive monosynaptic inputs from VIS (Fig. 6b–e). Such selective innervation of sensory cortex-projecting neurons was also found in the somatosensory → motor cortex connection (Supplementary Fig. 15)8, resulting in a recurrent loop between the sensory cortex and a subset of frontal cortical neurons. Among other cortical areas, inputs from PL and ILA were much more extensive for ACA→SC neurons, whereas those from PTLp were much more extensive for ACA→VIS neurons (Fig. 6c2,c5,d2,d5,e), matching the distributions of axonal projections of the two ACA subpopulations (Fig. 5c2,c5,d2,d5,e). Together, these findings suggest that the ACA→VIS neurons have enhanced reciprocal connections with the posterior sensory and association areas, whereas the ACA→SC neurons are preferentially connected to the medial prefrontal cortex (Fig. 7, Supplementary Movie 6 and Supplementary Table 1). Thus, within the visual network, there are two subnetworks involving separate populations of ACA neurons.
From the thalamus, the ACA→VIS neurons receive more inputs than ACA→SC neurons (Fig. 6e), opposite to the relative strengths of their projections to the thalamus (Fig. 5e). Thus the corticothalamic connections are much less reciprocal than the corticocortical connections (Fig. 7). Finally, we found inputs from the pallidum to ACA→VIS neurons but not to ACA→SC neurons.
DISCUSSION
Using a variety of virus-assisted circuit tracing techniques, we have mapped the long-range inputs and outputs of the frontal cortical regions that are directly connected to the visual, somatosensory, and auditory cortices. Both anterograde and retrograde tracing from the sensory areas indicate a clear spatial segregation between the frontal neurons connected to the visual cortex (ACA) vs. somatosensory and auditory cortices (MO) (Fig. 2). Anterograde and retrograde tracing from ACA and MO allowed us to delineate separate brain networks associated with different sensory modalities (Figs. 3,4). Furthermore, within the visual network, we identified two distinct subnetworks, involving subpopulations of ACA neurons that project to the visual cortex vs. the superior colliculus (Figs. 5–7).
Our anterograde tracing approach is similar to that used for generating the Allen Mouse Brain Connectivity Atlas10, but we focused on the brain networks for top-down modulation of sensory processing. In addition, we have complemented anterograde tracing of axonal projections with RV-mediated retrograde tracing of input neurons. The results of these different tracing strategies are highly consistent with each other. The distributions of thalamic inputs to ACA and MO mapped with RV tracing (Fig. 3h) are also broadly consistent with a recent mapping study using anterograde tracing from the thalamus15.
The visual and somatosensory/auditory networks we have identified using viral tracers generally correspond to the medial and somatic sensorimotor networks mapped in the Mouse Connectome Project using non-viral tracers13. However, there are some noticeable differences. While Zingg et al. (2014) grouped the visual and auditory areas into the same medial network and the somatosensory cortex in a separate network, we found that MOp and lateral MOs are connected to both AUD and SS whereas ACA and medial MOs are connected mainly to VIS (Fig. 2). This difference may reflect different emphases of the two studies; while Zingg et al. (2014) performed cluster analysis of all corticocortical connections, our study focused on the frontal-sensory cortex connections. In addition, preferential labeling of different neuronal subtypes by viral vs. non-viral tracers may also contribute to the difference between the two studies. Although the somatosensory and auditory networks overlap spatially in the frontal cortex, they are largely separate in other brain regions. For example, in the thalamus AUD receives extensive inputs from MG but SS mainly from VP, VAL and VM. Even in the frontal cortex, the neurons connected to AUD are likely to be distinct from those connected to SS at the level of single cells. Of course, it is also important to note that in addition to their distinct connections, the visual, somatosensory, and auditory networks also receive shared inputs and project to common targets in both the cortex (e.g., PTLp and ORB) and thalamus (e.g., MD), allowing cross talk between the different sensory modalities.
While ACA→VIS neurons make dense reciprocal connections with sensory and association areas, ACA→SC neurons are preferentially connected to PL and ILA (Figs. 5,6). These medial prefrontal areas are known to be important for the control of actions7,36, and the SC is crucial for controlling saccadic eye movement37,38. Thus the two subnetworks may be specialized in different functions, one for sensory perception (RSP&PTLp ↔ ACA ↔ VIS ↔ RSP&PTLp, the “perception subnetwork”) and the other for action control (RSP&PL/ILA ↔ ACA → SC, the “action subnetwork”). Interestingly, we found inputs from the pallidum to ACA→VIS but not ACA→SC neurons (Fig. 6e). Cholinergic and non-cholinergic pallidal neurons project widely to the cortex and play important roles in brain-state-dependent modulation of sensory processing39–43. Their selective innervation of ACA→VIS neurons should allow preferential regulation of the perception subnetwork in a brain state-dependent manner. In contrast, RSP provides dense inputs to and thus may regulate the activity of both subnetworks.
In both frontal and sensory cortices, intratelencephalic (IT) neurons provide extensive inputs to, but receive little innervation from, the pyramidal tract (PT) neurons24,32,44–46, suggesting a non-reciprocal connection from ACA→VIS to ACA→SC neurons. IT and PT neurons also differ in other cellular properties, e.g. with greater hyperpolarization-activated current (Ih) and faster action potentials in PT than IT neurons24. Interestingly, ACA→SC neurons provide much more projections to the thalamus (Fig. 5e), whereas ACA→VIS neurons receive more thalamic inputs (Fig. 6e). This points to a largely unidirectional thalamocortical loop for the interaction between ACA→VIS and ACA→SC neurons and between the two subnetworks (Fig. 7, blue lines). Note that the mouse LP is densely connected to VIS (Supplementary Fig. 3,5) and thought to be functionally analogous to the primate pulvinar10, which powerfully controls the responses in visual cortex47. The ACA→SC → LP projection (Fig. 5d2) may thus provide an additional pathway for top-down modulation of visual cortical processing. In the somatosensory network, the MO → PO → SS pathway (Fig. 3d and Supplementary Fig. 3d,f) may serve a similar function. However, deep-layer MO→AUD neurons were found to project to thalamus and brainstem motor regions3, suggesting a different organization of the auditory network.
The long-range projections from ACA to both VIS and SC suggest a strong analogy between the mouse ACA and the primate frontal eye field (FEF)13. Optogenetic activation of ACA markedly enhances visual performance of the mouse and neuronal responses in VIS5, similar to the effect of FEF stimulation on attentional modulation in primates9,48,49. In the rat, a frontal orienting field has also been identified, whose activation can bias the orientating response, and presumably attention, to the contralateral side50. The current study indicates that the mouse ACA is a point of convergence between the visual perception and action subnetworks. Such anatomical characterization provides a blueprint for future physiological investigation of how each subnetwork contributes to top-down attentional modulation and behavioral control.
METHODS
Virus preparation
AAV preparation followed previously reported protocol51. To construct AAV-EF1α-DIO-TVA-EGFP, TVA and EGFP linked by the 2A ‘self-cleaving’ peptides or rabies glycoprotein was cloned into pAAV-MCS in an antisense direction flanked by a pair of canonical loxP sites and a pair of lox2272 sites. TVA was subcloned from the AAV-TRE-HTG plasmid from L. Luo16. The AAV-CAG-DIO-Glycoprotein and AAV-CAG-DIO-TVA-mCherry vector was from Addgene (Plasmid #48333 and #48332)52. AAV particles (AAV2/2) were produced by co-transfection of packaging plasmids into HEK293T cells, and cell lysates were fractionated by iodixanol gradient ultracentrifugation. Viral particles were further purified from the crude fraction by heparin affinity column, desalted and concentrated with centrifugal filter (100K). The genomic titer of AAV2/2-EF1α-DIO-TVA-EGFP (4.4 × 1013 gc/mL), AAV2/2-CAG-DIO-TVA-mCherry (3.1 × 1012 gc/mL) and AAV2/2-CAG-DIO-Glycoprotein (8.7 × 1012 gc/mL) was determined by quantitative PCR. AAV2/2-CaMKIIα-mCherry (6.6 × 1012 gc/mL), AAV2/2-EF1α-mCherry-IRES-WGA-Cre (2.7 × 1012 gc/mL) and AAV2/2-EF1α-DIO-EYFP (4.2 × 1012 gc/mL) were purchased from the UNC Vector Core (Chapel Hill, NC). CAV-FLExloxP-Flp (5.0 × 1012 gc/mL) and AAV-DJ-hSyn1-FLExFRT-mGFP-2A-synaptophysin-mRuby (2.9 × 1013 gc/mL) were obtained from Stanford University (kind gift from Dr. Liqun Luo).
Glycoprotein-deleted (ΔG) and EnvA-pseudotyped rabies virus (RV-ΔG-tdTomato+EnvA) was used for retrograde monosynaptic tracing from sensory and frontal cortical pyramidal neurons53,54. RV-ΔG-tdTomato (1.5 × 109 IU/mL) and RV-ΔG-EGFP (5 × 108 IU/mL) were amplified in B7GG cells and pseudotyped using BHK-EnvA cells in a manner similar to that previously described by Osakada and Callaway (2013)55. EnvA pseudotyped rabies virus was titered using HEK293-TVA cells. RV-ΔG-tdTomato was a gift from B. Lim. B7GG cells, BHK-EnvA cells and HEK293-TVA cells were gifts from E. Callaway.
Animals and surgery
All experimental procedures were approved by the Animal Care and Use Committee at the University of California, Berkeley. Experiments were performed on wild-type (C57), CaMKIIα-Cre (Jackson lab stock #005359), PV-Cre (#008069), SOM-Cre (#013044) and VIP-Cre (#010908) mice.
Mice (>P60) were anesthetized with isoflurane (5% induction and 1.5% maintenance) and placed on a stereotaxic frame. Temperature was kept at 37 °C throughout the procedure using a heating pad. After asepsis, the skin was incised to expose the skull and the overlying connective tissue was removed. A craniotomy (~0.5 mm diameter) was made above the injection site. Viruses were loaded in a sharp micropipette mounted on a Nanoject II attached to a micromanipulator and then injected at a speed of 60 nL per min.
For retrograde monosynaptic tracing, TVA receptor and rabies glycoprotein, which are required for virus infection and trans-synaptic spread, respectively, were expressed in Cre-positive neurons by co-injection of AAV2/2-EF1α-DIO-TVA-EGFP / AAV2/2-CAG-DIO-TVA-mCherry and AAV2/2-CAG-DIO-Glycoprotein (200–500 nL) into VIS (Bregma -3 mm, lateral 2.5 mm, depth 0.5 mm), SS (Bregma -1 mm, lateral 3 mm, depth 0.8 mm), AUD (Bregma -2.5 mm, lateral 4 mm and depth 0.5 mm), RSP (Bregma -1.8 mm, lateral 0.3 mm, depth 0.5 mm), PTLp (Bregma -1.8 mm, lateral 1.2 mm, depth 0.5 mm), ACA (Bregma +0.5 mm, lateral 0.3 mm, depth 0.9 mm) or MO (Bregma +1.5 mm, lateral 1.2 mm, depth 0.8 mm) of CaMKIIα-Cre mice and into VIS, RSP, PTLp and ACA of PV-Cre, SOM-Cre and VIP-Cre mice. Two weeks later, RV-ΔG-tdTomato+EnvA or RV-ΔG-EGFP+EnvA (500 nL) was injected into the same site as AAV injection. The histology experiments were performed 7 days after rabies virus injection.
For anterograde tracing from cortical projection neurons, AAV2/2-CaMKIIα-mCherry (200–500 nL) was injected into VIS, SS, AUD, ACA or MO of wild type mice.
For anterograde tracing from subgroups of frontal cortical projection neurons, AAV2/2-EF1α-DIO-TVA-EGFP (4.4 × 1011 gc/mL, 500 nL) was injected into ACA or MO of CaMKIIα-Cre mice. Two weeks later, RV-ΔG-tdTomato+EnvA (500 nL) was injected into VIS or SC in ACA injected mice and SS in MO injected mice.
For axon arborization experiments, CAV-FLExloxP-Flp was injected into VIS or SC, and AAV-DJ-hSyn1-FLExFRT-mGFP-2A-synaptophysin-mRuby was injected into Cg of CaMKIIα-Cre mice. The histology experiments were performed 7–8 weeks after the injection.
For retrograde monosynaptic tracing from subgroups of frontal cortical projection neurons, AAV2/2-EF1α-DIO-TVA-EGFP and AAV2/2-CAG-DIO-Glycoprotein (500 nL) were co-injected into ACA or MO of wildtype mice. At the same time AAV2/2-EF1α-mCherry-IRES-WGA-Cre (500 nL) was injected into VIS or SC in ACA injected mice and SS in MO injected mice. Four weeks later, RV-ΔG-EGFP+EnvA (500 nL) was injected into ACA or MO.
For the control experiment on the distribution of WGA-Cre-labeled ACA→VIS and ACA→SC neurons, AAV2/2-EF1α-DIO-EYFP (500 nL) was injected into ACA, and AAV2/2-EF1α-mCherry-IRES-WGA-Cre (500 nL) were injected into VIS or SC (Supplementary Fig. 11).
For the control experiment testing the specificity of WGA-Cre-mediated labeling of ACA→VIS and ACA→SC neurons, AAV2/2-EF1α-DIO-EYFP (500 nL) was injected into ACA, and AAV2/2-EF1α-mCherry-IRES-WGA-Cre (500 nL) and red retrobeads (150 nL) were injected into VIS or SC (Supplementary Fig. 12–13).
For the control experiment measuring the spatial extent of RV-labeling in ACA without rabies glycoprotein18, AAV2/2-EF1α-DIO-TVA-EGFP (500 nL) was injected into ACA, and AAV2/2-EF1α-mCherry-IRES-WGA-Cre (500 nL) was injected into VIS or SC of wildtype mice. Four weeks later, RV-ΔG-EGFP+EnvA (500 nL) was injected into ACA (Supplementary Fig. 14).
Histology
The mice were deeply anesthetized with isoflurane and immediately perfused with chilled 0.1 M PBS followed by 4% paraformaldehyde (wt/vol) in PBS. The brain was removed and post-fixed overnight at 4 °C. After fixation, the brain was placed in 30% sucrose (wt/vol) in PBS solution for 1–2 d at 4 °C. After embedding and freezing, the brain was sectioned into 50 μm coronal slices using a cryostat. For fluorescence images, brain slices were washed with PBS for 0.5 hr and mounted with VECTASHIELD mounting medium with DAPI. For immunohistochemistry for tdTomato, brain slices were washed with PBS for 0.5 hr, quenched with 3% H2O2 for 0.5 hr, permeabilized using PBST (0.3% Triton X-100 in PBS) for 0.5 hr and then incubated with blocking solution (2% normal goat serum in PBST) for 1 hr followed by primary antibody incubation overnight at 4 ºC using anti-mCherry rat monoclonal antibody (M11217, Life Technologies; 1:1000). The next day, slices were washed three times with PBS, incubated with biotinylated secondary antibody (biotin-goat anti-rat IgG, 629540, Life Technologies; 1:1000) for 2 hrs and then incubated with VECTASTAIN→ ABC Reagent (PK-6100, Vector labs) overnight at 4 ºC. The next day, the slices were washed three times with PBS, incubated with DAB peroxidase substrate (SK-4100, Vector labs) for ~10 mins, washed with PBS again and then mounted with VECTASHIELD mounting medium. One out of every three sections were imaged using 20×/0.75 objective in a high-throughput slide scanner (Nanozoomer-2.0RS, Hamamatsu) for further processing. We also imaged selected example slices under a confocal microscope (Zeiss, LSM 710).
3D reconstruction and quantification
A software package was developed in Matlab to analyze the digitized brain images. The analysis software consists of three modules: image registration, signal detection, and quantification/visualization.
Registration module
The registration module is a reference point-based image alignment software used to align images of brain sections to the Allen Mouse Brain Atlas for further quantification and 3D reconstruction. First, we manually selected a set of reference points in both the atlas and the brain image. The module then applied several geometric transformations (translation, rotation and scaling) of the brain section to optimize the match of the reference points between the brain image and the atlas. Since histological sectioning can sometimes cause tissue compression, we allowed the scaling factors along the dorsal-ventral and medial-lateral axes to be optimized independently. Following the transformation, the match between the image and the atlas was inspected, and further adjustments were made manually if necessary.
Detection module
The detection module has two independent sub-modules designed for counting RV-labeled cells and detecting axons, respectively. The cell counting module records the position of manually identified tdTomato-labeled neurons in each digitized brain section image. For axon detection, the ridge detection method was used (http://en.wikipedia.org/wiki/Ridge_detection). The following steps were taken to maximize the detection accuracy: (1) Image ridges were computed across multiple scales to extract all possible axon-like signals from each image. In the resulting binary ‘ridge image’, the number of pixels occupied by each detected axon depends on the length but not the thickness of the axon. In addition to valid axons, the ridge image also contains many noise pixels. (2) To remove the noise pixels due to the general background in the fluorescence image, we set a threshold based on the intensity distribution of the original image, and use this as a mask to remove the noise pixels in the ridge image obtained from step (1). (3) To remove the discrete noise pixels with fluorescence intensities higher than the general background (thus not removed by step 2), we first identified pixels that are spatially contiguous in the ridge image (after a spatial convolution with a Gaussian kernel), computed the size of each contiguous region, and removed the regions (of the original ridge image) below a threshold size. Steps 2 and 3 were repeated until satisfactory detection results were achieved. (4) The results were then visually inspected and the remaining noise pixels, which were mostly artifacts introduced during brain tissue processing, were rejected manually.
Quantification/visualization module
After detection and registration, signals were quantified across the whole brain and projected to the 3D reference atlas for better visualization. The 3D viewer plug-in of the ImageJ software was used to animate the final 3D model. The atlas, 3D reference mouse brain, quantification ontology, and layouts for sunburst plot were obtained from the open online resource of Allen Institute for Brain Science, licensed under the Apache License (Version 2.0). The input from each region was quantified by dividing the number of labeled neurons in that region by the total number of labeled neurons detected in the whole brain (excluding the injection site). The output (axon projection) to each region was quantified as the number of pixels occupied by detected axons in the cleaned ridge image (see Detection module above) divided by the total number of axon-occupied pixels in the entire brain (excluding the injection site and locations with known major fiber tracks). In addition, the density of labeled neurons and axons (number of neurons/length of axons divided by volume) in each structure was computed (Supplementary Table 1).
To assess the data variability, we have computed the correlation coefficients (CCs) between individual brain samples for both input and output distributions. The mean CC between individual samples of the same group was 0.90±0.02 (s.e.m.). For the same brain sample, the CC between the whole-brain distributions of axons detected by two different observers was 0.96, and the CC between the whole-brain distributions of RV-labeled cells detected by two observers was >0.99.
Code availability
The atlas, 3D reference mouse brain, quantification ontology, and layouts for sunburst plot are freely available in the open online resource of Allen Institute for Brain Science, licensed under the Apache License (Version 2.0). The other computer codes used to generate the findings of this study are available from the corresponding author upon request.
Statistical analyses
A supplementary methods checklist is available summarizing statistical tests and results. Data collection and analysis were not performed blind to the conditions of the experiments. Data randomization was not applicable to our study, and no statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications16–17.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Movie S1. Whole-Brain Distributions of RV-Labeled Neurons Presynaptic to VIS, SS and AUD.
Movie S2. Whole-Brain Distributions of Axons from VIS, SS and AUD.
Movie S3. Whole-Brain Distributions of Axons from ACA and MO.
Movie S4. Whole-Brain Distributions of RV-Labeled Neurons Presynaptic to ACA and MO.
Movie S5. Whole-Brain Distributions of Axons from ACA→VIS and ACA→SC Neurons.
Movie S6. Whole-Brain Distributions of RV-Labeled Neurons Presynaptic to ACA→VIS and ACA→SC Neurons.
Acknowledgments
We thank B. Kim, S. Zhu and P. Kim for technical help. We thank L. Luo, K. Beier, B. Lim and E. Callaway for viral vectors and cell lines. We thank T. Kamigaki for helpful discussion. This work was supported by NIH R01 EY018861.
Footnotes
AUTHOR CONTRIBUTIONS
S.Z. and Y.D. conceived and designed the experiments. S.Z. performed and organized all the experiments. M.X. wrote the software for data analyses and analysed the data. W.-C.C., C.M. and J.D. prepared AAV and RV vectors for rabies virus based retrograde tracing. W.-C.C. also performed some viral injections. T.L. and D.J. performed the detection for labelled cells and axons in some brain samples. J.L.F. and D.J. performed the brain tissue sectioning with cryostat. S.Z., M.X. and Y.D. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Xu NL, et al. Nonlinear dendritic integration of sensory and motor input during an active sensing task. Nature. 2012;492:247–251. doi: 10.1038/nature11601. [DOI] [PubMed] [Google Scholar]
- 2.Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B. A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat Neurosci. 2013;16:1662–1670. doi: 10.1038/nn.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson A, et al. A circuit for motor cortical modulation of auditory cortical activity. J Neurosci. 2013;33:14342–14353. doi: 10.1523/JNEUROSCI.2275-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider DM, Nelson A, Mooney R. A synaptic and circuit basis for corollary discharge in the auditory cortex. Nature. 2014;513:189–194. doi: 10.1038/nature13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, et al. Selective attention. Long-range and local circuits for top-down modulation of visual cortex processing. Science. 2014;345:660–665. doi: 10.1126/science.1254126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desimone R, Duncan J. Neural mechanisms of selective visual attention. Annu Rev Neurosci. 1995;18:193–222. doi: 10.1146/annurev.ne.18.030195.001205. [DOI] [PubMed] [Google Scholar]
- 7.Fuster JM. The prefrontal cortex : anatomy, physiology, and neuropsychology of the frontal lobe. Lippincott-Raven; Philadelphia: 1997. [Google Scholar]
- 8.Mao T, et al. Long-range neuronal circuits underlying the interaction between sensory and motor cortex. Neuron. 2011;72:111–123. doi: 10.1016/j.neuron.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noudoost B, Chang MH, Steinmetz NA, Moore T. Top-down control of visual attention. Curr Opin Neurobiol. 2010;20:183–190. doi: 10.1016/j.conb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen CC. Cortical control of whisker movement. Annu Rev Neurosci. 2014;37:183–203. doi: 10.1146/annurev-neuro-062012-170344. [DOI] [PubMed] [Google Scholar]
- 12.Stanton GB, Bruce CJ, Goldberg ME. Topography of projections to posterior cortical areas from the macaque frontal eye fields. J Comp Neurol. 1995;353:291–305. doi: 10.1002/cne.903530210. [DOI] [PubMed] [Google Scholar]
- 13.Zingg B, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooks BM, et al. Organization of cortical and thalamic input to pyramidal neurons in mouse motor cortex. J Neurosci. 2013;33:748–760. doi: 10.1523/JNEUROSCI.4338-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunnicutt BJ, et al. A comprehensive thalamocortical projection map at the mesoscopic level. Nat Neurosci. 2014;17:1276–1285. doi: 10.1038/nn.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamichi K, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Sporns O, Burkhalter A. Network analysis of corticocortical connections reveals ventral and dorsal processing streams in mouse visual cortex. J Neurosci. 2012;32:4386–4399. doi: 10.1523/JNEUROSCI.6063-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones EG. The thalamus. 2. Cambridge University Press; 2007. [Google Scholar]
- 20.Wu H, Williams J, Nathans J. Complete morphologies of basal forebrain cholinergic neurons in the mouse. eLife. 2014;3:e02444. doi: 10.7554/eLife.02444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fishell G, Rudy B. Mechanisms of inhibition within the telencephalon: “where the wild things are”. Annu Rev Neurosci. 2011;34:535–567. doi: 10.1146/annurev-neuro-061010-113717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodato S, Shetty AS, Arlotta P. Cerebral cortex assembly: generating and reprogramming projection neuron diversity. Trends Neurosci. 2015;38:117–125. doi: 10.1016/j.tins.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepherd GM. Corticostriatal connectivity and its role in disease. Nat Rev Neurosci. 2013;14:278–291. doi: 10.1038/nrn3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino H, Komiyama T. Learning enhances the relative impact of top-down processing in the visual cortex. Nat Neurosci. 2015;18:1116–1122. doi: 10.1038/nn.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouget P, et al. Visual and motor connectivity and the distribution of calcium-binding proteins in macaque frontal eye field: implications for saccade target selection. Front Neuroanat. 2009;3:2. doi: 10.3389/neuro.05.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg ME, Wurtz RH. Activity of superior colliculus in behaving monkey. II Effect of attention on neuronal responses. J Neurophysiol. 1972;35:560–574. doi: 10.1152/jn.1972.35.4.560. [DOI] [PubMed] [Google Scholar]
- 28.Horwitz GD, Newsome WT. Separate signals for target selection and movement specification in the superior colliculus. Science. 1999;284:1158–1161. doi: 10.1126/science.284.5417.1158. [DOI] [PubMed] [Google Scholar]
- 29.Ignashchenkova A, Dicke PW, Haarmeier T, Thier P. Neuron-specific contribution of the superior colliculus to overt and covert shifts of attention. Nat Neurosci. 2004;7:56–64. doi: 10.1038/nn1169. [DOI] [PubMed] [Google Scholar]
- 30.Kustov AA, Robinson DL. Shared neural control of attentional shifts and eye movements. Nature. 1996;384:74–77. doi: 10.1038/384074a0. [DOI] [PubMed] [Google Scholar]
- 31.Zenon A, Krauzlis RJ. Attention deficits without cortical neuronal deficits. Nature. 2012;489:434–437. doi: 10.1038/nature11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N, Chen TW, Guo ZV, Gerfen CR, Svoboda K. A motor cortex circuit for motor planning and movement. Nature. 2015;519:51–56. doi: 10.1038/nature14178. [DOI] [PubMed] [Google Scholar]
- 33.Beier KT, et al. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lerner TN, et al. Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell. 2015;162:635–647. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Euston DR, Gruber AJ, McNaughton BL. The role of medial prefrontal cortex in memory and decision making. Neuron. 2012;76:1057–1070. doi: 10.1016/j.neuron.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson DA. Eye movements evoked by collicular stimulation in the alert monkey. Vision Res. 1972;12:1795–1808. doi: 10.1016/0042-6989(72)90070-3. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Liu M, Segraves MA, Cang J. Visual Experience Is Required for the Development of Eye Movement Maps in the Mouse Superior Colliculus. J Neurosci. 2015;35:12281–12286. doi: 10.1523/JNEUROSCI.0117-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, et al. A cortical circuit for gain control by behavioral state. Cell. 2014;156:1139–1152. doi: 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goard M, Dan Y. Basal forebrain activation enhances cortical coding of natural scenes. Nat Neurosci. 2009;12:1444–1449. doi: 10.1038/nn.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metherate R, Ashe JH. Nucleus basalis stimulation facilitates thalamocortical synaptic transmission in the rat auditory cortex. Synapse. 1993;14:132–143. doi: 10.1002/syn.890140206. [DOI] [PubMed] [Google Scholar]
- 42.Pinto L, et al. Fast modulation of visual perception by basal forebrain cholinergic neurons. Nat Neurosci. 2013;16:1857–1863. doi: 10.1038/nn.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson A, Mooney R. The Basal Forebrain and Motor Cortex Provide Convergent yet Distinct Movement-Related Inputs to the Auditory Cortex. Neuron. 2016;90:635–648. doi: 10.1016/j.neuron.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown SP, Hestrin S. Intracortical circuits of pyramidal neurons reflect their long-range axonal targets. Nature. 2009;457:1133–1136. doi: 10.1038/nature07658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiritani T, Wickersham IR, Seung HS, Shepherd GM. Hierarchical connectivity and connection-specific dynamics in the corticospinal-corticostriatal microcircuit in mouse motor cortex. J Neurosci. 2012;32:4992–5001. doi: 10.1523/JNEUROSCI.4759-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morishima M, Kawaguchi Y. Recurrent connection patterns of corticostriatal pyramidal cells in frontal cortex. J Neurosci. 2006;26:4394–4405. doi: 10.1523/JNEUROSCI.0252-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purushothaman G, Marion R, Li K, Casagrande VA. Gating and control of primary visual cortex by pulvinar. Nat Neurosci. 2012;15:905–912. doi: 10.1038/nn.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore T, Armstrong KM. Selective gating of visual signals by microstimulation of frontal cortex. Nature. 2003;421:370–373. doi: 10.1038/nature01341. [DOI] [PubMed] [Google Scholar]
- 49.Noudoost B, Moore T. Control of visual cortical signals by prefrontal dopamine. Nature. 2011;474:372–375. doi: 10.1038/nature09995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erlich JC, Bialek M, Brody CD. A cortical substrate for memory-guided orienting in the rat. Neuron. 2011;72:330–343. doi: 10.1016/j.neuron.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 52.Weissbourd B, et al. Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron. 2014;83:645–662. doi: 10.1016/j.neuron.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim BK, Huang KW, Grueter BA, Rothwell PE, Malenka RC. Anhedonia requires MC4R-mediated synaptic adaptations in nucleus accumbens. Nature. 2012;487:183–189. doi: 10.1038/nature11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osakada F, Callaway EM. Design and generation of recombinant rabies virus vectors. Nat Protoc. 2013;8:1583–1601. doi: 10.1038/nprot.2013.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Whole-Brain Distributions of RV-Labeled Neurons Presynaptic to VIS, SS and AUD.
Movie S2. Whole-Brain Distributions of Axons from VIS, SS and AUD.
Movie S3. Whole-Brain Distributions of Axons from ACA and MO.
Movie S4. Whole-Brain Distributions of RV-Labeled Neurons Presynaptic to ACA and MO.
Movie S5. Whole-Brain Distributions of Axons from ACA→VIS and ACA→SC Neurons.
Movie S6. Whole-Brain Distributions of RV-Labeled Neurons Presynaptic to ACA→VIS and ACA→SC Neurons.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.