Abstract
Background
Aberrant calcium signaling may contribute to arrhythmias and adverse remodeling in hypertrophic cardiomyopathy (HCM). Mutations in sarcomere genes may distinctly alter calcium handling pathways.
Methods
We analyzed gene expression, protein levels, and functional assays for calcium regulatory pathways in human HCM surgical samples with (n=25) and without (n=10) sarcomere mutations compared with control hearts (n=8).
Results
Gene expression and protein levels for calsequestrin, L-type calcium channel, sodium-calcium exchanger, phospholamban (PLN), calcineurin, and calcium/calmodulin-dependent protein kinase type II (CaMKII) were similar in HCM compared to controls. CaMKII protein abundance was increased only in sarcomere-mutation HCM (p<0.001). The CaMKII target, pT17-PLN, was 5.5-fold increased only in sarcomere-mutation HCM (p=0.01), as was auto-phosphorylated CaMKII (p<0.01) suggestive of constitutive activation. Calcineurin (PPP3CB) mRNA was not increased, nor was RCAN1 mRNA level, indicating lack of calcineurin activation. Further, MEF2 and NFAT transcription factor activity was not increased in HCM, suggesting that calcineurin pathway activation is not an upstream cause of increased CAMKII protein abundance or activation. SERCA2A mRNA transcript levels were reduced in HCM regardless of genotype, as was SERCA2/PLN protein ratio (45% reduced, p=0.03).45Ca SERCA uptake assay showed reduced uptake velocity in HCM regardless of genotype (p=0.01). The cardiac ryanodine receptor (RyR2) was not altered in transcript, protein, or phosphorylated (pS2808, pS2814) protein abundance, and [3H]ryanodine binding was not different in HCM, consistent with no major modification of RyR2.
Conclusions
Human HCM demonstrates calcium mishandling through both genotype-specific and common pathways. Post-translational activation of the CaMKII pathway is specific to sarcomere mutation-HCM, while SERCA2 abundance and SR Ca uptake are depressed in both sarcomere mutation-positive and negative HCM.
Keywords: Hypertrophy, cardiomyopathy, calcium, genetics
Ventricular arrhythmias can be a fatal consequence of hypertrophic cardiomyopathy (HCM), but whether this risk is due to structural remodeling, dysregulated calcium handling, or other electrophysiologic consequences is not clear. Extensive data from animal, in vitro, and human tissue studies have shown that mutations causative of HCM in both the thick and thin sarcomere filaments result in abnormal calcium handling, possibly contributing to the arrhythmia risk in HCM.1–5 In patients with HCM, arrhythmia risk has been linked to hypertrophy burden and scar, but the predictive value of these variables is low, raising the possibility that macroreentry from remodeling does not fully explain clinical arrhythmias.6, 7 Furthermore, resting electrocardiogram findings, such as prolonged QT interval, have not been demonstrated to be independent predictors of sudden death but do not necessarily reflect abnormal calcium handling.8 In human surgical samples, calcium dysregulation has been observed independent of genotype, and altered Ca2+/calmodulin-dependent protein kinase II (CaMKII) activity was proposed as a possible link between HCM-causal mutations and abnormal calcium signaling, implicating a potential single pathway for induction of both hypertrophy and arrhythmias.9 The implication of a single node, CaMKII, modulating diverse disease effects in HCM has important relevance for therapeutic development. Here, we find that sarcomere-mutation HCM is strongly associated with constitutive activation of CaMKIIδ protein in the lack of transcriptional dysregulation of CAMK2D or the calcineurin pathway. We do not find direct evidence of activation of MEF2 as a link between CaMKII-induced calcium mishandling and hypertrophic/pro-fibrotic signaling. In contrast, we find genotype-independent downregulation of SERCA2A gene expression and activity in HCM. Altered SERCA function has been demonstrated in mouse models of HCM, but has not been previously explored in human HCM.2
Methods
An expanded Methods section is available in the Online Data Supplement.
Human Heart Tissue Procurement
Myocardial tissue from the interventricular septum was obtained from unrelated subjects with HCM at the time of surgical myectomy for symptomatic left ventricular outflow tract obstruction (n=33) or at time of heart transplant for severe refractory heart failure (n=2). HCM was diagnosed based on standard criteria and in the absence of any cause for secondary hypertrophy.7 Summary subject clinical data is shown in the Table, and detailed per-subject clinical information and genetic testing results are shown in Supplemental Table 1. Genetic testing was performed on a clinical basis by CLIA-certified laboratories using standard HCM gene panels (including at least all standard sarcomere genes and infiltrative genetic causes). Due to limited tissue amount depending on extent of surgical myectomy, not all assays were performed in all patients. When tissue was limited, the number assayed for each group is listed in the text and/or figure legend. Patient demographic data were recorded at the time of tissue collection, including genotype status determined by clinical genetic testing performed in CLIA approved laboratories. Nomenclature for sequence variants conforms to the recommendations of the Human Genome Variation Society. Tissue was snap-frozen in liquid N2 immediately upon excision. Ventricular septal tissue was obtained from control donor hearts at time of explant (n=8). Donor hearts were perfused with cardioplegia solution prior to removal and tissue was snap frozen in liquid N2 immediately upon dissection. This study had the approval of the University of Michigan Institutional Review Board, and subjects gave informed consent.
Transcript and Protein Analysis
Reverse transcription polymerase chain reaction (RT-PCR), cDNA sequencing, and quantitative RT-PCR were performed by standard techniques (see Supplemental Table 2). Immunoblotting of protein isolated from human surgical samples was performed by established methods (see Data Supplement).
SERCA Uptake Assay
SERCA Ca2+ uptake was determined as previously described using 50 mg of homogenized frozen human heart tissue per sample.2 Both SERCA Ca2+ uptake velocity and total uptake capacity at one hour were determined for a range of pCa values from 8 to 5. Each measurement was performed in triplicate. Measurements were repeated for a subset of samples after pretreatment with lambda phosphatase inhibitor.
[3H]Ryanodine Binding
[3H]Ryanodine binding was performed as previously described.10 To maximize signal from limited quantity of surgical myectomy tissue, total binding was determined at an optimal calcium concentration (pCa 5) to reflect maximal [3H]ryanodine binding activity. Bindings were also performed in the presence of spermine NONOate and H2O2 to determine post-translational modification by nitrosylation or oxidation, respectively. Each measurement was repeated in triplicate.
Transcription Factor Studies
Activation of NFATc1 and MEF2 were assessed by an ELISA-based method per manufacturer’s recommended conditions (Active Motif, Carlsbad, CA). Nuclear extracts from frozen surgical samples were prepared and 10 µg of nuclear extracts were applied to wells coated with oligonucleotides containing a transcription factor consensus binding sites. After incubation with the primary antibody, an HRP-conjugated secondary antibody, and quantification was performed using a microplate reader at 450 nm with a reference wavelength of 655 nm. C2C12 nuclear extract was used as a positive control for MEF2 and nuclear extract from Jurkat T-cells were a positive control for NFATc1.
Statistical Analysis
Values are expressed as mean±SEM unless otherwise indicated. Normality was determined by the Shapiro-Wilk test. Normally distributed data was analyzed by 1-way ANOVA, and the Tukey post-hoc test was used for multiple comparisons between HCM subgroups and controls if the overall result was statistically significant. Western blot, qRT-PCR, and ryanodine binding data were not normally distributed and are presented as boxplots (boxes denoting 1st, 2nd, and 3rd quartiles; fences denoting the minimum and maximum data within 1.5 times the interquartile range of the 1st and 3rd quartile, respectively) and analyzed using the Kruskal-Wallis test for overall statistical significance, and pairwise significance was tested if the overall P value was <0.05. Where noted, distributions between groups with small sample sizes were compared using the Mann-Whitney test with exact P-value calcuations. Statistical analysis was performed using IBM SPSS Statistics. Statistical significance for all tests was defined by a 2-sided P value <0.05. MYBPC3- and MYH7-mutation carriers were combined as a single sarcomere-mutation HCM group to increase statistical power compared to sarcomere mutation-negative HCM and controls unless otherwise stated.
Results
Clinical Characteristics
Summary clinical characteristics of subjects are shown in the Table, and detailed clinical information is in Supplemental Table 1. MYBPC3 mutation carriers were younger than controls and had higher ejection fractions than HCM without sarcomere mutations. The ejection fraction was higher in both MYH7 and MYBPC3 mutation carriers compared to controls, and was also higher in MYBPC3 carriers compared to sarcomere negative patients. The magnitude of left ventricular outflow tract obstruction was similar among sarcomere negative and sarcomere positive patients. Calcium channel blocker and beta blocker prescriptions were also similar among groups (see Supplemental Table 1).
Shared and Divergent Calcium Handling Gene Expression and Protein Levelsin HCM
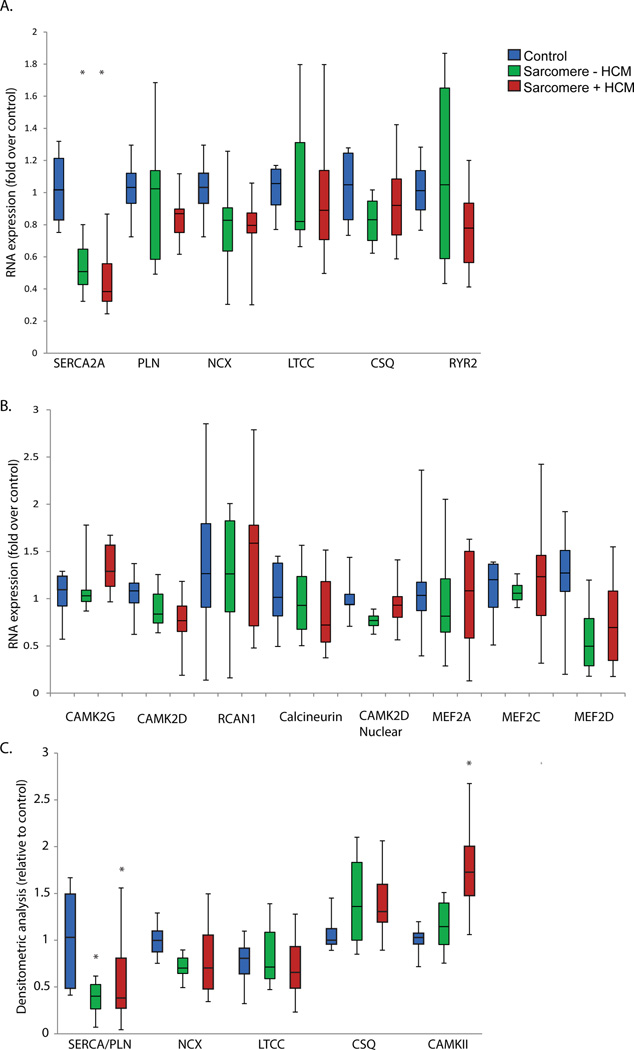
Analysis of most mRNA transcript levels of calcium handling and CaMKII pathway genes showed no statistically significant difference in HCM regardless of genotype (Figure 1A and 1B). However, SERCA2A (ATP2A2A) was decreased consistently across all samples in both HCM with MYBPC3 and MYH7 mutations (mean reduction 53%, p<0.001), as well as in sarcomere mutation-negative HCM (mean reduction 44%, p=0.001). No statistically significant difference was observed among HCM groups.
Figure 1.
A. mRNA transcript levels of calcium handling genes (control n=6, sarcomere mutation positive HCM n=20, sarcomere-negative HCM n=10). B. mRNA transcript levels of CaMKII-calcineurin pathway genes. C. Western blot analysis of calcium-handling protein abundance in HCM relative to controls, normalized to GAPDH (control n=6, sarcomere mutation positive HCM n=12, sarcomere-negative HCM n=7). *=p<0.05
Consistent with reduced gene expression, a significant reduction was observed in the SERCA2/phospholamban ratio in HCM, regardless of HCM genotype. Total protein abundance for most other calcium regulatory genes was not different in either sarcomere-mutation or sarcomere-negative HCM (Figure 1C). The exception was total CaMKIIδ protein level, which was increased in HCM due to MYBPC3 and MYH7 mutations, but not in sarcomere-negative HCM (no statistically significant difference between MYBPC3 and MYH7 groups).
Post-translational CaMKII Pathway Activation in Sarcomere Mutation HCM
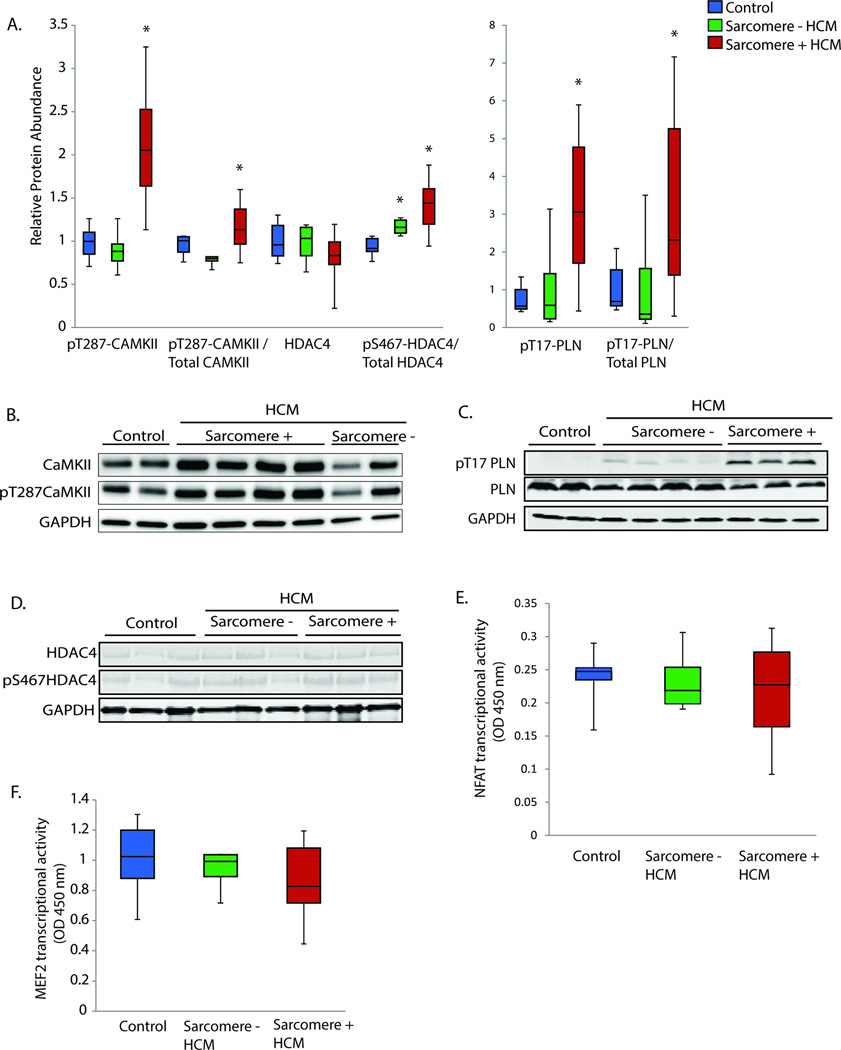
Since autophosphorylation of CaMKIIδ renders the enzyme constitutively active, we also quantified autophosphorylated CaMKIIδ (pT287- CaMKIIδ), which was increased only in HCM due to MYBPC3 and MYH7 mutations (Figure 2A). The CaMKII phosphorylation target pT17-phospholamban was also found to be significantly increased only in HCM due to MYBPC3 and MYH7 mutations (Figure 2A). Since calcineurin is implicated in CAMKII pathway activation11, we measured the mRNA transcript level of the calcineurin catalytic subunit beta isoform (PPP3CB), which has previously been demonstrated to be transcriptionally regulated in response to hypertrophic stimuli12, and found it to be unaltered. Since direct methods of measuring calcineurin activity in the absence of genetically-engineered model systems are limited, we quantified RCAN1 mRNA transcript level, which has been shown to be a reliable marker of calcineurin activity13, and also found no statistically significant difference (Figure 1B). Lastly, we assayed downstream NFAT transcription factor activity and again found no statistically significant difference in HCM regardless of genotype (Figure 2E). We also assessed activation of the CaMKII downstream nuclear phosphorylation target, HDAC4.14 pS467-HDAC4 was increased in HCM, but the increase was present across HCM genotypes, seemingly discordant with the evidence for sarcomere-HCM specific activation of CaMKII (Figure 2A). One possible explanation is that we did not find a significant increase in the nuclear transcript isoform of CAMK2D in any HCM group (Figure 1B), suggesting that increased nuclear HDAC4 phosphorylation may have occurred from an alternate kinase. Since HDAC4 and HDAC5 phosphorylation by CaMKII is known to increase transcriptional activation of MEF2, we also performed a transcription factor assay for MEF2 and found no statistically significant difference in HCM (Figure 2F). MEF2A, MEF2C, and MEF2D mRNA transcript levels were also not different in HCM (Figure 1B). Taken together, these results strongly implicate post-translational CaMKII pathway activation in sarcomere-mutation HCM, but do not support either calcineurin-NFAT signaling as the upstream driver or nuclear CaMKII driven induction of the HDAC4/5-MEF2pathway.
Figure 2.
A. Western blot quantification of autophosphorylated CaMKII (pT287-CaMKII), HDAC4, and the CaMKII phosphorylation targets pS467-HDAC4 and pT17-PLN. Quantification is relative to controls and normalized to GAPDH. For phosphorylation antibodies, quantification is also shown relative to the total antibody quantification. Control n=6, sarcomere mutation positive HCM n=16 (MYBPC3 n=10, MYH7 n=6), sarcomere-negative HCM n=7. B–D. Representative Western blots for CaMKII, HDAC4, and PLN. E–F. NFATc1 and MEF2 transcription factor activity measurement by absorbance at 450 nm wavelength, reference 655 nm. Control n=6, sarcomere mutation positive HCM n=12 (MYBPC3 n=6, MYH7 n=6), sarcomere-negative HCM n=5.. *=p<0.05
SERCA2 Abundance and Uptake is Diminished in HCM Regardless of Genotype
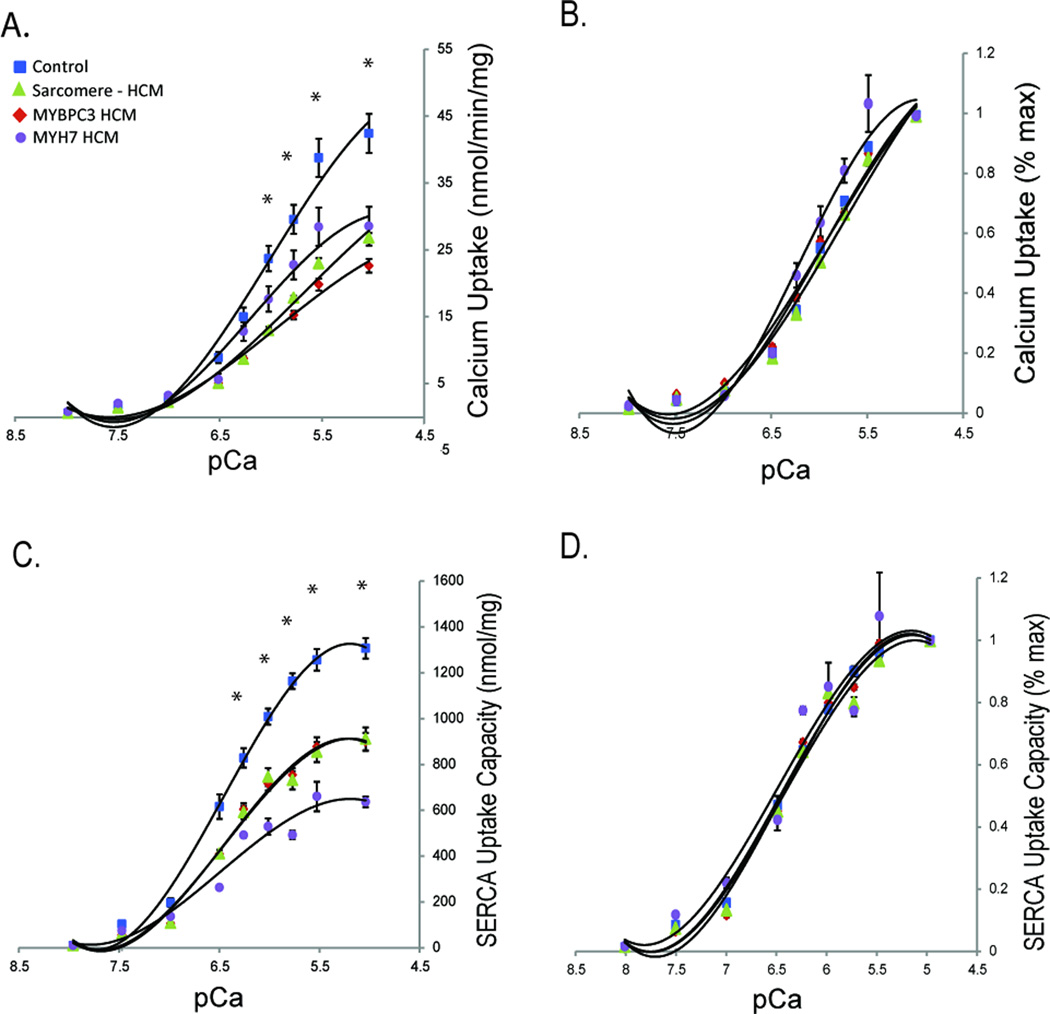
Although SERCA2 protein abundance relative to phospholamban was diminished in HCM (Figure 1C), increased phosphorylation of phospholamban at the Thr-17 CaMKII target site (Figure 2A) would be expected to increase SERCA2 calcium uptake activity. We performed a 45Ca uptake assay to determine the net balance of SERCA2 calcium uptake due to the potential for these opposing effects. We observed an overall decrease in both the velocity and the total capacity of SERCA calcium uptake in HCM, regardless of genotype (Figure 3A and 3C). When normalized to peak calcium uptake for each curve, no shift in the sensitivity of SERCA to calcium was observed, indicating no major net effect on SERCA activity by post-translational modifications (Figure 3B and 3D). Therefore, the major effect on SERCA uptake in HCM is most likely explained by the reduced ratio of SERCA2/phospholamban. To confirm lack of a major impact of the phospholamban phosphorylation by CaMKII, we repeated the SERCA2 uptake assay in the absence of phosphatase inhibitor and in the presence of lambda phosphatase to dephosphorylate phospholamban in a subset of samples. A similar reduction in SERCA calcium uptake velocity activity regardless of genotype in HCM was observed (Supplemental Figure 1A). When normalized to maximum uptake, a release of phospholamban phosphorylation would be expected to cause reduced sensitivity of SERCA to calcium and a rightward shift in the normalized uptake velocity curve. While the required experimental conditions of the SERCA uptake assay do not enable complete dephosphorylation (see Supplemental Material), no statistically significant difference existed, nor any trend toward rightward shift of SERCA calcium sensitivity, among the HCM groups, indicating a lack of major net effect from the CaMKII phosphorylation of phospholamban.
Figure 3.
A. SERCA uptake velocity measured by 45Ca SERCA uptake assay (control n=6, MYBPC3-mutation HCM n=7, MYH7-mutation HCM n=3, sarcomere-negative HCM n=5). B. Normalized SERCA uptake velocity. C. Total SERCA uptake measured by 45Ca SERCA uptake assay as maximum (saturating) uptake after one hour incubation. D. Normalized total SERCA uptake measured by 45Ca SERCA uptake assay as maximum (saturating) uptake after one hour incubation. *p<0.05 for all HCM groups vs control.
Ryanodine Receptors are Normally Regulated in HCM
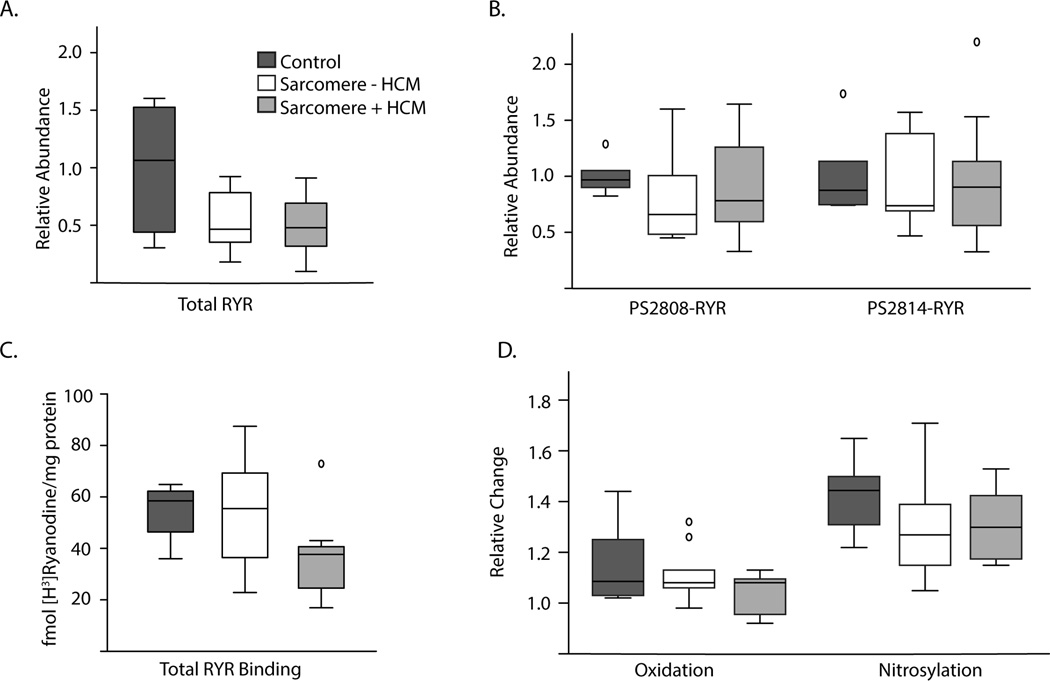
We hypothesized that ryanodine receptor quantity and/or post-translational modifications might contribute to abnormal calcium signaling in HCM. We did not find conclusive evidence of lowered ryanodine receptor abundance in HCM, though there was a trend in the sarcomere-mutation group (p=0.07, Figure 4A). We quantified the amount of ryanodine receptor phosphorylated at both the CaMKII-specific target site (pS2814-RyR2) and the protein kinase A/CaMKII target site (pS2808-RyR2) and found no statistically significant difference in the relative abundance of either (Figure 4B). We then performed [3H]ryanodine binding assays to measure ryanodine receptor abundance and function in the absence and presence of oxidizing and nitrosylating agents. At baseline, there was again no statistically significant difference in total [3H]ryanodine binding at pCa 5, but a trend toward reduced binding in sarcomere-mutation HCM (p=0.10, Figure 4C), consistent with the trend toward an absolute reduction in quantity by Western blot. There was no statistically significant difference in response to either oxidation or nitrosylation in HCM, suggesting no baseline difference in these post-translational modifications (p=0.5 and p=0.3, Figure 4D). These findings reduce the likelihood of significant post-translational modifications to RyR2 channels as a major mechanism for arrhythmias in HCM.
Figure 4.
A. Total RyR2 abundance measured by Western blot (control n=6, sarcomere-mutation HCM n=16, sarcomere-negative HCM n=7). B. Phosphorylated RyR2 abundance by Western blot for the CaMKII/protein kinase A phosphorylation site (pS2808-RyR2) and the CaMKII-specific phosphorylation site (pS2814-RyR2). C. Total ryanodine receptor binding following addition of [3H]ryanodine to cardiac tissue homogenates. Control n=6, sarcomere-mutation HCM n=11 (MYBPC3-mutation n=7, MYH7-mutation n=4), sarcomere-negative HCM n=9. D. Relative change in RyR2 binding specific counts with pre-incubation with either H2O2 (oxidation) or spermine NONOate (nitrosyl donor).
Discussion
We have investigated calcium mishandling in established human HCM, to determine whether ongoing dysregulation could present potential therapeutic targets and whether those targets differ by HCM genotype. We find that the nodal calcium regulatory protein CaMKII is specifically post-translationally dysregulated and auto-activated in sarcomere-mutation HCM, implicating a genotype-specific pathway for disease pathogenesis. Nonetheless, we do not find evidence linking CaMKII dysregulation to activation of the prohypertrophic/profibrotic calcineurin or MEF2 pathway. In contrast, we find genotype-independent reduction of SERCA2 activity in HCM that is not directly attributable to CaMKII and may represent a novel target for therapy in HCM.
Calcium mishandling is a common downstream consequence of sarcomere mutations in HCM, demonstrated across a range of experimental systems. The proximal upstream causes are likely diverse, depending on thick versus thin filament involvement, the specific mutated sarcomere protein, the particular domain carrying the mutation within the sarcomere protein, allosteric effects of the mutation, alteration of the calcium sensitivity of force generation, abnormal calcium buffering, and effects on sarcomere protein phosphorylation status.3–5, 15 The fact that these derangements lead to downstream alterations in calcium handling is highlighted by evidence that calcium channel blockade at least partially prevents adverse remodeling in both the MYH7 R403Q mutant mouse and the TNNT2 I79N mutant mouse.1, 16 Furthermore, a recent randomized clinical trial of diltiazem in preclinical HCM demonstrated evidence of attenuated early left ventricular remodeling specifically for MYBPC3 mutation carriers, highlighting the relevance of calcium handling as disease altering therapeutic target, at least in the early stage of disease before the development of overt hypertrophy.
The increase in constitutively activated CAMKII specific to sarcomere mutation HCM in this study strongly implicates calcium-sensing post-translational modifications as a downstream consequence of sarcomere mutations that may be a distinct disease mechanism that is not relevant to sarcomere negative HCM. CaMKII has been strongly linked to both pathologic hypertrophy and dilated cardiomyopathy.17 Coppini, et al. have proposed CaMKII as a central modulating disease node specifically in HCM, with postulated adverse downstream effects on both electrophysiologic and hypertrophic signaling.9 We found both the total protein level (CaMKIIδ) and the autophosphorylated (activated) protein level (pT287- CaMKIIδ) to be increased in HCM. Increased phosphorylation of phospholamban at the CaMKII specific site is also consistent with activated CaMKII.
In contrast to genotype-specific activation of CaMKII, we found reduced SERCA2 activity level in all HCM samples relative to controls, correlating with reduced transcription and protein levels across all samples, suggesting a common mechanism of calcium mishandling in HCM (see Supplemental Table 1 for per-subject summary). Despite increased CaMKII-phosphorylation of phospholamban, the net balance of SERCA2/phospholamban function appears to be driven more by the reduced abundance of SERCA2, since we found reduced total SERCA uptake velocity but no alteration in the calcium sensitivity of SERCA and no statistically significant difference in SERCA activity when pre-treated with phosphatase. SERCA2 abundance was also noted to be reduced in human HCM samples by Coppini and colleagues.9 Reduction in SERCA2 activity is not unique to HCM. Loss of SERCA2 abundance and activity has been well-established in heart failure, and clinical trials of gene therapy to replace SERCA2 function are ongoing.18, 19 Whether intracellular calcium levels or hypertrophic remodeling is the proximal upstream transcriptional repressor of SERCA2 levels, and whether direct intervention to increase SERCA2 levels would improve HCM pathogenesis merits further study. Alternatively, SERCA reduction may lead to high diastolic calcium levels, providing a potential link to CaMKII activation, but why this relationship would only be observed in sarcomere-gene mutation HCM is unclear.
We also evaluated potential post-translational modifications of RyR2 through [3H]ryanodine binding experiments and immunoblots, since RyR2 dysfunction is clearly a pro-arrhythmic mechanism and may be a consequence of CaMKII activation.20 We found no evidence of altered RyR2 phosphorylation, oxidation, or nitrosylation in HCM, reducing the likelihood that inherent RyR2 dysfunction is a major arrhythmia driver in HCM. Taken together, we do not find evidence of a major effect of CaMKII activation on either SERCA2 or RyR2 functional states, suggesting the relevance of multiple disease pathways in HCM beyond the CaMKII pathway.
A direct link between CaMKII activity and hypertrophic signaling through MEF2 was also not supported. This finding could be due to lack of increased activity of the nuclear CAMK2D isoform, which carries an additional nuclear localization signal that enhances its specificity for nuclear transcriptional targets, including HDAC4. Although we found an increased abundance of phosphorylated HDAC4 in HCM, this finding was not significantly different across HCM groups (incongruent with the sarcomere-mutation specific findings for CaMKII) and downstream activity on MEF2 was not increased. However, MEF2 receives input signals from numerous upstream molecules, so it is possible that opposing signals could mute a potential activation signal from CaMKII. In addition, our study does not rule out the possibility that early MEF2 activation could instigate hypertrophy and fibrosis, but then later reach equilibrium as hypertrophic remodeling plateaus. In the R403Q MYH7 mutant mouse model, increased MEF2 expression was shown only in focal areas of fibrosis, so it is possible that our assay of human surgical samples may not have detected this type of focal non-myocyte involvement.21
Calcineurin, a calcium-sensitive regulator of cardiac growth and hypertrophy, has been implicated in pathologic cardiac hypertrophy.22 However, the calcineurin inhibitor cyclosporine paradoxically was found to worsen the disease phenotype in the R403Q MYH7 mutant mouse, through unclear mechanisms.1 We find that the hypertrophy-regulating12 calcineurin catalytic subunit (PPP3CB) transcript level is not different in HCM compared to control hearts. Since calcineurin activity is not able to be robustly quantified in tissue samples, we measured RCAN1 mRNA level, which has been shown to strongly correlate with calcineurin activity13, and found no statistically significant difference, nor did we observe a difference in NFATc1 transcription factor activity. These findings counter the hypothesis that calcineurin is an active upstream driver of hypertrophy in HCM.
Approximately 50% of familial hypertrophic cardiomyopathy is due to mutations in sarcomere genes. A very small proportion of the remaining genetic causes (<2%) have been explained. Thus, the remaining 50% likely are associated with heterogenous causes that may or may not be directly associated with sarcomere function. Sarcomere mutations either directly or indirectly influence intracellular calcium, potentially explaining altered CaMKII activation. Of note, Coppini, et al. found a similar increase in action potential duration in HCM, regardless of genotype, in their study of human myectomy samples (n=6 for sarcomere-negative HCM, n=14 for sarcomere-mutation HCM), but did not specify mutational status in their other assays. While CaMKII activity may be one driver for prolonged action potential duration, other mechanisms common to both sarcomere-mutation HCM and sarcomere-negative HCM could also induce a prolonged action potential.
This study has several limitations. First, we investigated human surgical samples, which represent a mature form of the disease. Advanced remodeling may have distinct pathways from disease-originating pathways. In addition, in this cross-sectional study of human samples, causality of abnormally-regulated pathways cannot be proven. Also, human sample tissue quantity was limited and variable depending on the extent of surgical myectomy; therefore, not all assays were able to be performed for all subjects. Lastly, we limited our study of sarcomere-mutation HCM to samples with MYBPC3 and MYH7 mutations because of the lesser availability of surgical samples with thin filament mutations.
In conclusion, protein abundance and activity of CaMKII are increased in sarcomere-mutation HCM, highlighting the possibility that CaMKII could be an important genotype-specific therapeutic target. While evidence of persistent CaMKII-induced hypertrophic signaling through MEF2 was not present in the human HCM cardiac tissue, further study will be required to determine whether aberrant CaMKII signaling could link abnormal calcium handling with early development of hypertrophy. Lastly, altered SERCA2 activity in HCM likely is driven by an independent mechanism from CaMKII signaling, highlighting the likelihood of multiple disease pathways that may be independent targets for therapy in HCM.
Supplementary Material
Table 1.
Summary subject clinical data
| Age (y) | % male |
Septal thickness (mm) |
EF (%) | LVOT gradient (mmHg) |
|
|---|---|---|---|---|---|
| Controls (n=8) | 56 ± 6 | 13 | 11.6 ± 0.8 | 64 ± 2 | - |
| HCM patients | |||||
| No sarcomere mutation (n=10) | 49 ± 3 | 80 | 20.4 ± 0.9* | 67 ± 2 | 73 ± 18 |
| MYH7 mutations (n=8) | 44 ± 6 | 50 | 18.3 ± 0.8 | 73 ± 4* | 68 ± 8 |
| MYBPC3 mutations (n=17) | 39 ± 3* | 41 | 28.0 ± 3.3* | 75 ± 2*† | 77 ± 11 |
| Overall P-value | 0.038 | <0.001 | 0.006 | 0.9 |
P<0.05 vs controls
P<0.05 vs HCM patients with no sarcomere mutation
Clinical Perspective Summary.
What is new?
Intracellular calcium mishandling has been implicated in hypertrophic cardiomyopathy (HCM), but limited data are available in humans, and it is unclear whether calcium mishandling is specific to sarcomere mutation carriers.
In surgical myectomy samples from HCM patients, a marked reduction of SERCA2 abundance correlated with reduced SERCA2 function in HCM compared to control hearts regardless of the underlying genetic etiology.
However, CaMKII, a calcium-sensing kinase, was activated in HCM, but only in sarcomere gene mutation carriers.
Activation of CaMKII was associated with an increase in phospholamban phosphorylation at Thr17 in HCM but MEF2 activity was not increased.
What are the clinical implications?
Calcium mishandling may be a potential link between the primary genetic causes of HCM and the downstream signaling cascade that leads to hypertrophy and arrhythmias.
This study showed that two major calcium modulators, SERCA2 and CaMKII, are dysregulated in human HCM.
SERCA2 function is also reduced in dilated cardiomyopathy, and gene therapy to increase SERCA2 activity is currently being investigated in clinical trials.
CaMKII pathway inhibition may improve aberrant calcium cycling through modulation by multiple CaMKII phosphorylation targets.
However, since we found no evidence for CaMKII activation of transcriptional targets, pro-hypertrophic signaling may not be affected by CaMKII inhibition.
Acknowledgments
Funding Sources: Funding for this study was provided by NIH R01 HL093338-01, The Taubman Medical Research Institute, and the Cardiovascular Center Inaugural Grant Fund at the University of Michigan to SMD, and NIH R01-HL055438 and R01-120108 to HHV.
Footnotes
Disclosures: None.
References
- 1.Fatkin D, McConnell BK, Mudd JO, Semsarian C, Moskowitz IG, Schoen FJ, Giewat M, Seidman CE, Seidman JG. An abnormal ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. The Journal of clinical investigation. 2000;106:1351–1359. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guinto PJ, Haim TE, Dowell-Martino CC, Sibinga N, Tardiff JC. Temporal and mutation-specific alterations in ca2+ homeostasis differentially determine the progression of ctnt-related cardiomyopathies in murine models. American journal of physiology. Heart and circulatory physiology. 2009;297:H614–H626. doi: 10.1152/ajpheart.01143.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tardiff JC. Thin filament mutations: Developing an integrative approach to a complex disorder. Circulation research. 2011;108:765–782. doi: 10.1161/CIRCRESAHA.110.224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraysse B, Weinberger F, Bardswell SC, Cuello F, Vignier N, Geertz B, Starbatty J, Kramer E, Coirault C, Eschenhagen T, Kentish JC, Avkiran M, Carrier L. Increased myofilament ca2+ sensitivity and diastolic dysfunction as early consequences of mybpc3 mutation in heterozygous knock-in mice. Journal of molecular and cellular cardiology. 2012;52:1299–1307. doi: 10.1016/j.yjmcc.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sequeira V, Wijnker PJ, Nijenkamp LL, Kuster DW, Najafi A, Witjas-Paalberends ER, Regan JA, Boontje N, Ten Cate FJ, Germans T, Carrier L, Sadayappan S, van Slegtenhorst MA, Zaremba R, Foster DB, Murphy AM, Poggesi C, Dos Remedios C, Stienen GJ, Ho CY, Michels M, van der Velden J. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circulation research. 2013;112:1491–1505. doi: 10.1161/CIRCRESAHA.111.300436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hanlon R, Grasso A, Roughton M, Moon JC, Clark S, Wage R, Webb J, Kulkarni M, Dawson D, Sulaibeekh L, Chandrasekaran B, Bucciarelli-Ducci C, Pasquale F, Cowie MR, McKenna WJ, Sheppard MN, Elliott PM, Pennell DJ, Prasad SK. Prognostic significance of myocardial fibrosis in hypertrophic cardiomyopathy. Journal of the American College of Cardiology. 2010;56:867–874. doi: 10.1016/j.jacc.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW American College of Cardiology Foundation/American Heart Association Task Force on Practice G, American Association for Thoracic S, American Society of E, American Society of Nuclear C, Heart Failure Society of A, Heart Rhythm S, Society for Cardiovascular A, Interventions, Society of Thoracic S. 2011 accf/aha guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: Executive summary: A report of the american college of cardiology foundation/american heart association task force on practice guidelines. Circulation. 2011;124:2761–2796. doi: 10.1161/CIR.0b013e318223e230. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JN, Grifoni C, Bos JM, Saber-Ayad M, Ommen SR, Nistri S, Cecchi F, Olivotto I, Ackerman MJ. Prevalence and clinical correlates of qt prolongation in patients with hypertrophic cardiomyopathy. European heart journal. 2011;32:1114–1120. doi: 10.1093/eurheartj/ehr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coppini R, Ferrantini C, Yao L, Fan P, Del Lungo M, Stillitano F, Sartiani L, Tosi B, Suffredini S, Tesi C, Yacoub M, Olivotto I, Belardinelli L, Poggesi C, Cerbai E, Mugelli A. Late sodium current inhibition reverses electromechanical dysfunction in human hypertrophic cardiomyopathy. Circulation. 2013;127:575–584. doi: 10.1161/CIRCULATIONAHA.112.134932. [DOI] [PubMed] [Google Scholar]
- 10.Loaiza R, Benkusky NA, Powers PP, Hacker T, Noujaim S, Ackerman MJ, Jalife J, Valdivia HH. Heterogeneity of ryanodine receptor dysfunction in a mouse model of catecholaminergic polymorphic ventricular tachycardia. Circulation research. 2013;112:298–308. doi: 10.1161/CIRCRESAHA.112.274803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoo MS, Li J, Singh MV, Yang Y, Kannankeril P, Wu Y, Grueter CE, Guan X, Oddis CV, Zhang R, Mendes L, Ni G, Madu EC, Yang J, Bass M, Gomez RJ, Wadzinski BE, Olson EN, Colbran RJ, Anderson ME. Death, cardiac dysfunction, and arrhythmias are increased by calmodulin kinase ii in calcineurin cardiomyopathy. Circulation. 2006;114:1352–1359. doi: 10.1161/CIRCULATIONAHA.106.644583. [DOI] [PubMed] [Google Scholar]
- 12.Oka T, Dai YS, Molkentin JD. Regulation of calcineurin through transcriptional induction of the calcineurin a beta promoter in vitro and in vivo. Molecular and cellular biology. 2005;25:6649–6659. doi: 10.1128/MCB.25.15.6649-6659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank D, Kuhn C, van Eickels M, Gehring D, Hanselmann C, Lippl S, Will R, Katus HA, Frey N. Calsarcin-1 protects against angiotensin-ii induced cardiac hypertrophy. Circulation. 2007;116:2587–2596. doi: 10.1161/CIRCULATIONAHA.107.711317. [DOI] [PubMed] [Google Scholar]
- 14.Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, Bodine SC, Yao TP. The histone deacetylase hdac4 connects neural activity to muscle transcriptional reprogramming. The Journal of biological chemistry. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 15.Schober T, Huke S, Venkataraman R, Gryshchenko O, Kryshtal D, Hwang HS, Baudenbacher FJ, Knollmann BC. Myofilament ca sensitization increases cytosolic ca binding affinity, alters intracellular ca homeostasis, and causes pause-dependent ca-triggered arrhythmia. Circulation research. 2012;111:170–179. doi: 10.1161/CIRCRESAHA.112.270041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermann D, Knollmann BC, Steendijk P, Rutschow S, Riad A, Pauschinger M, Potter JD, Schultheiss HP, Tschope C. Diltiazem treatment prevents diastolic heart failure in mice with familial hypertrophic cardiomyopathy. European journal of heart failure. 2006;8:115–121. doi: 10.1016/j.ejheart.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Zhang T, Maier LS, Dalton ND, Miyamoto S, Ross J, Jr, Bers DM, Brown JH. The deltac isoform of camkii is activated in cardiac hypertrophy and induces dilated cardiomyopathy and heart failure. Circulation research. 2003;92:912–919. doi: 10.1161/01.RES.0000069686.31472.C5. [DOI] [PubMed] [Google Scholar]
- 18.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL. Hypertrophic cardiomyopathy in cardiac myosin binding protein-c knockout mice. Circulation research. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 19.Hayward C, Banner NR, Morley-Smith A, Lyon AR, Harding SE. The current and future landscape of serca gene therapy for heart failure: A clinical perspective. doi: 10.1089/hum.2015.018. [DOI] [PubMed] [Google Scholar]
- 20.Zhao YT, Valdivia CR, Gurrola GB, Hernandez JJ, Valdivia HH. Arrhythmogenic mechanisms in ryanodine receptor channelopathies. Science China. Life sciences. 2015;58:54–58. doi: 10.1007/s11427-014-4778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konno T, Chen D, Wang L, Wakimoto H, Teekakirikul P, Nayor M, Kawana M, Eminaga S, Gorham JM, Pandya K, Smithies O, Naya FJ, Olson EN, Seidman JG, Seidman CE. Heterogeneous myocyte enhancer factor-2 (mef2) activation in myocytes predicts focal scarring in hypertrophic cardiomyopathy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18097–18102. doi: 10.1073/pnas.1012826107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochemical and biophysical research communications. 2004;322:1178–1191. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.