Abstract
A previous report indicated that prototype chlorovirus PBCV-1 replicated in two Chlorella variabilis algal strains, NC64A and Syngen 2-3, that are ex-endosymbionts isolated from the protozoan Paramecium bursaria. Surprisingly, plaque-forming viruses on Syngen 2-3 lawns were often higher than on NC64A lawns from indigenous water samples. These differences led to the discovery of viruses that exclusively replicate in Syngen 2-3 cells, named Only Syngen (OSy) viruses. OSy-NE5, the prototype virus for the proposed new species, had a linear dsDNA genome of 327 kb with 44-nucleotide-long, incompletely base-paired, covalently closed hairpin ends. Each hairpin structure was followed by an identical 2,612 base-paired inverted sequence after which the DNA sequence diverged. OSy-NE5 encoded 357 predicted CDSs and 13 tRNAs. Interestingly, OSy-NE5 attached to and initiated infection in NC64A cells but infectious progeny viruses were not produced; thus OSy-NE5 replication in NC64A is blocked at some later stage of replication.
Keywords: Chloroviruses, algal viruses, Chlorella variabilis, OSy-NE5, permissive cells, non-permissive cells
Graphical abstract
Introduction
Large dsDNA-containing viruses that infect algae comprise the family Phycodnaviridae. They have genomes ranging from 160 to 485 kb that contain up to 500 protein-encoding genes (CDSs) (Van Etten et al., 2010; Wilson et al., 2009, 2012). These large viruses are found in aqueous environments throughout the world in both inland and marine waters. Currently, phycodnaviruses are classified into six genera. Members of one genus, Chlorovirus, are icosahedral, plaque-forming viruses that replicate in certain unicellular chlorella-like green algae. Chloroviruses are cosmopolitan residents of inland waters with titers occasionally reaching thousands of plaque forming units (PFU) per ml of indigenous water (Quispe et al., 2016; Reisser et al., 1988; Van Etten et al. 1985a, 1985b; Yamada et al., 1991; Zhang et al., 1988).
Chlorovirus hosts, which are normally endosymbionts, are often referred to as zoochlorellae (Karakashian & Karakashian, 1965; Karakashian, 1975; Kodama et al., 2014). They are associated with either the protozoan Paramecium bursaria, the coelenterate Hydra viridis or the heliozoon Acanthocystis turfacea (Van Etten & Dunigan, 2012). Zoochlorellae are resistant to viruses in their symbiotic state. Fortunately, some zoochlorellae grow independently of their partners in the laboratory, permitting plaque assay of the viruses and synchronous infection of their hosts, which allows one to study the virus life cycle in detail (Van Etten et al., 1983). Three such zoochlorellae are Chlorella variabilis NC64A [previously named Chlorella NC64A (Hoshina et al., 2010; Proschold et al., 2011) and its viruses are referred to as NC64A viruses], Chlorella heliozoae SAG 3.83 (previously named Chlorella SAG 3.83 and its viruses are called SAG viruses), and Micractinium conductrix (previously named Chlorella Pbi and its viruses are called Pbi viruses). However, little is known about the natural history of the chloroviruses and we suspect that more chlorovirus hosts and viruses exist in nature.
Paramecium bursaria chlorella virus 1 (PBCV-1) is the type member of the genus Chlorovirus. PBCV-1 infects and forms plaques on two Chlorella variabilis strains, NC64A and Syngen 2-3 (hereafter referred to as Syngen) (Van Etten et al. 1983a). Both C. variabilis strains are endosymbionts of the protozoan P. bursaria. At the time the plaque assay was developed in 1983, we assumed that Chlorella NC64A and Syngen strains were identical and consequently we focused our studies on PBCV-1 and NC64A for the past 35 years (Van Etten & Dunigan, 2012). However, a taxonomic study on rDNA (Kamako et al., 2005) as well as physiological analyses (Quispe et al., 2016a) of zoochlorellae established that NC64A and Syngen were similar, but not identical chlorella strains. The rDNA report prompted us to look for viruses in native water that would plaque on Syngen lawns. Surprisingly, as reported in this manuscript, viruses that formed plaques on Syngen were more common in indigenous waters of Nebraska than viruses that formed plaques on NC64A at certain times of the year. This observation led to the hypothesis that a yet unknown chlorovirus type might only replicate in Syngen cells but not in NC64A cells. The current manuscript describes this new group of chloroviruses, named Only Syngen viruses (OSy) because they replicate in Syngen cells. Furthermore, the OSy viruses can initiate infection in C. variabilis NC64A but they are unable to complete virus replication.
Results and Discussion
Difference in number of plaques from indigenous water samples on lawns of Syngen and NC64A cells
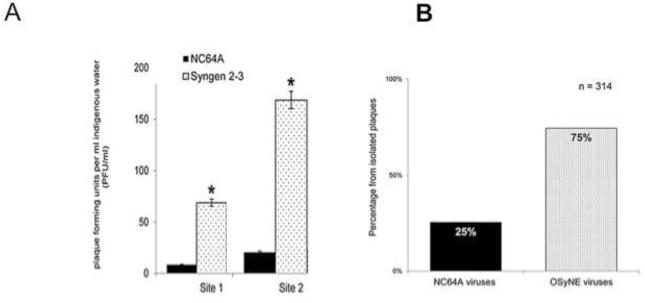
Chlorovirus PBCV-1 infects and forms plaques on both C. variabilis strains NC64A and Syngen. Indigenous water samples from an urban lake in Lincoln, Nebraska were collected and plaque assayed on lawns of these two cells as part of a three-year study of chloroviruses (Quispe et al., 2016). Unexpectedly, some water samples produced up to ten times more plaques when plated on Syngen lawns than when plated on NC64A lawns (Fig. 1A). Usually, samples collected earlier in the year (January to April) showed the highest differences in plaque forming units (PFU). This observation led to the prediction that an unknown chlorovirus type replicated in Syngen cells but not in NC64A cells. That is, Syngen might serve as a host for two distinct virus populations, viruses such as PBCV-1 that replicate in NC64A and Syngen cells and viruses that only replicate in Syngen cells.
Figure 1.
A) Two independent inland water samples collected from different sites in Holmes Lake in Lincoln, NE showed significantly higher plaque numbers on C. variabilis Syngen lawns compared to C. variabilis NC64A lawns. B) Distribution of isolates between NC64A and OSyNE viruses. Three hundred and fourteen plaques were isolated; 75% belonged to the OSy group and 25% to the NC64A genus highlighting the ubiquity and abundance of OSy viruses in inland waters.
Isolation of a new chlorovirus type
To investigate this unexpected difference in plaque-forming viruses in the water samples, 314 single plaques were isolated from the Syngen lawns that showed the greatest difference in plaque number when compared to NC64A lawns. The plaques were then inoculated on the two C. variabilis strains in both liquid and agar cultures. Seventy-five percent of the 314 viruses only formed plaques on Syngen cells, whereas 25% of the viruses formed plaques and lysed both chlorella strains in liquid culture (Fig. 1B). As noted above these new chloroviruses were named OSy viruses because they only formed plaques on Syngen cells. Subsequently, OSy viruses have been isolated from other in-land water sites in the continental United States leading us to believe they are widespread in nature. One isolate from Nebraska, called OSy-NE5, was selected and characterized as the type virus for the group.
Morphology of OSy-NE5 virus
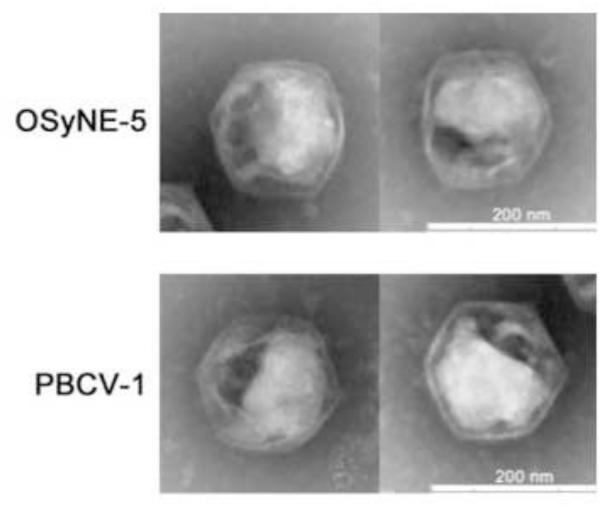
The OSy-NE5 has the same icosahedral morphology as chlorovirus PBCV-1 (Fig. 2) with a diameter of ~190 nm, which is identical to the diameter measured along the five-fold axis for PBCV-1 (Yan et al., 2000). Another characteristic of PBCV-1 is the presence of an internal, single bilayered membrane that is derived from the host endoplasmic reticulum located near the outer nuclear membrane (Milrot et al., 2016). The membrane is located underneath the outer capsid shell and it is required for virus infectivity because PBCV-1 is sensitive to a short exposure to chloroform (Skrdla et al., 1984). OSy-NE5 infectivity was also rapidly reduced by exposure to chloroform, indicating that it probably has an internal membrane (results not shown).
Figure 2.
Electron micrographs of OSy-NE5 and PBCV-1 after negative staining of purified viral particles. The micrographs reveal similarities in shape (icosahedral morphology) and size (~190 nm) between OSy-NE5 and PBCV-1.
Growth of OSy-NE5
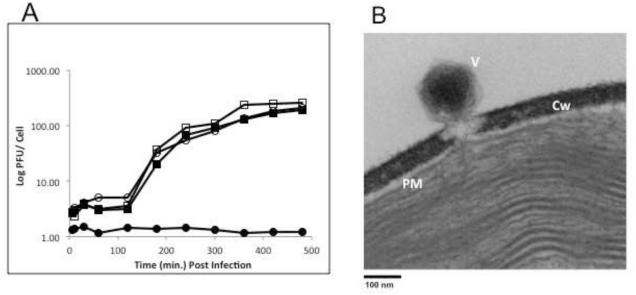
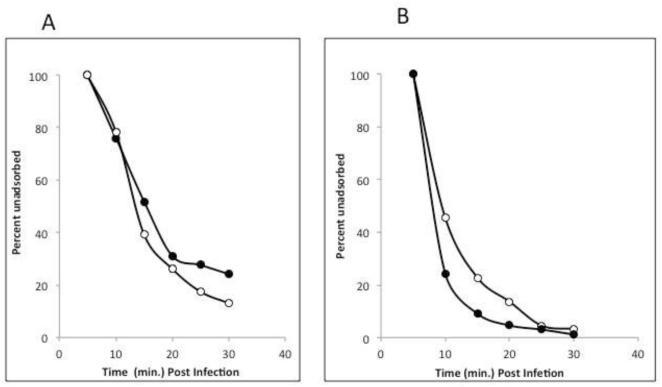
Figure 3A shows the kinetics of one step growth experiments of OSy-NE5 and PBCV-1 on both Syngen and NC64A cells. OSy-NE5 entered an eclipse stage for about 2 h on Syngen cells followed by a rise period of 3 to 4 h beginning at about 2 h post infection (PI). Lysis occurred by 8 h PI with a burst size of about 250 PFUs per cell. OSy-NE5 did not replicate in NC64A cells although occasionally 1 or 2 infectious OSy-NE5 particles were recovered from OSy-NE5 infected NC64A cells when assayed on Syngen cells at 24 h PI. However, these escaped OSy-NE5 viruses had not mutated to the extent that they could replicate normally in NC64A. In contrast to OSy-NE5, PBCV-1 infection of both Syngen and NC64A cells produced similar one step growth curves and burst sizes as OSy-NE5 on Syngen cells (Fig. 3A).
Figure 3.
A) One-step growth curves for chloroviruses OSy-NE5 (circles) and PBCV-1 (squares) on Syngen (open symbols) and NC64A (closed symbols) cells. B) Attachment and initial digestion of the cell wall of Syngen cells by OSy-NE5.
Like PBCV-1 (Meints et al., 1984), OSy-NE5 infects Syngen cells by attaching to the cell wall and digesting a hole at the point of attachment (Fig. 3B). We assume the OSy-NE5 internal membrane then fuses with the host plasma membrane allowing the viral DNA to enter the cell.
OSy-NE5 protein profile
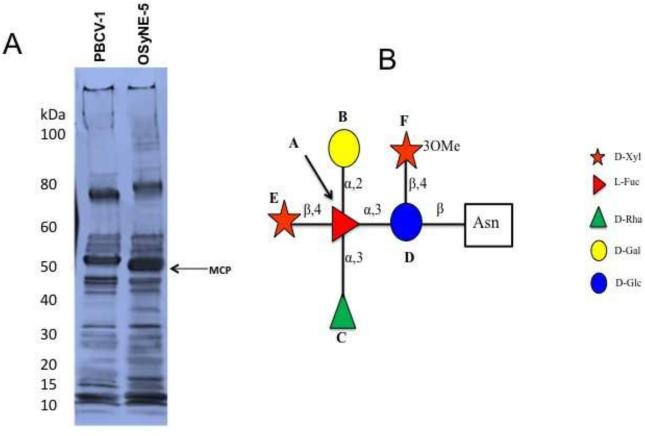
The PBCV-1 virus particle proteome consists of 148 viral-encoded proteins and at least one host-encoded protein (Dunigan et al., 2012). To determine if OSy-NE5 has comparable proteomic features, the protein profiles of OSy-NE5 and PBCV-1 were compared by SDS-PAGE. The OSy-NE5 protein profile resembled the PBCV-1 profile but with some differences (Fig. 4A). One difference was that the OSy-NE5 major capsid protein (MCP) migrated slightly faster than the PBCV-1 MCP (54 kDa) with a predicted molecular weight of ~50 kDa. The 50 kDa polypeptide is in the size range of MCPs from other chloroviruses, which range between 51 kDa and 55 kDa (DeCastro et al., 2016).
Figure 4.
A) SDS-PAGE profile of the virion protein compositions of OSy-NE5 and PBCV-1 purified particles. The OSy-NE5 MCP migrated slightly faster (~50 kDa) than the PBCV-1 MCP (54 kDa). B) Structure of the glycan of the MCP (OS5_219L) from chlorovirus OSy-NE5. The core structure that is present in the glycans of all the chlorovirus MCPs is boxed in.
The PBCV-1 MCP is a glycoprotein with four Asn-linked glycans, whose structures are very unusual and differ from previously described glycans in the three domains of life (De Castro et al., 2013). Some of their unusual structural features include: i) the most abundant glycan consists of 9 neutral monosaccharide residues organized in a highly branched fashion, ii) the glycans are attached to Asn in the MCP by a beta-glucose linkage, iii) a dimethylated rhamnose is the capping residue of the main glycan chain, iv) the glycan contains a hyperbranched fucose unit, and v) the glycan contains both D- and L-rhamnose residues. Another unusual feature is that the Asn-linked glycans are not located in a typical Asn-X-(Thr/Ser) sequon site that is characteristic of glycans synthesized via the endoplasmic reticulum/Golgi system.
Since Asn-linked glycans are associated with the MCPs from other chloroviruses (De Castro et al., 2016), we assumed that a similar situation occurred in the OSy-NE5 MCP. Therefore, the glycan structure(s) of its MCP (protein OS5_219L) was determined (Fig. 4B). The structure resembled that of six other chlorovirus MCPs, including a core structure of five residues that exists in all six chloroviruses (De Castro et al., 2016). Like the NC64A chloroviruses, the OSy-NE5 glycan has a D-rhamnose attached to the core structure, rather than an L-rhamnose typical of some of the other chloroviruses (De Castro et al., 2016). Methylation at position three of the proximal xylose was the only modification of this conserved motif that differed from all the other chloroviruses. Indeed, this modification was not present in any of the xylose units of the NC64A viruses, while viruses ATCV-1 and MT325 (SAG and Pbi viruses, respectively) had a methyl group exclusively at position four. The amino acid locations of the OSy-NE5 MCP glycan(s) were not determined but protein OS5_219L has four Asn in about the same locations as the four PBCV-1 glycan-linked Asn and we suspect that all four OSy-NE5 Asn are glycosylated.
OSy-NE5 genome
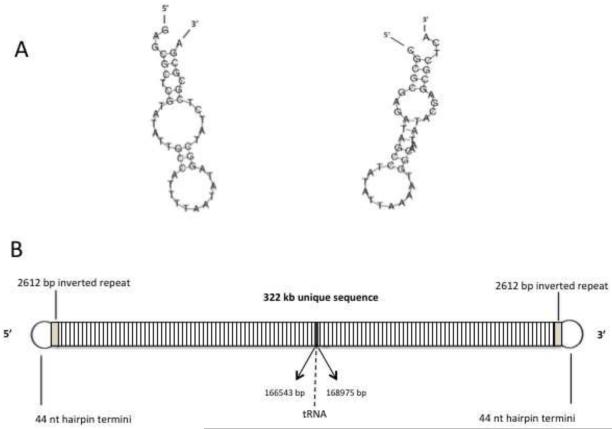
Pulsed field gel electrophoresis established that the OSy-NE5 genome is a linear molecule, like the other chlorovirus genomes, and it electrophoresed slightly faster than the 334 kb PBCV-1 genome indicating that it was slightly smaller (results not shown). Sequencing the OSy-NE5 genome established that it was 327,147 bp long. Like the PBCV-1 genome (Zhang et al., 1994), the OSy-NE5 genome has 44 nucleotide (nt) long, incompletely base-paired (bp), covalently closed hairpin ends that are complementary when the 44 nt sequences are inverted with respect to one another (Fig. 5A). Each hairpin loop structure is followed by an identical 2,612 bp inverted sequence after which the DNA sequences diverge (Fig. 5B).
Figure 5.
A) Sequence and predicted structure of the 44 nt single-strand hairpin termini of OSy-NE5. Note that the two hairpin ends are complementary to one another when they are inverted. B) The OSy-NE5 genome is a linear dsDNA molecule with 44 nt, incompletely base-paired, covalently closed hairpin ends. Each hairpin loop structure is followed by an identical 2,612 bp inverted sequence after which the DNA sequence diverges. Thirteen predicted tRNA genes are located near the center of the genome.
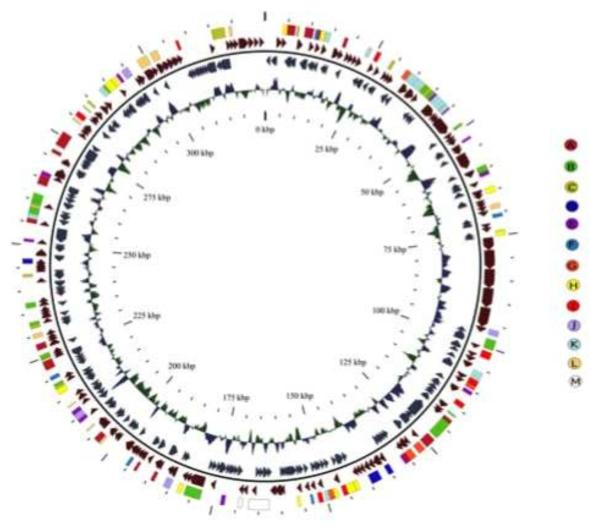
The OSy-NE5 virus is predicted to encode 357 CDSs (Sup. Table 1) and 13 tRNA genes (Sup. Table 2), located as a cluster near the center of the genome (Fig. 6). Of the 357 CDSs, all but 11 of them have homologs in one or more of the other sequenced chloroviruses (Jeanniard et al., 2013). Three of the 11 orphans have strong identity to known proteins: OS5_308R resembles ADP-ribosylglycohydrolase, OS5_333R resembles CTP synthetase and OS5_354R resembles a Ser/Thr protein phosphatase, which is the first time these 3 putative genes have been found in the chloroviruses.
Figure 6.
The OSy-NE5 genome represented as a circle. The 357 CDSs are represented as brown and blue-gray arrows showing predicted genes transcribed clockwise and counter clockwise, respectively, along the genome. Note that the diagram is circular, but there is a break at the 12 o’clock position because the viral genome is a linear molecule with terminal inverted repeats (Fig. 5B) and hairpin ends Fig. 5A). The polycistronic gene encoding 13 tRNAs is shown as a white rectangle at about 6 o’clock in the genome. The inner circle is the GC content of the genome. The outer circle of colored rectangles is used to classify protein-coding genes with predicted functions according to the color scheme in the figure. The specific colored coded genes in the various categories are listed in Sup. Table 3. The map was developed with CGView software.
An unusual feature of the OSy-NE5 genome is that it has 5 adjacent homologs (OS5_081R to OS5_85R) that resemble a putative viral surface glycoprotein referred to as Vp260 in PBCV-1 (Que et al., 1994). These 5 adjacent homologs also exist in a chlorovirus CVK2, as well as some other NC64A chloroviruses isolated in Japan (Chuchird et al., 2002).
The DNA polymerase gene has a 101 nt group I intron and the Tyr tRNA gene is predicted to have a small (12 nt) intron. The OSy-NE5 genome is 42% G+C. All of the NC64A viruses have a G+C content of about 40%, whereas the G+C content of the Pbi and SAG viruses are ~45% and 49%, respectively (Jeanniard et al., 2013). The percentage of the OSy-NE5 genome that is coding is 91.5%.
Synteny of OSy-NE5 with other chloroviruses
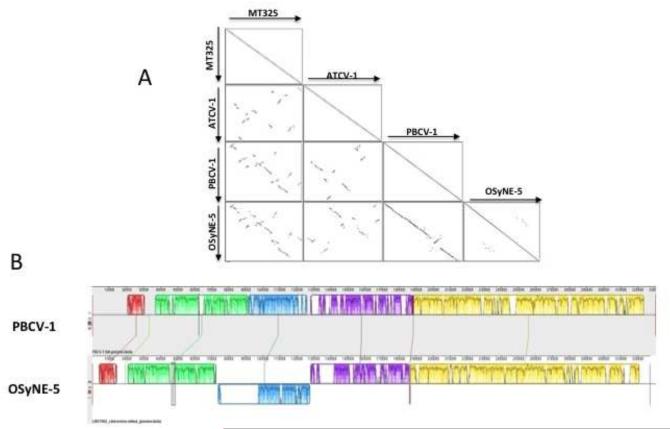
A previous report of 41 sequenced chloroviruses indicated that gene synteny, nucleotide conservation and phylogenetic affinity were highly conserved among viruses infecting the same algal host, with only a few localized rearrangements (Jeanniard et al., 2013). In contrast, low synteny was observed between chloroviruses that infected different hosts. Therefore, the OSy-NE5 gene order was compared to one representative from each of the 3 chlorovirus types; PBCV-1 represented the NC64A viruses, ATCV-1 represented the SAG viruses, and MT325 represented the Pbi viruses. As noted in (Fig. 7A), OSy-NE5 most closely resembled PBCV-1 (Fig. 7A), but there were differences between these viruses. There was an inverted section from 74 kb to 128 kb in the OSy-NE5 genome that corresponded to region 91.5 to 126 kb in the PBCV-1 genome. A 20 kb stretch from 76 kb to 96 kb within the inverted OSy-NE5 section was absent in the PBCV-1 genome. Also a 0 to 20 kb region existed at the left end of the PBCV-1 genome that contained sequences that did not resemble any OSy-NE5 sequences (Fig. 7B).
Figure 7.
A) Viral genome synteny. Dot-plot alignments between OSy-NE5 vs PBCV-1, vs ATCV-1, and vs MT325 viruses. Each dot represents a protein match between two viruses from genes in the same orientation (black) or in reverse orientation (gray). B) Alignment of OSy-NE5 and PBCV-1 genomes. Progressive Mauve alignment on default settings for the genomes of PBCV-1 (top) and OSy-NE5 (bottom). The degree of DNA sequence similarity is indicated by the height of each colored block. Homologous regions are connected by lines between genomes, and blocks below the center line in PBCV-1 indicate regions with inverse orientation in comparison to OSy-NE5. White spaces within blocks represent small-localized areas of the genome sequences that were not aligned. The largest of these is the stretch from around 76 kb to 96 kb within the inverted OSy-NE5 section, which is present in OSy-NE5 but not in the PBCV-1 genome. Blocks below the central line represent sequences that are inverted in comparison to the PBCV-1 arrangement.
Phylogenetic analysis
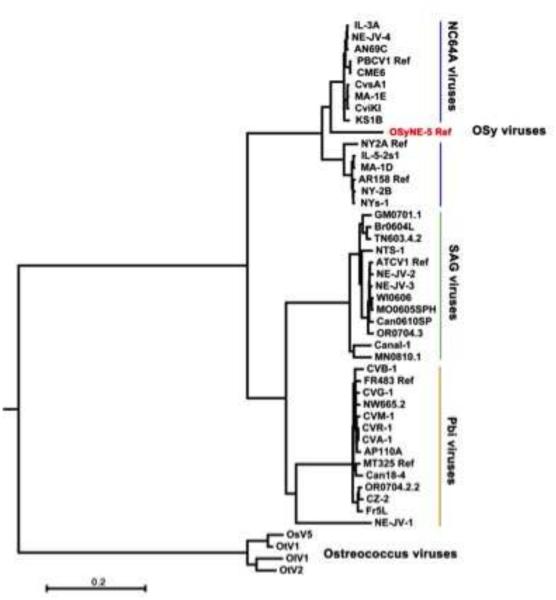
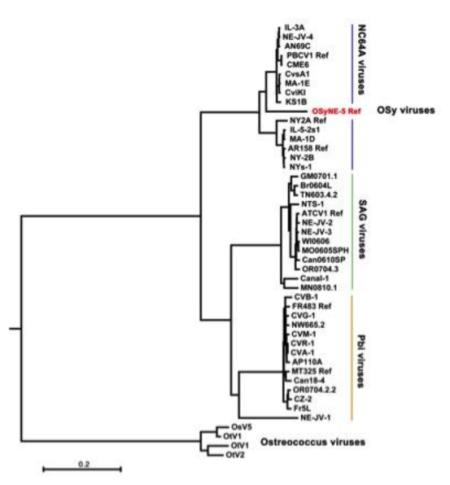
Phylogenetic relationships between OSy-NE5, 43 chloroviruses and 4 Ostreococcus viruses (outgroup) were determined using 29 of the 32 concatenated core chlorovirus genes used previously (Jeanniard et al., 2013). The resulting maximum likelihood (ML) phylogenetic tree is reported in Fig. 8, and is consistent with the alignments shown in Fig. 7A. While most of the OSy-NE5 genes resembled previously sequenced NC64A viruses, including PBCV-1, the isolated phylogenetic position of OSy-NE5 within the NC64A virus clade made it the first representative of a proposed new OSy species of chloroviruses. This new virus clade resides within two separate phylogenetic sub-groups of NC64A viruses – one contains PBCV-1 and the other NY-2A. Both of these NC64A subgroups have similar gene co-linearity (Jeanniard et al., 2013). The 29 core proteins identified in the OSy-NE5 genome share a high (average of 89%) amino acid identity with their PBCV-1 orthologs. For comparison, the protein sequence identity between clades of chlorovirus species ranged from 63% (NC64A vs. Pbi viruses) to 71% (Pbi vs. SAG viruses) (Jeanniard et al., 2013). Thus, the molecular phylogenetic analysis indicates a high phylogenetic affinity between the OSy and NC64A viruses.
Figure 8.
Phylogenetic tree shows the evolutionary relationships between 47 viral concatenated amino acid sequences (7762 gap-free sites). The Maximum Likelihood (ML) tree was constructed using the MEGA 6.0 software with the ML algorithm and default setting. The bar length represents 0.2 substitutions per amino acid site. The proposed new OSy chlorovirus species is indicated in red and resides between the two separate phylogenetic subclades of NC64A viruses – one subclade contains PBCV-1 while the other subclade contains chlorovirus NY-2A. Both subclades share almost perfect gene colinearity and they replicate in the same host. Branch support was estimated from 1000 bootstrap replicates. Four Ostreococcus virus sequences serve as outgroups to root the tree.
OSy-NE5 virus attaches to NC64A cells
The OSy viruses are interesting because they are unable to replicate in NC64A cells and yet NC64A viruses can replicate in both NC64A and Syngen cells. Previously, we have screened NC64A cells for resistant mutants that did not support PBCV-1 replication and such mutant cells are reasonably easy to isolate. However, in every instance where we investigated the nature of the resistance, the viruses were unable to attach to the resistant mutants, indicating a change in the host receptor (Van Etten et al., unpublished results). Therefore, we tested to see if OSy-NE5 could attach to NC64A cells. As noted in Fig. 9, OSy-NE5 attached to both Syngen and NC64A cells equally well. Therefore, the inability of OSy-NE5 to replicate in NC64A cells is not due to a failure in attachment.
Figure 9.
Attachment of OSy-NE5 (A) and PBCV-1 (B) to Syngen (open circles) and NC64A (closed circles) cells. The viruses were allowed to adsorb to the hosts and briefly centrifuged to pellet viruses attached to cells. The supernatant was removed and plaque assayed for unattached viruses.
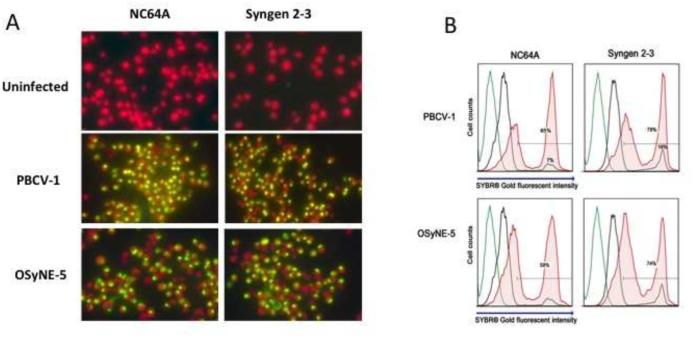
Previous experiments have established that PBCV-1 infection of NC64A cells results in rapid uptake of SYBR® Gold stain and consequently positive DNA fluorescent staining in the infected cells occurs (Dunigan et al., unpublished results). Therefore, fluorescent microscopy and flow cytometry analyses using SYBR® Gold stain were performed to determine if OSy-NE5 initiated infection in NC64A cells. PBCV-1 and an SAG virus (ATCV-1) served as positive and negative controls, respectively. Cultures were infected at a multiplicity of infection (MOI) of 10 and at 1 h PI cells were mixed with SYBR® Gold and analyzed by fluorescent microscopy and flow cytometry. The two chlorella strains, both alone or after mixing with chlorovirus ATCV-1, were negative for DNA staining. Syngen cells infected with either OSy-NE5 or PBCV-1 showed rapid SYBR® Gold uptake and consequently positive DNA fluorescent staining following viral infection. Surprisingly, NC64A cells were also positive for DNA staining after OSy-NE5 attachment (Fig. 10A).
Figure 10.
A. SYBR® Gold uptake analysis of infected and uninfected C. variabilis cells with chlorovirus types 1 h PI A) Epifluorescent microscopy observations. Chlorophyll fluoresces red while DNA fluoresces yellow under UV light. SYBR® Gold is able to enter the cell and fluoresce the DNA when the virus attaches and initiates infection. Chlorophyll stains red and DNA yellow under UV light. B) Infected and uninfected NC64A and Syngen cells mixed with OSy-NE5 and PBCV-1 (1 h PI). The histogram depicts the level of SYBR® Gold stain intensity of infected and uninfected cells. SYBR® Gold intensity increases when chlorella cells were infected by their respective chloroviruses. The gating was set on an uninfected control sample stained with SYBR® Gold (black line) and applied to the infected samples. Negative control cells are plotted in green (no dye) and black (plus dye). Experimental cells mixed with the DNA dye (SYBR® Gold) 1 h PI are plotted in red. Samples were run at 1 × 106 cells/ml, and approximately 1 × 104 cell events were collected per sample.
To corroborate and quantify these results, flow cytometry analysis was performed following a similar procedure. Samples of infected, uninfected and control cells were run at 1 × 106 cells/ml and approximately 1 × 104 cell events were collected per sample. Similar to the fluorescent microscopy analysis, OSy-NE5 significantly increased the population of NC64A cells with higher SYBR® Gold fluorescent intensity compared to uninfected cells (Fig. 10B). The increase in the OSy-NE5 induced fluorescent was similar to that observed in PBCV-1 on Syngen and NC64A cells. Background staining accounted for only 10% of the total cell population across the strains. Consequently, OSy-NE5 attached and initiated infection not only in Syngen, but also in NC64A cells. We also determined that OSy-NE5 did not attach and initiate infection in 2 other chlorella-like ex-symbiotic strains, SAG 3.83 and Pbi (results not shown).
Conclusions
This manuscript describes the characterization of a new group of chloroviruses, called OSy viruses that infect C. variabilis Syngen, that we propose form a new species of Chlorovirus. The OSy viruses are most closely related to the NC64A viruses at the genomic and phylogenetic levels. Interestingly, the OSy viruses can initiate infection in C. variabilis NC64A cells but they are unable to complete virus replication (non-permissive cells). In contrast, the NC64A viruses are able to infect and complete replication in two C. variabilis strains (NC64A and Syngen). Previous attempts to isolate chlorella cells that were resistant to chlorovirus infection always resulted in cells with an altered virus receptor such that the viruses could not attach. However, the finding in this report that OSy viruses can initiate infection in NC64A cells and not complete replication opens up a new area of investigation. That is, the OSy viruses are blocked at some later stage(s) of infection and the location of the block is under active investigation.
Methods
Cell cultures and virus purification
C. variabilis NC64A and Syngen 2-3 were maintained as slant stocks at 4°C. NC64A and Syngen cells were grown on Modified Bold’s Basal Medium (MBBM) with 1% thiamine (V/V) (complete MBBM) (Nichols & Bold, 1965; Van Etten et al., 1983). All experiments were performed with cells grown to early log phase (4 - 7 × 106 cells/ml). Cell cultures were shaken (200 rpm) at 26°C under continuous light. Procedures for producing and purifying chloroviruses have been described previously (Van Etten et al. 1983, 1983a; Agarkova et al., 2006).
Plaque assays
Indigenous water samples were analyzed by plaque assay on the two Chlorella variablis strains as previously described (Van Etten et al., 1983a) with minor modifications. The strains were grown to early log phase (4 - 7 × 106 cells/ml) and concentrated tenfold (4 - 7 × 107 cells/ml) by centrifugation for the plaque assays. Three ml of MBBM top agar (7.5 g/L agar) were mixed with 300 μl of concentrated actively growing cells and the water sample. Adequate amounts of 0.45 μm filtered water samples were plated to produce 25 - 120 plaques/plate when possible. The samples were poured over solidified MBBM containing agar (15 g/L). Plates were incubated for several days in constant light at 26°C and plaque averages were determined from 4 plates/sample/strain.
Isolation of OSyNE viruses
Indigenous water samples (collected in Jan., 2012 from Holmes Lake in Lincoln, NE) with significantly higher numbers of plaques on Syngen cells compared to NC64A were selected. A total of 314 single plaques were isolated from Syngen lawns and transferred to liquid cultures of Syngen and NC64A cells to amplify the viruses. After incubating in MBBM at 26°C in continuous light for seven days, tubes were centrifuged at 5000 rpm for 5 min to pellet fragments and whole cells. A green supernatant indicated lysis whereas a green pellet of intact algal cells suggested no lysis. Viruses that lysed Syngen cells without lysing NC64A cells were selected. These viruses were diluted and plaque assayed on the two strains. Viruses that formed plaques exclusively on Syngen lawns were selected and re-plaqued at least two times and then amplified in liquid culture for final virus purification. Lysate and purified viruses were stored at 4°C in glass vials.
One-Step growth experiments
One-step growth experiments were described previously (Van Etten et al., 1983) where a known number of chlorella cells were infected with viruses at a MOI of 1. After 5 min of adsorption, the infected cells were diluted 1/100 to reduce secondary infections. At appropriate times virus titers were determined by plaque assay (Van Etten et al., 1983a).
Transmission electron microscopy
For electron microscopic studies, virus particles were added to a strip of parafilm and transferred to 400-mesh copper grids (Electron Microscopy Sciences) supported by carbon-coated Formvar film. The grid was submerged in buffer suspension for 3 min and air-dried for 60 sec. Negative staining solution (2% w/v aqueous phosphate tungsten acid, pH 7.2) (1:1 ratio) was added for 5 min and air-dried for 1 h at room temperature. Virus particles were visualized in a Hitachi H7500 transmission electron microscope.
Cultures of Syngen were infected with OSy-NE5 at an MOI of 5 and samples were centrifuged in a Sorvall GSA rotor at 5,000 × g for 5 min at 4°C. Samples were fixed in 2% glutaraldehyde, 2% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) and further fixed in 1% osmium tetroxide in 0.1 M phosphate buffer (pH 7.4) for 1 h. Samples were dehydrated in a graduated ethanol series, and embedded in Epon 812 (Electron Microscopic Sciences, Fort Washington, PA). Thin sections (80 nm) were stained with uranyl acetate and lead citrate. The thin sections were observed under the transmission electron microscope. All electron micrograph experiments were conducted in the Morrison Microscopy Core facility at the University of Nebraska-Lincoln.
Protein and glycan analysis
SDS-PAGE profiles of OSy-NE5 and PBCV-1 proteins were performed using 15 μg of protein extracted from particles and run on a 4-20% Tris-Glycine PAGEr® Gold precast protein gel before silver staining (Morrissey, 1981). The isolation of the OSy-NE5 major capsid protein, digestion and glycopeptide purification were performed as described previously (De Castro et al., 2013). Details on the NMR characterization of the glycan structure are provided in the supplemental section.
DNA isolation and genomic library preparation and sequencing
DNA was isolated from virus preparations pretreated with DNAse I (to remove any exogenous DNA) using a phenol-chloroform procedure (Doyle, 1987). Isolated DNA was evaluated for quantity and quality with a Thermo Scientific NanoDrop 2000 spectrophotometer and stored at 4°C.
Sequencing for assembly of the OSy-NE5 genome was performed on the RSII platform as recommended by the manufacturer (Pacific Biosciences, Menlo Park, CA). Briefly, high molecular weight DNA was sheared using a G-tube device (Covaris Inc., Woburn MA) at 4400 and 4600 rpm in a microcentrifuge, then passed a second time through the G-tube at 4200 rpm. Sheared DNA was purified by binding to and eluting from 0.45 × volume AMPure PB beads and processed into a SMRTbell library as directed using the Template Prep Kit v1.0 instructions for “Greater than 10 kb Template Preparation” (Pacific Biosciences, Menlo Park, CA). Importantly, the first enzymatic step post-shearing is the “polishing” of single-stranded DNA to create blunt ends. In the case of the OSy-NE5 genome, sheared fragments containing either end of the linear double-stranded DNA genome were assumed to contain the single-strand hairpin termini, and therefore the hairpin was likely converted to double-stranded DNA, creating fragments that have read-through of the hairpin sequence that have the reverse-complement sequence artificially appended during the process. After creation of the SMRTbell library, size distribution and integrity of the library was verified on a Fragment Analyzer (Advanced Analytical Technology Inc., Ames IA). Sequencing was performed in a single SMRT cell on the RSII platform using P4/C6 chemistry, producing 68,554 post-filter polymerase reads and 172,448 subreads of average length 4,362 bases.
Sequence assembly and annotation
Assembly with the RSII data was performed using smrtanalysis v2.3. The sub-reads were assembled using the RS_HGAP_Assembly.3 protocol with 350 kb predicted genome size setting, in a de novo assembly (i.e., not guided by any existing virus reference genome). The length cutoff for error-corrected reads going into the overlap-layout-consensus step was 10,984 bases, and a total of 62,573,175 bases in 7,330 reads were used for assembly, resulting in a single 352,593 base initial contig having 1,808-fold average coverage. Remaining putative single-base insertions or deletions (the principal error mode of the platform) were polished by using the contig consensus sequence as reference for an additional round of error-correction with quiver in the RS_Resequencing.1 protocol and default settings. Self-self plot of the contig identified sequence at either end showing evidence that the hairpins had been converted to double-strand DNA as noted in the preceding section. The true end of the genome, and the hairpin sequence, was determined by manual identification of where the sequence became palindromic, i.e. the sequence reads extended through the hairpin and began to read back along the complementary strand of the genome. After trimming the excess self-complementary sequence, a genome of 327,235 nucleotides was produced that includes the hairpins as a 44 base extension at the beginning and end of the assembly. Due to the unusual structures at the ends, this representation begins with the first base that is actually connected by phosphodiester bonds to the 3’ end of the complementary strand recessed by 44 based from the start of the sequence, and ends with the base that is connected to the 5’ end of the complementary strand recessed 44 bases prior to the end of the sequence (see Results and Fig. 5).
The OSy-NE5 CDSs were predicted using the GeneMarkS web tool, which provides accesses to version 4.28 of the gene prediction program (Besemer et al., 2001). This version combines the original 2001 prokaryotic GeneMarkS with later developments to extend the unsupervised gene prediction to intron-less eukaryotes, eukaryotic viruses, phages, and EST/cDNA sequences. Prokaryotic sequence types and genetic code 11 were chosen for the analysis. The predicted genes were then annotated using an in house developed pipeline based on BLAST and NR databases from NCBI. The annotated genes were then manually curated and listed in Sup. Table 1.
tRNAs were predicted using tRNAscan-SE v.1.3.1 (Lowe & Eddy, 1997). This program used a modified, optimized version of tRNAscan v1.3 (Fichant & Burks, 1981), a new implementation of a multistep weight matrix algorithm for identification of eukaryotic tRNA promoter regions (Pavesi et al., 1994), and the RNA covariance analysis package Cove v.2.4.2. (Eddy & Durbin, 1994).
The genomic circular map was generated using GView (Petkau et al., 2010). COG groups were manually added in the outer track based on Sup Table 3. The genomic dotplots were generated using SynMap web tool from CoGe, which is a platform for performing comparative genomic research (Lyons et al., 2008).
The OSy-NE5 DNA sequence is in GeneBank under accession number KX857749.
Phylogenetic analysis
The evolutionary history was inferred by using the Maximum Likelihood (ML) method based on the JTT matrix-based model (Jones et al., 1992). The rooted tree with the highest log likelihood is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 29 of 32 core-viral concatenated amino acid sequences (Jeanniard et al., 2013). Forty-one viruses are chlorovirus sequences and four are Ostreococcus viral sequences (out group). All positions containing gaps and missing data were eliminated. There were a total of 7762 positions in the final dataset. Evolutionary analyses were conducted in MEGA 6.0 software (Tamura et al., 2013).
Epifluorescent microscopy
An Axio Imager A1 epifluorescent microscope was used with 20x and 40x objectives. SYBR® Gold stain was visualized by excitation with a mercury vapor short arc lamp HBO®.
Chlorella NC64A and Syngen cells were grown to early log phase (4 - 7 × 106 cells/ml) and concentrated ten fold. Three hundred μl of cells were mixed with 30 μl of SYBR® Gold stain (10X) following manufacturer's instructions. Cells were aliquoted in 30 μl amounts and infected with 10 μl of purified virus at a MOI of 20. After 30 min, cells were visualized under the microscope. Pictures were taken with a ProgRes® C14plus camera.
Flow cytometry analysis
Viral infection was analyzed by flow cytometry in the Morrison Core Research Facility at the University of Nebraska-Lincoln, using BD FACSCalibur for acquisition and FlowJo software for data analysis. Samples were run at 1 × 106 cells/ml, and approximately 1 × 104 cell events were collected per sample. SYBR® Gold stain was diluted in Tris buffer (50 mM Tris, 10 mM EDTA, pH 7.4) and mixed with uninfected and infected cells following the manufacturer’s instructions. Chlorella cells were gated based on light scatter properties and analyzed in the FL-1 channel for SYBR® Gold stain intensity (488 nm excitation, 530/30 nm emission). Histograms were representative of 3 independent experiments.
Supplementary Material
Acknowledgements
Funding for this study was partially provided by the NSF-EPSCoR grant EPS-1004094 (JVE), the COBRE program of the National Center for Research Resources Grant P20-RR15535 (JVE), the National Center for Research Resources grant 5P20RR016469 (GAD), and the National Institute for General Medical Science grant 8P20GM103427 (GAD). OS was supported by UNL UCARE and ARD scholarships. AE was supported by the Egyptian Ministry of Higher Education (Cultural Affairs and Missions Sector). We thank Danielle Shae and Han Chen in the Morrison Core Research Facility at UNL and William Thompson and Kristen Kuhn at USDA/ARS, for technical assistance. TPLS was supported by USDA/ARS appropriated project 5438-31320-012-00D. The USDA is an equal opportunity employer. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors declare no conflict of interest.
Author Contributions: C.F.Q., A.E., O.S. and J.L.V.E. designed research; C.F.Q., A.E., O.S., M.M., I.A., T.P.L.S., C.D., I.S. performed the experiments; T.P.L.S., G.A.D., A.E. and F.M. annotated the viral genome; C.F.Q., A.E., I.A., D.D.D. and J.L.V.E. analyzed data; and C.F.Q., A.E., O.S., M.B., C.D., and J.L.V.E. wrote the paper.
References
- Agarkova IV, Dunigan DD, Van Etten JL. Virion-associated restriction endonucleases of chloroviruses. J. Virol. 2006;80:8114–8123. doi: 10.1128/JVI.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuchird N, Nishida K, Kawasaki T, Fujie M, Usami S, Yamada T. A variable region on the chlorovirus CVK2 genome contains five copies of the gene for Vp260, a viral glycoprotein. Virology. 2002;295:289–298. doi: 10.1006/viro.2002.1408. [DOI] [PubMed] [Google Scholar]
- De Castro C, Molinaro A, Piacente F, Gurnon JR, Sturiale L, Palmigiano A, Lanzetta R, Parrilli M, Garosso D, Tonetti M, Van Etten JL. Structure of the N-linked oligosaccharides attached to virus PBCV-1 major capsid protein: an unusual class of complex N-glycans. Proc. Natl. Acad. Sci. USA. 2013;110:13956–13960. doi: 10.1073/pnas.1313005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro C, Speciale I, Duncan G, Dunigan DD, Agarkova I, Lanzetta R, Sturiale L, Palmigiano A, Garozzo D, Molinaro A, Tonetti M, Van Etten JL. Chloroviruses N-linked glycans share a new type of conserved core architecture unprecedented in any form of life. Angew. Chem. Int. Ed. 2016;55:654–658. doi: 10.1002/anie.201509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Dunigan DD, Cerny RL, Bauman AT, Roach JC, Lane LC, Agarkova IV, Wulser K, Yanai-Balser GM, Gurnon JR, Vitek JC, Kronschnabel BJ, Jeanniard A, Blanc G, Upton C, Duncan GA, McClung OW, Ma F, Van Etten JL. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J. Virol. 2012;86:8821–8834. doi: 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy ER, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichant GA, Burks C. Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol. 1991;220:659–671. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- Hoshina R, Iwataki, Mimamura N. Chlorella variabilis and Micractinium reisseri sp. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycol. Res. 2010;58:188–201. [Google Scholar]
- Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J, Duncan G, McClung OW, Larsen M, Claverie JM, Van Etten JL, Blanc G. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics. 2013;14:158. doi: 10.1186/1471-2164-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. BioSci. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kamako S, Hoshina R, Ueno S, Imamura N. Establishment of axenic endosymbiotic strains of Japanese Paramecium bursaria and the utilization of carbohydrate and nitrogen compounds by the isolated algae. Eur. J. Protistol. 2005;41:193–202. [Google Scholar]
- Karakashian MW. Symbiosis in Paramecium bursaria. Symp. Soc. Exp. Biol. 1975;29:145–173. [PubMed] [Google Scholar]
- Karakashian SJ, Karakashian MW. Evolution and symbiosis in the genus Chlorella and related algae. Evolution. 1965;19:368–377. [Google Scholar]
- Kodama Y, Suzuki H, Dohra H, Sugii M, Kitazume T, Yamaguchi K, Shigenobu S, Fujishima M. Comparison of gene expression of Paramecium bursaria with and without Chlorella variabilis symbionts. BMC Genomics. 2014;15:183. doi: 10.1186/1471-2164-15-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M. The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates rosids. Tropical Plant Biol. 2008;1:181–190. [Google Scholar]
- Meints RH, Lee K, Burbank DE, Van Etten JL. Infection of a chlorella-like alga with the virus, PBCV-1: Ultrastructural studies. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- Milrot E, Mutsafi Y, Fridmann-Sirkis Y, Shimoni E, Rechav K, Gurnon JR, Van Etten JL, Minsky A. Virus-host interactions: Insights from the replication cycle of the large Paramecium bursaria chlorella virus. Cellular Microbiol. 2016;18:3–16. doi: 10.1111/cmi.12486. [DOI] [PubMed] [Google Scholar]
- Morrissey JH. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Analytical Biochem. 1981;17:30731. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Nicols HW, Bold HC. Trichosarchina polymorpha gen.et. sp. nov. J. Phycol. 1965;1:34–38. [Google Scholar]
- Pavesi A, Conterio F, Bolchi A, Dieci G, Ottonello S. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res. 1994;22:1247–1256. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschold T, Darienko T, Silva PC, Reisser W, Krienitz L. The systematics of zoochlorella revisited employing an integrative approach. Environ. Microbiol. 2011;13:350–364. doi: 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- Que Q, Li Y, Wang IN, Lane LC, Chaney WG, Van Etten JL. Proteinglycosylation and myristylation in chlorella virus PBCV-1 and its antigenic variants. Virology. 1994;203:320–327. doi: 10.1006/viro.1994.1490. [DOI] [PubMed] [Google Scholar]
- Quispe CF, Sonderman O, Khasin M, Riekhof WR, Van Etten JL, Nickerson KW. Comparative genomics, transcriptomics, and physiology distinguish symbiotic from free-living Chlorella strains. Algal Res. 2016a;18:332–340. [Google Scholar]
- Quispe C, Sonderman O, Seng A, Rasmussen B, Webber G, Mueller C, Dunigan DD, Van Etten JL. Three-year survey of abundance, prevalence, and genetic diversity of Chlorovirus populations in a small urban lake. Arch. Virology. 2016;161:1839–1847. doi: 10.1007/s00705-016-2853-4. [DOI] [PubMed] [Google Scholar]
- Reisser W, Burbank DE, Meints SM, Meints RH, Becker B, Van Etten JL. A comparison of viruses infecting two different Chlorella-like green alga. Virology. 1988;167:143–149. doi: 10.1016/0042-6822(88)90063-3. [DOI] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33(sup 2):W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrdla MP, Burbank DE, Xia Y, Meints RH, Van Etten JL. Structural proteins and lipids in a virus, PBCV-1, which replicates in a chlorella-like alga. Virology. 1984;135:308–315. doi: 10.1016/0042-6822(84)90188-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Burbank DE, Kuczmarski D, Meints RH. Virus infection of culturable chlorella-like algae and development of a plaque assay. Science. 1983a;219:994–996. doi: 10.1126/science.219.4587.994. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Burbank DE, Schuster AM, Meints RH. Lytic viruses infecting a chlorella-like alga. Virology. 1985a;140:135–143. doi: 10.1016/0042-6822(85)90452-0. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Burbank DE, Xia Y, Meints RH. Growth cycle of a virus, PBCV-1, that infects Chlorella-like algae. Virology. 1983;126:117–125. doi: 10.1016/0042-6822(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Dunigan DD. Chloroviruses: not your everyday plant virus. Trends Plant Sci. 2012;17:1–8. doi: 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Lane LC, Dunigan DD. DNA viruses: The really big ones (giruses) Annu. Rev. Microbiol. 2010;64:83–99. doi: 10.1146/annurev.micro.112408.134338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Van Etten CH, Johnson JK, Burbank DE. A survey for viruses from fresh water that infect a eucaryotic chlorella-like green alga. Appl. Environ. Microbiol. 1985b;49:1326–1328. doi: 10.1128/aem.49.5.1326-1328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Alllen MJ. The Phycodnaviridae: the story of how tiny giants rule the world. Curr. Top. Microbiol. Immunol. 2009;328:1–42. doi: 10.1007/978-3-540-68618-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Schroeder DS, Nagasaki K, Brussaard C, Bratbak G, Suttle C. Phycodnaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy, IXth Report of the ICTV. Elsevier/Academic Press; Amsterdam: 2012. pp. 219–262. [Google Scholar]
- Yamada T, Higashiyama T, Fukuda T. Screening of natural waters for viruses which infect chlorella cells. Appl. Environ. Microbiol. 1991;57:3433–3437. doi: 10.1128/aem.57.12.3433-3437.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Olson NH, Van Etten JL, Bergoin M, Rossmann MG, Baker TS. Structure and assembly of large lipid-containin dsDNA viruses. Nat. Struc. Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Burbank DE, Van Etten JL. Chlorella viruses isolated in China. Appl. Environ. Microbiol. 1988;54:2170–2173. doi: 10.1128/aem.54.9.2170-2173.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Strasser P, Grabherr R, Van Etten JL. Hairpin loop structure at the termini of the chlorella virus PBCV-1 genome. Virology. 1994;202:1079–1082. doi: 10.1006/viro.1994.1444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.