Abstract
The prevalence of pathogenic multi-drug resistant (MDR) extended-spectrum β-lactamase (ESBL)-producing Escherichia coli is rapidly increasing, becoming a global concern. In a veterinary context, ESBL-producing E. coli are mostly reported in poultry and pigs. Here, we report on the prevalence and characterize ESBL-producing E. coli isolated from diverse dairy farms in China. Overall, 36 (23.53%) out of 153 E. coli isolates from mastitic milk samples (n = 1252) were confirmed as ESBL-producers by double-disc synergy testing and PCR. Nucleotide analysis of PCR amplicons revealed that blaCTX-M was the predominant ESBL gene detected in 28 (77.78%) isolates, with blaCTX-M-15 being the major (78.57%) allele encoding for ESBLs. Also, 20 (55.56%) and 6 (16.67%) of the ESBL isolates were carrying blaTEM and blaSHV genes, respectively, in singlet or in combination. The majority of these isolates belonged to phylo-group A (69.44%) and D (16.67%). Strikingly, all these isolates were found to be MDR showing high resistance to cephalosporins including the fourth generation cefepime and common non β-lactams. Additionally, class 1 integrons (intI1) were found in 30 (83.33%) isolates. Analysis of the class 1 integrons variable regions indicated that they were carrying up to five different gene cassettes conferring resistance to various drugs with a predominant combination of dfrA17-aadA5 genes in tandem, conferring resistance to aminoglycosides and trimethoprim. However, no ESBL encoding genes were found in the cassettes. Interestingly, 22 (66.11%) of the ESBL isolates were also carrying insertion sequence common region 1 (ISCR1) which was found to be associated with most of the CTX-M genes. Altogether, the current study reports on the high prevalence of ESBL-positive E. coli, particularly CTX-M-15, carrying clinical class 1 integrons and ISCR1 elements are likely indicative of their rapid and wider dissemination, posing threats to veterinary and public health. To the best of our knowledge, this is the first comprehensive study to report on the alarming high occurrence of ESBL-producing E. coli from mastitic cows in China.
Keywords: E. coli, ESBLs, CTX-M-15, integrons, gene cassettes, bovine mastitis
Introduction
Bovine mastitis, inflammation of the mammary gland, is the most prevalent and economically important disease of dairy animals (Halasa et al., 2007). Mastitis can be caused by a variety of bacterial pathogens, but Escherichia coli is one of the leading causes (Dahmen et al., 2013). Antimicrobial agents are used for therapeutic as well as preventive measures against bacterial infections including bovine mastitis in farm animals. Beta-lactams, such as ampicillin and amoxicillin, remain the first-line treatment in veterinary medicine but an increase in drug-resistance to these antibiotics has been observed. Therefore, extended-spectrum cephalosporins (ESC) such as ceftiofur have been approved in China for the treatment of animal diseases (MAO, 2010). Unfortunately, several recent studies have reported the increasing occurrence of highly resistant extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, mainly E. coli, isolated from food-producing animals from various countries including China (Rao et al., 2014; Xu et al., 2015; Seni et al., 2016).
Bacterial resistance to β-lactams, popular antibiotics due to their proven safety and efficiency, is increasing at an alarming rate. This resistance is mainly achieved through β-lactamases that can hydrolyse most β-lactam antibiotics including the third and fourth generation ESCs and monobactams (Bush and Jacoby, 2010). ESBLs are predominantly produced in gram negative bacteria, particularly in E. coli, and are considered a key mechanism conferring resistance to cephalosporins (Perez et al., 2007). Multi-drug resistance (MDR) has been commonly observed in most ESBL-producers and more alarmingly, co-resistance to other commonly used antibiotics like aminoglycosides, fluoroquinolones, tetracycline has been often reported (Chen et al., 2010; Timofte et al., 2014; Xu et al., 2015). This renders these organisms resistant to a wide range of antibiotics with limited therapeutic options. ESBL encoding genes have been categorized into three main types: blaCTX-M, blaSHV, and blaTEM. The blaCTX-M has been further categorized into five sub-groups (blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, blaCTX-M-25) and more than 150 variants have been documented (http://www.lahey.org/studies). In the past few years, CTX-M, especially CTX-M-15, has emerged as the most dominant type of ESBLs globally (D'Andrea et al., 2013). Recently, CTX-M-15 producing E. coli have been frequently documented from various sources including humans and food producing animals (Timofte et al., 2014; Liu et al., 2015; Xu et al., 2015), showing the broad spectrum of reservoirs carrying and spreading these genes. Food-animals are well established reservoirs of ESBL-producing E. coli, which can be transmitted from animals to humans by various direct and indirect means (Dahmen et al., 2013; Geser et al., 2015). This is also verified by Madec et al. (2012), they reported that the plasmids carrying CTX-M-15 genes in E. coli isolated from cattle were highly similar to those found in ESBL-producing E. coli isolates from human beings.
Integrons are genetic elements that play a vital role in the development and dissemination of MDR in clinical isolates due to their ability to capture, integrate and express gene cassettes (Vinue et al., 2008; Chen et al., 2010). Three main classes of integrons (1–3), carrying the gene cassettes encoding for antimicrobial resistance genes, are generally found to be associated with antibiotic resistance genes in pathogenic E. coli, Class 1 integrons are the most common in clinical E. coli, followed by less frequent class 2 integrons (Vinue et al., 2008; Xu et al., 2015). Class 1 integrons contain a 5′ conserved segment (CS) and 3′CS, followed by a variable region that contains one or more gene cassettes. The 5′CS consists of an integrase gene (intI1), a recombination-site (attI1), and the Pc promoter(s), and the 3′CS includes qacEΔ1 and sul1 genes which encode for quaternary ammonium compound and sulphonamide resistance (Hall and Stokes, 1993). Moreover, insertion sequences like ISCR1 (insertion sequence common region 1) as part of the complex class 1 integrons are found to be associated with ESBL and other resistance encoding genes and are probably involved in their mobilization and transposition. ISCR1 may mobilize the truncated 3′CS and nearby sequences from one integron to the 3′CS of another integron utilizing rolling circle transposition, thus facilitating dissemination of resistance elements (Eckert et al., 2006; Toleman et al., 2006).
Limited studies, particularly from China, have characterized ESBL-producing E. coli isolated from diseased food producing animals, mainly from mastitic cows (Lu et al., 2010; Timofte et al., 2014). Thus, we designed the current study to investigate the prevalence of pathogenic ESBL-producing E. coli and to characterize the ESBL genes and genetic elements which are likely to be responsible for their mobility and dissemination. To the best of our knowledge, this is the first comprehensive study into the molecular characterization of ESBL genes in E. coli isolated from dairy cows in China.
Materials and methods
Statement of ethics
The present study was conducted in accordance with the ethical guide lines of China Agricultural University (CAU), Beijing. Proper ethical approval was granted by the departmental committee of College of Veterinary Medicine, CAU. Sampling was carried according to the standard protocols and with prior consent of the dairy herd's authority.
Sample collection and location
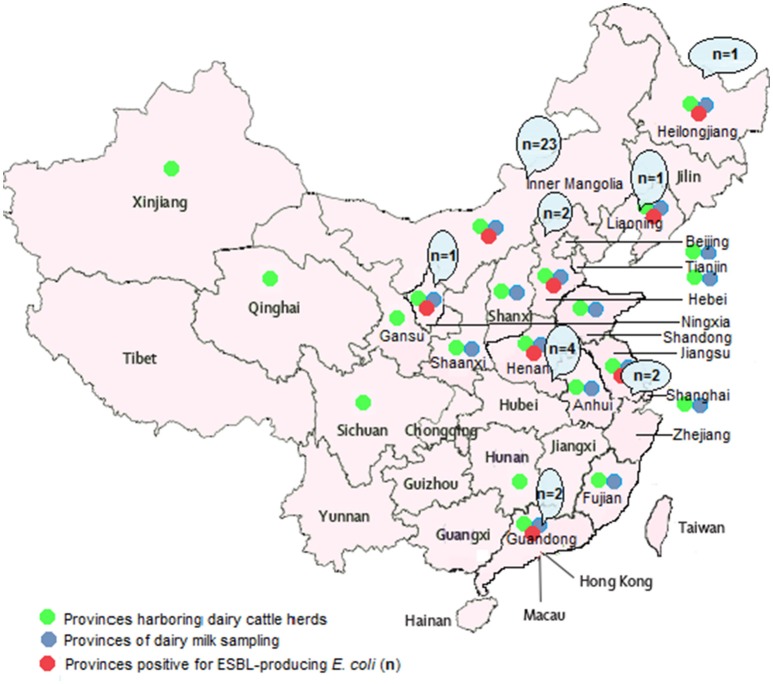
Milk samples of mastitic cows (n = 1252) were collected from 61 large commercial dairy herds (2000–40,000 cows/herd) located in 16 provinces of China during January 2015 to May 2016 (Figure 1 and Table 1). Sampling was carried out when the cows were suffering from mastitis and not according to a fixed schedule. The guidelines of the National Mastitis Council (NMC, 1999) were followed for the collection of milk samples from cows. Samples were taken in 50 mL sterile tubes and transported on ice to the laboratory for further processing.
Figure 1.
Map of China (mainland) showing 16 provinces from where samples were collected.
Table 1.
Occurrence of ESBL-producing E. coli isolated from dairy herds located in 16 provinces of China.
| Provinces of sampling | No. of dairy herdsa | No. of milk samplesb | E. coli isolatesc | ESBL E. coli from each herdd |
|---|---|---|---|---|
| Anhui | A | 63 | 4 | 0 |
| Beijing | B/B1/B2 | 26 (6/9/11) | 5 (2/2/1) | 0/0/0 |
| Fujian | F | 12 | 0 | 0 |
| Guangdong | G/G1 | 98 (23/75) | 3 (1/2) | 2 (0/2) |
| Hebei | Hb/ Hb1/ Hb2/ Hb3 | 220 (11/10/16/16 | 36 (2/0/0/5 | 2 (0/0/0/1 |
| /Hb4/Hb5/Hb6/Hb7 /Hb8/Hb9/Hb10 | /6/11/12/18/23 /24/12/38/14/9) | /1/0/0/4/5 /3/3/6/6/1) | /0/0/0/0/1 /0/0/0/0/0) | |
| /Hb11/Hb12/Hb13 | ||||
| Heilongjiang | H/H1/H2/H3 | 73 (10/13/10/40) | 7 (4/2/0/1) | 1 (1/0/0/0) |
| Henan | Hn/Hn1/Hn2/Hn3/Hn4 | 43 (12/6/7/12/6) | 5 (2/0/0/2/1) | 4 (2/0/0/2/0) |
| Inner-Mongolia | I/I1/I2/I3/I4/I5 | 425 (17/14/12/18 | 45 (1/2/0/1 | 23 (0/0/0/0 |
| /I6/I7/I8/I9/I10 | /42/37/49/22/24 | /6/8/7/5/3 | /5/7/6/3/0 | |
| /I11/I12/I13 | /61/33/53/22/21) | /6/0/3/2/1) | /2/0/0/0/0) | |
| Jiangsu | J | 9 | 4 | 2 |
| Liaoning | L | 20 | 4 | 1 |
| Ningxia | N/N1/N2/N3 | 97 (15/32/20/30) | 19 (2/7/5/5) | 1 (0/1/0/0) |
| Shaanxi | Sx-Bj | 13 | 2 | 0 |
| Shandong | S/S1/S2/S3 | 83 (14/14/15/40) | 8 (0/1/5/2) | 0/0/0/0 |
| Shanxi | Sx-Cz | 6 | 0 | 0 |
| Shanghai | S/S1/S2/S3 | 59 (16/16/10/17) | 10 (1/0/6/3) | 0/0/0/0 |
| Tianjin | T | 5 | 1 | 0 |
| Total | 61 | 1252 | 153 (12.22%) | 36 (23.53%) |
The letters in the column represent the farm number.
The numbers in parenthesis indicate no. of milk samples corresponding to the respective farm in second column.
The numbers in parenthesis show E. coli isolates from the respective farm.
The numbers in parenthesis indicate ESBL-producing E. coli isolated from the respective farm.
Isolation and identification of E. coli
Milk samples, shortly after arrival, were streaked (10 μL) onto MacConkey Agar (Difco™, Becton Dickinson, Sparks, MD USA) and incubated at 37°C for 18–24 h. Presumptive E. coli colonies with the dark pink to red colors, were further confirmed with the API-20E kit (bioMérieux, Marcy I'Etoile, France) as per instruction of the manufacturer. Biochemically confirmed E. coli isolates were further verified by PCR as described previously (Tantawiwat et al., 2005). Confirmed E. coli isolates were stored in brain heart infusion broth (BHI; Sigma-Aldrich) containing 30% glycerol at −80°C.
Phenotypic screening of ESBL-Producers
E. coli isolates were first screened for the phenotypic identification of ESBLs-producers on MacConkey agar containing cefotaxime (1 mg/L). These presumptive ESBL-producing E. coli were further confirmed by double-disc synergy testing in accordance with recommendations of the Clinical and Laboratory Standards Institute (CLSI, 2014), using antimicrobial discs (Becton Dickinson, Sparks, MD USA) of cefotaxime (30 μg), cefotaxime plus clavulanic acid (30/10 μg), ceftazidime (30 μg), and ceftazidime plus clavulanic acid (30/10 μg). The test was recorded positive when the zone of inhibition of cefotaxime plus clavulanic acid or ceftazidime plus clavulanic acid was ≥5 mm larger than their respective single discs (CLSI, 2014).
Genotypic screening of ESBL-Producing E. coli isolates
Bacterial DNA from ESBL-positive E. coli was isolated by the TIANamp Bacteria DNA Kit (TIANGEN, Beijing, China) according to the manufacturer's instructions. PCR assays were used for the detection of blaCTX-M,blaSHV, blaTEM genes as described previously (Chen et al., 2010). Details of the primers used in this study are shown in Table 2. All ESBL genes relevant PCR amplicons were purified by the TIANquick Midi Purification Kit (TIANGEN, Beijing, China), bi-directionally sequenced and aligned with sequences available in GenBank (Chen et al., 2010). Klebsiella pneumoniae ATCC 700603 (ESBLs-positive strain) and ddH2O, instead of template DNA, was used as positive and negative controls, respectively, in all PCR assays.
Table 2.
Details of primers used in this study.
| Primers | Sequence (5′ to 3′) | Target gene | Annealing temperature | Amplicons size | References |
|---|---|---|---|---|---|
| β-lactamases | |||||
| CTX-MA CTX-MB |
CGC TTT GCG ATG TGC AG ACC GCG ATA TCG TTG GT |
blaCTX-M | 54°C | 550-bp | Villegas et al., 2004 |
| SHV-F SHV-R |
GGG TTA TTC TTA TTT GTC GC TTA GCG TTG CCA GTG CTC |
blaSHV | 58°C | 567-bp | Chang et al., 2001 |
| TEM-F TEM-R |
ATA AAA TTC TTG AAG ACG AAA GAC AGT TAC CAA TGC TTA ATC |
blaTEM | 56°C | 1086-bp | Yao et al., 2007 |
| INTEGRONS | |||||
| intI1-F | CCT CCC GCA CGA TGA TC | intI1 | 54°C | 280-bp | Dillon et al., 2005 |
| intI1-R | TCC ACG CAT CGT CAG GC | ||||
| intI2-F | AAA TCT TTA ACC CGC AAA CGC | intI2 | 54°C | 439-bp | Dillon et al., 2005 |
| intI2-R | ATG TCT AAC AGT CCA TTT TTA AAT TCT A | ||||
| intI3-F | AGT GGG TGG CGA ATG AGT G | intI3 | 54°C | 599-bp | Dillon et al., 2005 |
| intI3-R | TGT TCT TGT ATC GGC AGG TG | ||||
| intI1-VR-F | TCA TGG CTT GTT ATG ACT GT | intI1variable region | 56°C | variable | White et al., 2000 |
| ntI1-VR-R | GTA GGG CTT ATT ATG CAC GC | ||||
| E. coli-SPECIFIC | |||||
| UAL | TGG TAA TTA CCG ACG AAA ACG GC | uidA | 62°C | 147-bp | Tantawiwat et al., 2005 |
| UAR | ACG CGT GGT TAC AGT CTT GCG | ||||
| PHYLO-GROUPS | |||||
| ChuA-F | GAC GAA CCA ACG GTC AGG AT | ChuA | 55°C | 279-bp | Clermont et al., 2000 |
| ChuA-R | TGC CGC CAG TAC CAA AGA CA | ||||
| YjaA-F | TGA AGT GTC AGG AGA CGC TG | YjaA | 55oC | 211-bp | Clermont et al., 2000 |
| YjaA-R | ATG GAG AAT GCG TTC CTC AAC | ||||
| TspE4C2-F | GAG TAA TGT CGG GGC ATT CA | TspE4C2 | 55°C | 152-bp | Clermont et al., 2000 |
| TspE4C2-R | CGC GCC AAC AAA GTA TTA CG | ||||
| ISCR1 | |||||
| ISCR1-F ISCR1-R |
CGC CCA CTC AAA CAA ACG GAG GCT TTG GTG TAA CCG |
ISCR1 | 55°C | 469-bp | Kiiru et al., 2013 |
F, forward; R, reverse.
Phylogenetic grouping
ESBL-positive E. coli isolates were placed in one of the four phylogenetic groups: phylo-group A, group B1, group B2 or group D. For this purpose, a triplex PCR assay targeting the chuA and yjaA genes and TspE4 was used as described previously by Clermont et al. (2000). The primers sequences and the annealing temperatures are listed in Table 2.
Antibiotic susceptibility testing
Antibiotic susceptibility of ESBL isolates was carried out on Mueller-Hinton agar (Difco™) against 16 different antibiotics discs (Becton Dickinson, Sparks, MD, USA), using the standard Kirby-Bauer disk diffusion method according to recommendations of the CLSI (2014). The panel of antimicrobial agents consisted of both β-lactam and non-β-lactam antibiotics as listed in Table 3. E. coli ATCC 25922 (ESBL-negative strain) and K. pneumoniae ATCC 700603 (ESBL-positive strain) were used as quality control strains (CLSI, 2014). The isolates were declared as multi-drug resistant (MDR) when found resistant to three or more categories of antimicrobial drugs.
Table 3.
Antibiotic susceptibility profiles of ESBL-producing E. coli isolates (n = 36) from milk of mastitic cows.
| Antimicrobial agents | Abbreviations | Conc.* (μg) | Susceptible (%) | Intermediate (%) | Resistance (%) |
|---|---|---|---|---|---|
| Ampicillin | AM | 10 | 11.11 (04/36) | 02.78 (01/36) | 86.11 (31/36) |
| Amoxicillin/clavulanic acid | AMX/CA | 20/10 | 25.00 (9/36) | 11.11 (04/36) | 63.89 (23/36) |
| Cefalexin | CX | 30 | 00.00 (00/36) | 00.00 (00/36) | 100 (36/36) |
| Cefaclor | CEC | 30 | 05.56 (02/36) | 00.00 (00/36) | 94.44 (34/36) |
| Cefoxatin | FOX | 30 | 83.34 (30/36) | 08.33 (03/36) | 8.33 (3/36) |
| Cefotaxime | CTX | 30 | 00.00 (00/36) | 00.00 (00/36) | 100.0 (36/36) |
| Ceftazidime | CAZ | 30 | 33.33 (12/36) | 00.00 (00/36) | 66.67 (24/36) |
| Cefepime | FEP | 30 | 41.67 (15/36) | 11.11 (04/36) | 47.22 (17/36) |
| Aztreonam | AZT | 30 | 13.89 (05/36) | 00.00 (00/36) | 86.11 (31/36) |
| Meropenem | MPN | 10 | 100.0 (36/36) | 00.00 (00/36) | 00.00 (00/36) |
| Tetracycline | TE | 30 | 16.67 (06/36) | 11.11 (04/36) | 72.22 (26/36) |
| Gentamicin | G | 10 | 27.78 (10/36) | 11.11 (04/36) | 61.11 (22/36) |
| Ciprofloxacin | CIP | 05 | 55.56 (20/36) | 08.33 (03/36) | 36.11 (13/36) |
| Chloramphenicol | C | 30 | 47.22 (17/36) | 11.11 (4/36) | 41.67 (15/36) |
| Nalidixic acid | NAL | 30 | 19.44 (07/36) | 02.78 (01/36) | 77.78 (28/36) |
| Trimethoprim/sulphamethoxazole | STX | 1.25/23.75 | 25.00 (09/36) | 02.78 (01/36) | 72.22 (26/36) |
Conc: concentrations.
Detection of integrons, gene cassettes and ISCR1
A PCR assay was used to detect Class 1, 2, and 3 integrons in all ESBL-producing E. coli using integron-integrase gene specific primers, intI1, intI2, and intI3, respectively (Dillon et al., 2005). Subsequently, the intI1 positive genotypes (n = 24) were determined by sequencing amplicons derived from PCR for the class 1 integron variable regions as described previously (White et al., 2000). The ISCR1 region was PCR amplified from ESBL-producing isolates using specific primers (Table 2). The sequenced amplicons of the ISCR1 elements were confirmed by BLAST analysis (see below). A combination of primers specific to the ISCR1 elements (Kiiru et al., 2013) elements and consensus primers of ESBL genes were used to verify their association.
PCR-RFLP genotyping of Class 1 integron variable region amplicons
A PCR-based restriction fragment length polymorphism (PCR-RFLP) assay was adopted to identify genetic variation in the amplified products using restriction enzyme HinfI (Takara, Shiga Japan) as published previously (Gu et al., 2008). PCR-RFLP products with similar band profiles were regarded as the same genotypes carrying identical gene cassette(s).
Nucleotide sequencing and data analysis
Regardless of the similar PCR-RFLP genotypes, all amplicons of gene cassettes, ISCR1 elements, and ESBL genes were bi-directionally sequenced using ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA). PCR amplicons of >1.8 kb were further sequenced using primer walking based on the sequenced amplicons. The obtained sequences were subjected to BLAST homology searches in the INTEGRALL database (http://integrall.bio.ua.pt). Other sequence analyses were compared with BLASTN software (http://www.ncbi.nlm.nih.gov/BLAST/). Clone Manager 7 (Sci-Ed software, Denver, USA) and ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) were also used for detailed analysis such as alignments and open reading frames.
Results
Prevalence and characterization of ESBL-Producing E. coli
Overall, 153 E. coli isolates were recovered from 1252 milk samples of mastitic dairy cows from 16 different provinces of China. Thirty six (23.53%) isolates were detected as ESBL-producing E. coli by phenotypic confirmatory tests and this was also verified by ESBL genotype specific PCR assay. The distribution of these isolates among different cattle herds is shown in Table 1. The highest occurrence of ESBL producers was observed in the Inner Mongolia province (23 isolates), followed by the Henan region (four isolates).
Figure 2 shows the frequency (%) of various ESBL encoding genes among 36 ESBL-producing E. coli isolated from mastitic milk. Overall, blaCTX-M was the most prevalent ESBL gene (77.78%; 28/36), while blaTEM and blaSHV genes were present in 55.56% (20/36) and 16.67% (6/36) of ESBL-positive isolates, respectively. The blaTEM and blaSHV genes were most frequently observed together with blaCTX-M, rather than alone (Figure 2). Notably, two of the isolates from Inner Mongolia carried three β-lactamase genes (blaCTX-M-15+blaTEM-1+blaSHV-12) in combination. Sequence analysis revealed that blaCTX-M-15 was the dominant (78.57%; 22/28) subtype. The other blaCTX-Msubtypes were: blaCTX-M-14 (10.71%; 3/28), blaCTX-M-1 (3.57%; 1/28), blaCTX-M-3(3.57%; 1/28), and blaCTX-M-55 (3.57%; 1/28). The phylo-group A was the most prevalent (69.44%; 25/36) among 36-ESBL-positive E. coli followed by group D (16.67%; 6/36), B1 (8.33%; 3/36), and B2 (5.56%; 2/36) as depicted in Table 4.
Figure 2.
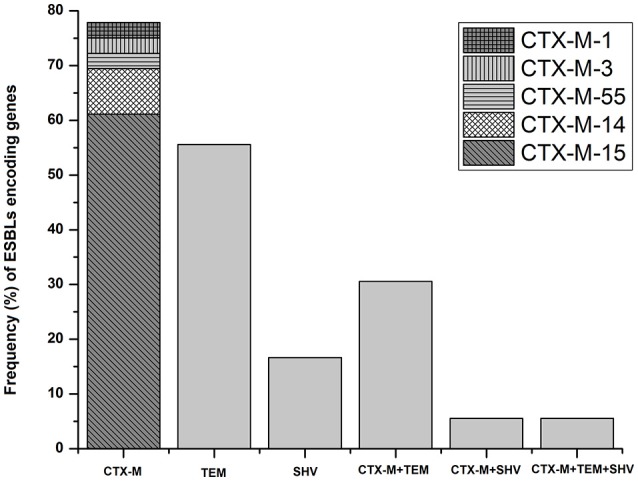
Distribution of ESBLs encoding genes and CTX-M subtypes among ESBL-producing E. coli (n = 36) isolated from bovine mastitis.
Table 4.
Characteristics of ESBL-producing E. coli strains (n = 36) isolated from mastitic cows.
| E. coli isolates | Place of isolation | Phlyo-groups | β-lactamase genes | ISCR1* | ISCR1 association with bla genes | Integron class 1 | IntI1-VR** amplicons (bp) | Gene Cassettes 5′CS -3′CS | GenBank accession numbers | R/I* phenotypes to other non β-lactam antibiotics |
|---|---|---|---|---|---|---|---|---|---|---|
| I-3 | Inner Mongolia | D | TEM-1+SHV-1 | + | + | + | 2200 | dfrA1-aacA4 | KY114582 | Cip; C; NAL; SXT; TE |
| I-4 | Inner Mongolia | A | CTX-M-15 | + | + | + | 1700 | dfrA17-aadA5 | KY114583 | Cip; C; G; NAL; SXT; TE |
| I-5 | Inner Mongolia | D | CTX-M-15+TEM-1+SHV-1 | − | − | + | 2200 | dfrA1- aacA4 | KY114584 | Cip; G; NAL; SXT; TE |
| I-6 | Inner Mongolia | A | TEM-1 | + | + | + | 1700 | dfrA17-aadA5 | KY114585 | Cip; C; G; NAL; SXT; TE |
| I-12 | Inner Mongolia | B1 | CTX-M-15+TEM-1 | + | + | − | − | − | − | Cip; G; NAL; SXT; TE |
| I-14 | Inner Mongolia | A | CTX-M-15+ SHV-1 | + | + | + | 1700 | dfrA17-aadA5 | KY114586 | Cip; C; G; NAL; SXT; TE |
| I-17 | Inner Mongolia | A | CTX-M-15 | − | − | + | 1700 | dfrA17-aadA5 | KY114587 | Cip; G; NAL; SXT; TE |
| I-18 | Inner Mongolia | A | CTX-M-15 | + | + | − | − | − | − | Cip; G; NAL; SXT; TE |
| I-22 | Inner Mongolia | D | CTX-M-15+SHV-1 | + | + | + | − | − | − | Cip; G; NAL; SXT; TE |
| I-25 | Inner Mongolia | A | CTX-M-15+TEM-1+SHV-1 | − | − | + | 1700 | dfrA17-aadA5 | KY114588 | Cip; G; NAL; SXT; TE |
| I1-1 | Inner Mongolia | A | CTX-M-14 | + | + | + | 1800 | dfrA17-aada4 | KY114589 | − |
| I1-2 | Inner Mongolia | A | TEM-1 | + | + | + | − | − | − | G; TE |
| I1-3 | Inner Mongolia | A | TEM-1 | + | − | − | − | − | − | − |
| I1-4 | Inner Mongolia | A | CTX-M-15 | − | − | + | 1700 | dfrA17-aadA5 | KY114590 | − |
| I1-5 | Inner Mongolia | A | CTX-M-15 | _ | _ | _ | _ | _ | _ | NAL |
| I1-7 | Inner Mongolia | B1 | CTX-M-55+TEM-1 | _ | _ | + | 1700 | dfrA17-aadA5 | KY114591 | Cip; C; G; NAL; SXT; TE |
| I1-8 | Inner Mongolia | A | CTX-M-14 | − | − | + | 1800 | dfra17-aada4 | KY114592 | C; G; NAL; SXT; TE |
| I1-11 | Inner Mongolia | B1 | TEM-1 | − | − | + | 1700 | dfrA17-aadA5 | KY114593 | C; G; NAL; SXT; TE |
| I2-1 | Inner Mongolia | A | CTX-M-15+TEM-1 | − | − | + | 1700 | dfrA17-aadA5 | KY114594 | Cip; C; G; NAL; SXT; TE |
| I2-2 | Inner Mongolia | A | TEM-1 | − | − | + | 1700 | dfrA17-aadA5 | KY114595 | C; G; NAL; SXT; TE |
| I2-3 | Inner Mongolia | A | CTX-M-14+TEM-1 | − | − | + | 1800 | dfra17-aada4 | KY114596 | C; G; NAL; SXT; TE |
| I3-1 | Inner Mongolia | B2 | CTX-M-15 | + | + | + | 1700 | dfrA17-aadA5 | KY114597 | C; G; NAL; SXT; TE |
| I3-2 | Inner Mongolia | B2 | CTX-M-15 | + | + | + | 1700 | dfrA17-aadA5 | KY114598 | C; G; NAL; SXT; TE |
| G-1 | Guangdong | A | CTX-M-15 | + | + | + | 1700 | dfrA17-aadA5 | KY114599 | Cip; C; NAL; SXT; TE |
| G-2 | Guangdong | A | CTX-M-15+TEM-1 | + | + | + | 1700 | dfrA17- aadA5 | KY114600 | C; NAL; SXT; TE |
| J-4 | Jiangsu | D | CTX-M-15+TEM-1 | + | + | + | − | − | − | C; G; NAL; SXT; TE |
| J-5 | Jiangsu | D | CTX-M-15+TEM-1 | + | + | + | − | − | − | C; G; NAL; SXT; TE |
| Hb2-4 | Hebei | D | TEM-1 | − | − | + | 2000 | dfrA17-aadA5 | KY114601 | C; G; NAL; SXT; TE |
| Hb3-1 | Hebei | A | CTX-M-15 | + | + | + | 1000 | aadA1 | KY114602 | Cip; C; G; NAL; SXT; TE |
| L-1 | Liaoning | A | CTX-M-15+TEM-1 | + | + | + | 1700 | dfrA17-aadA5 | KY114603 | C; G; NAL; SXT; TE |
| N-1 | Ningxia | A | CTX-M-3+TEM-1 | − | − | + | 1300 | aadA5 | KY114604 | TE |
| H-5 | Heilojinag | A | SHV-12 | + | + | − | − | − | − | − |
| Hn-6 | Henan | A | CTX-M-15 | − | − | − | − | − | − | C; TE |
| Hn1-2 | Henan | A | CTX-M-1 | + | + | + | 1700 | dfrA17-aadA5 | KY114605 | G; NAL |
| Hn1-6 | Henan | A | CTX-M-15+TEM-1 | + | − | + | 1700 | dfrA17-aadA5 | KY114606 | G; NAL; SXT; TE |
| Hn1-7 | Henan | A | CTX-M-15+TEM-1 | + | + | + | 1700 | dfrA17-aadA5 | KY114607 | G; NAL; SXT; TE |
R/I, Resistance/Intermediary; aacA4, aminoglycoside 6′-N-acetyltransferase; aadA, aminoglycoside adenyltransferase; dfrA1, dihydrofolate reductase type A; dfrA17, dihydrofolate reductase DHFRXVII; Cip, ciprofloxacin; C, chloramphenicol; G, gentamicin; NAL, nalidixic acid; SXT, trimethoprim/sulphamethoxazole; TE, tetracycline.
ISCR1: Insertion sequence common region 1.
IntI1-VR-: class 1 integrons variable regions, approximate size of base pairs deduced from running the amplicons on 1% agarose gel and sequencing the amplicon.
Antibiotic susceptibility profiles
All 36 ESBLs-producing E. coli isolates were found to be multiple-drug resistant (MDR). However, different isolates exhibited slight variation in their antibiotic susceptibility profiles against the 16 tested antibiotics (Table 3). The majority of the isolates were resistant to first (cephalexin, 100%), second (cefaclor, 94.4%), third (cefotaxime and ceftazidime, 100% and 66.67%, respectively), and fourth (cefepime, 58.33%) generation cephalosporins. However, a high rate of susceptibility was observed toward cefamycin (cefoxatin, 83.34%) and carbapenem (meropenem, 100%), but susceptibility to monobactams (aztreonam, 13.89%) was low. The isolates were also resistant to other β-lactam and non-β-lactam antibiotics including ampicillin (88.89%), amoxicillin/clavulanic acid (75.00%), chloramphenicol (52.78%), ciprofloxacin (44.44%), gentamicin (72.22%), nalidixic acid (80.56%), tetracycline (83.33%) and trimethoprim/sulphamethoxazole (75%).
Detection of integrons, gene cassettes and ISCR1
Thirty (83.33%) of the ESBL-producing E. coli carried clinical class 1 integrons but class 2 and class 3 integrons were not detected in any of the isolates. Among the intI1+ESBL-producing E. coli, 24 (80.00%) isolates tested positive for the presence of variable regions, while six of the isolates could not be amplified (Table 4). Furthermore, these 24 isolates were also positive for qacEΔ1/sul1 indicating a complete clinical class 1 integron. Integrons lacking 3′CS were not PCR amplified for qacEΔ1/sul1 (results not shown).
The PCR-amplicon sizes of the inserted gene cassettes ranged between ~1.0 and ~2.2 kb with the most predominant being ~1.7 kb amplicons (Figure 3). Most of the PCR amplicons of the variable regions of gene cassette arrays were a single band. However, two of the isolates produced a double band (of ~1.7 and ~ 0.2 kb). Subsequent sequence analysis of the gel extracted amplicons indicated that the smaller band was nonspecific amplification. Different band profiles of PCR-RFLP products indicated five distinct genotypic configurations (Figure 4). The most predominant PCR-RFLP genotype produced a profile of ~0.6, ~0.4, ~0.45, ~0.2, and ~0.22 kb restriction fragments consistent with digestion of 1.7 kb PCR amplicon of the variable regions. Amplicons sequence analysis of the variable regions revealed five gene cassettes carrying single or two genes in tandem. The predominant combination was dfrA17-aadA5 in tandem that conferred resistance to aminoglycosides and trimethoprim. Interestingly, all the CTX-15-positive isolates, except two, carried dfrA17-aadA5 genes in combination. This was consistent with the antibiotic susceptibility profile of these isolates reflecting resistance to the relevant drugs (Table 4). Surprisingly, no ESBL genes were found encoded in the variable region of the gene cassette array of these isolates. Therefore, ISCR1 elements were investigated by targeted-PCR. The PCR amplicons of ISCR1 elements were sequenced and confirmed by homology. Results indicated that ISCR1 was found in 22 (66.11%) ESBL positive isolates (Table 4). Moreover, ISCR1 (Accession number KY095113) was found associated with blaCTX-M, blaTEM and blaSHV in 16, 3, and 4 isolates, respectively. Interestingly, all blaCTX-M-15 positive isolates, except one (Hn1-6), have been always found associated with ISCR1 elements. However, blaTEM, when found alone or in combination with others, except blaCTX-M-15, was mainly negative for ISCR1 elements. The amplicon size resulting from PCR using primer combinations specific to ISCR1 elements and ESBL genes revealed that the ESBL genes were oriented downstream of ISCR1 elements. Altogether, these results indicated that ISCR1 elements are associated with ESBL genes. The detailed characterizations of all ESBLs-producing E. coli are elaborated in Table 4.
Figure 3.
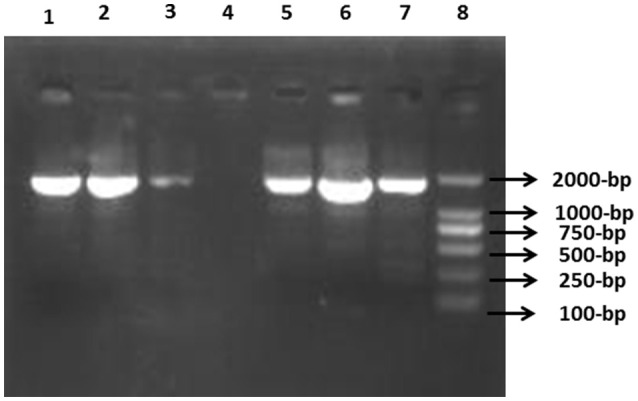
Detection of class 1 integrons variable regions in ESBL-producing E. coli. PCR product was separated on 1% agarose gel. Lane 1, I-3 (intI1+) isolate; Lane 2, I1-8strain; Lane 3, G-2 isolate; Lane 4, H-5 (intI1-ve) isolate; Lane 5, I2-3 E. coli; Lane 6, Hn1-2 strain; Lane 7, positive control strain; Lane 8, 2K molecular marker (Transgen, Beijing, China).
Figure 4.
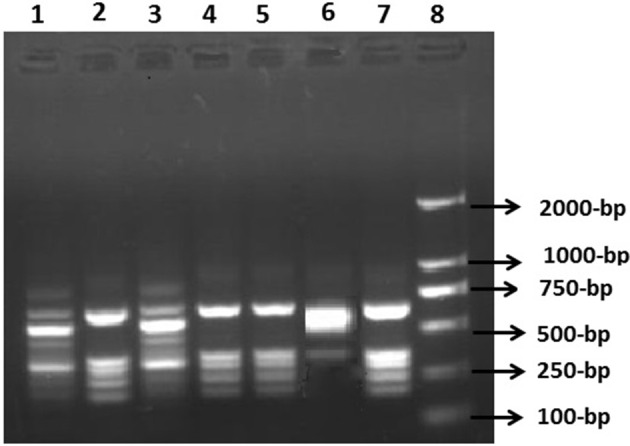
Restriction fragment length polymorphism (RFLP) analysis of intI1 variable region amplicons using Hinf I enzyme. RFLP product was analyzed on 1.5% agarose gel. Lane 1, I-3 isolate; Lane 2, I-25 E. coli; Lane 3, Hn1-2 isolate; Lane 4, I2-3 isolate; Lane 5, G-2 strain; Lane 6, Hb3-1isolate; Lane 7, Hn1-7 E. coli strain; Lane 8, 2K molecular marker.
Discussion
In the past few years, ESBL-producing E. coli have been increasingly isolated from food-producing animals raising global concerns for veterinary and public health (Seiffert et al., 2013). The current study reports on the higher occurrence (23.53%) of ESBL-producing E. coli (n = 36) among 153 E. coli isolates from mastitic cows, as compared to previous reports from China (Yu et al., 2015) and other countries (Dahmen et al., 2013; Geser et al., 2015; Freitag et al., 2016). Interestingly, the majority of ESBL-producing E. coli (23 isolates) were recovered from herds in Inner Mongolia province possibly linked to dense farming (2.37 million dairy cows) with the largest dairy herds in this province (Dou, 2014). Dense farming is significantly correlated with the incidence of bovine mastitis (Ali et al., 2014), and mastitis is the main reason for frequent and prolonged use of antibiotics that exert selective pressure for emergence and dissemination of resistant isolates (Berge et al., 2005). Our results revealed that CTX-M, mainly the CTX-M-15, was the most prevalent genotype, followed by TEM and SHV. These findings agree with other contemporary studies from China and around the world that also reported CTX-M as the dominant ESBL genotype (Locatelli et al., 2009; Dahmen et al., 2013; Geser et al., 2015; Kar et al., 2015). This goes along with the recent detection of CTX-M-15 producing E. coli from cattle and other food-animals in east Asia (Ohnishi et al., 2013; Yu et al., 2015), India (Upadhyay et al., 2015), the United Kingdom (Timofte et al., 2014), Germany (Freitag et al., 2016) and Tanzania (Seni et al., 2016). A national resistance surveillance study in China reported that the prevalence of ESBL-producing E. coli in humans has persisted above 50% since 2000 (Xiao et al., 2011), and recently Liu et al. reported even higher prevalence (68.2%) of ESBLs in clinical E. coli isolates, mainly the blaCTX-M-15 (Liu et al., 2015). Similarly, in animals the prevalence of ESBL-producing E. coli in China has considerably increased in recent years with CTX-M being the major prevailing gene encoding for ESBLs (Rao et al., 2014). It is known that, generally, ESBL genes are located on plasmids that could spread easily among commensal and pathogenic bacteria in the herd and the environment. Due to limited resources, in the present study, we could not investigate the prevalence of ESBL genes in other bacteria, in other healthy cows, or in cows with subclinical mastitis which is usually 30–40% higher than clinical mastitis (Halasa et al., 2007). We presume that the actual prevalence of ESBL-producers, particularly E. coli, may be much higher than the reported.
In ESBL-positive E. coli, phylogenetic group A represented the most prevalent group followed by virulent extra-intestinal group D; however, this was in contrast to our previous study (Liu et al., 2014), which reported that the pathogenic E. coli associated with mastitis mainly belonged to phylo-group B1 (58.6%) rather than group A (35.7%). Nevertheless, similar phylogenetic distributions were observed in some ESBL-producing E. coli isolated from animals (Abraham et al., 2014; Xu et al., 2015). Our results indicated that all the ESBL-producing E. coli isolates were MDR. The majority of these isolates (54–100%) were found resistant to cephalosporins. In addition, low susceptibility was also observed against the common β-lactam and non-β-lactam antibiotics such as ampicillin, aminoglycosides, tetracycline and fluoroquinolones. Recently, many studies have reported MDR ESBL-producing E. coli isolated from cattle (Timofte et al., 2014), poultry (Kar et al., 2015), pigs (Xu et al., 2015), and humans (Gu et al., 2008). Fluoroquinolones, following ciprofloxacin, are the second important antimicrobial drug in veterinary and human medicine (Coque et al., 2008). Quinolone resistance is traditionally caused by chromosomal mutations in gyrase or topoisomerase encoding genes or efflux pump expression regulating genes (Hopkins et al., 2005); nonetheless, plasmid mediated quinolone resistance is also increasingly reported in ESBL-producing E. coli (Xu et al., 2015).
Integrons play an important role in the emergence of MDR bacteria and in the dissemination of resistance genes. Published reports on the characterization of integrons in ESBLs-positive E. coli from dairy cows are scarce, but previous studies have been conducted in other food-animals, humans and the environment (Gu et al., 2008; Chen et al., 2010; Xu et al., 2015). In accordance to these studies, clinical class 1 integrons were found in the majority of ESBL-positive E. coli (83.33%). The gene cassette arrays of the class 1 integron variable regions contained five different gene combinations that likely impart additional resistance features to our isolates (see Table 3). Six of the intI1 positive amplicons were failed to generate gene cassettes which may be related to the absence of 3′CS in these integrons (Lu et al., 2010). The dfrA17-aadA5 was the predominant gene array that corroborates with the previous studies in China (Gu et al., 2008; Xu et al., 2015). Strikingly, we determined that majority of blaCTX-M genes were associated with ISCR1 elements, but no ESBL genes were found in the class 1 integron cassettes. It agrees with other published reports that also did not detect ESBL genes in the cassettes (Kiiru et al., 2013; Kar et al., 2015; Xu et al., 2015). Notably, our findings of the most pre-dominant CTX-M type (blaCTXM-15) and its association with the ISCR1 elements rather than gene cassette arrays indicated that they are more likely mobilized by ISCR1 elements. Conversely, blaTEM, when found alone or not associated with blaCTXM-15, was not often found linked to ISCR1, and therefore, was comparatively less prevalent. It has been proposed that antibiotic resistance gene elements are added to the 3′-CS of class 1 integrons by co-mobilization with the nearby ISCR1 from the neighbor integron, implying rolling circle transposition and homologous recombination mechanisms, thus facilitating the formation of complex class 1 integrons (Toleman et al., 2006). Taken together, the current high occurrence of multi-resistant ESBL-producing E. coli carrying clinical class 1 integrons and its association with ISCR1 is worrisome. This may suggest these bacteria are armouring against the antibiotics by devising various tools to render antibiotics useless. Fear exists that this co-existence of ESBL genes along with class 1 integrons as gene cassettes and ISCR1 mobile elements would more robustly disseminate resistance elements within bacterial populations. This calls for an efficient control policy with restriction on the consumption of extended spectrum cephalosporins for long term use.
Conclusions
Here, we report on the high occurrence of ESBL-producing E. coli from bovine mastitis. Genotypic characterization indicated a dominance of blaCTX-M-15 genes harboring clinical class 1 integrons associated with ISCR1 elements, indicative rapid and wider dissemination potential and posing threats to veterinary and public health. To the best of our knowledge, this is the first comprehensive study to report on the alarming high prevalence of blaCTX-M-15 and class 1 integron resistance conferring elements in ESBL-producing E. coli from mastitic cows in China.
Author contributions
BH, TA, and SR, conceived and designed the experiment. TA, MS, and SZ, performed the research. JG, GL, and LZ, contributed in reagents/materials/analysis. BH, TA, and SR, wrote the manuscript.
Funding
This research was supported by the Chinese Twelfth “Five-year” National Science and Technology Support Project (No. 2012BAD12B03), Ministry of Education in China major project (No. 313054), Specialized Research Fund for the Doctoral Program of Higher Education (SRFDP) State Education Ministry (No. 20120008110042), and the National Natural Science Foundation of China (No. 3151101034) and (NO. 31572587).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Elizabeth Rettedal, Cork Cancer Centre, University College of Cork for editing and reviewing the manuscript.
References
- Abraham S., Trott D. J., Jordan D., Gordon D. M., Groves M. D., Fairbrother J. M., et al. (2014). Phylogenetic and molecular insights into the evolution of multidrug-resistant porcine enterotoxigenic Escherichia coli in Australia. Int. J. Antimicrob. Agents 44, 105–111. 10.1016/j.ijantimicag.2014.04.011 [DOI] [PubMed] [Google Scholar]
- Ali T., Rahman A., Qureshi M. S., Hussain M. T., Khan M. S., Uddin S., et al. (2014). Effect of management practices and animal age on incidence of mastitis in Nili Ravi buffaloes. Trop. Anim. Health Prod. 46, 1279–1285. 10.1007/s11250-014-0641-2 [DOI] [PubMed] [Google Scholar]
- Berge A. C., Atwill E. R., Sischo W. M. (2005). Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69, 25–38. 10.1016/j.prevetmed.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Bush K., Jacoby G. A. (2010). Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54, 969–976. 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. Y., Siu L. K., Fung C. P., Huang M. H., Ho M. (2001). Diversity of SHV and TEM beta-lactamases in Klebsiella pneumoniae: gene evolution in Northern Taiwan and two novel beta-lactamases, SHV-25 and SHV-26. Antimicrob. Agents Chemother. 45, 2407–2413. 10.1128/AAC.45.9.2407-2413.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shu W., Chang X., Chen J. A., Guo Y., Tan Y. (2010). The profile of antibiotics resistance and integrons of extended-spectrum beta-lactamase producing thermotolerant coliforms isolated from the Yangtze River basin in Chongqing. Environ. Pollut. 158, 2459–2464. 10.1016/j.envpol.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2014). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standard Institute. CLSI document M100–S124. [Google Scholar]
- Coque T. M., Baquero F., Canton R. (2008). Increasing prevalence of ESBl-producing Enterobacteriaceae in Europe. Euro Surveill. 13, 1–11. [PubMed] [Google Scholar]
- Dahmen S., Métayer V., Gay E., Madec J. Y., Haenni M. (2013). Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet. Microbiol. 162, 793–799. 10.1016/j.vetmic.2012.10.015 [DOI] [PubMed] [Google Scholar]
- D'Andrea M. M., Arena F., Pallecchi L., Rossolini G. M. (2013). CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317. 10.1016/j.ijmm.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Dillon B., Thomas L., Mohmand G., Zelynski A., Iredell J. (2005). Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods 62, 221–232. 10.1016/j.mimet.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Dou M. (2014). China Dairy Big Data. Chaoyang, Beijing, China: A609 Times Fortune, No.6 Shuguang Xili, yearbook@21dairy.com. [Google Scholar]
- Eckert C., Gautier V Arlet G. (2006). DNA sequence analysis of the genetic environment of blaCTX-M genes. J. Antimicrob. Chemother. 57, 14–23. 10.1093/jac/dki398 [DOI] [PubMed] [Google Scholar]
- Freitag C., Michael G. B., Kadlec K., Hassel M., Schwarz S. (2016). Detection of plasmid-borne extended-spectrum beta-lactamase (ESBL) genes in Escherichia coli isolates from bovine mastitis. Vet. Microbiol. [Epub ahead of print]. 10.1016/j.vetmic.2016.08.010 [DOI] [PubMed] [Google Scholar]
- Geser N. S., Stephan R., Hächler H. (2015). Occurrence and characteristics of extended-spectrum b-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 8:21 10.1186/1746-6148-8-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Pan S., Wang T., Zhao W., Mei Y., Huang P., et al. (2008). Novel cassette arrays of integrons in clinical strains of Enterobacteriaceae in China. Int. J. Antimicrob. Agents 32, 529–533. 10.1016/j.ijantimicag.2008.06.019 [DOI] [PubMed] [Google Scholar]
- Halasa T., Huijps K., Østerås O., Hogeveen H. (2007). Economic effects of bovine mastitis and mastitis management: a review. Vet. Q 29, 18–31. 10.1080/01652176.2007.9695224 [DOI] [PubMed] [Google Scholar]
- Hall R. M., Stokes H. W. (1993). Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90, 115–132. 10.1007/BF01435034 [DOI] [PubMed] [Google Scholar]
- Hopkins K. L., Davies R. H, Threlfall E. J. (2005). Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25, 358–373. 10.1016/j.ijantimicag.2005.02.006 [DOI] [PubMed] [Google Scholar]
- Kar D., Bandyopadhyay S., Bhattacharyya D., Samanta I., Mahanti A., Nanda P. K., et al. (2015). Molecular and phylogenetic characterization of multidrug resistant extended spectrum beta-lactamase producing Escherichia coli isolated from poultry and cattle in Odisha, India. Infect. Genet. Evol. 29, 82–90. 10.1016/j.meegid.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Kiiru J., Butaye P., Goddeeris B. M., Kariuki S. (2013). Analysis for prevalence and physical linkages amongst integrons, ISEcp1, ISCR1, Tn21 and Tn7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992-2011). BMC Microbiol. 13:109. 10.1186/1471-2180-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang Y., Wang G., Xing Q., Shao L., Dong X., et al. (2015). The prevalence of Escherichia coli strains with extended spectrum beta-lactamases isolated in China. Front. Microbiol. 6:335. 10.3389/fmicb.2015.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu G., Liu W., Liu Y., Ali T., Chen W., et al. (2014). Phylogenetic group, virulence factors and antimicrobial resistance of Escherichia coli associated with bovine mastitis. Res. Microbiol. 165, 273–277. 10.1016/j.resmic.2014.03.007 [DOI] [PubMed] [Google Scholar]
- Locatelli C., Caronte I., Scaccabarozzi L., Migliavacca R., Pagani L., Moroni P. (2009). Extended-spectrum beta-lactamase production in E. coli strains isolated from clinical bovine mastitis. Vet. Res. Commun. 33(Suppl. 1), 141–144. 10.1007/s11259-009-9263-y [DOI] [PubMed] [Google Scholar]
- Lu L., Dai L., Wang Y., Wu C., Chen X., Li L., et al. (2010). Characterization of antimicrobial resistance and integrons among Escherichia coli isolated from animal farms in Eastern China. Acta Tropica 113, 20–25. 10.1016/j.actatropica.2009.08.028 [DOI] [PubMed] [Google Scholar]
- Madec J. Y., Poirel L., Saras E., Gourguechon A., Girlich D., Nordmann P., et al. (2012). Non-ST131 Escherichia coli from cattle harbouring human-like blaCTX-M-15-carrying plasmids. J. Antimicrob. Chemother. 67, 578–581. 10.1093/jac/dkr542 [DOI] [PubMed] [Google Scholar]
- MAO. (2010). Ministry of Agriculture of the People's Republic of China. Announcement No.1435 of the Ministry of Agriculture. Available online at: www.moa.gov.cn/govpublic/SYJ/201008/t20100823_1622639.html (Online in Chinese).
- NMC (1999). Laboratory Handbook on Bovine Mastitis. National Mastitis Council. Madison, WI: NMC Inc. [Google Scholar]
- Ohnishi M., Okatani A. T., Harada K., Sawada T., Marumo K., Murakami M., et al. (2013). Genetic characteristics of CTX-M-type extended-spectrum-beta-lactamase (ESBL)-producing Enterobacteriaceae involved in mastitis cases on Japanese dairy farms, 2007 to 2011. J. Clin. Microbiol. 51, 3117–3122. 10.1128/JCM.00920-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Hujer A. M., Hujer K. M., Decker B. K., Rather P. N., Bonomo R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. 10.1128/AAC.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L., Lv L., Zeng Z., Chen S., He D., Chen X., et al. (2014). Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet. Microbiol. 172, 534–541. 10.1016/j.vetmic.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Seiffert S. N., Hilty M., Perreten V., Endimiani A. (2013). Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: an emerging problem for human health? Drug Resist. Updat. 16, 22–45. 10.1016/j.drup.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Seni J., Falgenhauer L., Simeo N., Mirambo M. M., Imirzalioglu C., Matee M., et al. (2016). Multiple ESBL-Producing Escherichia coli Sequence Types Carrying Quinolone and Aminoglycoside Resistance Genes Circulating in Companion and Domestic Farm Animals in Mwanza, Tanzania, Harbor Commonly Occurring Plasmids. Front. Microbiol. 7:142. 10.3389/fmicb.2016.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantawiwat S., Tansuphasiri U., Wongwit W., Wongchotigul V., Kitayaporn D. (2005). Development of multiplex PCR for the detection of total coliform bacteria for Escherichia coli and Clostridium perfringens in drinking water. Southeast Asian J. Trop. Med. Public Health 36, 162–169. [PubMed] [Google Scholar]
- Timofte D., Maciuca I. E., Evans N. J., Williams H., Wattret A., Fick J. C., et al. (2014). Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 beta-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob. Agents Chemother. 58, 789–794. 10.1128/AAC.00752-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Walsh T. R. (2006). Common regions e.g., orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 58, 1–6. 10.1093/jac/dkl204 [DOI] [PubMed] [Google Scholar]
- Upadhyay S., Hussain A., Mishra S., Maurya A. P., Bhattacharjee A., Joshi S. R. (2015). Genetic Environment of Plasmid Mediated CTX-M-15 Extended Spectrum Beta-Lactamases from Clinical and Food Borne Bacteria in North-Eastern India. PLoS ONE 10:e0138056. 10.1371/journal.pone.0138056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas M. V., Correa A., Perez F., Zuluaga T., Radice M., Gutkind G., et al. (2004). CTX-M-12 beta-lactamase in a Klebsiella pneumoniae clinical isolate in Colombia. Antimicrob. Agents Chemother. 48, 629–631. 10.1128/AAC.48.2.629-631.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinué L., Sáenz Y., Somalo S., Escudero E., Moreno M. A., Ruiz-Larrea F., et al. (2008). Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 62, 934–937. 10.1093/jac/dkn331 [DOI] [PubMed] [Google Scholar]
- White P. A., McIver C. J., Deng Y., Rawlinson W. D. (2000). Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182, 265–269. 10.1111/j.1574-6968.2000.tb08906.x [DOI] [PubMed] [Google Scholar]
- Xiao Y. H., Giske C. G., Wei Z. Q., Shen P., Heddini A., Li L. J. (2011). Epidemiology and characteristics of antimicrobial resistance in China. Drug Resist. Updat. 14, 236–250. 10.1016/j.drup.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Xu G., An W., Wang H., Zhang X. (2015). Prevalence and characteristics of extended-spectrum beta-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front. Microbiol. 6:1103. 10.3389/fmicb.2015.01103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F., Qian Y., Chen S., Wang P., Huang Y. (2007). Incidence of extended-spectrum beta-lactamases and characterization of integrons in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Shantou, China. Acta Biochim. Biophys. Sin. (Shanghai) 39, 527–532. 10.1111/j.1745-7270.2007.00304.x [DOI] [PubMed] [Google Scholar]
- Yu T., He T., Yao H., Zhang J. B., Li X. N., Zhang R. M., et al. (2015). Prevalence of 16S rRNA Methylase Gene rmtB Among Escherichia coli Isolated from Bovine Mastitis in Ningxia, China. Foodborne Pathog. Dis. 12, 770–777. 10.1089/fpd.2015.1983 [DOI] [PubMed] [Google Scholar]