Abstract
MyoD is a master regulator of myogenesis with a potent ability to redirect the cell fate of even terminally differentiated cells. Hence, enhancing the activity of MyoD is an important step to maximising its potential utility for in vitro disease modelling and cell replacement therapies. We have previously shown that the reprogramming activity of several neurogenic bHLH proteins can be substantially enhanced by inhibiting their multi-site phosphorylation by proline-directed kinases. Here we have used Xenopus embryos as an in vivo developmental and reprogramming system to investigate the multi-site phospho-regulation of MyoD during muscle differentiation. We show that, in addition to modification of a previously well-characterised site, Serine 200, MyoD is phosphorylated on multiple additional serine/threonine sites during primary myogenesis. Through mutational analysis, we derive an optimally active phospho-mutant form of MyoD that has a dramatically enhanced ability to drive myogenic reprogramming in vivo. Mechanistically, this is achieved through increased protein stability and enhanced chromatin association. Therefore, multi-site phospho-regulation of class II bHLH proteins is conserved across cell lineages and germ layers, and manipulation of phosphorylation of these key regulators may have further potential for enhancing mammalian cell reprogramming.
Keywords: MyoD, Myogenesis, Phosphorylation, bHLH, Xenopus, Reprogramming
Abbreviations: bHLH, basic-Helix-Loop-Helix; cdk, cyclin-dependent-kinase; ISH, In situ hybridisation; MRF, Muscle Regulatory Factor; SP, serine-proline; TP, threonine-proline; WT, wild-type
Highlights
-
•
MyoD is phosphorylated on multiple S/T-P sites in muscle differentiation.
-
•
Myogenic activity of MyoD is regulated by up to five C terminal phospho-sites.
-
•
Our optimal phospho-mutant form of MyoD dramatically enhances myogenesis in vivo.
-
•
MyoD multi-site phosphorylation controls protein stability and chromatin association.
-
•
Multi-site phospho-regulation is a conserved mechanism across cell lineages.
1. Introduction
The basic-Helix-Loop-Helix (bHLH) protein MyoD is an extensively studied Muscle Regulatory Factor (MRF) that plays a pivotal role during muscle development, and has also received much attention for its ability to direct a myogenic program even in terminally differentiated somatic cells [1], [2]. Using MyoD to direct trans-differentiation has potential utility in cellular reprogramming efforts directed at in vitro modelling of muscle diseases for drug screening [3] and in cell replacement therapies [4]. However, refining of these reprogramming methods requires optimising the myogenic activity of MyoD.
We have previously studied other master regulatory proneural bHLH transcription factors that drive differentiation in the neural lineage, namely Neurogenin2, Ascl1 and NeuroD4 [5], [6], [7], [8]. We see that phosphorylation of these factors on multiple serine-proline and threonine-proline (SP and TP) sites suppressed both their ability to drive neuronal differentiation during development [5], [7] and during cellular reprogramming of mammalian cells [6].
MyoD phosphorylation on Serine 200 (S200) and S5, sites both found as part of an SP motif, has long been recognised to underlie the characteristic MyoD protein fluctuations and activity that vary with cell cycle phase in proliferating myoblasts [9], [10], [11], [12], [13]. MyoD also contains multiple additional SP and TP sites which may, by analogy with proneural factors, play an additional role in regulation of the ability of MyoD to promote myogenic conversion and differentiation. Here, using the Xenopus tadpole as a model to test MyoD's myogenic conversion potential, we show that myogenic activity of MyoD is regulated by up to five S/TP kinase sites specifically located in the C terminus of the protein. Furthermore, we see that multi-site phosphorylation regulates both MyoD protein stability and chromatin association. Finally, we identify an optimal phospho-mutant form of MyoD that dramatically enhances myogenic conversion of embryonic ectoderm in vivo.
2. Materials and methods
2.1. Cloning
Wild-type (WT) mouse MyoD in pCS2 has been described [14]. A single C terminal HA tag was added by PCR using the primers: 5′GATCGGATCCACCATGGAGCTTCTATCGCCG-3′; 5′GATCCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTAAAGCACCTGATAAATCGCATT-3′. All single or multiple site mutations were performed using the QuikChange II or QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies) respectively (PCR primers available on request). Nucleotide and protein sequence alignments were conducted using ClustalW software [15].
2.2. Xenopus laevis embryo manipulation
Acquisition of Xenopus laevis embryos, preparation and injection of synthetic mRNA, staging of embryos and in situ hybridisation (ISH) were conducted as described previously [6], [16], [17]. Dig-oxigenin-labelled anti-sense probes were synthesised from linearised plasmid Actc1 in pSP64T (Xenbase; EcoR1, SP6). Semi-quantitative scoring was conducted for Muscle Actin staining on the injected side of the embryo relative to the uninjected side; grades were assigned 0–3, where 0 indicates no increase in myogenesis, through to 3 indicating extensive myogenesis.
2.3. Quantitative real-time PCR (qRT-PCR)
Whole embryo RNA was extracted using the Qiagen RNeasy® Mini Kit and cDNAs were prepared with the QuantiTect® Reverse Transcription Kit (Qiagen). qPCR analysis was conducted using the Quantifast® SYBR Green PCR Kit (Qiagen) in a LightCycler® 480 (Roche). Thermal cycling parameters are described in Ref. [5], and primer sequences (5′–3′): EF1αF = CACCATGAAGCCCTTACTGAG; EF1αR = TGATAACCTGTGCGGTAAATG; xMyh4F = GAACAAGGACCCACTGAACG; xMyh4R = TCCACCTTTACCAGCAGCAT.
2.4. Western blotting
Protein extracts were prepared from whole embryos and incubated with or without lambda protein phosphatase (NEB) as described [7], prior to separation on an 18% Tris/Glycine/SDS gel by standard methods. For assay of chromatin association, cross linking, cytoplasmic and chromatin fractionation, western blot and protein quantification were conducted as described [7].
2.5. Statistical analysis
For western blotting, experiments were performed in independent duplicate with representative results shown. For qPCR data, mRNA expression was normalised to expression of the housekeeping gene Elongation Factor 1α (EF1α), and for analysis, mRNA levels in injected embryos were calculated relative to stage-matched uninjected controls. Mean values and the standard error of the mean (s.e.m.) were calculated from at least three independent experiments (n = 3). Statistical significance was determined using a paired two-tailed Student's t-test with (p < 0.05) = *; (p < 0.025) = **; (p < 0.0125) = ***. For in situ hybridisation data, experiments were conducted in independent duplicate or triplicate and the n numbers reported refer to the range of total numbers of embryos in each injection category. Statistical significance was determined between categories using a Fisher's Exact Test for Count Data, and p values are as described above.
3. Results
3.1. MyoD is phosphorylated on sites in addition to S200 during Xenopus primary myogenesis
Early studies investigating phospho-regulation of MyoD largely focused on its modification in proliferating cells in culture; either C2C12 myoblasts or C3H10T1/2 fibroblasts transfected with MyoD [10], [12]. To investigate phosphorylation of MyoD and its reprogramming activity in vivo, we chose the highly versatile Xenopus embryo system, which can be used to study mammalian MyoD activity both in the endogenous myotome and also for reprogramming activity in the non-myogenic ectoderm tissue [14]. Furthermore, the first cleavage of the fertilised egg bisects the embryos into future left and right sides, so unilateral mRNA injection allows MyoD activity to be directly compared between injected and uninjected embryo sides.
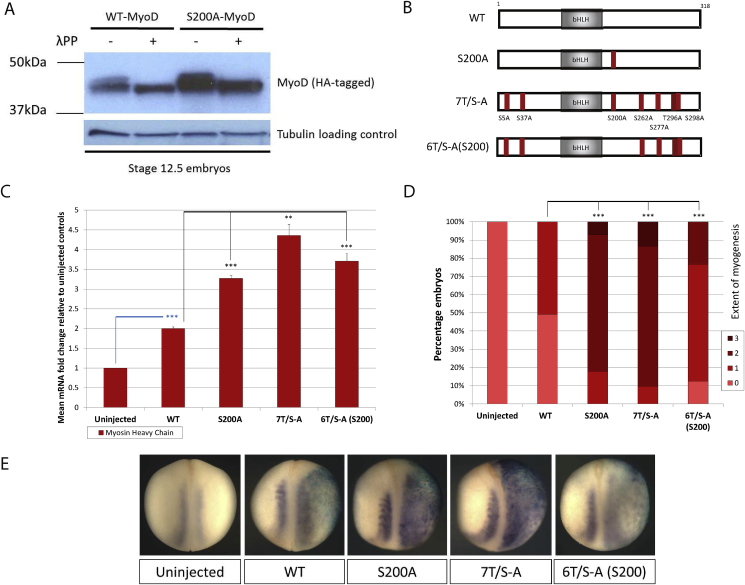
Firstly, we explored whether MyoD is phosphorylated during primary myogenesis. Extracts were prepared from stage 12.5 embryos injected with mRNA encoding HA-tagged Wild-Type (WT) MyoD, or S200A MyoD, a mutant version with S200 mutated to alanine to prevent phosphorylation at this established site. Western blot analysis reveals a broad protein band for both WT and S200A proteins (Fig. 1A), with more S200A protein than WT, consistent with previous reports that demonstrate a longer half-life for this mutant protein [10], [11], [12]. Incubation with a broad spectrum protein phosphatase enhances the migration of both WT and S200A, indicating phosphorylation on at least one additional site during differentiation in vivo.
Fig. 1.
MyoD is phospho-regulated on sites in addition to S200 during primary myogenesis. (A) Western blot analysis of protein extracts from stage 12.5 embryos injected with 200 pg mRNA encoding HA-tagged WT or S200A-MyoD. Samples were incubated with or without lambda protein phosphatase. Tubulin provided a loading control. (B) Schematic representation of WT mouse MyoD protein and phospho-mutant variants, showing approximate locations of SP/TP sites that are mutated to AP in each. (C–E) Embryos were unilaterally injected at the 2-cell stage with 100 pg mRNA encoding the respective MyoD constructs. At stage 18, embryos were assayed by RT-qPCR for Myosin Heavy Chain expression (C [n = 3]) or by ISH with semi-quantitative scoring for Muscle Actin expression (D [n = 52–73]) as described in the methods. Representative embryos are shown in (E); injected side to the right. * = (p < 0.05); ** = (p < 0.025); *** = (p < 0.0125).
3.2. A multi-site phospho-mutant MyoD promotes enhanced myogenesis in vivo
Mouse MyoD protein contains seven potential proline-directed kinase sites (six serine-proline (SP), and one threonine-proline (TP)) that are highly conserved with human MyoD (Supplementary Fig. 1). To further explore a potential regulatory role of S/TP sites additional to S200 during myogenesis, a panel of phospho-mutant forms of MyoD were created (Fig. 1B): 7T/S-A MyoD contains all seven SP/TP sites mutated to alanine-proline (AP), while 6T/S-A(S200) has the S200 site restored but the remaining six sites mutated. In order to compare their myogenic activity, mRNA encoding these proteins was injected unilaterally into two-cell stage Xenopus embryos, and myogenesis was assayed in stage 18 embryos by RT-qPCR and in situ hybridisation (ISH) for muscle structural genes Myosin Heavy Chain or Muscle Actin respectively (Fig. 1C–E). At this level of over-expression, WT MyoD induces a small increase in myogenesis, largely confined to expansion of the myotome on the injected side. By comparison, both S200A and full phospho-mutant 7T/S-A induce a marked increase in myogenesis spreading laterally, and demonstrating reprogramming of ectoderm tissue. Interestingly, 6T/S-A(S200) also induces a significant increase in myogenesis relative to WT MyoD, albeit with less activity than S200A or 7T/S-A MyoDs. This demonstrates that sites in addition to S200 can regulate myogenic activity of MyoD.
3.3. Regulatory activity resides in C terminal phosphorylation sites
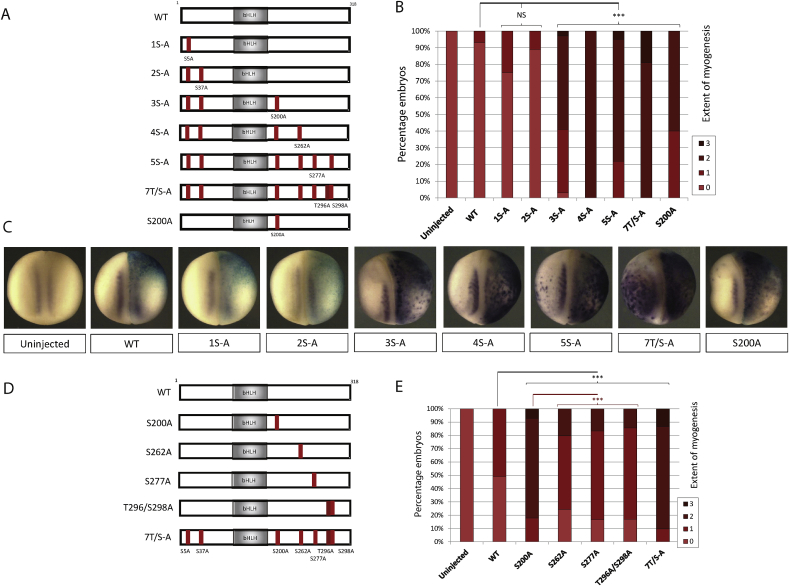
We have previously described a model for phospho-regulation of proneural protein activity, where the number, rather than location of available phospho-sites is the key determinant of bHLH activity; mutation of increasing numbers of phosphorylation sites additively increases the neurogenic activity of the mutant proteins [5], [7]. To explore if a similar phenomenon may control regulation of MyoD, a panel of MyoD phospho-mutants were made with SP/TP sites cumulatively mutated from the N terminus to generate 1S-A, 2S-A etc (Fig. 2A). Due to the immediate proximity of T296/S298, these sites were mutated simultaneously.
Fig. 2.
Mutational analysis shows regulatory activity of S200 and additional C terminal phosphorylation sites. Schematic representation of phospho-mutant constructs with mutation of cumulative (A) or individual (D) sites. Two-cell stage embryos were unilaterally injected with 100 pg mRNA of the respective MyoD construct and assayed at stage 18 for expression of Muscle Actin by ISH: (B) Cumulative mutant series [n = 68–74] with representative images shown in (C). (D) Single site mutants [n = 52–83]. NS = Not significant; * = (p < 0.05); ** = (p < 0.025); *** = (p < 0.0125).
Embryos were injected with mRNA as before and scored for myogenesis at stage 18 by ISH (Fig. 2B–C). Mutation of the N terminal SP sites in MyoD has no significant effect on its activity relative to the wild-type protein. Similarly, mutation of S200 in addition to the two N terminal sites (i.e. 3S-A) produces a level of myogenesis comparable to mutation of the single S200 site alone, suggesting that the N terminal sites do not contribute to regulation of MyoD myogenic activity. However, mutation of C terminal residues in addition to S200 results in a more active MyoD than the single phosphorylation site mutant S200A MyoD. Thus, for MyoD, both number and location of phospho-sites contribute to its regulation.
3.4. S200 is the most important individual C terminal site for regulating myogenic activity
The five C terminal phospho-sites in mouse MyoD are also the phospho-sites that are most conserved with both human and Xenopus MyoD (Supplementary Fig. 1); S200 and S262 are highly conserved between all three species, suggesting that these may have enhanced functional significance. We therefore sought to address whether myogenic activity depends on any particular C terminal phospho-site.
Each C terminal site was mutated individually and myogenic activity was assayed as described above (Fig. 2D–E). Mutation of any individual C terminal phospho-site results in significantly enhanced myogenesis compared to WT MyoD. Furthermore, the three most C terminal sites when mutated individually (S262, S277 and T296/S298) are very similar to each other in inducing a moderate increase in myogenesis. By comparison, mutation of the S200 site alone results in myogenic activity that is significantly greater than any other single site mutant, yet this is still not as active as the full 7T/S-A phospho-mutant. Therefore, S200 confers the most significant regulatory activity but its mutation alone is not sufficient to maximally activate MyoD; additional C terminal residues must also be mutated.
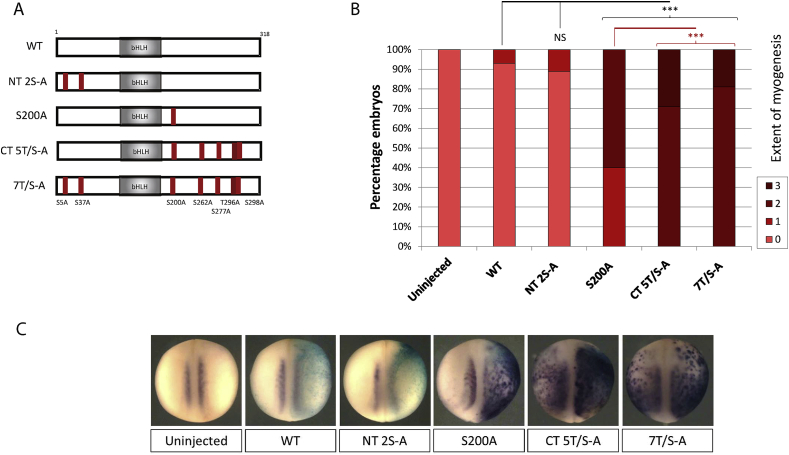
3.5. Identification of an optimal phospho-mutant MyoD with maximal myogenic activity
For the purposes of cellular reprogramming, an optimal form of MyoD would have maximal activity in driving myogenic conversion of non-muscle tissue. Therefore, we finally sought to derive an optimal phospho-mutant MyoD in our Xenopus assay. For the proneural proteins, maximal activity is achieved by mutation of all available phospho-sites [5], [6], [7], but for MyoD, we see that N terminal sites do not contribute to regulation (Fig. 2). S200 is the most important single C terminal site (Fig. 2), so additional constructs were made to mutate S200 in combination with other C terminal sites (data not shown). In all assays conducted, the most extensive myogenesis is induced by CT 5T/S-A MyoD, in which all five C terminal phospho-sites are mutated but the two N-terminal phospho-sites are intact. These results are summarised in Fig. 3, where NT 2S-A MyoD shows equivalent activity to WT protein, and CT 5T/S-A MyoD is more active than the full 7T/S-A phospho-mutant. We note that both of these multi-site mutants are significantly more active than the single S200A mutant, inducing extensive conversion of the lateral ectoderm to a myogenic cell fate, often seen bilaterally over the embryo. Thus, while S200 shows the greatest regulatory role of any individual site, phosphorylation must be prevented on all five C-terminal serine/threonines for maximal differentiation activity.
Fig. 3.
Maximal myogenic activity is achieved by mutation of multiple C terminal sites. (A) Schematic representation of phospho-mutant constructs. (B) Two-cell stage embryos were unilaterally injected with 100 pg mRNA of the respective MyoD construct and assayed at stage 18 as before [n = 32–41] with representative images shown in (C). NS = Not significant; * = (p < 0.05); ** = (p < 0.025); *** = (p < 0.0125).
3.6. CT 5T/S-A MyoD has both increased protein accumulation and enhanced chromatin binding relative to WT and S200A MyoD
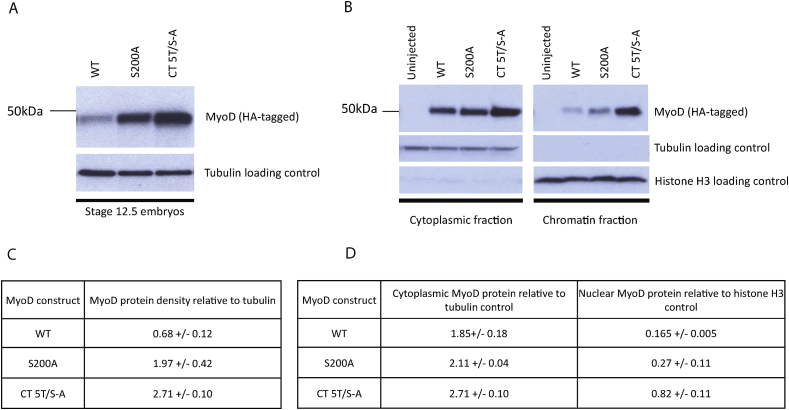
Having established an optimal phospho-mutant form of MyoD that has superior myogenic activity in vivo, we investigated the mechanisms by which this enhanced differentiation is achieved. One mechanism may be by enhancing MyoD protein stability. We have already shown that injection of equal amounts of WT and S200A MyoD mRNA produces a greater accumulation of S200A protein relative to WT MyoD (Fig. 1) so we next determined whether the optimal phospho-mutant CT 5T/S-A MyoD showed further enhancement of stability. HA-tagged mRNA was injected into Xenopus embryos, and protein extracts were prepared at stage 12.5 for western blot analysis; the density of each MyoD protein band was calculated relative to the respective tubulin loading control (Fig. 4A, C). While injection of the same amount of mRNA results in an almost three-fold increase in S200A protein relative to WT MyoD, CT 5T/S-A MyoD protein accumulates to a level that is more than four-fold higher than WT MyoD. Thus, whilst S200 is an established phospho-site that regulates MyoD stability [10], [12], further enhancement of MyoD protein stability accompanies mutation of additional C-terminal SP sites.
Fig. 4.
CT 5T/S-A MyoD shows enhanced protein stability and enhanced chromatin binding relative to both WT and S200A MyoD. Embryos were injected with 200 pg mRNA encoding HA-tagged MyoD constructs as indicated, and western blot analysis was performed on whole embryo extracts at stage 12.5 (A) or cross linking and cytoplasmic/chromatin extracts at stage 13 (B). MyoD protein density was quantified relative to tubulin loading control for whole embryo extracts (C) and cytoplasmic fractions (D), or relative to histone H3 for chromatin fractions (D). Mean values are shown from independent duplicate samples with standard error of the mean.
Phospho-mutant proneural proteins show both an enhanced stability compared to their wild-type counterparts, but also exhibit enhanced DNA binding affinity, and both of these attributes contribute to superior reprogramming activity [5], [7]. To test whether MyoD phospho-status influences protein binding to embryonic chromatin, embryos were injected as before, and cross-linking was performed in stage 13 embryos prior to nuclear extraction, chromatin isolation and western blot analysis (Fig. 4B, D). Cytoplasmic samples were also collected and MyoD protein accumulation was calculated relative to tubulin or histone H3 loading controls in cytoplasmic and chromatin fractions respectively. The absence of tubulin protein in the chromatin fraction confirms successful chromatin isolation, and no MyoD protein was detected in the uninjected embryos, confirming specificity of the anti-HA antibody.
Comparing the relative amounts of chromatin bound MyoD protein (Fig. 4D) with the relative amounts of whole embryo MyoD protein (Fig. 4C), S200A MyoD is only 1.6-fold higher than WT MyoD in chromatin samples, suggesting that S200A displays greater protein stability than WT MyoD, but no greater chromatin binding affinity. Therefore, protein stability alone contributes to the enhanced activity of this mutant S200A protein. In contrast, chromatin-bound CT 5T/S-A MyoD accumulates to five-fold higher than WT and three-fold higher than S200A MyoD, demonstrating an increased chromatin binding affinity in addition to increased protein accumulation. Thus, our optimal phospho-mutant CT 5T/S-A dramatically enhances myogenesis in vivo by a combination of increased protein accumulation and enhanced chromatin binding; this latter feature being unique to the CT 5T/S-A MyoD phospho-mutant and not shared with S200A MyoD.
4. Discussion
The bHLH transcription factor MyoD is a master regulator of myogenesis, and manipulating its activity both for in vitro modelling of muscle disease and for cell replacement therapy has huge potential in translation medicine and cellular reprogramming [18]. We set out to engineer a maximally active form of MyoD for in vivo reprogramming in the Xenopus frog system. Neurogenin2, Ascl1 and NeuroD4, bHLH transcription factors regulating neurogenesis, undergo multi-site phosphorylation that coordinates neuronal differentiation with the cellular kinase environment in vivo [5], [6], [7], [8]. As such, multi-site bHLH protein phosphorylation in rapidly dividing cells suppresses their ability to drive neuronal differentiation; a multiply phospho-mutant form of these proneural proteins substantially enhances their ability to reprogram both Xenopus ectoderm and mammalian fibroblasts into neurons [5], [6], [7], [8]. Here, we show here that multi-site phosphorylation also controls the ability of MyoD to regulate cellular reprogramming to muscle in vivo, demonstrating conservation of a similar regulation of bHLH proteins across cell lineages and germ layers.
Analogous to our findings for proneural bHLH proteins [5], [6], [7], [8], we see that MyoD is phospho-regulated on multiple proline-directed kinase sites during developmental myogenesis. Moreover, by combining mutational analysis with this in vivo assay system, we have identified an optimal phospho-mutant form of MyoD, CT 5T/S-A MyoD, that displays markedly enhanced ability to drive myogenic differentiation compared to any previously described phospho-mutant [9], [10], [11], [12], [13]. Our optimal C terminal phospho-mutant protein displays increased protein stability relative to both WT MyoD and S200A MyoD, a mutant previously shown to have enhanced stability and activity compared to the WT protein [10], [11], [12]. In addition, we also see that CT 5T/S-A MyoD has increased chromatin affinity that is not shared by S200A MyoD. The data we present here, in combination with previous work on proneural proteins [5], [6], [7], [8], support the conservation of a phospho-regulatory model of bHLH protein action, both functionally and mechanistically, across both nerve and muscle cell lineages.
However, in contrast to proneural proteins where only the number of available phospho-sites is key to regulating protein function [5], [6], [7], [8], we find that both number and location of phospho-sites are important for MyoD, with regulatory phospho-sites located solely in the C terminus of the protein. In contrast to MyoD N terminal phospho-sites, C terminal sites show strong sequence conservation between species. Indeed, even in proliferating myoblasts, four major isoforms of MyoD were found to be subsequently resolved by phosphatase treatment [10], and an early in vitro assay found increased reporter transactivation when other individual C terminal proline-directed kinase sites were mutated [12]. These were found not to be phosphorylated by cdks in dividing culture cells, but they may be targeted by other proline-directed kinases such as MAPKs in the environment of the differentiating myotome.
The distinct C terminal location of regulatory phospho-sites may also relate to the complexity of MyoD function that is being revealed by genome-wide analysis. MyoD temporally orchestrates expression of multiple subsets of genes during the specification and differentiation stages of myogenesis [19]. Additionally, MyoD has been found to bind at sites throughout the genome where it induces histone modifications rather than gene expression [20]. Thus, for MyoD, DNA binding affinity does not always correlate with transcriptional activation and instead, the timing and location of cofactor association may be the critical determinant for temporal and spatial activity. For example, genome-wide analysis has shown that MyoD transcriptional activity is temporally regulated through a direct repression of promoter-bound MyoD [19]. A comparison of the genome-wide promoter occupancy of both WT and phospho-mutant MyoD may reveal clues as to potential differential phospho-regulation of distinct downstream MyoD targets. Furthermore, consistent with a role in cofactor association, the ability of MyoD to activate transcription in inaccessible chromatin has been mapped to the C terminal domain [21], and partly attributed to the helix 3 domain [22] that enables binding to pioneer factors Pbx/Meis [23]; spanning resides 218–269, this includes the S262 phospho-site. It will be interesting to determine whether co-factor binding is regulated by MyoD phosphorylation.
In conclusion, we have characterised multi-site phospho-regulation of MyoD on C-terminal S/TP sites that controls its ability to drive myogenesis in vivo: Whilst there are differences between the myogenic and neurogenic bHLH factors, the essence of multi-site phospho-regulation is both functionally and mechanistically conserved in muscle and nerve. Using an in vivo developmental and reprogramming assay system in Xenopus embryos, we have derived a novel C terminal phospho-mutant MyoD protein that has significantly enhanced myogenic activity compared to any previously described mutants. This has the potential for enhancing trans-differentiation of mammalian cells into muscle for translational applications, and this should form the basis of further investigations.
Acknowledgements
We thank Yea Ji Yeong for initial assistance with cloning. This work was supported by UK Medical Research Council (MRC) Research Grants MR/L021129/1 and MR/K01329/1, and core support from Wellcome Trust and MRC Cambridge Stem Cell Institute. LH was supported by an MRC Doctoral Training Award.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2016.11.009.
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2016.11.009.
Contributor Information
Laura J.A. Hardwick, Email: ljah2@cam.ac.uk.
John D. Davies, Email: jdd24@cam.ac.uk.
Anna Philpott, Email: ap113@cam.ac.uk.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary Fig. 1Protein sequence alignment of human, mouse and Xenopus MyoD. The bHLH domain is underlined and SP/TP sites are highlighted in red. A consensus line is also shown below the alignment to indicate the degree of conservation at each position: Residues may be identical (*), strongly conserved (:) or weakly conserved (.).
Transparency document
References
- 1.Buckingham M., Rigby P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 2.Moncaut N., Rigby P.W., Carvajal J.J. Dial M(RF) for myogenesis. FEBS J. 2013;280:3980–3990. doi: 10.1111/febs.12379. [DOI] [PubMed] [Google Scholar]
- 3.Abujarour R., Bennett M., Valamehr B., Lee T.T., Robinson M., Robbins D., Le T., Lai K., Flynn P. Myogenic differentiation of muscular dystrophy-specific induced pluripotent stem cells for use in drug discovery. Stem Cells Transl. Med. 2014;3:149–160. doi: 10.5966/sctm.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muir L.A., Nguyen Q.G., Hauschka S.D., Chamberlain J.S. Engraftment potential of dermal fibroblasts following in vivo myogenic conversion in immunocompetent dystrophic skeletal muscle. Mol. Ther. Methods & Clin. Dev. 2014;1:14025. doi: 10.1038/mtm.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali F., Hindley C., McDowell G., Deibler R., Jones A., Kirschner M., Guillemot F., Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali F.R., Cheng K., Kirwan P., Metcalfe S., Livesey F.J., Barker R.A., Philpott A. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development. 2014;141:2216–2224. doi: 10.1242/dev.106377. [DOI] [PubMed] [Google Scholar]
- 7.Hardwick L.J., Philpott A. Multi-site phosphorylation regulates NeuroD4 activity during primary neurogenesis: a conserved mechanism amongst proneural proteins. Neural Dev. 2015;10:15. doi: 10.1186/s13064-015-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindley C., Ali F., McDowell G., Cheng K., Jones A., Guillemot F., Philpott A. Post-translational modification of Ngn2 differentially affects transcription of distinct targets to regulate the balance between progenitor maintenance and differentiation. Development. 2012;139:1718–1723. doi: 10.1242/dev.077552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitzmann M., Fernandez A. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell. Mol. Life Sci. 2001;58:571–579. doi: 10.1007/PL00000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitzmann M., Vandromme M., Schaeffer V., Carnac G., Labbe J.C., Lamb N., Fernandez A. cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life and myogenic activity. Mol. Cell Biol. 1999;19:3167–3176. doi: 10.1128/mcb.19.4.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tintignac L.A., Leibovitch M.P., Kitzmann M., Fernandez A., Ducommun B., Meijer L., Leibovitch S.A. Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp. Cell Res. 2000;259:300–307. doi: 10.1006/excr.2000.4973. [DOI] [PubMed] [Google Scholar]
- 12.Song A., Wang Q., Goebl M.G., Harrington M.A. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell Biol. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tintignac L.A.J., Sirri V., Leibovitch M.P., Lecluse Y., Castedo M., Metivier D., Kroemer G., Leibovitch S.A. Mutant MyoD lacking Cdc2 phosphorylation sites delays M-Phase entry. Mol. Cell. Biol. 2004;24:1809–1821. doi: 10.1128/MCB.24.4.1809-1821.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupp R.A., Snider L., Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes & Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 15.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernon A.E., Devine C., Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130:85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- 17.Almeida A.D., Wise H.M., Hindley C.J., Slevin M.K., Hartley R.S., Philpott A. The F-box protein Cdc4/Fbxw7 is a novel regulator of neural crest development in Xenopus laevis. Neural Dev. 2010;5:1. doi: 10.1186/1749-8104-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong A.P., Tapscott S.J. Skeletal muscle programming and re-programming. Curr. Opin. Genet. Dev. 2013;23:568–573. doi: 10.1016/j.gde.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho O.H., Mallappa C., Hernandez-Hernandez J.M., Rivera-Perez J.A., Imbalzano A.N. Contrasting roles for MyoD in organizing myogenic promoter structures during embryonic skeletal muscle development. Dev. Dyn. Off. Publ. Am. Assoc. Anatomists. 2015;244:43–55. doi: 10.1002/dvdy.24217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L., Gentleman R.C., Tapscott S.J. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishibashi J., Perry R.L., Asakura A., Rudnicki M.A. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J. Cell Biol. 2005;171:471–482. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerber A.N., Klesert T.R., Bergstrom D.A., Tapscott S.J. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes & Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 23.Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1Protein sequence alignment of human, mouse and Xenopus MyoD. The bHLH domain is underlined and SP/TP sites are highlighted in red. A consensus line is also shown below the alignment to indicate the degree of conservation at each position: Residues may be identical (*), strongly conserved (:) or weakly conserved (.).