Abstract
Data presented in this Data in Brief article correspond to the article "in vivo" silencing of CD40 reduces progression of experimental atherogenesis through a NFκB/miR-125b axis and reveals new potential mediators in the pathogenesis of atherosclerosis" (M. Hueso, L. De Ramon, E. Navarro, E. Ripoll, J.M. Cruzado, J.M. Grinyo, J. Torras, 2016) [1]. Here, we describe the validation of the silencing of CD40 expression with a specific siRNA in ApoE−/− mouse aortas, and its systemic effects on splenic lymphocytic subpopulations as well as on the infiltration of aortic intima by F4/80+, galectin-3+ macrophages or by NF-κB+ cells. We also show the output of a Gene Ontology and TLDA analysis which allowed the detection of potential mediators of atherosclerosis progression. We provide the scientific community with a set of genes whose expression is increased during atherosclerosis progression but downregulated upon CD40 silencing.
Keywords: CD40, siRNA, Macrophages, Atherosclerosis, GO analysis, NF-kB, miR-125b, Clec/Klr
Specifications Table
| Subject area | Molecular Biology |
| More specific subject area | Cardiovascular gene expression |
| Type of data | Tables and Figures |
| How data was acquired | By flow cytometry, immunohistochemistry, microarray profiling, Taqman low density array profiling |
| Data format | Analyzed |
| Experimental factors | ApoE−/− intraperitoneally-treated with an anti-CD40 specific siRNA |
| Experimental features | Expression of mRNAs/miRNAs in the ascending aorta of ApoE−/− was compared with that of mice treated with a scrambled siRNA as control |
| Data source location | n.a |
| Data accessibility | Data are available with this article |
Value of the Data
-
1.
CD40 was silenced "in vivo" with a specific siRNA in ApoE−/− mice. Silencing was confirmed by measuring CD40 expression by qPCR, IHC and flow-cytometry.
-
2.
Infiltrating macrophages were detected in atherosclerotic aortas of ApoE−/− mice and its number decreased upon CD40 silencing.
-
3.
Gene Ontology (GO) analysis targeted a number of components of the NF-kB pathway, as well as members of the Clec/Klr gene families as potentially involved in disease progression. Furthermore, TLDA profiling detected a number of murine miRNAs also potentially involved in disease progression.
-
4.
This data provide evidences of the potency of the specific siRNA used.
1. Data
The dataset of this article provides information on the validation of the silencing of the CD40 gene in the aorta from ApoE-deficient mice upon treatment with a specific siCD40 [Fig. 1], the description of their systemic effects in spleen cell subpopulations [Table 1], and the quantification of the infiltration of macrophages and of NF-κB+ cells in their aortic plaques [Fig. 2]. We also present a Gene Ontology (GO) analysis [Fig. 3], centered in components of the NF-κB pathway [Table 2], or in the Clec and Klr families [Table 3], as well as a miRNA gene expression data analysis [Table 4] during disease progression and upon siCD40 treatment. Finally we present the demographic data for the patients included in the accompanying study [Table 5].
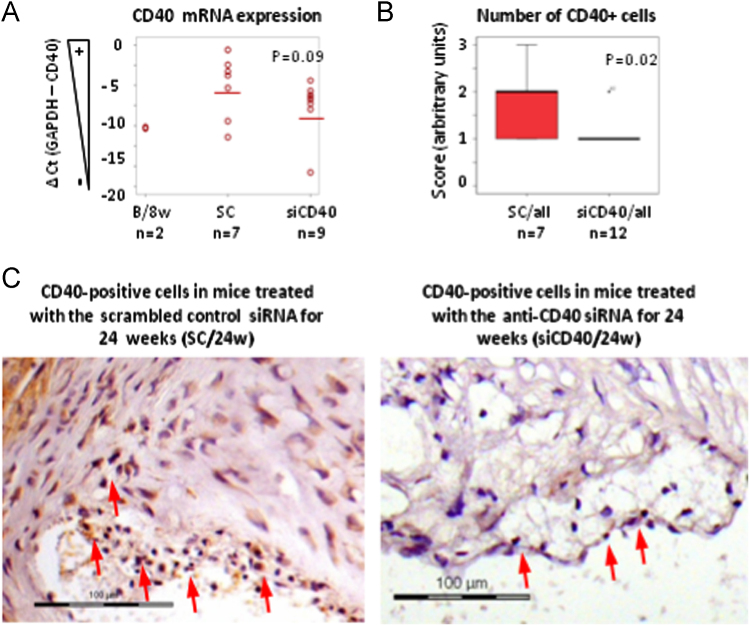
Fig. 1.
Efficiency of CD40 expression silencing with the siCD40. (A). Dot plot of CD40 expression in the aortas of ApoE−/− mice at basal conditions (B/8w), or treated with the anti-CD40 siRNA (siCD40) or with the SC control siRNA (SC). Mean is represented by a line. (B). Box plot quantifying CD40+ cells in ascending aortas of mice treated with siCD40 (n=12 mice) or SC (n=7). (C). Representative staining of CD40+ cells in the aortic sinus (arrows). Scale bars are 100 µm. Kruskall–Wallis test.
Table 1.
Immunophenotype data analysis of spleen cell sub-populations from CD40-silenced in ApoE-deficient mice.
| B/8w | SC/24w | siCD40/24w | p | |
|---|---|---|---|---|
| n | 5 | 4 | 8 | |
| CD3+ | 17±6% | 28±8% | 33±2% | 0.044 |
| CD3+CD40+ | 17±6% | 16±10% | 7±2%* | 0.008 |
| CD4+/CD8+ | 1.8±0.5% | 2.7±1.1% | 3.2±1.2% | 0.040 |
| CD19+ | 48±7% | 29±4% | 19±9% | 0.005 |
| CD19+CD40+ | 45±6% | 28±4% | 20±14% | 0.01 |
| CD11b+ | 12±2% | 13±11% | 9±4% | 0.32 |
| CD11b+CD40+ | 18±2% | 31±4% | 18±6%* | 0.029 |
| CD11b+CD86+CD40+ | 5±2% | 4±1% | 3±1% | 0.15 |
| CD11b+CD206+CD40+ | 4±2% | 2±0.4% | 2±2% | 0.08 |
| CD11c+CD40+ | 7±2% | 8±1% | 6±2% | 0.39 |
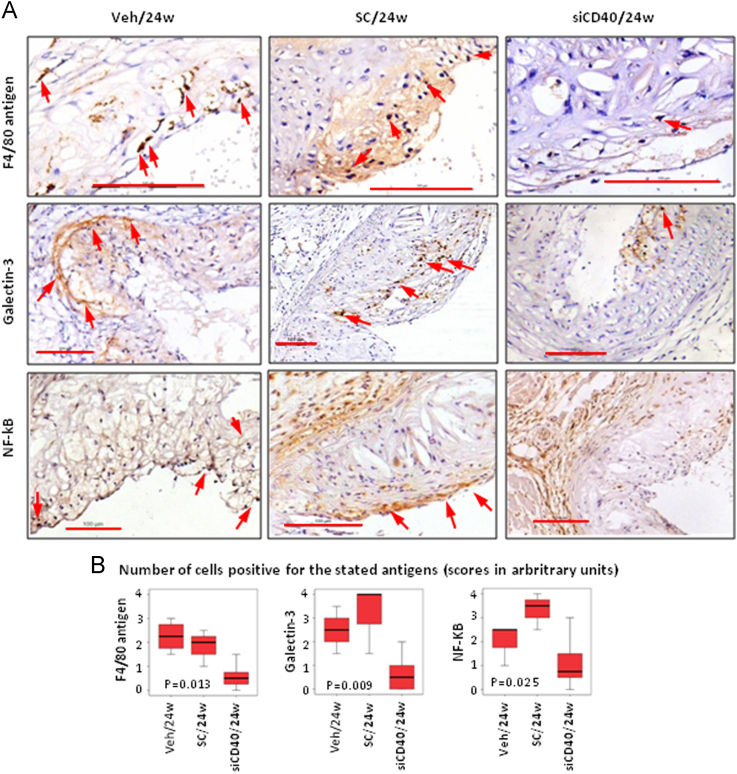
Fig. 2.
CD40 silencing results in a reduced macrophage infiltration and NF-κB+cells in plaques ofApoE−/−mice treated with siCD40. (A). Representative images of F4/80, galectin-3 or NF-κB staining at 24w. In all cases, arrows show positive cells for the stated antigen. (B). Box plot quantifying neointimal macrophages (as F4/80+ or galectin-3+ cells) and NF-κB+ cells in ascending aortas of mice treated with siCD40 (n=8 mice), SC (n=3) and Veh (n=4) at 24w. Scale bars are 100 µm. Kruskall–Wallis test.
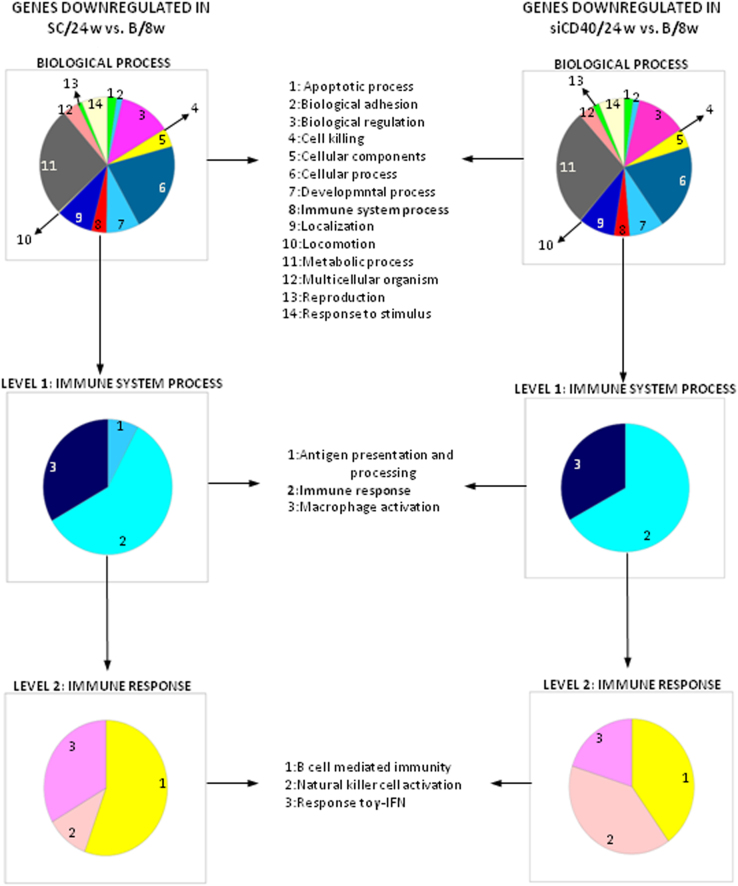
Fig. 3.
GO charts for genes down-regulated in the stated experiments. Every pie portion stands for a functional group of genes and its size is proportional to the number of genes that belong to that group.
Table 2.
Gene Ontology data analysis (GO=Biological process/Immune system process/Immune response) centered on the genes encoding components of the NF-κB pathway.
| Gene |
SC/10w vs. B/8w UP |
SC/24w vs. B/8w UP |
SC/24w vs. B/8w DOWN |
siCD40/24w vs. SC/24w UP |
siCD40/24w vs. SC/24w DOWN |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| log2FC | -log10(PVal) | log2FC | -log10(PVal) | log2FC | -log10(PVal) | log2FC | -log10(PVal) | log2FC | -log10(PVal) | |
| IKKA-CHUK | 1.13 | 1.83 | n.s. | n.s. | n.s. | n.s. | 1.09 | 1.50 | n.s. | n.s. |
| IKKB | 3.31 | 3.05 | 2.79 | 2.75 | n.s. | n.s. | n.s. | n.s. | 1.29 | 1.72 |
| IKKE1 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | 1.16 | 3.28 | n.s. | n.s. |
| SIKE | n.s. | n.s. | n.s. | n.s. | 2.05 | 3.38 | n.s. | n.s. | n.s. | n.s. |
| IKBA | n.s. | n.s. | n.s. | n.s. | 2.33 | 2.05 | n.s. | n.s. | n.s. | n.s. |
Table 3.
Gene Ontology data analysis (GO=Biological process/Immune system process/Immune response) centered on the genes encoding components of the Clec and Klr families.
| Gene |
SC/10w vs. B/8w UP |
SC/10w vs. B/8w DOWN |
SC/24w vs. B/8w UP |
SC/24w vs. B/8w DOWN |
||||
|---|---|---|---|---|---|---|---|---|
| log2FC | −log10(PVal) | log2FC | −log10(PVal) | log2FC | −log10(PVal) | log2FC | −log10(PVal) | |
| Clec1a | n.s. | n.s. | n.s. | n.s. | 1.90 | 3.07 | n.s. | n.s. |
| Clec2e | n.s. | n.s. | n.s. | n.s. | 1.90 | 1.69 | n.s. | n.s. |
| Clec2g | n.s. | n.s. | n.s. | n.s. | 3.86 | 1.62 | n.s. | n.s. |
| Clec2i | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. |
| Clec4a2⁎ | n.s. | n.s. | n.s. | n.s. | 1.04 | 2.64 | n.s. | n.s. |
| Clec4a2⁎⁎ | 1.47 | 5.57 | n.s. | n.s. | 1.01 | 1.91 | n.s. | n.s. |
| Clec4d | n.s. | n.s. | 1.20 | 1.37 | n.s. | n.s. | n.s. | n.s. |
| Clec4e | n.s. | n.s. | n.s. | n.s. | 1.37 | 2.50 | n.s. | n.s. |
| Clec4f | n.s. | n.s. | n.s. | n.s. | 2.02 | 2.10 | n.s. | n.s. |
| Clec11a | n.s. | n.s. | n.s. | n.s. | 2.23 | 1.82 | n.s. | n.s. |
| Clec18a | 2.42 | 3.42 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. |
| D21Rik⁎⁎⁎ | n.s. | n.s. | n.s. | n.s. | 3.06 | 1.40 | n.s. | n.s. |
| Klra1 | n.s. | n.s. | n.s. | n.s. | 3.01 | 1.41 | n.s. | n.s. |
| Klra2 | n.s. | n.s. | n.s. | n.s. | 1.41 | 2.20 | n.s. | n.s. |
| Klra9 | 1.02 | 1.64 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. |
| Klra18 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. |
| Klra21 | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | 2.01 | 1.37 |
| Klrb1a | n.s. | n.s. | n.s. | n.s. | 1.66 | 1.30 | n.s. | n.s. |
| Klrc2 | n.s. | n.s. | 1.10 | 1.62 | n.s. | n.s. | n.s. | n.s. |
| Klre1 | n.s. | n.s. | n.s. | n.s. | 1.39 | 1.65 | n.s. | n.s. |
| Klri2 | n.s. | n.s. | 1.47 | 1.85 | n.s. | n.s. | n.s. | n.s. |
Clec4a2 Transcript variant 2.
Clec4a2 Transcript variant 3.
D21Rik stands for RIKEN cDNA 4922502D21, also known as Clec2m.
Table 4.
TLDA data analysis of miRNA expression.
| miRNA |
SC/24w vs. B/8w |
siCD40/24w vs. SC/24w |
||
|---|---|---|---|---|
| log2FC | −log10(PVal) | log2FC | −log10(PVal) | |
| miR-let7i | 2,96 | 1,79 | −0,71 | 0,53 |
| miR-10a | 4,77 | 2,69 | −0,26 | 0,08 |
| miR-26a | 2,51 | 1,82 | −1,28 | 1,30 |
| miR-27a | 1,53 | 1,85 | −0,64 | 0,52 |
| miR-27b | 4,69 | 1,49 | −0,83 | 0,69 |
| miR-30a | 2,35 | 2,69 | −1,15 | 1,34 |
| miR-122 | 6,28 | 1,35 | 0,99 | 1,07 |
| miR-125b-5p | 4,29 | 2,39 | −1,73 | 2,52 |
| miR-130a | 4,10 | 1,60 | −1,15 | 0,88 |
| miR-132 | −0,15 | 0,22 | −0,51 | 0,63 |
| miR-324-5p | 4,25 | 1,45 | −1,68 | 0,86 |
| miR-363 | N.D. | N.D. | −2,55 | 2,39 |
| miR-465a-5p | 1,88 | 1,52 | N.D. | N.D. |
| miR-491 | 2,29 | 1,69 | −0,41 | 0,34 |
| miR-543 | 4,43 | 1,60 | −0,47 | 0,33 |
Table 5.
Clinical characteristics of patients from which aortic tissue (advanced plaque and normal aorta) was extracted.
| ID | Age | Gender | Cause of death | Diabetes | Hypertension | Dyslipidemia |
|---|---|---|---|---|---|---|
| 4 | 60 | Female | Cardiovascular | No | Yes | No |
| 5 | 83 | Female | Cardiovascular | No | Yes | No |
| 12 | 60 | Female | Cardiovascular | Yes | Yes | Yes |
| 21 | 76 | Male | Infection | Yes | Yes | No |
| 136 | 68 | Female | Cancer | No | No | No |
| 149 | 86 | Female | Cardiovascular | No | Yes | No |
ID: Our identification Number.
2. Experimental design, materials and methods
2.1. Study design
CD40 was silenced with a specific siRNA (siCD40) in ApoE−/− mice. Global patterns of expression of mRNAs/miRNAs in the ascending aorta were compared among siCD40 and SC-control treated mice.
2.2. Validation of CD40 silencing efficiency
CD40-silencing was validated by qPCR and IHC (Fig. 1). Sections were evaluated by hematoxylin/eosin staining and antigen-specific IHC using standard procedures.
2.3. Analysis of macrophage cell infiltration in plaques of ApoE-deficient mice treated with siCD40
F4/80+, galectin-3+, and NF-kB+ cells were detected and quantified by antigen-specific IHC using standard procedures (Fig. 2).
2.4. Evaluation of the systemic effect of CD40 silencing
Changes in splenic lymphoid cell subsets were characterizaed by using a BD FACS Canto II Cytometer after double or triple staining with monoclonal antibodies (Table 1). Splenocytes were isolated as previously described [2] and incubated with anti-CD19APC (clone 1D3), anti-CD3APC (clone 145-2C11), anti-CD4PECY7 (clone RM4-5), anti-CD8PERCP-CY5.5 (clone 53-6.7), anti-CD11cPE (clone HL3), anti-CD11bAPC-CY7 (cloneM1/70), anti-F4/80PE (clone BM8), antiCD40FITC (clone HM40-3), anti-CD86FITC (Clone GL1), and anti-CD206FITC (clone C068C2), all from BD Biosciences (BD Biosciences, San Jose, CA, USA). For each marker, results are expressed as percentage of the total number of cells acquired. Kruskal–Wallis test. *p<0.05 by Bonferroni test to compare SC/24w vs siCD40/24w.
2.5. Gene Ontology (GO) analysis
RNA extraction, microarray hybridization and analysis were performed on a commercial basis at Arraystar Inc. (Rockville, MD, USA). Differentially expressed mRNAs were identified in Volcano plots by using the standard thresholds of log2 (Fold Change)>1 and −log10 (P-Value)>1.30 [1]. The Gene Ontology (GO) analysis (www.pantherdb.org) [3], [4], [5] was used to classify differentially expressed mRNAs by their functional roles. Genes which passed the volcano plot thresholds were arranged in pie charts (Fig. 3) and then classified in “GO Biological process” categories. Transcripts belonging to the “Immune System Process” category (level 1, GO:0002376) were subsequently studied (Level 0=Biological process, Level 1=Immune system process, (GO:0002376), and Level 2=Immune Response (GO:0006955) [6]. Every pie portion stands for a functional group of genes and its size is proportional to the number of genes that belong to that group. Shown are genes of the NF-kB pathway (Table 2), and of the Clec/Klr family (Table 3).
2.6. Gene expression miRNA profiling of mouse aortas using TLDA cards
Total RNA from frozen aortic samples was studied by TLDA cards. MiRNA expression data from the TLDAs were analyzed with the ExpressionSuite Software v1.0.3 (Life Technologies) by using the ΔΔCt method after global normalization [7]. Differentially expressed miRNAs were identified in “Volcano plots. Expression changes are shown as log2 Fold Change (log2FC) after comparing normalized expression for each experiment. Gene name (Gene), changes in expression (log2 FC), and the statistical significance of the replicates (−log10 (PVal) are shown. In all cases the thresholds used were: log2 FC>1 and −log10 (PVal) >1.30. n.s.=either log2FC or −log10 (PVal) were below the thresholds. Table 4 shows the most significant hits found.
2.7. Demographics of human samples
Abdominal human aortas were collected from autopsy material from patients deceased in the HUB from November/2009-February/2010. Confidentiality was protected following national guidelines. The study was performed conform the declaration of Helsinki and approved by the Clinical Research Ethics Committee of HUB (PR163/13). Demographics of the patients involved in the study are shown in Table 5.
Acknowledgments
The authors thank Instituto de Salud Carlos III through the project (PI11/00556, PI14/00762 and PI13/00969) co-funded by European Union (ERDF/ESF, “Investing in your future”), Shire Pharmaceutical Spain and Societat Catalana de Transplantament for the financial support of this project. We thank Cristian Varela for his excellent technical assistance and Nuria Bolaños, both from Laboratori de Nefrologia Experimental-IDIBELL, who contributed in mice experiments and in histological studies; Benjamin Torrejón from Centre Cientific y Tecnològic of Universitat de Barcelona for helping with the histomorphometric analysis.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2016.11.045.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.M. Hueso, L. De Ramon, E. Navarro, E. Ripoll, J.M. Cruzado, J.M. Grinyo, J. Torras, "in vivo" silencing of CD40 reduces progression of experimental atherogenesis through a NFκB/miR-125b axis and reveals new potential mediators in the pathogenesis of atherosclerosis, 2016, in press [DOI] [PubMed]
- 2.Ripoll E., Merino A., Herrero-Fresneda I., Aran J.M., Goma M., Bolanos N., de Ramon L., Bestard O., Cruzado J.M., Grinyo J.M., Torras J. CD40 gene silencing reduces the progression of experimental lupus nephritis modulating local milieu and systemic mechanisms. PLoS One. 2013;8:e65068. doi: 10.1371/journal.pone.0065068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. Gene Ontol. Consort. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium T.G.O. The Gene Ontology: enhancements for 2011. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1028. (D559-564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium T.G.O. Gene Ontology annotations and resources. Nucleic Acids Res. 2013;41:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D׳Haene B., Mestdagh P., Hellemans J., Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol. Biol. 2012;822:261–272. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material