Abstract
Novel molecular targets are being searched to aid in prostate cancer diagnosis and therapy. Recently, ZFP91 zinc finger protein has been found to be upregulated in prostate cancer cell lines. It is a potentially important oncogenic protein; however only limited data regarding its biological function and expression patterns are available. To date, ZFP91 has been shown to be a key factor in activation of noncanonical NF-κB signaling pathway as well as to be involved in HIF-1α signaling in cancer cells. The present study aimed to characterize ZFP91 expression in prostate cancer specimens. Furthermore, since our earlier reports showed discrepancies between ZFP91 mRNA and protein levels, we studied this interrelationship in LNCaP and PC-3 prostate cancer cell lines using siRNA mediated knockdown. QPCR analysis revealed marked upregulation of ZFP91 mRNA in the majority of prostate cancer specimens. Transfection of prostate cancer cells with ZFP91 siRNA resulted in a 10-fold decrease in mRNA levels. On a protein level, however, no inhibitory effect was observed over the time of the cell culture. We conclude that ZFP91 is overexpressed in prostate cancer and that potential accumulation of the ZFP91 protein in studied cells may be of importance in prostate cancer biology.
1. Introduction
Prostate cancer is worldwide the second most common cancer in men with over million of new cases diagnosed every year and is the fifth leading cause of death of cancer in men (WHO International Agency for Research on Cancer, http://www.iarc.fr/). With the advent of whole genome expression analyses and progress in molecular biology techniques several molecular pathways involved in prostate cancer pathogenesis have been discovered. Still novel targets are being searched to aid in diagnosis and treatment of this complex and heterogeneous disease [1]. In 2014 our team described changes in expression of little-studied ZFP91 zinc finger protein gene (ZFP91) in benign prostate hyperplasia (BPH) and in prostate cancer cell lines [2]. Recently found oncogenic properties of this gene prompted us to further study its significance in prostate cancer pathogenesis.
ZFP91 gene was discovered in 1995 by Saotome et al. [3]. In 2003, using cDNA microarray screening, Unoki et al. found its overexpression in mononuclear cells from patients with acute myelogenous leukemia (AML) and in several neoplastic blood cell lines [4]. Subsequently, in 2006 an interaction of ZFP91 with ARF tumor suppressor protein (cyclin-dependent kinase inhibitor 2A, isoform 4) was discovered. This protein serves important antioncogenic functions based on p53-dependent cell death or cell cycle arrest in response to oncogenes activation [5]. To date the most important findings regarding ZFP91 gene functions come from works of Lee and Jin et al. This team patented concepts of ZFP91-based therapies and published a series of papers providing valuable insight into ZFP91 role in human biology and cancer pathogenesis [6–9].
ZFP91 expression is positively regulated by agonists of the nuclear factor kappa B (NF-κB) signaling pathway through a promotor sequence located in ZFP91 gene's 5′ upstream region. On the other hand, ZFP91 overexpression results in increased NF-κB activity in a dose dependent manner. This effect is also dependent on presence of MAP3K14 protein, known also as NIK (NF-κB-inducing kinase). This is a key kinase in activation of noncanonical (alternative) NF-κB signaling pathway [7, 10]. It has been shown that ZFP91 protein functions as an atypical E3 ubiquitin-protein ligase. Ubiquitination of NIK by ZFP91 leads to its stabilization and activation of the noncanonical NF-κB pathway and this pathway's target genes expression [7, 8]. What is important, available data imply oncogenic activity of the NIK and its overexpression has been associated with cancer pathogenesis in, for example, melanoma, pancreatic, breast, and lung cancer [11].
Oncogenic properties of ZFP91 gene are not only limited to NIK stabilization and NF-κB signaling pathway activation. Most recently, it has been shown that ZFP91 is overexpressed in human colon cancer and promotes this cancer progression. Through interaction with NF-κB/p65, ZFP91 protein upregulates hypoxia inducible factor-1α (HIF-1α). HIF-1α is a subunit of a key transcription factor responsible for cellular response to hypoxia and implicated on many levels in cancer pathogenesis and biology [12, 13]. With regard to prostate cancer, HIF-1α is overexpressed in actively growing prostate tissues: BPH and prostate cancer [14]. Under hypoxic conditions HIF-1α dependent signaling promotes epithelial to mesenchymal transition (EMT) in prostate cancer cells which is proven to play a role in cancer progression and invasiveness [15, 16].
In the current study, the ZFP91 gene expression was examined in prostate cancer specimens and found to be markedly upregulated. To study further ZFP91 expression in prostate cancer cells, LNCaP and PC-3 prostate cancer cell lines were transfected with ZFP91 targeting siRNA. In the result a significant discrepancy between ZFP91 mRNA level changes and protein levels in these cells was observed. This indicates that ZFP91 protein may be stabilized and accumulated in prostate cancer cells and this effect may be connected with oncogenic properties of ZFP91.
2. Materials and Methods
2.1. Prostate Cancer cDNA Samples
Prostate Cancer cDNA Array III (Origene, HPRT103) was utilized providing samples from 48 prostate cancer patients. Description of every sample includes relevant clinical information, full pathology report, and RNA quality data (data available at http://www.origene.com/qPCR/Tissue-qPCR-Arrays.aspx). In 9 cases tissue samples came from parts of prostate without pathological changes and in remaining 39 cases from parts of prostate with cancer. Each sample was evaluated for mRNA expression of ZFP91 and of three reference genes: tubulin alpha 1b (TUBA1B), 5′-aminolevulinate synthase 1 (ALAS1), and β2-microglobulin (B2M). Out of these three, TUBA1B and ALAS1 were selected using geNorm method as a reference to normalize data. Of note, selected genes were proven to be among the most stable and useful ones for normalization purposes in gene profiling studies of prostate tissues, both malignant and not [17].
2.2. Prostate Cancer Cell Lines
Prostate cancer cell lines, LNCaP and PC-3, were obtained from American Type Culture Collection (ATCC, Manassas, USA) and maintained in RPMI-1640 Medium (LNCaP) or F12K Medium (PC-3). Media were purchased from ATCC and supplemented with 10% fetal bovine serum. The cells were grown in 75 cm2 flasks at 37°C in a humidified atmosphere of 5% CO2. The culture medium was changed every 2 days. When cells reached approximately 80% confluence, they were either subcultured or harvested by 0.25% trypsin-EDTA. Harvested cells were frozen in −80°C for further analyses.
2.3. Transfection
LNCaP and PC-3 cells transfection conditions were optimized using siGLO Green Transfection Indicator (Dharmacon, GE Healthcare, Lafayette, USA) and Fluoview FV10i-LIV confocal microscope (Olympus, Melville, USA) for image acquisition. Cells were transfected with ZFP91 siRNA (ON-TARGETplus SMARTpool, Dharmacon) or negative control siRNA (ON-TARGETplus Nontargeting control Pool, Dharmacon) or left untreated. DharmaFECT Transfection Reagents 2 and 3 (Dharmacon) were used as transfection agents. The procedure was performed on logarithmically growing LNCaP and PC-3 cells according to manufacturer's recommendations with several modifications of the procedure tested. Viability of cells was determined by microscopic evaluation and trypan blue exclusion test.
2.4. Total RNA and Protein Extraction
Total RNA and protein were extracted by means of NucleoSpin RNA/Protein and NucleoSpin RNA Clean-Up XS (Macherey-Nagel Ltd., Oensingen, Switzerland). RNA concentration and purity were determined spectrophotometrically (NanoDrop, Thermo Scientific, Waltham, USA). For each sample 1 μg of total RNA was reversely transcribed using MMLV reverse transcriptase kit (Novozym, Poznan, Poland) with Oligo dT (PE Biosystems, Warrington, UK) as primers. The reaction was performed at 42.8°C for 60 min (UNO II thermocycler, Biometra, Goettingen, Germany).
2.5. QPCR Analysis
Analyses were performed as described earlier [2, 18, 19]. Briefly, primers were designed by Primer 3 software (Whitehead Institute for Biomedical Research, Cambridge, USA) and purchased from the Laboratory of DNA Sequencing and Oligonucleotide Synthesis (Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland). Primers sequences are listed in Table 1. Real-time PCR was carried out in a LightCycler 2.0 thermocycler (Roche Diagnostics, Basel, Switzerland) with software version 4.05. SYBR Green detection system was used based on LightCycler FastStart DNA Master SYBR Green I mix (Roche). PCR reactions were carried out in 20 μL mixtures, containing 4 μL template cDNA, 0.2 μM of each gene specific primer, and 3.5 mM of Mg2+ ions. The real-time PCR program included a 10 min denaturation step to activate the Taq DNA polymerase, followed by a three-step amplification program: denaturation at 95.0°C for 9 s, annealing at 58.0°C for 5 s, and extension at 72.0°C for 5 s. Specificity of the reaction products was routinely checked by determination of melting points (0.1°C/s transition rate) and random sample separation in a 2.5% ethidium bromide/agarose gel (Figure 1). PCR efficiency was assessed by a serial dilution method. Briefly, products of PCR reactions were separated in a 2.5% agarose gel and specific bands were extracted using a DNA gel extraction kit (Millipore, Billerica, USA). The amount of extracted DNA was estimated spectrophotometrically. Extracted DNA was diluted (10-fold serial dilutions) in order to generate a standard curve for efficiency calculation (LightCycler software version 4.05.).
Table 1.
Oligonucleotide sequences of sense (S) and antisense (A) primers are shown for ZFP91 zinc finger protein (ZFP91), two primer pairs, tubulin alpha 1b (TUBA1B), 5′-aminolevulinate synthase 1 (ALAS1), and β2-microglobulin (B2M).
| cDNA | GenBank accession number | Primer | Primer sequence (5′-3′) | Position | PCR product size (bp) |
|---|---|---|---|---|---|
| ZFP91 | NM_053023 | S | TGTCCTTGCCCATCCTCGCTA | 1128–1148 | 190 |
| A | ACTCTTGAAGGCCCGAGCAC | 1298–1317 | |||
| S | GAAACCCCAAAGCCACGGAG | 892–911 | 227 | ||
| A | CCTTCCATCTCACAACGGACA | 1098–1118 | |||
| TUBA1B | NM_006082 | S | TGGAACCCACAGTCATTGATGA | 430–451 | 135 |
| A | TGATCTCCTTGCCAATGGTGTA | 543–564 | |||
| ALAS1 | NM_000688 | S | AGACATAACATCTACGTGCAA | 2031–2051 | 167 |
| A | GAATGAGGCTTCAGTTCCA | 2179–2197 | |||
| B2M | NM_004048 | S | CAGCCCAAGATAGTTAAGTG | 385–404 | 262 |
| A | CCCTCCTAGAGCTACCTGT | 628–646 |
Figure 1.
Ethidium bromide-stained 2.5% agarose gel showing random samples out of prostate cancer cDNA Array amplified with ZFP91 primers. Note presence of reaction products with the expected size of 190 bp. As a DNA size standard O'RangeRuler 50 bp DNA Ladder (Fermentas) was used.
2.6. Western Blot Analysis
For each sample, 20 μg of protein was separated in a 4–20% gradient SDS-polyacrylamide electrophoretic gel and transferred onto a PVDF membrane. Transferred proteins were stained with Ponceau S. Membranes were incubated in a blocking buffer consisting of 5% nonfat dry milk in TBST for 1 h, followed by primary antibody incubation overnight at 4°C with rabbit anti-ZFP91 at 1 : 200 (sc-102172; Santa Cruz Biotechnology, Dallas, USA) or (to further validate the results) with rabbit anti-ZFP91 at 1 : 200 (ab30970; Abcam, Cambridge, UK) and with rabbit anti-β-Actin at 1 : 2000 (#5142; Cell Signaling Technology, Danvers, USA). Afterwards, membranes were thoroughly washed and incubated with an anti-rabbit HRP-linked antibody at 1 : 2000 (#7074; Cell Signaling Technology) for 1 h at room temperature. After washing, membranes were incubated with enhanced chemiluminescence (ECL Plus, Amersham, GE Healthcare) detection reagents (5 min, room temperature) and visualized on GelDoc-It Imaging System (UVP, Upland, USA) with use of VisionWorks LS Software. ECL DualVue Western Blotting Markers (GE Healthcare) served as a protein size standard. Band detection and quantification of band intensity were performed using TotalLab (Nonlinear dynamics, Newcastle upon Tyne, England).
2.7. Statistical Analysis
GraphPad Prism version 5.00 (GraphPad Software, San Diego, USA) was used to perform statistical analyses. Differences were considered significant at p < 0.05. For a more detailed description of particular experiment statistics, see description below each figure.
3. Results
3.1. ZFP91 mRNA Expression in Prostate Cancer Specimens and Its Correlation with Gleason Score
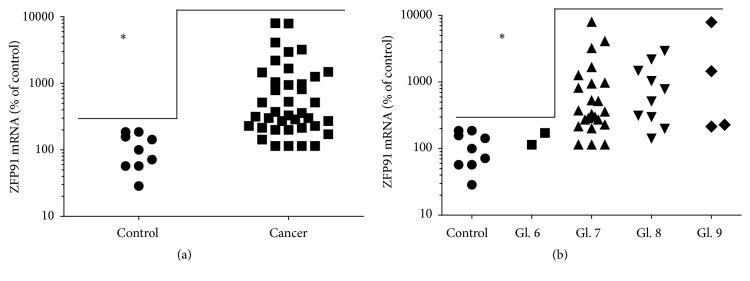
Samples with cDNA from 48 patients with pathologically confirmed prostate cancer were examined. Samples came from tissue fragments evaluated microscopically as normal prostate or prostate cancer. Using QPCR method ZFP91 expression was analyzed in above-mentioned samples finding its significant overexpression in prostate cancer (Figure 2(a)). In some cases the difference was over 10-fold. On the other hand, the range of ZFP91 expression in cancer samples was very wide and specimens with similar expression as in control group were also present.
Figure 2.
QPCR analysis of ZFP91 gene expression in normal prostates (control n = 9) compared to prostate cancer specimens (Cancer n = 39) (a) and compared to prostate cancer specimens grouped according to Gleason score (Gl. 6 n = 2, Gl. 7 n = 23, Gl. 8 n = 10, and Gl. 9 n = 4) (b). Results are presented as a scatter plot and median expression in control group was assigned a value of 100. Statistical comparison by Mann-Whitney test (a) and by Kruskal-Wallis test followed by Dunn's test (b); ∗ p < 0.05.
In subsequent analysis, samples taken from prostate cancer fragments were divided according to the Gleason score (Figure 2(b)). No significant differences in ZFP91 expression were noted between tissues with Gleason scores 7, 8, and 9. Samples with Gleason score 6 prostate cancer had ZFP91 expression similar to control group, that is, significantly lower. However, this interesting result requires further studies due to limited number of samples in Gleason score 6 group.
3.2. ZFP91 siRNA Mediated Knockdown in LNCaP and PC-3 Prostate Cancer Cells
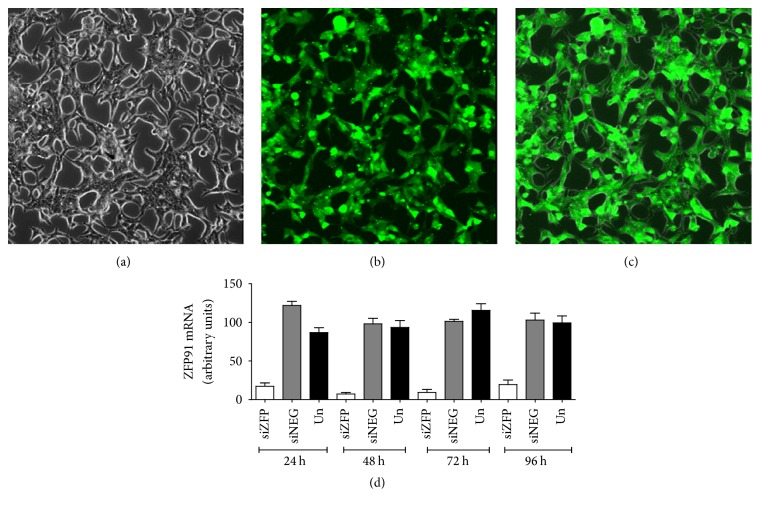
In order to optimize conditions for ZFP91 gene knockdown in prostate cancer cells, LNCaP and PC-3 cell lines were firstly transfected with fluorescent indicator of transfection efficiency (siGLO Green Transfection Indicator, Dharmacon). After a series of experiments a protocol was established allowing successfully transfecting almost all cells in a culture (a representative image for LNCaP cells is shown in Figure 3). In a next step, using a pool of predesigned ZFP91 siRNA (ON-TARGETplus SMARTpool, Dharmacon) transfection conditions were further optimized in order to achieve maximal ZFP91 knockdown while maintaining unaffected cell viability. ZFP91 mRNA downregulation to the levels between 5 and 20 percent of control groups was achieved in LNCaP cells (Figure 3). Similar results were obtained in PC-3 cells (data not shown). What is important, the silencing effect of RNA interference was stable over at least 4 days of cell culture (Figure 3).
Figure 3.
Transfection of LNCaP cell line with ZFP91 siRNA. Top. cells transfected with siGLO Green Transfection Indicator showing good transfection efficiency. Representative cell images under bright field (a), fluorescence field with a FITC filter (b), and superimposed images (c). (d) QPCR analysis of ZFP91 gene RNA interference. Cells transfected with ZFP91 siRNA pool (siZFP), nontargeting siRNA control pool (siNEG), and untreated (Un). ZFP91 mRNA levels at different time points were examined. Bars represent mean ZFP91 expression ± SE; median value in untreated group was assigned a value of 100. Each experiment was performed in triplicate.
3.3. Discrepancy between ZFP91 mRNA and Protein Levels
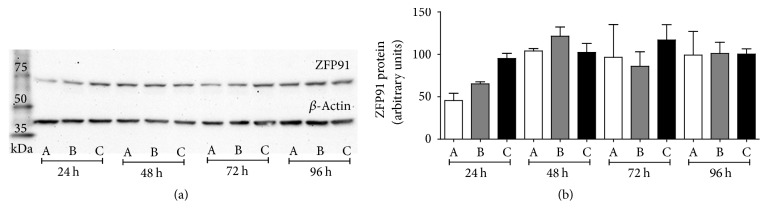
Protein levels of ZFP91 were examined in prostate cancer cells treated with ZFP91 siRNA. Despite marked ZFP91 mRNA downregulation no effect on the protein level was observed. Figure 4 shows representative results of ZFP91 immunoblotting in LNCaP cells. Similar results were obtained for PC-3 cells (data not shown). What is important, this discrepancy between mRNA and protein levels was maintained over time of the cell culture. This finding was further validated by a set of experiments using, among others, different culture and transfection conditions and different ZFP91 primers and antibodies.
Figure 4.
Representative experiment of ZFP91 protein immunoblotting in LNCaP cells transfected with ZFP91 siRNA pool (A), nontargeting siRNA control pool (B), and untreated (C) at indicated time points. Bars represent protein expression relative to β-Actin levels. Median expression in untreated group was assigned a value of 100. Results are presented as means ± SE; each experiment was at least duplicated.
4. Discussion
The results of the present study show significantly elevated expression of ZFP91 mRNA in prostate cancer samples compared with normal prostate, in some cases over 10-fold. These results are in accordance with an observation of Lee et al. of increased ZFP91 mRNA staining in prostate cancer specimens using in situ hybridization method [6]. What is important, the hereby presented results come from a significant number of samples: 48, with full pathology reports available. As found in our previous work, similar ZFP91 mRNA overexpression was not noted in prostate cancer cell lines, despite upregulated ZFP91 protein levels in these cells. This difference, although surprising, could be explained by genome differences between cancer cell lines and cancer specimens [20–23].
Analysis of ZFP91 gene expression in prostate cancer samples subdivided according to Gleason score did not reveal significant differences between Gleason scores 7, 8, and 9. Lower ZFP91 expression in samples with Gleason score 6 is an interesting result as it represents a low-risk prostate cancer and efforts are still being made to distinguish low- and high-risk disease [24]. However, the sample number in Gleason score 6 group is insufficient to draw any conclusions and this issue requires further study. Overall, no evidence suggests that observed ZFP91 overexpression in prostate cancer is correlated with Gleason score or disease staging (data not shown). In this regard, it would be of interest to study potential differences between patients with high ZFP91 expression in cancer samples and those with its expression unchanged. In the field of prostate cancer studies, genes whose expression is an independent risk factor of the disease course have been discovered [25]. Whether ZFP91 gene expression could be such a factor remains to be answered. It is possible that it only accompanies other cellular processes (e.g., NF-κB pathway activation) and as such does not represent an independent risk factor. Abundance and complexity of factors influencing NF-κB pathway activation cause problems in selecting key compounds in the studied processes [26]. It would be important to test significance of the observed changes at the protein level, especially as marked discrepancies between mRNA and protein levels of ZFP91 have been described. Marked overexpression of ZFP91 in many samples of prostate cancer remains unclear and requires further investigation.
Studies on ZFP91 gene's potential role in prostate cancer pathogenesis seem important not only due to its oncogenic properties. In recent years one of the thoroughly investigated molecular targets in prostate cancer therapy is factors involved in the NF-κB pathway activation. Its role as a key regulator of immune processes is well documented; still an increasing line of evidence implies its influence on cancer initiation and propagation. With regard to prostate cancer biology, particularly underlined are multidimensional relations between NF-κB pathway activation and activation of signaling pathways dependent on androgen receptor [27]. Interaction between androgen receptor and p52 factor (engaged in noncanonical NF-κB pathway activation) promotes prostate cancer cell growth, protects from apoptotic cell death, and influences expression of androgen receptor dependent genes [28–31]. Taking this into consideration, ZFP91 may potentially influence androgen receptor signaling in prostate cancer cells and therefore affect the biology of prostate cancer.
With regard to NF-κB pathway function in prostate cancer cells, the influence of the canonical NF-κB pathway activation agonists such as TNF and IL-1β is relatively well characterized. Less is known in this field regarding its noncanonical pathway. Functions of this additional and more specific NF-κB activation pathway are not thoroughly studied; however available data suggest its involvement in prostate cancer biology [10, 27]. In vitro stimulation of cancer cells with TNF and LIGHT (peptide activating both canonical and alternative NF-κB signaling pathway) stimulates ZFP91 expression (in MCF-7 cells for TNF and HeLa cells for LIGHT) in a dose dependant manner [6, 8]. TNF influence on growth and function of prostate cancer cells in vitro has been well documented and, among other things, it is proven to induce apoptosis in LNCaP cells [32]. As for LIGHT, it enhances the proliferation rate of LNCaP cells [33]. Another agonist of both NF-κB signaling pathways, RANKL, stimulates migration of PC-3 cells and expression of genes associated with metastatic potential in these cells [34]. ZFP91 protein as a factor specifically regulating NF-κB noncanonical pathway activation [7, 8] may play an important role in above-mentioned relationships.
The present study is the first to our knowledge that directly tests the inhibition of ZFP91 expression in prostate cancer cell lines using RNA interference. In the work of Unoki et al. ZFP91 antisense oligonucleotides induced cell growth arrest and apoptosis in endometrial and colon cancer cell lines [4]. Lee and Jin et al. found that siRNA knockdown of ZFP91 inhibited activity of HIF-1α and its dependent genes in colon cancer cell lines and induced apoptosis in breast cancer and stomach cancer cell lines [6, 9]. In our hands successful ZFP91 mRNA knockdown was established in both LNCaP and PC-3 prostate cancer cell lines. On a protein level, however, no inhibitory effect was noted. Such phenomenon has been described in the literature and stems from only transient character of siRNA induced knockdown in cell culture [35, 36]. In cases of proteins with long turnover rate or proteins accumulated/stabilized in certain cell types siRNA mediated transfection does not result in phenotypic response. As found in our previous study, both LNCaP and PC-3 cell lines have elevated ZFP91 protein levels compared to normal prostate epithelial cells [2]. Taking this into consideration, together with a fact that ZFP91 protein knockdown was established in other cell lines, it may be hypothesized that in case of prostate cancer cells ZFP91 protein is accumulated or stabilized. Such posttranscriptional regulation of protein abundance has been described for other oncogenic proteins, for example, protein p53, where its accumulation involves enhanced translation of its mRNA and decreased proteolytic degradation [37]. It seems that in case of ZFP91 expression no direct correlation exists between its mRNA and protein level, as described in our previous report and noted also in works of Lee and Jin et al. [2, 6]. Available data imply that ZFP91 expression is subjected, at least partially, to a potent posttranscriptional regulatory mechanism. What is important, ZFP91 relative abundance in prostate cancer cells may play a role in this cancer biology.
A recent interesting study of Huang et al. presented effects of RNA-mediated knockdown of ZFP91 pseudogene on biology of pancreatic cancer cells [38]. Such a silencing resulted in decreased pancreatic cancer cells proliferation rate, inhibition of their migratory ability, and a reversal, at least partial, of the epithelial to mesenchymal transition process in these cells. Long noncoding RNA (lncRNA) expressed by ZFP91 pseudogene would be therefore another one in an emerging group of pseudogene lncRNAs involved in cellular development and cancer pathogenesis. Although no direct relation with ZFP91 gene expression or function could be assigned to this, it is a relevant information as lncRNAs are capable of acting, for example, through generation of endogenous siRNAs and influencing mRNAs in the cell [39]. Whether results observed by Huang et al. are somewhat connected with interference in ZFP91 expression requires further study.
In conclusion, recently discovered oncogenic properties of ZFP91 should draw more attention to this subject and propel research regarding expression patterns and functions of this little-studied gene. The results presented in this study indicate that ZFP91 is overexpressed in prostate cancer. A siRNA mediated knockdown of the ZFP91 gene revealed a potential accumulation of the ZFP91 protein in prostate cancer cells. Taking into consideration known ZFP91 functions, this could play a role in NF-κB and HIF-1α signaling in prostate cancer. In order to explore the significance of ZFP91 in prostate cancer biology further studies are definitely needed.
Acknowledgments
This work was supported by a grant from Ministry of Science and Higher Education of the Republic of Poland (no. DI2011 0219 41 to Lukasz Paschke).
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Shtivelman E., Beer T. M., Evans C. P. Molecular pathways and targets in prostate cancer. Oncotarget. 2014;5(17):7217–7259. doi: 10.18632/oncotarget.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paschke L., Rucinski M., Ziolkowska A., et al. ZFP91—a newly described gene potentially involved in prostate pathology. Pathology and Oncology Research. 2014;20(2):453–459. doi: 10.1007/s12253-013-9716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saotome Y., Winter C. G., Hirsh D. A widely expressed novel C2H2 zinc-finger protein with multiple consensus phosphorylation sites is conserved in mouse and man. Gene. 1995;152(2):233–238. doi: 10.1016/0378-1119(94)00717-7. [DOI] [PubMed] [Google Scholar]
- 4.Unoki M., Okutsu J., Nakamura Y. Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. International Journal of Oncology. 2003;22(6):1217–1223. [PubMed] [Google Scholar]
- 5.Van Tompkins, Hagen J., Zediak V. P., Quelle D. E. Identification of novel ARF binding proteins by two-hybrid screening. Cell Cycle. 2006;5(6):641–646. [PubMed] [Google Scholar]
- 6.Lee J. J., Lee J.-H., Lee K., Hong Y.-S., Jin X. Therapeutic agent for cancer, inflammation, and auto-immune disease containing inhibitor of Zinc Finger Protein 91. US Patent 20,080,248,024, 2008.
- 7.Jin X., Jin H. R., Jung H. S., Lee S. J., Lee J.-H., Lee J. J. An atypical E3 ligase zinc finger protein 91 stabilizes and activates NF-κB-inducing kinase via Lys63-linked ubiquitination. The Journal of Biological Chemistry. 2010;285(40):30539–30547. doi: 10.1074/jbc.m110.129551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin H. R., Jin X., Lee J. J. Zinc-finger protein 91 plays a key role in LIGHT-induced activation of non-canonical NF-κB pathway. Biochemical and Biophysical Research Communications. 2010;400(4):581–586. doi: 10.1016/j.bbrc.2010.08.107. [DOI] [PubMed] [Google Scholar]
- 9.Ma J., Mi C., Wang K. S., Lee J. J., Jin X. Zinc finger protein 91 (ZFP91) activates HIF-1α via NF-κB/p65 to promote proliferation and tumorigenesis of colon cancer. Oncotarget. 2016;7(24):36551–36562. doi: 10.18632/oncotarget.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S.-C. Non-canonical NF-κB signaling pathway. Cell Research. 2011;21(1):71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao G., Fu J. NF-kappaB and cancer: a paradigm of Yin-Yang. American Journal of Cancer Research. 2011;1(2):192–221. [PMC free article] [PubMed] [Google Scholar]
- 12.Mabjeesh N. J., Amir S. Hypoxia-inducible factor (HIF) in human tumorigenesis. Histology and Histopathology. 2007;22(5):559–572. doi: 10.14670/HH-22.559. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y., Liu J., Huang H. Recent agents targeting HIF-1α for cancer therapy. Journal of Cellular Biochemistry. 2013;114(3):498–509. doi: 10.1002/jcb.24390. [DOI] [PubMed] [Google Scholar]
- 14.Wu F., Ding S., Li X., et al. Elevated expression of HIF-lα in actively growing prostate tissues is associated with clinical features of benign prostatic hyperplasia. Oncotarget. 2016;7(11):12053–12062. doi: 10.18632/oncotarget.7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan M. I., Hamid A., Adhami V. M., Lall R. K., Mukhtar H. Role of epithelial mesenchymal transition in prostate tumorigenesis. Current Pharmaceutical Design. 2015;21(10):1240–1248. doi: 10.2174/1381612821666141211120326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M., Wang Y. X., Luo Y., et al. Hypoxia inducible factor-1α-dependent epithelial to mesenchymal transition under hypoxic conditions in prostate cancer cells. Oncology Reports. 2016;36(1):521–527. doi: 10.3892/or.2016.4766. [DOI] [PubMed] [Google Scholar]
- 17.Ohl F., Jung M., Xu C., et al. Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization? Journal of Molecular Medicine. 2005;83(12):1014–1024. doi: 10.1007/s00109-005-0703-z. [DOI] [PubMed] [Google Scholar]
- 18.Paschke L., Zemleduch T., Rucinski M., Ziolkowska A., Szyszka M., Malendowicz L. K. Adiponectin and adiponectin receptor system in the rat adrenal gland: ontogenetic and physiologic regulation, and its involvement in regulating adrenocortical growth and steroidogenesis. Peptides. 2010;31(9):1715–1724. doi: 10.1016/j.peptides.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Szyszka M., Paschke L., Tyczewska M., Rucinski M., Grabowska P., Malendowicz L. K. Lack of expression of preproorexin and orexin receptors genes in human normal and prostate cancer cell lines. Folia Histochemica et Cytobiologica. 2016;53(4):333–341. doi: 10.5603/fhc.a2015.0035. [DOI] [PubMed] [Google Scholar]
- 20.Ertel A., Verghese A., Byers S. W., Ochs M., Tozeren A. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Molecular Cancer. 2006;5(1, article 55) doi: 10.1186/1476-4598-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein W. D., Litman T., Fojo T., Bates S. E. A Serial Analysis of Gene Expression (SAGE) database analysis of chemosensitivity: comparing solid tumors with cell lines and comparing solid tumors from different tissue origins. Cancer Research. 2004;64(8):2805–2816. doi: 10.1158/0008-5472.can-03-3383. [DOI] [PubMed] [Google Scholar]
- 22.Gillet J.-P., Calcagno A. M., Varma S., et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(46):18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandberg R., Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI) Proceedings of the National Academy of Sciences of the United States of America. 2005;102(6):2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggener S. E., Badani K., Barocas D. A., et al. Gleason 6 prostate cancer: translating biology into population health. The Journal of Urology. 2015;194(3):626–634. doi: 10.1016/j.juro.2015.01.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapointe J., Li C., Higgins J. P., et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoesel B., Schmid J. A. The complexity of NF-κB signaling in inflammation and cancer. Molecular Cancer. 2013;12(1, article 86) doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain G., Cronauer M. V., Schrader M., Möller P., Marienfeld R. B. NF-κB signaling in prostate cancer: a promising therapeutic target? World Journal of Urology. 2012;30(3):303–310. doi: 10.1007/s00345-011-0792-y. [DOI] [PubMed] [Google Scholar]
- 28.Nadiminty N., Chun J. Y., Lou W., Lin X., Gao A. C. NF-κB2/p52 enhances androgen-independent growth of human LNCaP cells via protection from apoptotic cell death and cell cycle arrest induced by androgen-deprivation. Prostate. 2008;68(16):1725–1733. doi: 10.1002/pros.20839. [DOI] [PubMed] [Google Scholar]
- 29.Nadiminty N., Lou W., Lee S. O., Lin X., Trump D. L., Gao A. C. Stat3 activation of NF-κB p100 processing involves CBP/p300-mediated acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(19):7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadiminty N., Lou W., Sun M., et al. Aberrant activation of the androgen receptor by NF-κB2/p52 in prostate cancer cells. Cancer Research. 2010;70(8):3309–3319. doi: 10.1158/0008-5472.CAN-09-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessard L., Saad F., Le Page C., et al. NF-κB2 processing and p52 nuclear accumulation after androgenic stimulation of LNCaP prostate cancer cells. Cellular Signalling. 2007;19(5):1093–1100. doi: 10.1016/j.cellsig.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Chopra D. P., Menard R. E., Januszewski J., Mattingly R. R. TNF-α-mediated apoptosis in normal human prostate epithelial cells and tumor cell lines. Cancer Letters. 2004;203(2):145–154. doi: 10.1016/j.canlet.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Nadiminty N., Chun J. Y., Hu Y., Dutt S., Lin X., Gao A. C. LIGHT, a member of the TNF superfamily, activates Stat3 mediated by NIK pathway. Biochemical and Biophysical Research Communications. 2007;359(2):379–384. doi: 10.1016/j.bbrc.2007.05.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armstrong A. P., Miller R. E., Jones J. C., Zhang J., Keller E. T., Dougall W. C. RANKL acts directly on RANK-expressing prostate tumor cells and mediates migration and expression of tumor metastasis genes. Prostate. 2008;68(1):92–104. doi: 10.1002/pros.20678. [DOI] [PubMed] [Google Scholar]
- 35.Wennemers M., Bussink J., van den Beucken T., Sweep F. C. G. J., Span P. N. Regulation of TRIB3 mRNA and protein in breast cancer. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049439.e49439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu W., Hodges E., Redelius J., Höög C. A novel approach for evaluating the efficiency of siRNAs on protein levels in cultured cells. Nucleic Acids Research. 2004;32(2, article e17) doi: 10.1093/nar/gnh010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koumenis C., Alarcon R., Hammond E., et al. Regulation of p53 by Hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Molecular and Cellular Biology. 2001;21(4):1297–1310. doi: 10.1128/mcb.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W., Li N., Hu J., Wang L. Inhibitory effect of RNA-mediated knockdown of zinc finger protein 91 pseudogene on pancreatic cancer cell growth and invasion. Oncology Letters. 2016;12(2):1343–1348. doi: 10.3892/ol.2016.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X., Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500(1):10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]