Abstract
There exist many tools and methods for construction of co-expression network from gene expression data and for extraction of densely connected gene modules. In this paper, a method is introduced to construct co-expression network and to extract co-expressed modules having high biological significance. The proposed method has been validated on several well known microarray datasets extracted from a diverse set of species, using statistical measures, such as p and q values. The modules obtained in these studies are found to be biologically significant based on Gene Ontology enrichment analysis, pathway analysis, and KEGG enrichment analysis. Further, the method was applied on an Alzheimer’s disease dataset and some interesting genes are found, which have high semantic similarity among them, but are not significantly correlated in terms of expression similarity. Some of these interesting genes, such as MAPT, CASP2, and PSEN2, are linked with important aspects of Alzheimer’s disease, such as dementia, increase cell death, and deposition of amyloid-beta proteins in Alzheimer’s disease brains. The biological pathways associated with Alzheimer’s disease, such as, Wnt signaling, Apoptosis, p53 signaling, and Notch signaling, incorporate these interesting genes. The proposed method is evaluated in regard to existing literature.
The volume of data obtained by microarray or other new high throughput technologies have been increasing enormously, and as a result, the biological databases are growing enormously. The key challenge is to analyze these large amount of data and extract meaningful information. Therefore, it is an interesting and challenging task to represent these data in an easy way, so that researchers can communicate and interpret data efficiently. A biological network graph allows interactive visual representation and analysis of data. In a biological network, nodes represent bio-entities (genes or proteins) and edges represent associations between the bio-entities. The biological networks can be categorized broadly based on their constituent bio-entities, such as, protein-protein interaction network, gene co-expression network (CEN), and metabolic network. These biological networks encode the relationships, such as, correlation, co-expression, and co-regulation, among the bio-entities.
A gene CEN is defined as an undirected graph, where two genes are connected to each other if their expression levels are similar across conditions. Information related to physical-interaction, genetic interaction, shared protein domains, co-localization, pathway, and predicted functions can be inferred from a gene CEN1. A gene CEN primarily shows correlation or association in a pair of genes in terms of standard correlation measures and mutual information. These networks encode the relationships of each entity with its neighbors, which helps in geometrical interpretations for extraction of functionally related genes or network modules.
There are many established methods for construction of CEN based on various approaches and correlation measures. CEN serves in analysis of functionally related groups of genes, termed as network modules. Therefore, along with the construction of CEN, it is important to extract the functionally correlated network modules. Researchers have also introduced several tools and methods to support extraction of network modules from co-expression networks2,3,4,5.
Medical applications of analysis of co-expressed modules are many. One of such applications is to find interesting genes related to a disease. Most of the network based approaches extract co-expressed network modules from a CEN. Analysis of such network modules facilitates the study of molecular differences across different stages of a disease6. Methods presented in refs 7, 8, 9, 10, 11 suggest the application of CEN module extraction techniques in the analysis of genes related to diseases, such as Alzheimer’s Disease (AD). These studies consider the co-expressed genes in network modules based on expression similarity.
Gene expression data assess only experimental information, but not the functional associations among genes. However, for biological relevance of results, Gene Ontology (GO) derived information must be incorporated, which encode additional information, such as, functional similarity and co-membership of genes or proteins in a biological pathway12. Functional similarity between two genes with annotations can be measured using their semantic similarity. Semantic similarity is based on GO database and describes the topological distance between two terms in the hierarchical taxonomy. Semantic similarity between two genes can be measured on the basis of information content (Ic). Ic is defined as shown in Equation (1).
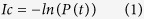 |
where,
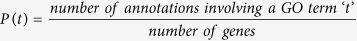 |
This paper considers the important issue of analysis of CEN using both gene expression similarity and semantic similarity. The proposed method analyzes CEN considering not only the highly co-expressed genes, but also the genes with less expression similarity, yet high semantic similarity, which are termed as border genes in this paper. The border genes obtained are found to be involved in biological pathways, and therefore, are highly functionally related.
The proposed method is applied to find the potential biomarkers related to the progression of AD. Alzheimer’ disease, mostly seen in elderly persons, is related to degeneration of neurons, leading towards dementia. With the growth of data from genomics, proteomics, and interactomics, it has become easy for researchers to understand the dysfunctionalities at molecular level during progression of a disease. In case of AD, pathways, such as Wnt signaling, Alzheimer’s disease, Apoptosis signaling, and Glycolysis, are related to the progression of the disease. Therefore, understanding of the genes participating in a network module with Pathway analysis, KEGG enrichment analysis, and GO terms helps to identify the biomarkers related to the disease.
Existing methods9,11,13,14,15, use CEN module extraction techniques that prioritize the genes and study them at molecular level during progression of AD and other neurodegenerative diseases. Alzbase11 is an integrated approach, which reveals links between upstream genetic variations and downstream endo-features and prioritizes genes in relation to AD. Method proposed by Yue et al.9 combines gene pair scores of four existing methods to provide robust and useful results enriched with pathways, such as AD, Parkinson’s disease, and Huntington’s disease.
In this paper, a method named THD-Module Extractor is introduced, which can extract biologically relevant modules from CEN and identify border genes related to AD with less expression similarity and high semantic similarity. The method accepts a microarray gene expression data M and two thresholds, namely, expression similarity threshold (δ) and minimum neighborhood threshold (ρ). The method constructs CEN from gene expression data and extracts highly co-expressed modules.
The proposed method uses SSSim16 as the expression similarity measure and Lin17 as semantic similarity measure. SSSim finds similarity between genes exhibiting shifting, scaling, and shifting-and-scaling patterns. The SSSim measure is chosen over other correlation measures since SSSim is robust to noisy values. For semantic similarity, the Lin measure is chosen as it is a normalized relative measure that finds the differences in information content of two GO terms being compared12.
The effectiveness of the proposed method is validated using various datasets, including AD dataset. In case of AD dataset, the method identifies the border genes involved in pathways related to the progression of the disease. It is observed that the highly differentially expressed border genes share some important pathways, GO terms, and KEGG pathways, known to be related to AD. The border genes are assessed to find their significance in progression of AD. The work flow of THD-Module Extractor, is illustrated in Fig. 1.
Figure 1. A schematic diagram of THD-Module Extractor framework.
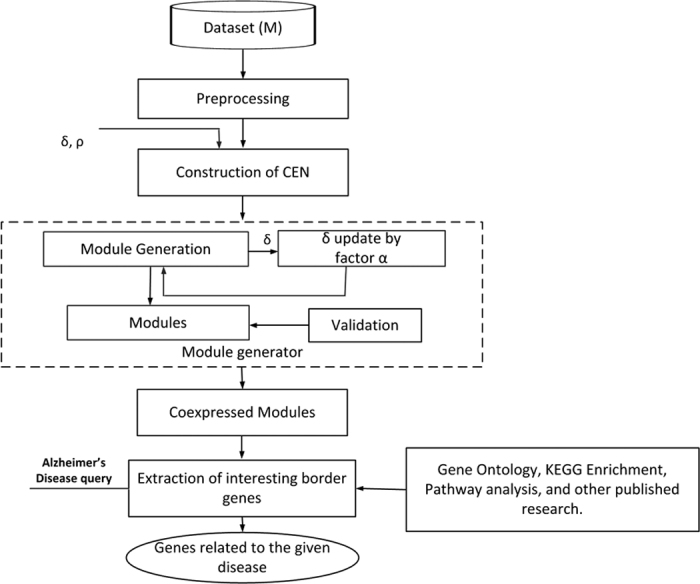
The proposed method accepts a microarray gene expression dataset, M. After preprocessing, it accepts two thresholds, namely, expression similarity threshold (δ) and minimum neighborhood threshold (ρ) to construct CEN and to extract modules with high expression similarity using similarity measure, SSSim. THD-Module Extractor updates δ by a factor α in each iteration of the module extraction process. The method identifies disease related genes by analyzing the border gene set extracted from the highly co-expressed modules and validates them using GO, KEGG Enrichment, pathway analysis, and existing literature.
This work provides a comprehensive solution towards identifying the biomarkers of diseases, such as AD, based on expression and semantic similarity of genes, obtained from module extraction method. The following are the major contributions of this work.
In this paper, a CEN module extraction method named THD-Module Extractor is introduced, which identifies not only the highly co-expressed genes, but also extracts the genes with less expression similarity and high semantic similarity. In contrast, these genes are not considered by any of the existing related methods.
The method is validated using datasets pertaining to various species, in terms of both statistical and biological significance measures. For the AD dataset, the genes with less expression similarity and high semantic similarity are assessed using GO, pathway analysis, and KEGG enrichment analysis to prioritize the genes related to the progression of the disease.
Correlation analysis of each pair of genes in a large dataset is computationally expensive. This work leverages the parallel computing capabilities of the Graphics Processing Units (GPU) to find the SSSim16 correlation matrix, implemented using the NVIDIA CUDA library.
Results
Construction of CEN and extraction of network modules
THD-Module Extractor accepts a microarray gene expression dataset and two thresholds, the expression similarity threshold (δ) and the minimum neighborhood threshold (ρ), to construct CEN and to extract modules with high expression similarity using a similarity measure, SSSim. The method identifies border genes with low expression similarity and high semantic similarity from the highly co-expressed network modules. These border genes are analyzed to identify disease related genes from AD dataset.
The threshold (δ) of the module extraction process is gradually updated from 0.9 to 0.5 by a factor α in each iteration. The minimum neighborhood threshold (ρ) is set to 3 in the conducted experiments. The generated co-expressed modules are validated in terms of p and q values.
For a list of genes, the p value represents the association of query genes with different GO terms. A lower value of p implies more biological significance of the network module. Similarly, q value is a statistical measure, which gives the minimum false discovery rate (FDR). In Tables 1 and 2, it is shown that the network modules obtained from Dataset 1 and Dataset 2 have less p and q values, respectively, which implies the biological significance of our results. Table 1 shows that the modules are biologically enriched with GO terms namely, cell cycle process, mitotic cell cycle process, chromosomal part, cell cycle, nuclear chromosome part with 3.801e-24, 5.760e-22, 2.351e-21, 2.188e-20, and 3.413e-18 p values, respectively. Table 2 shows that the modules extracted from Dataset 2 are enriched with functions like gluthathione peroxidate activity, response to wounding, monocarboxylic acid metabolic process, jasmonic acid biosynthetic process, perioxidase activity with q values 2.87e-16, 4.38e-15, 2.10e-12, 5.47e-10, 1.22e-9, respectively. Table 3 gives a comparison between FUMET4 and the proposed method in terms of p values obtained for Dataset 3. Additionally, Tables 4 and 5 depict that GO terms obtained using the proposed method for Dataset 1 have lower p values and q values than that of FUMET and Qcut, respectively. This signifies the ability of the method to detect biologically enriched modules with low p and q values. As shown in Tables 3, 4 and 5, THD-Module Extractor performs better in terms of p values and q values in comparison to FUMET and Qcut.
Table 1. p values of network modules for Dataset 1 (Subset of yeast cell cycle).
| Module No. | Number of genes | Gene Ontology ID | GeneOntology Attribute | p value |
|---|---|---|---|---|
| Module 1 | 284 genes | GO:0022402 | cell cycle process | 3.801e-24 |
| GO:1903047 | mitotic cell cycle process | 5.760e-22 | ||
| GO:0044427 | chromosomal part | 2.351e-21 | ||
| GO:0007049 | cell cycle | 2.188e-20 | ||
| GO:0044454 | nuclear chromosome part | 3.413e-18 | ||
| Module 2 | 97 genes | GO:0022402 | cell cycle process | 2.227e-14 |
| GO:1903047 | mitotic cell cycle process | 6.703e-14 | ||
| GO:0007049 | cell cycle | 2.438e-13 | ||
| GO:0000280 | nuclear division | 3.322e-9 | ||
| Module 3 | 89 genes | GO:0022402 | cell cycle process | 3.875e-14 |
| GO:0007049 | cell cycle | 1.537e-13 | ||
| GO:103047 | mitotic cell cycle process | 3.998e-13 | ||
| GO:0000280 | nuclear division | 8.879e-10 | ||
| GO:0048285 | organelle fission | 2.103e-9 |
Table 2. q values of network modules for Dataset 2 (Arabidopsis thaliana).
| Module No. | Number of genes | Functions | q value (FDR) |
|---|---|---|---|
| Module 1 | 70 genes | gluthathione peroxidate activity | 2.87e-16 |
| response to wounding | 4.38e-15 | ||
| monocarboxylic acid metabloic process | 2.1e-12 | ||
| jasmonic acid biosynthetic process | 5.47e-10 | ||
| perioxidase activity | 1.22e-9 | ||
| Module 2 | 16 genes | apoplast | 2.40e-7 |
| transferase activity transfering alkyl or aryl groups | 1.56e-3 | ||
| response to cold | 5.57e-3 | ||
| cellular response to water stimulus | 6.64e-3 | ||
| copper ion binding | 6.64e-3 | ||
| Module 3 | 27 genes | aromatic amino acid family biosynthetic process | 2.35e-9 |
| aromatic amino acid family metabolic process | 5.38e-9 | ||
| cellular amino acid biosynthetic process | 8.36e-6 | ||
| monocarboxylic acid metabolic process | 1.09e-5 | ||
| lyase activity intramolecular transferase activity | 2.91e-5 | ||
| indolalkylamine biosynthetic process | 2.94e-5 |
Table 3. Comparison of THD-Module Extractor with FUMET in terms of p values for Dataset 3 (Yeast sporulation)4.
| GO_ID | GO attribute | THD-Module Extractor | FUMET |
|---|---|---|---|
| GO:0048646 | Anatomical structure formation | 7.771e-36 | 3.38e-31 |
| GO:0043934 | Sporulation | 2.478e-34 | 1.060e-25 |
| GO:0030154 | Cell Differentiation | 1.757e-33 | 1.06e-25 |
| GO:0030435 | Sporulation resulting in formation of a cellular spore | 1.757e-33 | 1.43e-25 |
| GO:0032505 | Developmental process | 4.132e-28 | 2.06e-22 |
| GO:0048869 | Cellular developmental process | 4.002e-27 | 3.13e-23 |
| GO:0005628 | Prospore membrane | 8.597e-21 | 5.72e-16 |
Table 4. Comparison of THD-Module Extractor with FUMET and Qcut in terms of p values for Dataset 14.
| GO_ID | GO attribute | THD-Module Extractor | FUMET | Qcut |
|---|---|---|---|---|
| GO:0022402 | Cell Cycle process | 3.801e-24 | 1.482e-13 | 6.585e-8 |
| GO:0044427 | Chromosomal part | 2.351e-21 | 7.860e-10 | 2.274e-9 |
| GO:0007049 | Cell Cycle | 2.188e-20 | 5.388e-14 | 1.165e-7 |
| GO:0044454 | Nuclear chromosome part | 3.413e-18 | 7.860e-10 | 2.274e-9 |
| GO:0006281 | DNA repair | 1.949e-16 | 3.422e-9 | 2.970e-8 |
| GO:0006974 | Response to DNA damage stimulus | 9.465e-16 | 1.461e-9 | 2.983e-7 |
| GO:0051301 | Cell division | 2.508e-15 | 2.619e-7 | 1.547e-6 |
| GO:0051716 | Cellular response to stimulus | 1.662e-10 | 6.375e-8 | 1.872e-7 |
| GO:0006260 | DNA replication | 2.153e-13 | 2.124e-8 | 1.082e-7 |
| GO:0006270 | DNA replication initiation | 1.688e-8 | 2.360e-8 | 1.081e-7 |
| GO:0048523 | Negative regulation of cellular process | 5.185e-8 | 8.852e-9 | 7.970e-7 |
| GO:0048519 | Negative regulation of biological process | 4.719e-8 | 1.308e-8 | 1.073e-6 |
| GO:0005634 | Nucleus | 8.278e-8 | 4.693e-9 | 3.967e-8 |
Table 5. Comparison of results obtained from THD-Module Extractor with FUMET and Qcut in terms of q values for Dataset 14.
| GO Annotation | THD-Module Extractor | FUMET | Qcut |
|---|---|---|---|
| DNA repair | 1.97e-20 | 7.87e-14 | 5.25e-13 |
| Nuclear chromosome part | 3.76e-17 | 1.65e-13 | 1.02e-12 |
| Nuclear chromosome | 5.19e-18 | 1.05e-14 | 2.41e-13 |
| Cellular bud | 2.65e-7 | 6.22e-9 | 4.07e-3 |
| Replication fork | 8.43e-19 | 8.852e-11 | 1.78e-10 |
| Mitosis | 3.86e-8 | 4.12e-8 | 1.49e-5 |
Application of THD-Module Extractor on AD dataset
Application of THD-Module Extractor on AD dataset shows that the method can extract biologically and statistically enriched network modules. As shown in Table 6 and 7, the genes in network modules obtained from AD dataset have lower p and q values, respectively. For example, in case of Module 1 in Table 6, some of the GO terms, namely, intracellular organelle part, cytoplasm part organelle, organelle part, intracellular part, membrane bounded organelle have p values 4.920e-47, 8.199e-45, 1.106e-43, 7.388e-40, and 2.785e-39, respectively. Moreover, Module 1 is rich in functions like cystolic ribosome, translation elongation, nuclear transcribed mRNA catabolic process nonsense mediated decay with lower q values 8.21e-16, 8.80e-16, 1.11e-15, respectively. Therefore, it can be inferred that the network modules extracted from THD-Module Extractor are biologically enriched with GO terms, which have low p values and low q values with high biological significance.
Table 6. p values and q values of the network modules for Dataset 4 (AD).
| Module no. | Size of module | GO_ID | GO attribute | p value | Function | q value |
|---|---|---|---|---|---|---|
| 1 | 3652 genes | GO:0044446 | intracellular organelle part | 4.920e-47 | cystolic ribosome | 8.21e-16 |
| GO:0044444 | cytoplasm part organelle | 8.199e-45 | translational termination | 8.21e-16 | ||
| GO:0044422 | organelle part | 1.106e-43 | translational elongation | 8.8e-16 | ||
| GO:0044424 | intracellular part | 7.388e-40 | nuclear transcribedmRNA catabolic process, nonsense-mediated decay | 1.11e-15 | ||
| GO:0043227 | membrane-bounded organelle | 2.785e-39 | viral gene expression | 1.11e-15 | ||
| GO:0043229 | intracellular organelle | 1.767e-34 | viral trascription | 2.81e-15 | ||
| GO:0010467 | gene expression | 3.941e-26 | ribosomal subunit | 2.81e-15 | ||
| GO:0044403 | symbiosis, encompassing mutualism through parasitism | 2.279e-20 | multi-organism metabolic process | 7.27e-15 | ||
| GO:0043604 | amide biosynthetic process | 1.951e-18 | SRP-dependent cotranslational protein targeting to membrane | 1.33e-14 | ||
| GO:0006614 | SRP-dependent cotranslation protein targeting to membrane | 2.059e-18 | cotranslational protein targeting to membrane | 1.40e-14 |
Table 7. p values and q values of the network modules for Dataset 4 (continued).
| Module no. | Size of module | GO_ID | GO attribute | p value | Function | q value |
|---|---|---|---|---|---|---|
| Module 2 | 442 genes | GO:0003723 | RNA Binding | 3.226e-10 | translational initiation | 3.81e-8 |
| GO:0044444 | intracellular membrane-bounded organelle | 3.357e-8 | translational elongation | 8.05e-7 | ||
| GO:0043231 | negative regulation of organelle organization | 4.805e-8 | protein localization to endoplasmic reticulum | 4.44e-6 | ||
| GO:0005829 | cytosol | 6.028e-8 | viral life cycle | 7.37e-6 | ||
| GO:0032991 | macromolecular complex | 7.931e-8 | nuclear transcribed mRNA catabolic process, nonsense-mediated decay | 1.46e-5 | ||
| GO:0051129 | negative regulation of cellular component organization | 2.187e-7 | SRP-dependent cotranslational protein targeting to membrane | 2.43e-5 | ||
| Module 3 | 209 genes | GO:0031982 | vesicle | 1.950e-7 | protein polyubiquitination | 2.19e-2 |
| GO:0031968 | memebrane-bounded vesicle | 2.300e-7 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest | 2.19e-2 | ||
| GO:0065010 | extracellular memebrane-bounded organelle | 3.840e-7 | signal transduction involved in mitotic cell cycle checkpoint | 2.19e-2 | ||
| GO:0070062 | extracellular exosome | 3.840e-7 | signal transduction involved in mitotic DNA damage checkpoint | 2.19e-2 | ||
| GO:0043230 | extracellular organelle | 4.521e-7 | cell cycle arrest | 2.19e-2 | ||
| GO:1903561 | extracellular vesicle | 4.521e-7 | signal transduction involved in DNA integrity checkpoint | 2.22e-2 | ||
| GO:0002684 | positive regulation of immune system process | 3.55e-5 | G1 DNA damage checkpoint | 2.72e-2 | ||
| GO:0002757 | immune response activating signal transduction | 9.184e-4 | antigen processing and presentation of exogenous peptide antigen | 2.72e-2 |
Analysis of modules to find genes related to AD
Each network module are extracted from a CEN to find those genes that have less expression similarity with the core gene, yet high semantic similarity with all genes. A gene or a node in CEN vi ∈ V is a core gene iff it is the starting node of a module mj and satisfies the following conditions.
1. vi ∉ any other module mk where j ≠ k.
2.
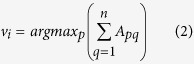 |
where A is the Adjacency matrix, p and q denote the rows and columns in A, and n is the total number of genes.
3. nNeighbor (vi) ≥ ρ, where nNeighbor (vi) is the total number of k genes directly connected to a gene vi, i.e. SSSim (vi, vk) ≥ δ.
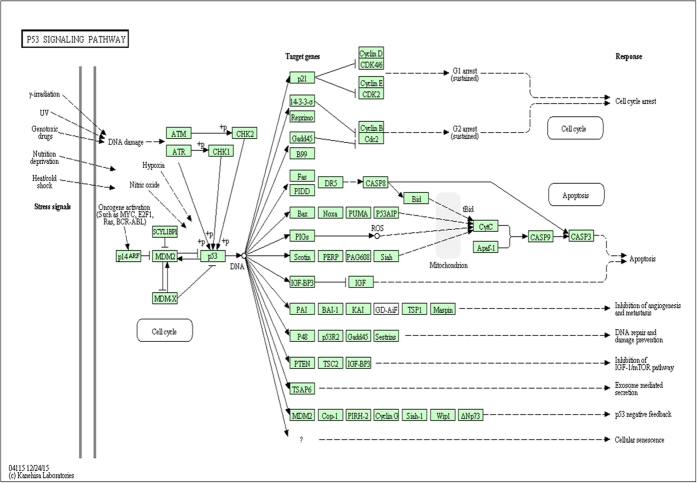
On the other hand, a gene vb within a module is a border gene iff ∀vj ∈ neighbor (vb), SSSim(vb, vj) ≥ δ and nNeighbor (vb) < ρ. From each biologically enriched modules extracted from the CEN for AD dataset, the border genes are found to have low expression similarity with respective core genes and high semantic similarity among each other. These border genes are then analyzed to find the differentially expressed genes across normal as well as disease samples. The 2000 differentially expressed genes with highest variance across normal and disease samples are considered. These differentially expressed genes are then analyzed using queryMany() of mygene package in R library to find the entrez IDs of the corresponding genes. The semantic correlations among these entrez IDs are found using mgeneSim of GOSemSim package in R using Lin’s semantic correlation measure17 and Biological Process (BP) structure. In GO-BP structure, genes with annotations are involved with multiple functions12. Figure 2 shows an evidence of genes connected in p53 pathway. From the semantic correlation matrix, the genes for which semantic correlations are greater or equal to β are extracted, where β is the mean semantic correlation score. From this analysis, 912 genes are found to be interesting, with semantic correlation ≥ β.
Figure 2. An evidence of p53 signaling pathway obtained using the DAVID tool from KEGG database.
32,33, under CC BY. Some of the genes, such as PIGs, CDK4/6, CASP9, IGF-BP3, GADD45 invovled in the pathway are found to be related to Alzheimer’s disease.
These 912 interesting genes are analyzed using PANTHER and validated using GeneCards. Table 8 provides description of some of the interesting border genes, namely, APBB2, CASP2, CSNK1D, CDK5, HSD17B10, MAPT, PSEN2, and RCAN1. Some of the pathways associated with these border genes are depicted in Table 9 and their description is given in Table 10. A few of these pathways, namely, Wnt signaling pathway, Alzheimer disease-amyloid secretase pathway, Apoptosis signaling pathway, and Glycolysis are found to be related to AD, which are further validated using GeneCards and existing literature18,19.
Table 8. Some Interesting Border Genes and their Description.
| Gene name | Description | Gene name | Description |
|---|---|---|---|
| APBB2 | Mutation in APBB2 (Amyloid Beta (A4) Precursor Protein-Binding, Family B, Member 2) causes Alzheimer’s disease1 | CASP2 | Caspase 2 is involved in activation of cascade of caspases responsible for apoptosis, which increases the cell death1 |
| CSNK1D | Casein kinase 1 delta protein is involved in apoptosis, which regulates degeneration or cell death during progresssion of AD34 | CDK5 | Cyclin-dependent kinase 5 enhances Aβ production or Aβ amyloidgenesis, which is seen in AD pateints during progression stages35 |
| HSD17B10 | Mutation in Hydroxysteroid (17-Beta) Dehydrogenase 10 is responsible for mental retardness in male36 | MAPT | Microtubule-Associated Protein Tau (MAPT) is associated with Pick disease type taupathy, which is linked with frontal lobe dementia29 |
| PSEN2 | Mutations in Presenilin 2 (PSEN2) is a key initiator for formation of amyloid plagues (nlm.nih.gov) | RCAN1 | Regulator Of Calcineurin 1 activates CASP3 and induces apoptosis37 |
Table 9. Pathways associated with interesting border genes obtained from Dataset 4.
| Pathway Accession Number | Pathways |
|---|---|
| P00004 | Alzheimer disease-presenilin pathway |
| P00059 | p53 pathway |
| P05912 | Dopamine receptor mediated signaling pathway |
| P00045 | Notch signaling pathway |
| P06587 | Nicotine pharmacodynamics pathway |
| P00003 | Alzheimer disease-amyloid secretase pathway |
| P00049 | Parkinson disease |
| P00015 | Circadian clock system |
| P06959 | CCKR signaling map |
| P00007 | Axon guidance mediated by semaphorins |
| P00057 | Wnt signaling pathway |
Table 10. Analysis of the pathways related to AD1.
| Pathway name | Description |
|---|---|
| Apoptosis Pathway | This pathway is responsible for death of cells. Since Alzheimer’s disease is associated with damage of neurons, so Apoptosis has a major role in Alzheimer’s disease38. Genes like MAPT, CASP2 catalyzes Apoptosis. |
| Notch Pathway | This pathway is responsible for maturation, cell division, and functioning of immune system. Genes like PSEN1, MYC, PSEN2 are associated with Notch pathway. In patients with Alzheimer’s disease, there is mutation in PSEN1 or PSEN2, which leads to deposition of longer form of amyloid beta in AD brains. |
| Wnt Pathway | Wnt plays an important role in development of neurons in adult brain and this implicate in neurodegenerative disease30. Genes like CSNK1D, DKK1, PIN1, PSEN1, PSEN2 are related to Wnt signaling pathways. |
| p53 signaling pathway | The p53 tumor suppressor gene is a key gene for apoptosis and activates cell growth. Since death of neuron is associated with Alzheimer’s disease, so p53 signaling pathway is an important regulator of Alzheimer’s disease. Genes like CASP3, CDK5, CREB1 are related to p53 signaling pathway. |
Further, these 912 interesting genes are analyzed to find GO terms related to AD. The GO terms that are found to be related to AD are: membrane-bounded organelle, negative regulation of apoptotic process, and antigen processing and presentation of peptide antigen via MHC class, which are validated using GeneCards.
Moreover, a Web-based tool called TopoGSA20 is used to study the topological structures. TopoGSA compares the interesting genes against KEGG database and finds KEGG enrichment scores for the given genes. Table 11 depicts that the interesting border genes extracted from the modules, obtained using THD-Module Extractor, are enriched with KEGG pathways. The KEGG pathways Wnt signaling, Apoptosis, p53 signaling pathway, Notch signaling pathway, Alzheimer’s disease signaling, associated with the interesting border genes and validated using GeneCards and existing literature18,19, are found to be related to AD. A brief description of the pathways related to AD and the interesting border genes are given in Table 9.
Table 11. Quality scores of KEGG pathways associated with interesting gene sets of Dataset 4.
| Identifier | Median degree | Median CC | Median SPL | Median BW | Median EVC | No. of genes | Score |
|---|---|---|---|---|---|---|---|
| hsa05220:Chronic myeloid leukemia | 36 | 0.08 | 3.3 | 49218 | 0.1 | 76 (0) | 0.97 |
| hsa05212:Pancreatic cancer | 30 | 0.07 | 3.35 | 45788 | 0.07 | 75 (0) | 0.93 |
| hsa05223:Non-small cell lung cancer | 29 | 0.07 | 3.35 | 38253 | 0.08 | 55 (0) | 0.93 |
| hsa04320:Dorso-ventral axis formation | 30 | 0.06 | 3.32 | 32814 | 0.1 | 22 (0) | 0.92 |
| hsa05221:Acute myeloid leukemia | 27 | 0.09 | 3.39 | 32521 | 0.07 | 57 (0) | 0.92 |
| hsa05215:Prostate cancer | 28 | 0.06 | 3.37 | 35660 | 0.07 | 84 (0) | 0.9 |
| hsa05040:Huntington’s disease | 31 | 0.03 | 3.35 | 59658 | 0.05 | 28 (0) | 0.85 |
| hsa04210:Apoptosis | 21 | 0.09 | 3.51 | 21503 | 0.04 | 81 (0) | 0.84 |
| hsa04115:p53 signaling pathway | 21 | 0.07 | 3.57 | 18855 | 0.02 | 59 (0) | 0.78 |
| hsa04330:Notch signaling pathway | 15 | 0.08 | 3.59 | 11534 | 0.03 | 40 (0) | 0.74 |
| hsa04370:VEGF signaling pathway | 16 | 0.04 | 3.5 | 15764 | 0.04 | 63 (0) | 0.73 |
| hsa04510:Focal adhesion | 15 | 0.06 | 3.55 | 14237 | 0.03 | 181 (1) | 0.72 |
| hsa04910:Insulin signaling pathway | 14 | 0.07 | 3.58 | 14485 | 0.03 | 123 (2) | 0.72 |
| hsa05010:Alzheimer’s disease | 16 | 0.05 | 3.63 | 24871 | 0.02 | 28 (1) | 0.71 |
| hsa05020:Parkinson’s disease | 9 | 0.03 | 3.74 | 13520 | 0.01 | 20 (2) | 0.55 |
| hsa04310:Wnt signaling pathway | 10 | 0.04 | 3.76 | 9607 | 0.01 | 123 (1) | 0.54 |
Discussion
In the past two decades, a good number of methods, such as, FCM21, SYNCLUS22, FLAME2, FUZZY-EWKM3,23, FUMET4, and Qcut5 have been introduced for construction of CEN. These methods are based on various correlation measures. Some other CEN methods24,25,26 consider gene CEN, formed by merging networks of heterogenous experiments, specific to plants. Leal et al.24 introduced a multivariate approach based on Principal Component Analysis (PCA) to study the plant immune responses, by considering gene expression similarity of various plants, namely rice, Arabidopsis thaliana, soybean, cassava and tomato. However, the method proposed herein analyzes genes based on both expression and semantic similarity. The experiments conducted show that the border genes with low expression similarity and high semantic similarity are differentially expressed between normal and disease set of samples with high variance.
The key consideration is that genes or proteins function in co-ordination and not in isolation. The genes or gene products interact in biomolecular network or pathways, which are ultimately associated with the functions of living organisms and other various traits. Therefore, analysis of genes involved in molecular pathway is important for identifying disease-related genes or biomarkers, responsible for dysregulations of functions in disease patients.
AD is a common cause of dementia. In AD patients, it is found that there are some differences in brain tissues in the form of misfolded proteins, known as senile plagues and neurofibrillary tangles. The senile plagues or amyloid plagues are some clumps of β amyloid proteins, which block communication between neurons, and implicate neuronal death. The formation of β amyloid clumps at early onset of AD is initiated by mutations in proteins, known as Amyloid Beta (A4) Precursor Protein (APP)27, Presenilin 1 (PSEN1), and Presenilin 2 (PSEN2) (nlm.nih.gov). On the other hand, neurofibrillary tangles are formed due to mutations in a protein called Tau, which is responsible for ensuring clear passage for food molecules inside microtubule. Mutation in the protein Tau causes disintegration of microtubule and formation of tangles, which obstructs food passage to the cell neurons, resulting in death of neurons28. Involvement of genes or proteins in molecular pathways related to AD, such as, Wnt signaling, p53 signaling, Alzheimer disease-amyloid secretase, Apoptosis signaling, and Glycolysis, show a close relation with pathogenesis of AD.
In the proposed method, the interesting border genes are extracted from network modules using PANTHER and validated using GeneCards. The border genes are semantically similar as they share some common functionalities in the GO-BP structure, and therefore are found to be involved in molecular pathways. The pathways associated with the border genes related to AD are Wnt signaling, p53 signaling, Alzheimer disease-amyloid secretase, Apoptosis signaling, and Glycolysis. Some of the interesting border genes that are found to be related to AD and validated against the existing literature1,29 are AATF, APBA2, APBB1, APOD, BACE2, CASP2, CAST, CDK5, CSNK1D, GAP43, HSD17B10, KCNIP3, KLK6, MAPT, NQO1, NRGN, OGT, PADI2, PSEN2, and RCAN1. Microtubule-Associated Protein Tau (MAPT) is linked with frontal lobe dementia29. PSEN2 is associated with deposition of longer form of amyloid-beta in AD brains, and Caspase 2 (CASP2) is involved in activation of cascade of caspases responsible for apoptosis, which increases cell death1. Gene CDK5 is associated with p53 signaling pathway. The p53 signaling pathway is a suppressor of cell growth, i.e., death of neurons in AD patients. Genes, such as, CSNK1D and PSEN2 related to Wnt signaling pathways are found to play an important role in neurodegenerative diseases30.
Method and datasets
The proposed THD-Module Extractor method is implemented using Matlab R2015a on a machine with an Intel(R) core I3-2120 CPU@3.30Ghz processor, a 2.00 GB RAM, and a 64-bit Windows 7 operating system. The algorithm is evaluated on four datasets. The description of the datasets is given in Table 12. Figure 1 depicts the work flow of the method. The various steps of execution are detailed below.
Table 12. Datasets and their description.
| Serial no. | Dataset | Size of dataset | Source |
|---|---|---|---|
| 1 | Subset of yeast cell cycle | 384 × 17 | http://faculty.washington.edu/kayee/cluster |
| 2 | Arabidopsis thaliana | 138 × 8 | http://homes.esat.kuleuven.be/sistawww/bioi/thijs/Work/Clustering.html |
| 3 | Yeast sporulation | 474 × 17 | http://cmgm.stanford.edu/pbrown/sporulation |
| 4 | Homo sapiens (AD) | 9084 × 28 | http://agnigarh.tezu.ernet.in/~dkb/resources.html |
Preprocessing
Preprocessing of datasets is important before implementation of any scientific algorithm. Many datasets contain noisy values and NULL values. The following preprocessing tasks are performed on the considered datasets using Matlab, before storing them in tab delimited form.
The expression values with unlabeled or unformatted28 genes are removed.
Genes having only undefined (NaN) expression value(s) are removed.
Redundant or duplicate genes are removed.
The AD dataset GSE4226 consists of blood mononuclear cells collected from AD patients and age-gender matched normal individuals with 9600 probes and 28 samples. This work considers perturbation of gene expression values during progression of the disease, from normal to disease stage. Therefore, expression values of probes for AD patients and normal individuals are considered and rest are discarded.
After preprocessing of the datasets, the SSSim score of each gene pair is computed. However, computation of SSSim score for each gene pair takes lots of execution time. The datasets considered are large, often containing several thousands of genes, each with many expression values for multiple varying conditions. In our case, the preprocessed AD dataset contains expression values of more than 9000 genes under 28 different conditions, even after removing the unnecessary entries. This requires computation of more than 81 million getSSSim values. Traditional implementation of the measure on a standard CPU requires a huge amount of processing time. Therefore, this work leverages the parallel processing capabilities of GPU to speed up the computation of the large getSSSim correlation matrix.
NVIDIA CUDA implementation for SSSim computation
The NVIDIA CUDA 7.5 API and its C/C++ based compiler (NVCC) are used to implement the getSSSim correlation matrix computation on GPU. An NVIDIA GeForce 980 GPU with compute capability 5.231 is used in this implementation. The computation of the getSSSim correlation matrix is distributed into a logical 2-D grid of thread blocks, where each thread processes one getSSSim value. The dimension of each thread block is 4 × 4 × 28, which specifies the number of threads per block. The dimension of the grid is 2271 × 2271, which specifies the number of blocks in a grid. Figure 3 shows the distribution of threads into the grid. The GPU used in our implementation contains 2048 cores and can process equal number of threads simultaneously. In case of the AD dataset, the speedup obtained using the GPU for the correlation matrix computation is more than 1200 times, in comparison to a Matlab based implementation using a standard Intel-i7 CPU.
Figure 3. GPU grid configuration for getSSSim.
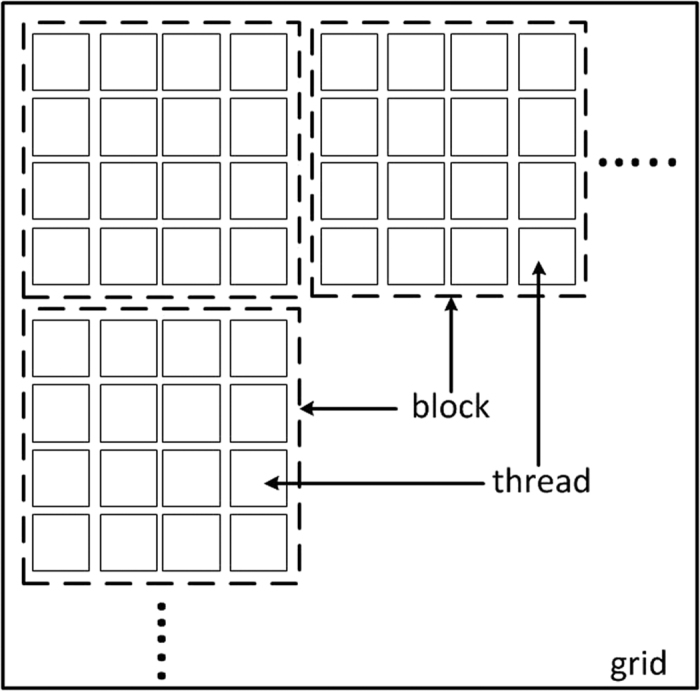
Computation of the getSSSim matrix is expressed as a large grid in the GPU kernel. The grid contains GPU threads equal to the number of getSSSim values to be calculated. The organization of the grid is hierarchical. The grid is a 2D array of blocks and each block is a 3D array of threads. For simplicity, the blocks are shown as 2D arrays of threads.
Once the score matrix has been computed, the CEN is constructed from the adjacency matrix, A. From the CEN, co-expressed modules are extracted for different values of δ and a static ρ. Each module contains a core gene that has maximum number of neighbor genes, satisfying the expression similarity threshold δ within the module. These co-expressed modules are then validated in terms of external validation metrics. A Web-based tool FuncAssociate 2.0 is used to find p values of each GO term associated with the genes of the co-expressed modules. Another Web-based tool called GeneMania1 is used to find the q values of each GO term. From each co-expressed module extracted from AD dataset, the border genes that satisfy the expression similarity threshold δ with their neighbors, but not the minimum neighborhood threshold ρ, are found.
THD-Module Extractor in finding interesting genes for AD
In addition to biologically relevant network module extraction, the proposed THD-Module Extractor allows identification of disease related genes by analyzing the border genes, extracted from the highly co-expressed modules. These border genes are analyzed using R library. Among the border genes, the k differentially expressed genes with highest variance between normal and disease samples are considered. Further, the entrez ID of each differentially expressed gene is found using mygene package of R. Thereafter, the semantic similarity of each of these border genes among each other is computed using GOSemSim package of R. On the basis of a static threshold, the border genes with high semantic similarity are filtered out. These interesting border genes are found to have low expression similarity with the core genes, yet high semantic similarity among each other. These interesting border genes are then validated using pathway analysis, KEGG enrichment analysis, GO terms enrichment analysis, and existing literature.
Additional Information
How to cite this article: Kakati, T. et al. THD-Module Extractor: An Application for CEN Module Extraction and Interesting Gene Identification for Alzheimer's Disease. Sci. Rep. 6, 38046; doi: 10.1038/srep38046 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Author Contributions T.K. and D.K.B. formulated the problem. T.K. and H.K. designed the method. T.K. implemented the method using Matlab and R, and wrote the manuscript. H.K. contributed the CUDA implementation of the measure used. H.K. and D.K.B. reviewed the manuscript.
References
- Warde-Farley D. et al. The genemania prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research 38, W214–W220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. & Medico E. Flame, a novel fuzzy clustering method for the analysis of dna microarray data. BMC bioinformatics 8, 3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ye Y., Huang J. & Feng S. Fuzzy soft subspace clustering method for gene co-expression network analysis. In Bioinformatics and Biomedicine Workshops (BIBMW), 2010 IEEE International Conference on 47–50 (IEEE, 2010). [Google Scholar]
- Mahanta P., Ahmed H. A., Bhattacharyya D. K. & Ghosh A. Fumet: A fuzzy network module extraction technique for gene expression data. Journal of biosciences 39, 351–364 (2014). [DOI] [PubMed] [Google Scholar]
- Ruan J., Dean A. K. & Zhang W. A general co-expression network-based approach to gene expression analysis: comparison and applications. BMC systems biology 4, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Liu Z.-P., Honda M., Kaneko S. & Chen L. Coexpression network analysis in chronic hepatitis b and c hepatic lesions reveals distinct patterns of disease progression to hepatocellular carcinoma. Journal of molecular cell biology 4, 140–152 (2012). [DOI] [PubMed] [Google Scholar]
- Liu Z.-P., Wang Y., Zhang X.-S. & Chen L.-n. Network-based analysis of complex diseases. Systems Biology, IET 6, 22–33 (2012). [DOI] [PubMed] [Google Scholar]
- Kikuchi M. et al. Network-based analysis for uncovering mechanisms underlying alzheimer’s disease. Systems Biology of Alzheimer’s Disease 479–491 (2016). [DOI] [PubMed] [Google Scholar]
- Yue H., Yang B., Yang F., Hu X.-L. & Kong F.-B. Co-expression network-based analysis of hippocampal expression data associated with alzheimer’s disease using a novel algorithm. Experimental and Therapeutic Medicine 11, 1707–1715 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q. et al. Inter-tissue coexpression network analysis reveals dpp4 as an important gene in heart to blood communication. Genome medicine 8, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Z. et al. Alzbase: an integrative database for gene dysregulation in alzheimer’s disease. Molecular neurobiology 53, 310–319 (2016). [DOI] [PubMed] [Google Scholar]
- Kustra R. & Zagdanski A. Data-fusion in clustering microarray data: Balancing discovery and interpretability. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB) 7, 50–63 (2010). [DOI] [PubMed] [Google Scholar]
- Ibáñez K., Guijarro M., Pajares G. & Valencia A. A computational approach inspired by simulated annealing to study the stability of protein interaction networks in cancer and neurological disorders. Data Mining and Knowledge Discovery 30, 226–242 (2016). [Google Scholar]
- Chandrasekaran S. & Bonchev D. Network analysis of human post-mortem microarrays reveals novel genes, micrornas, and mechanistic scenarios of potential importance in fighting huntington’s disease. Computational and Structural Biotechnology Journal 14, 117–130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiernan C. T. et al. Protein homeostasis gene dysregulation in pretangle-bearing nucleus basalis neurons during the progression of alzheimer’s disease. Neurobiology of aging 42, 80–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed H., Mahanta P., Bhattacharyya D. & Kalita J. Shifting-and-scaling correlation based biclustering algorithm. IEEE/ACM Transactions on Computational Biology and Bioinformatics 11, 1239–1252 (2014). [DOI] [PubMed] [Google Scholar]
- Lin, An information-theoretic definition of similarity. In ICML 98, 296–304 (1998). [Google Scholar]
- Liu C.-C. et al. Deficiency in lrp6-mediated wnt signaling contributes to synaptic abnormalities and amyloid pathology in alzheimer’s disease. Neuron 84, 63–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S. et al. Alzpathway: a comprehensive map of signaling pathways of alzheimer’s disease. BMC systems biology 6, 52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaab E., Baudot A., Krasnogor N. & Valencia A. Topogsa: network topological gene set analysis. Bioinformatics 26, 1271–1272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezdek J. C., Ehrlich R. & Full W. Fcm: The fuzzy c-means clustering algorithm. Computers & Geosciences 10, 191–203 (1984). [Google Scholar]
- DeSarbo W. S., Carroll J. D., Clark L. A. & Green P. E. Synthesized clustering: A method for amalgamating alternative clustering bases with differential weighting of variables. Psychometrika 49, 57–78 (1984). [Google Scholar]
- Jing L., Ng M. K. & Huang J. Z. An entropy weighting k-means algorithm for subspace clustering of high-dimensional sparse data. Knowledge and Data Engineering, IEEE Transactions on 19, 1026–1041 (2007). [Google Scholar]
- Leal L. G., López C. & López-Kleine L. Construction and comparison of gene co-expression networks shows complex plant immune responses. PeerJ 2, e610 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atias O., Chor B. & Chamovitz D. A. Large-scale analysis of arabidopsis transcription reveals a basal co-regulation network. BMC systems biology 3, 1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Kleine L., Leal L. & López C. Biostatistical approaches for the reconstruction of gene co-expression networks based on transcriptomic data. Briefings in functional genomics elt003 (2013). [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin M.-C. et al. Early-onset alzheimer’s disease caused by mutations at codon 717 of the β-amyloid precursor protein gene. Nature 353, 844–846 (1991). [DOI] [PubMed] [Google Scholar]
- Ballatore C., Lee V. M.-Y. & Trojanowski J. Q. Tau-mediated neurodegeneration in alzheimer’s disease and related disorders. Nature Reviews Neuroscience 8, 663–672 (2007). [DOI] [PubMed] [Google Scholar]
- Tanzi R. E. & Bertram L. Twenty years of the alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 120, 545–555 (2005). [DOI] [PubMed] [Google Scholar]
- Inestrosa N. C. & Varela-Nallar L. Wnt signaling in the nervous system and in alzheimer’s disease. Journal of molecular cell biology 6, 64–74 (2014). [DOI] [PubMed] [Google Scholar]
- Cook S. CUDA Programming: A Developer’s Guide to Parallel Computing with GPUs (Newnes, 2012). [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M. & Tanabe M. Kegg as a reference resource for gene and protein annotation. Nucleic Acids Research 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. & Goto S. Kegg: kyoto encyclopedia of genes and genomes. Nucleic acids research 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasojima K., Kuret J., DeMaggio A. J., McGeer E. & McGeer P. L. Casein kinase 1 delta mrna is upregulated in alzheimer disease brain. Brain research 865, 116–120 (2000). [DOI] [PubMed] [Google Scholar]
- Sheng Y., Zhang L., Su S. C., Tsai L.-H. & Zhu J. J. Cdk5 is a new rapid synaptic homeostasis regulator capable of initiating the early alzheimer-like pathology. Cerebral Cortex bhv032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-Y., He X.-Y. & Miller D. Hsd17b10: a gene involved in cognitive function through metabolism of isoleucine and neuroactive steroids. Molecular genetics and metabolism 92, 36–42 (2007). [DOI] [PubMed] [Google Scholar]
- Harris C. D., Ermak G. & Davies K. J. Rcan1-1l is overexpressed in neurons of alzheimer’s disease patients. FEBS Journal 274, 1715–1724 (2007). [DOI] [PubMed] [Google Scholar]
- LeBlanc A. C. The role of apoptotic pathways in alzheimer’s disease neurodegeneration and cell death. Current Alzheimer Research 2, 389–402 (2005). [DOI] [PubMed] [Google Scholar]