Abstract
Tristetraprolin (TTP) binds to mRNA transcripts to promote their degradation. The TTP protein family in humans includes two other proteins, TIS11b and TIS11d. All three proteins contain a highly homologous RNA binding domain (RBD) that consists of two CCCH zinc fingers (ZFs). Both ZFs are folded in the absence of RNA in TIS11d and TIS11b. In TTP, however, only ZF1 adopts a stable fold. The focus of this study is to understand the origin and biological significance of the structural differences of the RBD. We identified three residues that affect the affinity for the structural Zn2+ and determine the folding of ZF2 in the absence of RNA. We observed that the mRNA destabilizing activity of TTP was increased when the partially disordered RBD of TTP was replaced with the fully structured RBD of TIS11d, indicating that differences in the folded state of the RBD affect the activity of the proteins in the cell.
Graphical abstract
INTRODUCTION
Tristetraprolin (TTP), also known as TIS11, Nup475, GOS24, and ZFP361-7, is an RNA binding protein that binds to the 3’ UTR of transcripts at AU-rich elements (AREs)8. Binding of TTP to these transcripts promotes their fast degradation, as TTP recruits and activates mRNA decay enzymes9-12. The function of TTP became apparent with the observation that TTP knockout mice quickly develop severe inflammatory phenotypes after birth, a result of the accumulation of several transcripts13. The most abundant of these transcripts encodes the proinflammatory cytokine tumor necrosis factor α (TNFα)7.
In humans, the TTP protein family includes TIS11b and TIS11d, also known as ZFP36L1 and ZFP36L214,15, respectively. Based on their primary sequences, these three proteins are predicted to have low structural complexity and high flexibility. They each contain an RNA binding domain (RBD), surrounded by less-structured N- and C-terminal domains. The highly conserved RNA binding domain is a tandem zinc finger (TZF) motif with two CX8CX5CX3H zinc fingers, where X represents amino acids between each zinc coordinating residue. The two zinc fingers are separated by an 18 amino acid linker. A well conserved sequence, (R/K)YKTEL, precedes each finger and has been shown to be important for RNA binding16-18.
In 2004, Wright and coworkers determined the structure of TIS11d bound to the RNA sequence 5’-UUAUUUAUU-3’ (PDB code:1RGO) using NMR spectroscopy19. This structure depicts two folded zinc fingers where the zinc ions are coordinated by residues C159, C168, C174, and H178 in the first finger (ZF1) and C197, C206, C212, and H216 in the second finger (ZF2). The protein-RNA complex shows that the structure of the TZF domain of TIS11d is symmetrical, both ZF1 and ZF2 contain a short α-helix between the first two zinc coordinating cysteine residues (spanning residues 160-165 in ZF1 and 198-203 in ZF2). Additionally, both fingers contain a turn of 310-helix between the second and third cysteine coordinating resides (amino acids 171-173 in ZF1 and 209-211 in ZF2). The symmetry observed between the structures of the two zinc fingers is not surprising since their primary sequence is ~50% identical. The two zinc fingers are held together by a flexible linker, which contains a short 310-helix at residues 179-183 and is involved in forming a small hydrophobic cluster with residues of the first finger.
NMR and computational studies have shown that in the absence of RNA, the structure of the two zinc fingers of TIS11d is maintained although their orientation changes, as the linker region is highly flexible17,19. Despite their high sequence identities, TTP and TIS11d present striking differences in their unliganded structures. The structural symmetry observed for ZF1 and ZF2 in TIS11d is not observed in unliganded TTP. Previous NMR spectroscopic studies of TTP indicated that only ZF1 of TTP may adopt a stable fold in its RNA free state20,21.
The focus of this study is to understand the origin and biological significance of the structural differences observed between the RNA binding domains of TTP and TIS11d. We used TTP/TIS11d chimeras to identify the regions responsible for these differences. After testing that these chimeras are still active and capable of binding RNA, we used NMR spectroscopy to determine if ZF2 is folded in the free state. Finally, we determined how a fully folded TZF domain affects the activity of TTP using a luciferase reporter assay, where luciferase was placed under the control of the TNFα 3’UTR. The results reveal that the degree of structure of the RBD affects the activity of these proteins in the cell.
RESULTS AND DISCUSSION
Despite the importance of RNA-binding proteins to gene regulation, our understanding of how their structure and dynamics contribute to their biological activity is limited. In this study we focus on two related RNA-binding proteins—TTP and TIS11d—that regulate the stability of mRNA transcripts encoding key cancer-related proteins, such as tumor necrosis factor-α and vascular endothelial growth factor. TTP and TIS11d display differential folding propensity in the absence of RNA, despite sharing a high sequence identity. To understand the origin of the different folding propensities and how the activities of the two proteins are affected, we used an interdisciplinary approach that included biochemical and biophysical methods, cell-based assays and phylogenetic analysis.
The TZF domains of TTP and TIS11d share high sequence identity but differ in their RNA-free structures
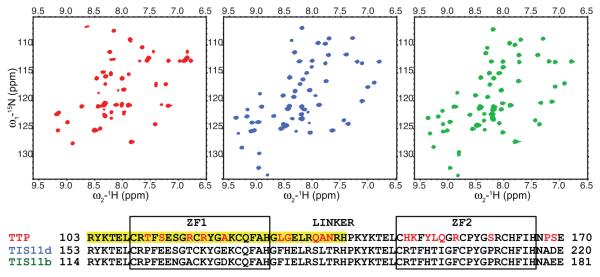
Although the TZF domains of TTP and TIS11d share 70% sequence identity, we observe that their solution structures differ dramatically in the apo state. The well-dispersed 15N-1H HSQC spectra of the TZF domains of TIS11d (residues 151-220) and TIS11b (residues 200-270) indicate that the structures of both ZF1 and ZF2 are preserved in the absence of RNA (Fig. 1). In contrast, the 15N-1H HSQC spectrum of the TTP TZF domain (residues 102-174) contains only cross peaks from residues of ZF1 and from the first nine residues of the linker (Fig. 1 and Supplementary Fig. 1). The lack of backbone amide (N-HN) cross peaks for ZF2 from the spectrum of TTP in the absence of RNA is the result of a chemical exchange process that is intermediate on the NMR chemical shift time scale, likely due to the flexibility of ZF2 that samples different conformations22-24. Cross-peaks from the C-terminal part of the linker and from ZF2 appear in the 15N-1H HSQC spectrum of TTP only upon addition of RNA (Fig. 2A). The overall increase in chemical shift dispersion observed upon addition of RNA results from the increased number of cross-peaks (cross-peaks from the linker and ZF2 are now present in the 15N-1H HSQC spectrum) and from the chemical shift changes of ZF1, due to RNA-binding. These data suggest that in the absence of RNA, ZF2 of TTP is in a molten globule state and that RNA is needed to stabilize ZF2 in a folded state, in agreement with the findings of Brewer et al.25. In a previous study, Berg and coworkers have shown that the TZF domain of murine TTP could bind 1.7 ± 0.4 equivalents of Co2+ 26 which suggests that both ZF1 and ZF2 of murine TTP can bind Zn2+ in the absence of RNA. In the same report, however, NMR spectroscopic studies of the TZF domain of murine TTP containing a single point mutation introduced to enhance solubility (Y143K), showed that only ZF1 could stably bind Zn2+ while ZF2 was unstructured26, in agreement with our observations in human TTP.
Figure 1.
The C-terminal ZF of TTP is unstructured in the free state. A) 15N-1H HSQC spectra of the TZF domains of TTP (red), TIS11d (blue) and TIS11b (green). B) Sequence alignments of TZF domains of TTP, TIS11d, and TIS11b. Sequence differences between TTP, TIS11d and TIS11b are highlighted in red. Residues with cross-peaks present in the 15N-1H HSQC spectrum of TTP collected in the absence of RNA are highlighted in yellow.
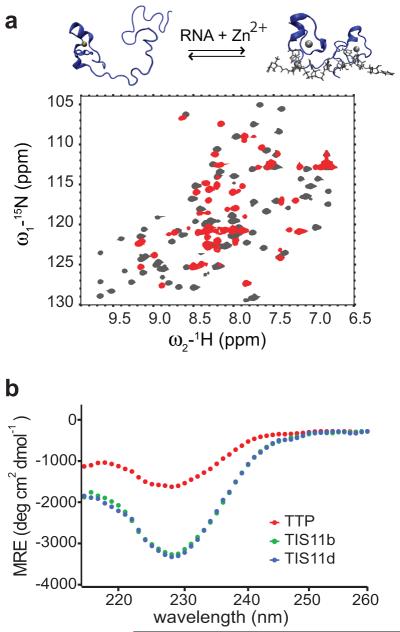
Figure 2.
In TTP ZF2 is unstructured in the unbound state but folds upon RNA-binding. a) 15N-1H HSQC spectra of RNA free (red)/RNA bound TTP (gray). New cross-peaks, corresponding to residues of the linker and ZF2 of TTP, appear upon addition of RNA. The structural transition occurring upon RNA-binding is depicted on top. The 3D structure of TTP was modeled from that of TIS11d (PDB id: 1RGO)19. b) Far-UV circular dichroism (CD) spectra of TTP (red), TIS11d (blue) and TIS11b (green). The CD spectra of TIS11b and TIS11d are almost entirely overlapped.
Due to chemical exchange line broadening, cross-peaks from ZF2 are absent in the 15N-1H HSQC spectrum of TTP, thus no structural information can be determined for this part of the domain using NMR spectroscopy. In order to test the hypothesis that ZF2 is in a molten globule state, we used several experimental approaches, including mutagenesis, CD spectroscopy and metal analysis, where we estimated the [Zn2+]/[TZF] ratio for both TTP and TIS11d.
NMR spectroscopy was used to determine the effect of a single point mutation, the third zinc-coordinating Cys to Ser, on the structure of TTP and TIS11d: C162S and C212S in TTP and TIS11d, respectively. These mutant proteins are lacking one of the zinc-coordinating Cys residues in ZF2, thus ZF2 cannot stably bind Zn2+ and fold (Supplementary Fig. 2). The 15N-1H HSQC spectra of wild-type (WT) TTP and C162S mutant TTP are nearly identical (Supplementary Fig. 2), with the exception of a few cross-peaks originating from presence of four extra residues at the C-terminus of the C162S mutant construct used. The 15N-1H HSQC spectrum of C212S TIS11d resembles that of wild-type TTP, as cross-peaks from the unstructured ZF2 are missing (Supplementary Fig. 2). These results support the hypothesis that in the absence of RNA, ZF2 of TTP cannot stably bind Zn2+. As a result, ZF2 of TTP samples many partially structured states on a time-scale that is intermediate relative the chemical shift differences, thus cross-peaks from ZF2 are broadened beyond detection24. In addition, these mutant proteins showed that in both TTP and TIS11d, ZF1 can independently adopt a stable fold when ZF2 is unfolded (Supplementary Fig. 2).
The CD spectra of TTP, TIS11b and TIS11d show that the TZF domain of TTP is less structured than that of TIS11d and TIS11b, as evidenced by the difference in CD signal peak intensities between 220-240 nm (Fig. 2B). In addition, Zn2+ titrations of TIS11d and TTP, monitored by CD spectroscopy, show that TIS11d coordinates two equivalents of Zn2+, one ion for each finger. TTP, however, only coordinates one zinc cation, indicating that ZF2 is disordered in solution (Supplementary Fig. 3). The same result was obtained when the [Zn2+]/[TZF] ratio was determined for both TTP and TIS11d using inductively coupled plasma-optical emission spectrometry (ICP-OES), [Zn2+]/[TTP] = 1.1 ± 0.1 and [Zn2+]/[TIS11d] = 1.8 ± 0.1.
Taken together, these findings demonstrate that only the N-terminal ZF, ZF1, of TTP adopts a stable fold in the unbound state, and that RNA binding, in the presence of Zn2+, induces the folding of ZF2. While TIS11d is fully folded, TTP is partially unfolded and requires RNA-binding to promote a transition to a fully folded state. Therefore, RNA recognition by TTP is achieved through an RNA-dependent conformational adaptation of the TZF domain.
The affinity for Zn2+ of different types of zinc fingers is known to differ over a range that spans about 7 orders of magnitude, with zinc fingers coordinating the zinc cation with four cysteine residues (Cys4-type) binding Zn2+ with the highest affinity27,28. In an attempt to increase the affinity of TTP ZF2 for Zn2+ the last Zn2+ coordinating residue of ZF2 was mutated from a His to a Cys, to make it into a Cys4-type zinc finger. The 15N-1H HSQC spectrum of the mutated TZF domain was still missing all the cross-peaks from the C-terminal half of the domain, as in the WT, suggesting that the mutated ZF2 (now Cys4-type) is still unable to stably bind Zn2+. This result is not surprising considering that engineered zinc fingers constructed from the same amino acid sequence except for the Zn2+ coordinating residues (Cys4, Cys3His and Cys2His2 were tested) have similar affinity for Zn2+ 27,29. These and our results indicate that differences in Zn2+ binding affinity arise not primarily from the particular atoms that coordinate the Zn2+, the first solvation shell, but mostly from the second coordination sphere27. Thus, the particular primary, secondary and tertiary structures of a protein determine the affinity for Zn2+ of the zinc finger.
The predicted secondary structure for ZF1 of TTP is the same as that observed for ZF1 of TIS11d
Small backbone chemical shift differences are observed for ZF1 and the linker of TTP and TIS11d suggesting that ZF1 of these two proteins have similar structures (Supplementary Fig. 4). This is not surprising since ZF1 of TTP and TIS11d (159-179 TIS11d/109-129 TTP) share 81.5% sequence identity. To support this conclusion, the backbone resonances of TTP (C , C , CO, N, and HN) were used to predict the secondary structure elements of ZF1 using δ2D software30. The predicted secondary elements are the same as those observed in the solution NMR structure for TIS11d. To understand how the primary sequence of ZF1 affects the observed chemical shifts, the residues that differ between TIS11d and TTP were mutated in TIS11d and chemical shift perturbations were calculated (Supplementary Fig. 4). From this data we determined that many differences observed between the spectra of TTP and TIS11d ZF1 arise from the identity of one residue, P161 in TIS11d and T111 in TTP. In TIS11d, this proline is in a short α-helix (residues 160-165) located between the first two zinc coordinating cysteine residues of the first finger. Mutation of residue P161 to T in TIS11d alone can capture many of the differences observed between the spectra of TIS11d and TTP and recapitulate most of the changes observed in the HSQC spectrum when we mutated all the residues of TIS11d ZF1 to those found in TTP (P161T, E163S, T167R, K169R, E172A), Supplementary Fig. 4. In TIS11d, F162 forms a hydrogen bond with the side chain of C159 that stabilizes the conformation of ZF1. We hypothesize that mutation of the preceding residue, P161T, affects the conformation of F162 and the hydrogen bond that it forms with C159, thus affecting the structure of many residues in ZF1 and their chemical shifts.
Identifying the origin of the difference in structures of TTP and TIS11d using chimeras
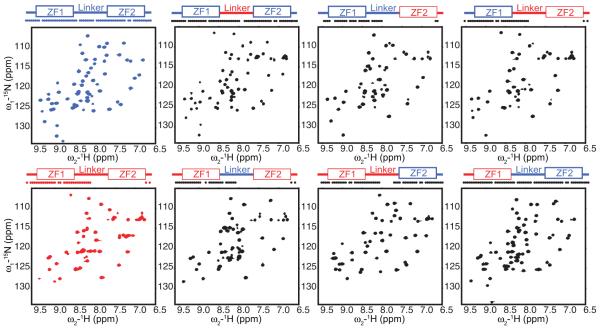
Many of the residues that differ between the TZF motif of TTP and TIS11d span the length of the motif, thus it is not easy to identify the residues that stabilize the structure of ZF2 from a simple analysis of the primary sequence. For this reason, chimeric proteins were constructed by combining the linker and/or zinc fingers of TTP and TIS11d to identify the role that different residues have in determining the structure of the zinc fingers (Fig. 3). Folding of each chimera was monitored using NMR spectroscopy to detect structural changes from the wild-type proteins with residue resolution. The absence of N-HN cross peaks for the residues of ZF2 and part of the linker from the 15N-1H HSQC spectrum, due to intermediate exchange line broadening, is an indication of lack of structure or partial folding of ZF2 (Fig. 3). From the data we can conclude that switching the linkers of TTP and TIS11d does not affect the structure of ZF2. However, the primary sequence of ZF2 affects its structure. Replacing the primary sequence of ZF2 of TTP with that of TIS11d increases the number of resonance peaks observed in the spectrum, with these cross-peaks corresponding to the C-terminal half of the domain that now folds into a stable structure. The converse is also true: changing the primary sequence of TIS11d ZF2 into that of TTP increases the propensity for intrinsic disorder of the chimera, and cross peaks from ZF2 are broadened beyond detection in the 15N-1H HSQC spectrum. Altogether, these findings show that the primary sequence of ZF2 alone determines its structure or lack thereof.
Figure 3.
The amino acid sequence of ZF2 determines the structure of ZF2. Backbone amide cross peaks represented for TTP (red), TIS11d (blue), and chimeras (black). On top, a schematic representation of the TZF domain shows the zinc fingers as rectangles and the linker as line (sequences from TIS11d and TTP shown in blue and red, respectively). The dots indicate residues along the sequence with a cross-peak in the 15N-1H HSQC spectrum.
All TTP/TIS11d chimeras bind to the ARE13 sequence with similar affinities
To determine if the chimeras retain RNA binding activity, we determined the equilibrium dissociation constant of each chimera using fluorescent electrophoretic mobility shift assays 31. All of the chimeras bind to a model ARE sequence (ARE13: 5'-UUUUAUUUAUUUU-3') with similar affinity, indicating that the folded/unfolded character of the second zinc finger does not strongly affect the RNA binding activity of the protein, Table 1. Analysis of the primary sequence shows that TTP has a net charge of 8.2, while TIS11d has a net charge of 3.3 at pH 7 (Innovagens - peptide properties calculator, http://www.innovagen.com/custom-peptide-synthesis/peptide-property-calculator/peptide-property-calculator-notes.asp). The more basic nature of TTP may result in enhanced attractive Coulomb interaction with the RNA and a more favorable enthalpy of binding thus compensating for the entropic penalty associated to folding of ZF2 upon RNA binding. This entropy-enthalpy compensation in the free energy of RNA-binding was observed for TIS11d and a mutant of TIS11d where residues D219 and E220 were deleted, thus changing the total net charge of the TZF domain32.
Table 1.
RNA binding activity of TTP/TIS11d chimeras Electrophoretic mobility shift assays (EMSA) with labeled ARE13 were used to determine the RNA binding activity of each chimera. Each chimera was made by fusing ZF1, the linker or ZF2 from either TTP (red) or TIS11d (blue).
| TZF Chimera | Kd apparent (nM) |
|---|---|
|
|
9 ± 2 |
|
|
9 ± 1 |
|
|
42 ± 5 |
|
|
15 ± 9 |
|
|
15 ± 3 |
|
|
19 ± 10 |
|
|
15 ± 5 |
|
|
23 ± 6 |
Understanding how the amino acid composition of ZF2 determines the unfolded/folded character of the zinc finger
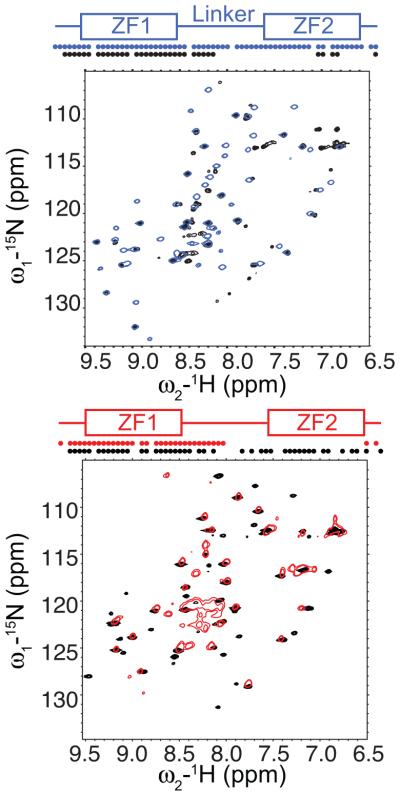
We found that the primary sequence of ZF2 determines the structure of ZF2. In order to dissect what accounts for a stably folded ZF2, we made mutations within the second zinc finger where residues of TIS11d were mutated to the equivalent residues in TTP (Fig. 4 and Supplementary Fig. 5). Most of the differences in amino acid sequence between TTP and TIS11d ZF2 are observed in the region that is helical in TIS11d19 (Fig. 1). Mutation of the residues of this region of TTP (residues 148-155) into those of TIS11d (residues 198-205) results in the presence of cross-peaks from ZF2 and the second half of the linker in the 15N-1H HSQC spectrum, indicating that ZF2 is now stably folded (Fig. 4). Furthermore, mutation of the residues in the second half of the α-helix of TIS11d (residues H201, T202 and I203) to those of TTP (residues Y151, L152, Q153) results in loss of signal from almost all residues of ZF2 and the second part of the linker (Fig. 4), indicating loss of structure in ZF2. Mutating the first two residues of the α-helix (R198 and T199) of TIS11d to those of TTP does not affect the structure of ZF2 (Supplementary Fig. 5).
Figure 4.
The most important region to stabilize the structure of ZF2 comprises residues 151-153 in TTP (residues 201-203 in TIS11d). (Left) Overlay of the15N-1H HSQC spectrum of WT TIS11d (blue) with that of the mutant protein containing the TTP amino acid sequence at the second half of the α-helical region of ZF2, residues 201-203, (black). (Right) Overlay of the 15N-1H HSQC spectrum of WT TTP (red) with that of the mutant protein containing the TIS11d amino acid sequence at the α-helical region of ZF2, residues 148-155, (black). On top, a schematic representation of the TZF domain shows the zinc fingers as rectangles and the linker as line. The dots indicate residues along the sequence with a cross-peak in the 15N-1H HSQC spectrum.
There are two other residues that are different between ZF2 of TIS11d and TTP (Fig. 1): F205 and P210. Mutation of F205 to R and of P210 to S in TIS11d did not change the structure of the finger indicating that these two residues do not affect the stability of ZF2 (Supplementary Fig. 5).
We also investigated how the C-terminal tail of the TZF domain (Fig. 1) affects the overall stability of ZF2. Previous experimental and molecular dynamics (MD) studies of TIS11d have shown how the two conserved negatively charged residues flanking ZF2 (D219 and E220) contribute to the stabilization of the TZF structure through their electrostatic interaction with the positively charged residues of the linker and that disruption of these interactions results in a more flexible but still folded ZF217,32. Here we show that mutation of A218 and D219 in TIS11d to the corresponding residues of TTP (P168 and S169, respectively) result in the disappearance of a few cross-peaks in the linker (188-189) and in ZF2 (216-219), Supplementary Fig. 5. In agreement with our previous studies17,32, this result indicates that the residues of the C-terminal tail affect the dynamics of the linker region and those of ZF2 but do not affect its folding.
From these experiments, we conclude that the most important region to stabilize the structure of ZF2 comprises residues 151-153 in TTP (residues 201-203 in TIS11d). In TIS11d these three residues are located on the second half of the α-helix between the first two zinc coordinating cysteine residues19. Secondary structure prediction (data not shown) for TTP bound to RNA (ARE13: 5'-UUUUAUUUAUUUU-3'), from the backbone and Cβ chemical shifts using δ2D and SPARTA+33 software, shows that the α-helix between the first two zinc coordinating cysteine residues in ZF2 is shorter in TTP (residues 148-150) than what is observed in TIS11d (residues 198-203). Our experimental results support the recent computational model of TTP bond to RNA bound developed by Blackshear and coworkers34. These computational studies suggested that while TIS11d forms two alpha helical turns in this region, TTP only forms one helical turn. Taken together these studies indicate that the ability to form a longer α-helix affects the ability of the finger to stably coordinate the Zn2+ ion and fold into a unique structure in the absence of RNA.
Comparative Genomics
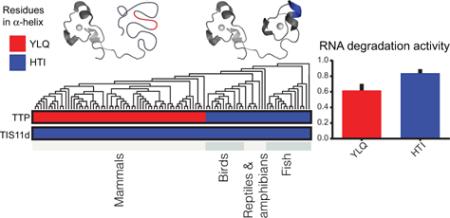
Comparison of the amino acid sequences of TIS11d and TTP from vertebrates from 100 species35-38 (Supplementary Fig. 6) indicates that the TZF domain of TIS11d is likely always folded, as the three residues of the α-helix that alone stabilize the structure of ZF2 (residues 201-203, HTI) are absolutely conserved. In TTP homologs, however, the corresponding three residues (res. 151-153, YLQ, associated with the disorder of the TZF domain) are only conserved in mammals (Supplementary Fig. 6). In fish, reptiles, amphibians and birds these three residues are the same as in TIS11d, thus, from our studies (Fig. 4), we predict that ZF2 is folded in these vertebrates. Comparative genomics suggests that TTP has evolved more recently in mammals to modulate its folding through Zn2+ and RNA binding. In addition, we observed that the presence of the HTI amino acids, which stabilize the structure of ZF2 (Fig. 4), correlates with the presence of two acidic amino acid residues at the tail of the TZF domain (Supplementary Fig. 6). Our previous studies have shown that these two acidic residues are important to stabilize the structure of the folded TZF domain of TIS11d in an RNA-binding competent state32. These data suggest that they are not conserved when ZF2 is unstructured.
Determining the effect of the structure of the TZF domain on the cellular activity of TTP
To determine whether the structure of the TZF domain of TTP affects its mRNA destabilizing activity in cells, we employed a reporter assay where luciferase was placed under the control of the TNFα 3’UTR. HEK293 cells (which do not express TTP constitutively) were co-transfected with a plasmid containing full length TTP or chimeric variants and a plasmid encoding both firefly and Renilla luciferase. The Renilla luciferase gene was fused to the 3’UTR sequence of TNFα, while the firefly luciferase was fused to a control vector 3’UTR. The ratio of Renilla to firefly luciferase reveals TTP variant activity normalized by transfection efficiency. Differences in luciferase activity observed between cells that expressed wild-type TTP or TTPTZF_TIS11d fusion protein, where the TZF domain of TTP has been replaced with that of TIS11d, assess the degree to which intrinsic disorder of the TZF domain affects the regulatory activity of the protein. The TTPTZF_TIS11d fusion protein, which has a stably folded ZF2, led to a two-fold reduction in luciferase activity compared to wild-type TTP (Fig. 5). Having a structured ZF2 decreased the expression of the protein in cells (decreased luciferase activity), therefore, the TTPTZF_TIS11d fusion protein has higher activity than WT TTP (Fig. 5). As expected, high luciferase activity was observed when the cells were transfected with a plasmid containing the mutant protein C162S of TTP, where the third zinc-coordinating cysteine has been mutated to a serine in ZF2 and thus cannot fold even in the presence of RNA (Fig. 5). Although we expected C162S TTP to be completely inactive, a residual mRNA destabilizing activity was observed for this mutant protein. This residual activity has been previously observed39 suggesting that mutation of the zinc-coordinating residues in ZF1 might be necessary to completely inactivate the protein. Together, the results show that the mRNA destabilizing activity of TTP in the cell is affected by the structure of ZF2 in the RNA binding domain, increasing the structure of the TZF domain of TTP increases the RNA-destabilizing activity of the protein in the cell.
Figure 5.
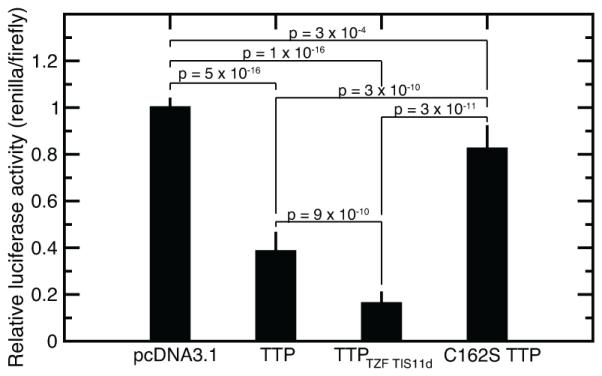
Measurements of the cellular activity of TTP and TTP/TIS11d chimeras. The plot shows the relative luciferase activity measured for HEK cells incubated for 48 hours with empty pcDNA3.1HISC plasmid, WT TTP, TTP chimera (TTP containing the TZF domain of TIS11d, TTPTZF TIS11d), and TTP C212S mutation.
There are several possible explanations for this observation. 1) Disorder in the TFZ domain may trigger TTP degradation, lowering its cytoplasmic concentration40. 2) The cytoplasmic concentration of TTP may also be decreased by an increased import into the nucleus. The nuclear transport protein importin has been shown to bind to the basic residues R134 and R137 of the linker region of the TZF41, thus the structure of ZF2 might affect the recognition and transport of TTP into and out of the nucleus. 3) The structure of the TZF domain may affect how the protein interacts with other enzymes of the RNA degradation machinery9. 4) A fully structured TZF domain may increase the RNA-binding affinity in cells. Any or all of these explanations may account for the increased cellular activity associated with a fully folded RNA binding domain.
Differential folding propensities of ZFs may explain observations of TZF proteins in other species, as well. The CCCH-type TZF domain of the TTP family of protein was first identified in the murine TTP2, and has since been found in more than 3600 proteins42,43. Analysis of the C. elegans genome has revealed the presence of an unusually high number of proteins (16) containing a TZF domain homologous to TTP. Several of these proteins (OMA-1, OMA-2, MEX-5, MEX-6, PIE-1, POS-1 and MEX-1) have a known function as mRNA regulators at the onset and during the early stages of embryogenesis44. Experimental evidence indicates that the two zinc fingers of these TZF domains play different functions during embryogenesis45. While the N-terminal zinc finger of POS-1, PIE-1 and MEX-1 has been demonstrated to be necessary and sufficient for degradation in somatic blastomeres, the C-terminal zinc finger is necessary and sufficient to target these proteins to P granules, which are germline cytoplasmic structures rich in RNA45. These findings suggest that the two fingers in the CCCH-type TZF domain may function independently and that the structure, or lack thereof, of each finger might affect the activity and/or localization of the protein within the cell. This independence may apply to the human TZF domains of the TTP family, thus the increased propensity for intrinsic disorder of TTP relative to TIS11b and TIS11d may indicate that the proteins have evolved to be able to modulate their activity through their thermodynamic stability.
Summary
In conclusion, our studies reveal 1) that the three residues located in the α-helical region of ZF2 alone determine if the TZF domain is fully or partially folded, 2) that the degree of structure of the TZF domain affects the activity of the protein in the cell and 3) this family of proteins has only recently evolved to be able to regulate its activity through modulation of its folding. It will be interesting to determine whether the structural disorder of the TZF domain affects binding partner recognition in the cell, cellular stability and localization, or all the above.
METHODS
Protein expression and purification
The RBD of human TTP (residues 102-170) and TIS11d (residues 152-220) was synthesized by Genscript and cloned into the pet21b vector between Nde1 and Xho1 restriction sites. Chimeras of TTP and TIS11d were generated via Quikchange mutagenesis. TTP, TIS11d and chimeras were expressed within BL21(DE3) E. coli competent cells. Isotopic labeling with 15N was performed by growing the cells in M9 enriched with 1 g of 15NH4Cl per liter. Carbon labeling was performed by growing cells in 13C-glucose. The cells were grown at 37° C to an OD600 of 0.8 and then induced for 4 hours with 1 mM Isopropyl β-D-1 thiogalactopyranoside (IPTG) and 0.1 mM ZnSO4 at the same temperature. Harvested cells were passed through a cell disrupter in 50 mL lysis buffer containing 50mM Tris HCl, pH 8.0, 50 mM NaCl, 2 mM dithiothreitol (DTT) and 1 EDTA free Complete protease inhibitor tablet (Roche). Lysates were centrifuged at 19500 RPM for 1 hr at 4° C and passed through a 20 mL DEAE column pre-equilibrated with 50 mM Tris HCl, pH 8.0, 50 mM NaCl and 2 mM DTT. The flow through was dialyzed overnight in 25 mM sodium phosphate, pH 6.5, 50 mM NaCl and 2 mM DTT and passed through a HiTRAP SP 10mL column (GE healthcare Life Science) preequilibrated with 25 mM sodium phosphate, pH 6.5, 50 mM NaCl and 2 mM DTT. The peptides were eluted with a gradient of NaCl from 0-1 M; the peptides typically eluted at 0.2-0.6 M NaCl. Fractions with peptide were combined and concentrated to 2 mL using a 3 KDa Centriprep (Millipore) and further purified using a 1.6 × 60 cm Superdex-75 size exclusion column (Amersham Bio\sciences) equilibrated with 50 mM Tris HCL pH 7, 300 mM NaCl, 2 mM DTT. For NMR analysis fractions with TTP, TIS11d or chimeras were buffer exchanged into 10 mM Tris pH 6.2, 20 mM KCl, 2 mM DTT, 0.1 mM ZnSO4. Protein concentrations were determined via Ninhydrin assay.
Measurements of RNA-binding affinity
The RNA-binding activity of the samples were determined using fluorescent electrophoretic mobility shift assays (EMSA) with fluorescein end labeled RNA as previously described4. Briefly, the affinities of TTP, TIS11d, and chimeras for the sequence 5’-UUUUAUUUAUUUU-3’ (ARE13) were measured by direct titration of 3 nM labeled RNA with increasing concentrations of protein. Varying concentrations of the protein were incubated for 3 hours at room temperature with 2 nM fluorescently labeled RNA in a buffer solution containing 10 mM TRIS (pH=8), 100 μM Zn(OAc)2, 100 mM NaCl, 0.01 mg tRNA, 0.01% (v/v) IGEPAL, and 2 mM DTT. After addition of 10 μL of 0.005%(w/v) bromocresol green in 30% glycerol to 100 μL of each sample, 50 μL of each mixture were loaded onto a 1% agarose, 1Å~TB gel and run for 35 min at 120 V at room temperature to separate bound from free RNA. To detect the fluorescently labeled RNA, the gel was imaged using a Fuji FLA-5000 laser imager. The apparent dissociation constant Kd,app was determined by fitting to the quadratic equation31,
where m and b are the maximum and base signals, Kd is the dissociation constant, and P and R are the concentrations of protein and labeled RNA in the sample, respectively.
NMR spectroscopy
Three dimensional triple resonance (1H-13C-15N) experiments, including HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH, were collected at 14.1 T and 298 K to assign the backbone 1H, 13C, and 15N resonances of [U-13C, U-15N] TTP and TIS11d in 92% H2O/8% D2O buffer solution (10 mM Tris, 20 mM KCl, 2 mM DTT, 0.1 mM ZnSO4 at pH 6.2). Additional 3D 15N edited 1H–1H NOESY and HMQC-NOESY-HSQC experiments were collected at 14.1 T and 298 K and used to aid in the backbone resonance assignment of the chimeras.
Folding of each chimera was monitored via NMR spectroscopy. 15N-1H HSQC spectra were collected at 298 K for each chimera. All NMR triple-resonance and 15N-1H HSQC experiments were collected on a Varian Inova spectrometer operating at 600 MHz equipped with a triple-resonance cold probe. The temperature was calibrated using a sample of 100% methanol. Data processing was performed using NMRPipe 46 and Sparky software 47.
Sample Preparation for Trace Metal Analysis
The RBD of human TIS11d (residues 152-220) and TTP (residues 102-170) were cloned into the pHMTC vector between BamHI and HindIII restriction sites. Constructs were expressed in BL21(DE3) E. coli competent cells. The cells were grown in Luria Broth (LB) media at 37°C to an OD600 of 0.6 and then induced for 3 hours with 1 mM IPTG and 0.1 mM ZnSO4 at the same temperature. Harvested cells were then passed through a cell disrupter in 50 ml lysis buffer containing 50 mM Tris HCl, pH 8.0, 100 mM NaCl, 100 μM ZnOAc, 2 mM DTT and 1 EDTA free Complete protease inhibitor tablet (Roche). Lysate was centrifuged at 19500 RPM for 1hr at 4°C and passed through a 20 ml Amylose column pre-equilibrated with wash buffer (50 mM Tris HCl, pH 8.0, 100 mM NaCl, 100 μM ZnOAc and 2 mM DTT). The column was washed with 10 column volumes of wash buffer. The protein was eluted with 10 mM Maltose in wash buffer. The eluent was then placed over a 5 ml Hi TRAP Q column (GE healthcare Life Science) pre-equilibrated with the same wash buffer used above. The column was washed with 3 column volumes of wash buffer. The protein was eluted with a gradient of NaCl from 0-1 M; the peptides typically eluted at 0.3-0.5 M NaCl. Fractions containing our protein were combined, concentrated and dialyzed in 50 mM Tris HCl, pH 8.0, 150 mM NaCl, trace amounts of ZnOAc and 0.1% β-mercaptoethanol (BME) and protein concentrations were determined by UV absorption. The concentration of Zn2+ in the samples was determined using inductively coupled plasma-optical emission spectrometry (ICP-OES) at the Center for Applied Isotope Studies at the University of Georgia.
CD spectroscopy
Far-UV circular dichroism (CD) spectra were recorded for TTP (50 μM) and TIS11d (18 μM) in 50 mM HEPES pH 7.0, 20 mM KCl and 1 mM TCEP using a Jasco-810 spectropolarimeter (Jasco Inc., Easton, MD). Curves were monitored from 200-260 nm in a 0.1 cm path length quartz cuvette using a scan rate of 20 nm min−1 and a response time of 8 s. The sample temperature for all CD measurements was maintained at 293 K.
Luciferase reporter assay
The pcDNA3.1 His-C-TTP (TTP) expression construct was kindly given to us by Dr. Seth Brooks48. The cDNA for the fusion protein containing the N- and C- terminal domain of TTP and the TZF domain of TIS11d was generated by Genscript and contained flanking EcoR1 restriction sites for subsequent cloning into pcDNA3.1 His-C-TTP. C162S mutant of TTP was generated using Q5-mutagenesis kit (NEB). Luciferase reporter constructs harboring cDNA of TNFα 3’UTR (nucleotides 1,231-1,506) was generated by gblock synthesis (IDT) and was inserted into the psicheck™-2 construct at restriction sites XhoI and NotI.
Human embryonic kidney (HEK) 293 cells were plated at a density of 300,000 cells/well in 6 well plates, with Dulbecco’s Modified Eagle’s Medium (DMEM) containing 100 units/mL penicillin and 100 μg/mL streptomycin and 10% fetal bovine serum (FBS). The plates were incubated for 24 hours in a humidified 5% CO2 incubator set at 37° C. When the cells were 60-70% confluent each well was cotransfected with 0.25 μg psicheck2_TNFalpha and 0.5 μg of pcDNA3.1-His-C (empty plasmid), pcDNA3.1 TTP, pcDNA3.1 TTPTZF_TIS11d, or pcDNA3.1 TTP (C162S) using Dharmafect Duo protocol (Thermo-Scientific). Transfections were performed in triplicate. 48 hours after transfection, cells were harvested and Luciferase activity was monitored using a Dual-luciferase Reporter Assay System (Promega) and normalized by the internal control values given by firefly luciferase. Luciferase activity was monitored in each well 3 times.
Supplementary Material
ACKNOWLEDGMENT
We thank S. P. Ryder, N. R. Zearfoss and C. C. Clingman for helpful discussion and technical assistance on the luciferase reporter assay, J. A. Zitzewitz and B. R. Morgan for helpful discussion, and C. R. Matthews for sharing equipment.
Funding Sources
This work was supported by National Institutes of Health Grant GM098763.
Abbreviations
- 3’ UTR
3’ untranslated region
- HEK
Human embryonic kidney
- HMQC
heteronuclear multiple-quantum coherence
- HSQC
heteronuclear single-quantum coherence
- MD
molecular dynamics
- NOESY
nuclear Overhauser effect spectroscopy
- NMR
nuclear magnetic resonance
- PDB
Protein Data Bank
- RBD
RNA-binding domain
- TNFα
tumor necrosis factor α
- TTP
tristetraprolin
- TTPTZF TIS11d
TTP containing the TZF domain of TIS11d
- TZF
tandem zinc finger
- ZF
zinc finger
- ZF1
N-terminal zinc finger
- ZF2
C-terminal zinc finger
- WT
wild type
Footnotes
Author Contributions
LMD performed all experiments. LMD and FM designed and interpreted the experiments and wrote the manuscript.
Supporting Information
A figure showing the assigned peaks in 15N-1H HSQC spectra of TTP and TIS11d, a figure showing the 15N-1H HSQC spectra of TTP, C162S TTP mutant, TIS11d and C212S TIS11d mutant, a figure CD spectra of TIS11d and TTP at different concentration of Zn2+, a figure illustrating the chemical shift perturbation of TIS11d mutant proteins, a figure of the 15N-1H HSQC spectra of TIS11d mutants designed to identify the residues important for stabilizing the structure of ZF2 and a figure showing the sequence alignment of the TZF domain of TIS11d and TTP from 100 vertebrate species. This material is available free of charge via the Internet.
REFERENCES
- (1).Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- (2).DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. Journal of Biological Chemistry. 1990;265:19185–19191. [PubMed] [Google Scholar]
- (3).Siderovski DP, Blum S, Forsdyke RE, Forsdyke DR. A set of human putative lymphocyte G0/G1 switch genes includes genes homologous to rodent cytokine and zinc finger protein-encoding genes. DNA Cell Biol. 1990;9:579–587. doi: 10.1089/dna.1990.9.579. [DOI] [PubMed] [Google Scholar]
- (4).Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. Journal of Biological Chemistry. 1990;265:16556–16563. [PubMed] [Google Scholar]
- (5).Stoecklin G, Gross B, Ming X-F, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- (6).Essafi-Benkhadir K, Onesto C, Stebe E, Moroni C, Pages G. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol Biol Cell. 2007;18:4648–4658. doi: 10.1091/mbc.E07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Carballo E, Lai WS, Blackshear PJ. Feedback Inhibition of Macrophage Tumor Necrosis Factor-α Production by Tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- (8).Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & Development. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sandler H, Kreth J, Timmers HTM, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Research. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol. 2013;20:735–739. doi: 10.1038/nsmb.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes & Development. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Carballo E, Gilkeson GS, Blackshear PJ. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J Clin Invest. 1997;100:986–995. doi: 10.1172/JCI119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- (15).Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- (16).Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- (17).Morgan BR. A computational study of RNA binding and specificity in the tandem zinc finger domain of TIS11d. Protein Sci. 2010;19:1222–1234. doi: 10.1002/pro.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Pagano JM, Farley BM, McCoig LM, Ryder SP. Molecular Basis of RNA Recognition by the Embryonic Polarity Determinant MEX-5. Journal of Biological Chemistry. 2007;282:8883–8894. doi: 10.1074/jbc.M700079200. [DOI] [PubMed] [Google Scholar]
- (19).Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- (20).Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry. 2003;42:217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- (21).Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J. Biol. Chem. 2003;278:19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- (22).Kosol S, Contreras-Martos S, Cedeño C, Tompa P. Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules. 2013;18:10802–10828. doi: 10.3390/molecules180910802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Barbar E. NMR characterization of partially folded and unfolded conformational ensembles of proteins. Biopolymers. 1999;51:191–207. doi: 10.1002/(SICI)1097-0282(1999)51:3<191::AID-BIP3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- (24).Schulman BA, Kim PS, Dobson CM, Redfield C. A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nat. Struct. Biol. 1997;4:630–634. doi: 10.1038/nsb0897-630. [DOI] [PubMed] [Google Scholar]
- (25).Brewer BY, Ballin JD, Fialcowitz-White EJ, Blackshear PJ, Wilson GM. Substrate dependence of conformational changes in the RNA-binding domain of tristetraprolin assessed by fluorescence spectroscopy of tryptophan mutants. Biochemistry. 2006;45:13807–13817. doi: 10.1021/bi061320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Worthington MT, Amann BT, Nathans D, Berg JM. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proceedings of the National Academy of Sciences. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sénèque O, Latour J-M. Coordination properties of zinc finger peptides revisited: ligand competition studies reveal higher affinities for zinc and cobalt. J. Am. Chem. Soc. 2010;132:17760–17774. doi: 10.1021/ja104992h. [DOI] [PubMed] [Google Scholar]
- (28).Velyvis A, Qin J. Zinc Finger Proteins. Springer US; Boston, MA: 2005. LIM Domain and Its Binding to Target Proteins; pp. 99–105. [Google Scholar]
- (29).Reddi AR, Guzman TR, Breece RM, Tierney DL, Gibney BR. Deducing the energetic cost of protein folding in zinc finger proteins using designed metallopeptides. J. Am. Chem. Soc. 2007;129:12815–12827. doi: 10.1021/ja073902+. [DOI] [PubMed] [Google Scholar]
- (30).Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- (31).Pagano JM, Clingman CC, Ryder SP. Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA. 2011;17:14–20. doi: 10.1261/rna.2428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Morgan BR, Deveau LM, Massi F. Probing the structural and dynamical effects of the charged residues of the TZF domain of TIS11d. Biophys. J. 2015;108:1503–1515. doi: 10.1016/j.bpj.2015.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- (34).Lai WS, Perera L, Hicks SN, Blackshear PJ. Mutational and structural analysis of the tandem zinc finger domain of tristetraprolin. Journal of Biological Chemistry. 2014;289:565–580. doi: 10.1074/jbc.M113.466326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Siepel A, Haussler D. Combining phylogenetic and hidden Markov models in biosequence analysis. J. Comput. Biol. 2004;11:413–428. doi: 10.1089/1066527041410472. [DOI] [PubMed] [Google Scholar]
- (37).Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief. Bioinformatics. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, Haussler D, Miller W. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gomperts M, Pascall JC, Brown KD. The nucleotide sequence of a cDNA encoding an EGF-inducible gene indicates the existence of a new family of mitogen-induced genes. Oncogene. 1990;5:1081–1083. [PubMed] [Google Scholar]
- (40).Ngoc LV, Wauquier C, Soin R, Bousbata S, Twyffels L, Kruys V, Gueydan C. Rapid proteasomal degradation of posttranscriptional regulators of the TIS11/tristetraprolin family is induced by an intrinsically unstructured region independently of ubiquitination. Molecular and Cellular Biology. 2014;34:4315–4328. doi: 10.1128/MCB.00643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Frederick ED, Ramos SBV, Blackshear PJ. A unique C-terminal repeat domain maintains the cytosolic localization of the placenta-specific tristetraprolin family member ZFP36L3. J. Biol. Chem. 2008;283:14792–14800. doi: 10.1074/jbc.M801234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A. The Pfam protein families database. Nucleic Acids Research. 2007;36:D281–D288. doi: 10.1093/nar/gkm960. %U http:––nar.oxfordjournals.org–content–36–suppl_1–D281.long. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: the protein families database. Nucleic Acids Research. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Lee M-H, Schedl T. RNA-binding proteins. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.79.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Molecular Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- (46).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- (47).Goddard TD, Kneller DG. SPARKY 3 [Google Scholar]
- (48).Brooks SA, Connolly JE, Diegel RJ, Fava RA, Rigby WFC. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 2002;46:1362–1370. doi: 10.1002/art.10235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.