Abstract
Background
Visceral fat plays a central role in the development of metabolic syndrome and atherosclerotic cardiovascular diseases. The association of visceral fat accumulation with cardio-metabolic diseases has been reported, but the impact of visceral fat on the gene expression profile in peripheral blood cells remains to be determined. The aim of this study was to determine the effects of visceral fat area (VFA) and subcutaneous fat area (SFA) on the gene expression profile in peripheral blood cells of obese subjects.
Methods
All 17 enrolled subjects were hospitalized to receive diet therapy for obesity (defined as body mass index, BMI, greater than 25 kg/m2). VFA and SFA were measured at the umbilical level by computed tomography (CT). Blood samples were subjected to gene expression profile analysis by using SurePrint G3 Human GE Microarray 8 × 60 k ver. 2.0. The correlation between various clinical parameters, including VFA and SFA, and peripheral blood gene expression levels was analyzed.
Results
Among the 17 subjects, 12 had normal glucose tolerance or borderline diabetes, and 5 were diagnosed with type 2 diabetes without medications [glycated hemoglobin (HbA1c); 6.3 ± 1.3%]. The mean BMI, VFA, and SFA were 30.0 ± 5.5 kg/m2, 177 ± 67 and 245 ± 131 cm2, respectively. Interestingly, VFA altered the expression of 1354 genes, including up-regulation of 307 and down-regulation of 1047, under the statistical environment that the parametric false discovery rate (FDR) was less than 0.1. However, no significant effects were noted for SFA or BMI. Gene ontology analysis showed higher prevalence of VFA-associated genes than that of SFA-associated genes, among the genes associated with inflammation, oxidative stress, immune response, lipid metabolism, and glucose metabolism.
Conclusions
Accumulation of visceral fat, but not subcutaneous fat, has a significant impact on the gene expression profile in peripheral blood cells in obese Japanese subjects.
Keywords: Obesity, Visceral fat, Subcutaneous fat, Fat distribution, Gene expression, Microarray, Metabolic syndrome, Diabetes, Adiponectin, KLF
Background
Increasing evidence demonstrates that excess visceral fat locates upstream of the metabolic syndrome, a cluster of diabetes, dyslipidemia, and hypertension, which is associated with atherosclerotic cardiovascular diseases [1]. In a series of clinical studies, we have shown that visceral fat area (VFA), but not subcutaneous fat area (SFA), correlates significantly and strongly with cardio-metabolic diseases [2, 3]. Various groups, including ours, have focused on the underlying molecular mechanism and links between visceral fat accumulation and cardio-metabolic diseases [4, 5]. Some of the discussed molecular pathological links between visceral adiposity and cardio-metabolic diseases include dysregulation of adipocytokines [1], chronic low-grade inflammation of visceral fat tissue [6], and harmful changes in gut microbiota [7]. However, the exact mechanism(s) remains unresolved.
We have also examined the role of gene expression profile in peripheral blood cells, and reported that visceral adiposity can alter the expression profiles of various genes in peripheral blood cells, including those involved in circadian rhythm and inflammation [8, 9]. However, in these studies, visceral adiposity, including VFA and SFA, was not assessed by modern precision technology such as computed tomography (CT). In addition, impact of SFA on gene expressions in peripheral blood cells was not determined. Moreover, most of the enrolled subjects were overt type 2 diabetes patients (HbA1c; 8.1 ± 2.2%) in our previous study [8, 9], suggesting that gene expression profile in peripheral blood cells influenced by these parameters. Other groups also investigated the impact of VFA and/or SFA on the expression of various genes in peripheral blood cells. For example, Lee et al. [10] found a significant association between VFA, but not SFA, and sirtuin 1 (SIRT1) mRNA level in peripheral blood mononuclear cells.
The aim of the present study was to define the association of VFA and SFA determined by CT, with the gene expression profile in peripheral blood cells in obese subjects free of overt diabetes.
Methods
Study population
The enrolled subjects were hospitalized at Sumitomo Hospital between February 2012 and April 2014 to receive calorie-restricted diet therapy for obesity. Subjects with type 1 diabetes mellitus, cancer, autoimmune diseases, and infectious diseases were excluded from the present study. Patients treated with glucose-lowering agents were also excluded. Written informed consent was obtained from each patient after explaining the purpose of study. The study protocol was approved by the human ethics committees of Sumitomo Hospital and Osaka University. The study was also registered with the University Hospital Medical Information Network (UMIN #000001663).
Clinical parameters
Obesity was defined as body mass index (BMI) greater than 25 kg/m2 according to the criteria of the Japan Society for the Study of Obesity [11]. VFA and SFA were measured on the cross-sectional CT slice at the umbilical level [12]. Waist circumference was measured with a tape at the umbilical level in standing position. Serum adiponectin concentration was measured by a latex particle-enhanced turbidimetric immunoassay with a human adiponectin latex kit (Otsuka Pharmaceutical Co., Tokyo, Japan). The homeostasis model − assessment of insulin resistance (HOMA-IR) was calculated by the equation: [HOMA-IR = fasting insulin (µU/mL) × fasting glucose (mg/dL)/405]. Type 2 diabetes mellitus and borderline diabetes were defined according to the criteria of the Japan Diabetes Society [13]. Briefly, diabetes was defined as fasting glucose of ≥126 mg/dL, casual glucose of ≥200 mg/dL, or HbA1c of ≥6.5%. Hypertension was defined as systolic blood pressure (SBP) of ≥140 mm Hg, diastolic BP (DBP) of ≥90 mm Hg, or treatment with anti-hypertensive agents. Dyslipidemia was defined as fasting triglycerides (TG) of ≥150 mg/dL, high-density lipoprotein cholesterol (HDL-C) of <40 mg/dL, or low-density lipoprotein cholesterol (LDL-C) of ≥140 mg/dL, or treatment with lipid-lowering agents. LDL-C was calculated using the Friedewald formula, except in cases with TG of >400 mg/dL. The estimated glomerular filtration rate (eGFR) was calculated by using the following formula: [eGFR = 194 × (serum creatinine−1.094) × (age−0.287) × F (male, F = 1; female, F = 0.739)] [14]. Intima-media thickness (IMT) of common carotid artery was measured by echography (HI VISION Preirus; Hitachi, Tokyo).
Microarray analysis
Blood samples were collected into PaxGene Blood RNA tubes (PreAnalytiX, Qiagen Inc., Valencia, CA) before breakfast and left to stand for 2 h at room temperature. The tubes were kept at −20 °C for 2 days and then stored at −80 °C. Total RNA was extracted from the blood sample by using PaxGene Blood RNA Kit (PreAnalytiX, Qiagen). After RNA was qualified by Agilent 2100 Bioanalyzer, 100 ng of total RNA was converted to cDNA, amplified, and labeled with Cy3-labeled CTP using the Quick Amp Labeling kit (Agilent Technologies, Santa Clara, CA). The amplified cRNA and dye incorporation were quantified using ND-1000 Spectrophotometer (Nano Drop Technologies, Wilmington, DE) and hybridized to SurePrint G3 Human GE Microarray 8 × 60 k ver. 2.0 (Design ID: 039494, Agilent Technologies). After hybridization, arrays were washed consecutively by using Gene Expression Wash Pack (Agilent Technologies). Fluorescence images of the hybridized arrays were generated using the Agilent DNA Microarray Scanner, and the intensities were extracted with Agilent Feature Extraction software ver. 10.7.3.1. The raw microarray data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO Series GSE85226).
Microarray data analyses
The raw microarray intensities were processed by the percentile shift method (75th percentile) with GeneSpring GX 13.0 (Agilent Technologies) so as to normalize the range of expression intensities for inter-microarray. Genes found to be expressed in more than 50% of the hybridizations were subjected to further analyses. The normalized data were exported from the GeneSpring GX software. The univariate correlation between clinical parameters, including VFA and SFA, and peripheral blood gene expression levels was examined by Pearson’s correlation under the R environment (http://cran.at.r-project.org). Gene ontology (GO) information was retrieved from the annotations in GeneSpring GX 13.0.
Results
Characteristics of the enrolled subjects
The clinical characteristics of the participating subjects are listed in Table 1. The mean BMI and waist circumference were 30.0 kg/m2 (range, 24.0–44.0 kg/m2) and 101.2 cm (range, 85–127 cm), respectively. The mean VFA and SFA were 177.3 cm2 (range, 78–318 cm2) and 244.7 cm2 (range, 80–558 cm2), respectively. The mean serum adiponectin concentration was 4.2 μg/mL (range, 2.3–9.8 μg/mL) and the mean HbA1c was 6.3% (range, 5.3–10.9%). Among the 17 subjects, 5 had type 2 diabetes, 6 had borderline diabetes, and 6 subjects had normal glucose tolerance. All 5 diabetic patients were not treated with any anti-diabetic agents. Atherosclerotic plaque in the carotid artery (IMT ≥1.1 mm) was observed in 7 subjects. Among the 17 subjects, dyslipidemia and hypertension were found in 15 and 8 subjects, respectively. Seven patients were treated with statins and four patients were treated with angiotensin converting enzyme inhibitor (ACE-I) or angiotensin II receptor blocker (ARB).
Table 1.
Characteristics of subjects
| N | 17 |
| Sex (male/female) | 14/3 |
| Age (years) | 54.6 ± 14.6 |
| BMI (kg/m2) | 30 ± 5.5 |
| Waist circumference (cm) | 101 ± 11 |
| Visceral fat area (cm2) | 177 ± 67 |
| Subcutaneous fat area (cm2) | 245 ± 131 |
| Adiponectin (μg/mL) | 4.2 ± 1.7 |
| Systolic blood pressure (mm Hg) | 132 ± 17 |
| Diastolic blood pressure (mm Hg) | 82 ± 13.7 |
| Fast plasma glucose (mg/dL) | 102 ± 21 |
| Hemoglobin A1c (%) | 6.3 ± 1.3 |
| Diagnosis (T2DM/B/N) | 5/6/6 |
| HOMA-IR | 3.2 ± 2.3 |
| Total cholesterol (mg/dL) | 206 ± 40 |
| Triglyceride (mg/dL) | 196 ± 129 |
| HDL-C (mg/dL) | 56.3 ± 18.1 |
| LDL-C (mg/dL) | 114 ± 40 |
| Uric acid (mg/dL) | 6.4 ± 0.8 |
| Urinary albumin (μg/day) | 12.3 ± 10.6 |
| eGFR (mL/min/1.73 m2) | 77.9 ± 19.8 |
| mean IMT ≥1.1 mm | 7/10 |
| Statin use (±) | 7/10 |
| ACE-I/ARB use (±) | 4/13 |
Data are mean ± SD
T2DM type 2 diabetes mellitus, B borderline diabetes, N normal glucose tolerance, HOMA-IR homeostasis model assessment of insulin resistance, HDL-C high density lipoprotein-cholesterol, LDL-C low density lipoprotein-cholesterol, eGFR estimated glomerular filtration rate, IMT intima-media thickness, ACE-I angiotensin converting enzyme inhibitor, ARB angiotensin II receptor blocker
Gene expression profiles
Peripheral blood RNA samples were subjected to microarray analysis. The target probes were selected under the condition that significant signals were detected in more than 7 cases among 17 subjects and thus 23,197 probes were extracted for gene expression analysis. Table 2 lists the number of probes that showed significant changes according to various clinical variables under the statistical environment that the parametric false discovery rate (FDR) was less than 0.1. Sex and age had impacts on 52 and 625 probes, respectively. Surprisingly, VFA had a great impact on peripheral blood cells gene expression, i.e., 1354 probes consisting of 307 up-regulated and 1047 down-regulated probes. However, no significant gene probes were detected with SFA or BMI. Serum adiponectin, diabetes, HbA1c, and HOMA-IR also had no impact on the gene expression in peripheral blood cells. Likewise, statins and ACE-I/ARB had no effect. Figure 1 illustrates the number of upregulated/downregulated probes according to various clinical parameters. Table 3 lists the top 30 genes that correlated significantly with VFA positively and negatively. Among these genes, Krüppel-like factor 10 (KLF10) was the most significant (Table 3).
Table 2.
Changes in probes according to various clinical parameters
| FDR < 0.1 | Up | Down | |
|---|---|---|---|
| Categorical | |||
| Sex | 52 | 20 | 32 |
| Diagnosis of diabetes | 0 | 0 | 0 |
| Mean IMT | 0 | 0 | 0 |
| Statin use | 0 | 0 | 0 |
| ACE-I/ARB use | 0 | 0 | 0 |
| Continuous | |||
| Age | 625 | 206 | 419 |
| Body mass index | 0 | 0 | 0 |
| Visceral fat area | 1354 | 307 | 1047 |
| Subcutaneous fat area | 0 | 0 | 0 |
| Adiponectin | 0 | 0 | 0 |
| Hemoglobin A1c | 0 | 0 | 0 |
| HOMA-IR | 0 | 0 | 0 |
Data represent number of probes
FDR false discovery rate, IMT intima-media thickness, ACE-I angiotensin converting enzyme inhibitor, ARB angiotensin II receptor blocker, HOMA-IR homeostasis model assessment of insulin resistance
Fig. 1.
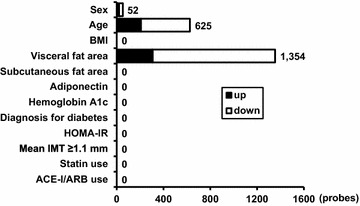
Changes in the number of genes according to various clinical parameters. The target 23,197 probes were selected under the condition that significant signals were detected in more than 7 cases among 17 subjects. Data represent the number of probes that showed significant upregulation and downregulation according to the listed clinical parameters under the statistical environment that the parametric false discovery rate (FDR) was less than 0.1. Parameters such as sex, diagnosis for diabetes, mean IMT, statin use, and ACE-I/ARB use were adopted as categorical variables. Age, BMI, visceral and subcutaneous fat areas, adiponectin, hemoglobin A1c, and HOMA-IR were adopted as continuous variables. BMI body mass index; HOMA-IR homeostasis model assessment of insulin resistance; IMT intima-media thickness; ACE-I angiotensin converting enzyme inhibitor; ARB angiotensin II receptor blocker
Table 3.
Top 30 genes that correlated positively and negatively with visceral fat area
| Probe name | Gene symbol | Gene name | R | p value | FDR |
|---|---|---|---|---|---|
| Positive correlation | |||||
| A_21_P0013668 | SPATA31C2 | SPATA31 subfamily C, member 2 | 0.830 | 3.72E−05 | 0.08430563 |
| A_19_P00803850 | LOC100505474 | Uncharacterized LOC100505474 | 0.828 | 4.01E−05 | 0.08430563 |
| A_33_P3238410 | SBF1 | SET binding factor 1 | 0.814 | 6.93E−05 | 0.08430563 |
| A_23_P325676 | ZNF653 | Zinc finger protein 653 | 0.803 | 1.03E−04 | 0.08430563 |
| A_23_P384532 | CCDC11 | Coiled-coil domain containing 11 | 0.802 | 1.07E−04 | 0.08430563 |
| A_33_P3311956 | FEZ2 | Fasciculation and elongation protein zeta 2 (zygin II) | 0.801 | 1.13E−04 | 0.08430563 |
| A_23_P430670 | CHST5 | Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 | 0.797 | 1.29E−04 | 0.08430563 |
| A_33_P3253653 | GPR155 | G protein-coupled receptor 155 | 0.779 | 2.29E−04 | 0.08430563 |
| A_33_P3314974 | PARD6G-AS1 | PARD6G antisense RNA 1 | 0.778 | 2.33E−04 | 0.08430563 |
| A_33_P3402773 | 0.775 | 2.61E−04 | 0.084956675 | ||
| A_24_P117942 | TOMM20L | Translocase of outer mitochondrial membrane 20 homolog (yeast)-like | 0.771 | 2.94E−04 | 0.084956675 |
| A_33_P3772937 | KRT8P12 | Keratin 8 pseudogene 12 | 0.768 | 3.15E−04 | 0.084956675 |
| A_33_P3404889 | 0.768 | 3.20E−04 | 0.084956675 | ||
| A_33_P3379436 | FAM74A4 | Family with sequence similarity 74, member A4 | 0.767 | 3.29E−04 | 0.084956675 |
| A_23_P34066 | IL9R | Interleukin 9 receptor | 0.765 | 3.50E−04 | 0.084956675 |
| A_23_P417415 | ACOT11 | Acyl-CoA thioesterase 11 | 0.758 | 4.19E−04 | 0.084956675 |
| A_33_P3410093 | LTA4H | Leukotriene A4 hydrolase | 0.758 | 4.22E−04 | 0.084956675 |
| A_33_P3334895 | GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | 0.750 | 5.26E−04 | 0.084956675 |
| A_23_P72697 | GPIHBP1 | Glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 | 0.749 | 5.40E−04 | 0.084956675 |
| A_33_P3378531 | AS3MT | Arsenite methyltransferase | 0.748 | 5.57E−04 | 0.084956675 |
| A_24_P75190 | HBD | Hemoglobin, delta | 0.747 | 5.76E−04 | 0.084956675 |
| A_23_P26457 | HBA2 | Hemoglobin, alpha 2 | 0.744 | 6.08E−04 | 0.084956675 |
| A_33_P3265866 | 0.744 | 6.09E−04 | 0.084956675 | ||
| A_21_P0004859 | BTN2A1 | Butyrophilin, subfamily 2, member A1 | 0.741 | 6.63E−04 | 0.084956675 |
| A_21_P0005185 | DKFZp686L13185 | Uncharacterized LOC401287 | 0.739 | 6.95E−04 | 0.084956675 |
| A_21_P0012204 | XLOC_014512 | 0.739 | 7.00E−04 | 0.084956675 | |
| A_21_P0009476 | XLOC_012670 | 0.737 | 7.45E−04 | 0.084956675 | |
| A_33_P3365932 | WASH1 | WAS protein family homolog 1 | 0.734 | 7.91E−04 | 0.084956675 |
| A_19_P00812257 | LINC01191 | Long intergenic non-protein coding RNA 1191 | 0.732 | 8.26E−04 | 0.084956675 |
| A_23_P209564 | CYBRD1 | Cytochrome b reductase 1 | 0.732 | 8.36E−04 | 0.084956675 |
| Negative correlation | |||||
| A_23_P168828 | KLF10 | KRUPPEL-like factor 10 | −0.856 | 1.16E−05 | 0.08430563 |
| A_32_P54544 | CCT6A | Chaperonin containing TCP1, subunit 6A (zeta 1) | −0.832 | 3.40E−05 | 0.08430563 |
| A_23_P389919 | WHSC1 | Wolf-Hirschhorn syndrome candidate 1 | −0.830 | 3.74E−05 | 0.08430563 |
| A_23_P44139 | PRIM2 | Primase, DNA, polypeptide 2 (58 kDa) | −0.827 | 4.21E−05 | 0.08430563 |
| A_19_P00331853 | LOC100131564 | uncharacterized LOC100131564 | −0.826 | 4.34E−05 | 0.08430563 |
| A_23_P501877 | ZFP64 | ZFP64 zinc finger protein | −0.826 | 4.36E−05 | 0.08430563 |
| A_24_P3973 | HNRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | −0.826 | 4.46E−05 | 0.08430563 |
| A_23_P215088 | ZC3HC1 | Zinc finger, C3HC-type containing 1 | −0.826 | 4.47E−05 | 0.08430563 |
| A_23_P7679 | NUP155 | Nucleoporin 155 kDa | −0.816 | 6.50E−05 | 0.08430563 |
| A_33_P3381483 | ZNF331 | Zinc finger protein 331 | −0.811 | 7.73E−05 | 0.08430563 |
| A_23_P115149 | WDR77 | WD repeat domain 77 | −0.808 | 8.71E−05 | 0.08430563 |
| A_23_P151093 | YARS2 | Tyrosyl-tRNA synthetase 2, mitochondrial | −0.807 | 8.92E−05 | 0.08430563 |
| A_23_P251421 | CDCA7 | Cell division cycle associated 7 | −0.807 | 8.94E−05 | 0.08430563 |
| A_21_P0008290 | LINC00641 | Long intergenic non-protein coding RNA 641 | −0.807 | 9.17E−05 | 0.08430563 |
| A_33_P3262665 | MAP7D3 | MAP7 domain containing 3 | −0.805 | 9.59E−05 | 0.08430563 |
| A_33_P3213557 | CCZ1 | CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) | −0.805 | 9.60E−05 | 0.08430563 |
| A_23_P202143 | NOLC1 | Nucleolar and coiled-body phosphoprotein 1 | −0.804 | 1.01E−04 | 0.08430563 |
| A_23_P46924 | BUB3 | BUB3 mitotic checkpoint protein | −0.803 | 1.04E−04 | 0.08430563 |
| A_24_P925635 | SEPT7P2 | Septin 7 pseudogene 2 | −0.803 | 1.06E−04 | 0.08430563 |
| A_24_P345822 | TFG | TRK-fused gene | −0.802 | 1.08E−04 | 0.08430563 |
| A_23_P85180 | TMEM187 | Transmembrane protein 187 | −0.801 | 1.10E−04 | 0.08430563 |
| A_33_P3221234 | IPP | Intracisternal A particle-promoted polypeptide | −0.801 | 1.13E−04 | 0.08430563 |
| A_33_P3415037 | VDAC2 | Voltage-dependent anion channel 2 | −0.799 | 1.20E−04 | 0.08430563 |
| A_33_P3309929 | HDAC3 | Histone deacetylase 3 | −0.799 | 1.21E−04 | 0.08430563 |
| A_23_P214798 | SYNCRIP | Synaptotagmin binding, cytoplasmic RNA interacting protein | −0.796 | 1.34E−04 | 0.08430563 |
| A_21_P0012709 | XLOC_014512 | −0.795 | 1.38E−04 | 0.08430563 | |
| A_24_P116909 | MALT1 | Mucosa associated lymphoid tissue lymphoma translocation gene 1 | −0.794 | 1.42E−04 | 0.08430563 |
| A_23_P69437 | YEATS2 | YEATS domain containing 2 | −0.793 | 1.45E−04 | 0.08430563 |
| A_33_P3251538 | MAPKAP1 | Mitogen-activated protein kinase associated protein 1 | −0.793 | 1.46E−04 | 0.08430563 |
| A_23_P102202 | MSH6 | Muts homolog 6 | −0.793 | 1.47E−04 | 0.08430563 |
Gene ontology
Gene ontology (GO) analysis was also performed to further determine the impact of VFA on gene expression profile in peripheral blood cells. As shown in Table 4, visceral fat adiposity correlated significantly with genes related to the metabolic process, oxygen transport, and nucleotide binding. Genes involved in inflammation (GO: 0006954), oxidative stress (GO: 0006979), immune response (GO: 0006955), lipid metabolism (GO: 0006629), and glucose metabolism (GO: 0006006), were finally examined. Figure 2 shows the percentage of genes (among all genes) that correlated significantly with SFA and VFA (p < 0.05). VFA correlated with 17.6, 26.8, 18.4, 25.5, and 26.4% of genes involved in inflammation, oxidative stress, immune response, lipid metabolism, and glucose metabolism, respectively, while the respective percentages for SFA were only 4.2, 2.6, 2.7, 3.4, and 3.2%.
Table 4.
Significant GO terms based on genes that correlated positively and negatively with visceral fat area
| GO | GO term | Corrected p value |
|---|---|---|
| Positive correlation | ||
| Biological process | Oxygen transport | 8.821E−04 |
| Gas transport | 3.707E−03 | |
| Molecular function | Oxygen transporter activity | 5.310E−04 |
| Cellular component | Hemoglobin complex | 3.428E−04 |
| Negative correlation | ||
| Biological process | RNA processing | 6.559E−21 |
| Heterocycle metabolic process | 1.252E−20 | |
| Cellular nitrogen compound metabolic process | 8.348E−20 | |
| Nucleobase-containing compound metabolic process | 1.702E−19 | |
| Organic cyclic compound metabolic process | 2.892E−19 | |
| Cellular aromatic compound metabolic process | 6.642E−19 | |
| Cellular metabolic process | 4.940E−18 | |
| Nitrogen compound metabolic process | 9.293E−18 | |
| Nucleic acid metabolic process | 1.052E−16 | |
| Cellular macromolecule metabolic process | 1.780E−16 | |
| Metabolic process | 1.122E−14 | |
| Primary metabolic process | 1.551E−14 | |
| Gene expression | 3.068E−14 | |
| Organic substance metabolic process | 7.768E−14 | |
| Macromolecule metabolic process | 1.529E−11 | |
| RNA metabolic process | 3.440E−11 | |
| mRNA processing | 5.194E−11 | |
| ncRNA metabolic process | 1.859E−10 | |
| RNA splicing | 5.494E−10 | |
| Molecular function | RNA binding | 1.470E−10 |
| Nucleotide binding | 2.106E−07 | |
| Nucleoside phosphate binding | 2.143E−07 | |
| Heterocyclic compound binding | 1.144E−06 | |
| Nucleic acid binding | 2.081E−06 | |
| Aminoacyl-tRNA ligase activity | 2.924E−06 | |
| Ligase activity, forming aminoacyl-tRNA and related compounds | 2.924E−06 | |
| Ligase activity, forming carbon–oxygen bonds | 2.924E−06 | |
| Organic cyclic compound binding | 3.035E−06 | |
| Small molecule binding | 3.265E−06 | |
| Catalytic activity | 1.736E−04 | |
| ATP-dependent helicase activity | 3.046E−04 | |
| Purine NTP-dependent helicase activity | 3.046E−04 | |
| Structure-specific DNA binding | 3.201E−04 | |
| ATPase activity | 9.197E−04 | |
| ATPase activity, coupled | 1.738E−03 | |
| Adenyl nucleotide binding | 1.738E−03 | |
| ATP binding | 1.799E−03 | |
| Adenyl ribonucleotide binding | 2.557E−03 | |
| Cellular component | Nuclear part | 2.984E−28 |
| Intracellular part | 6.115E−28 | |
| Intracellular | 1.984E−27 | |
| Intracellular membrane-bounded organelle | 8.017E−27 | |
| Membrane-enclosed lumen | 6.105E−24 | |
| Intracellular organelle lumen | 6.201E−24 | |
| Nuclear lumen | 6.172E−23 | |
| Organelle lumen | 8.194E−23 | |
| Intracellular organelle part | 2.042E−22 | |
| Intracellular organelle | 2.087E−22 | |
| Membrane-bounded organelle | 1.535E−21 | |
| Organelle part | 3.743E−21 | |
| Nucleus | 1.105E−20 | |
| Organelle | 1.383E−17 | |
| Nucleolus | 2.194E−16 | |
| Nucleoplasm | 4.535E−15 | |
| Mitochondrion | 1.779E−13 | |
| Mitochondrial part | 1.863E−09 | |
| Cytoplasm | 2.197E−09 | |
Fig. 2.
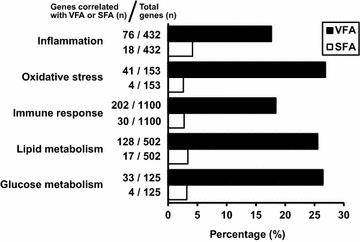
Percentages of obesity-related genes that correlated significantly with visceral and subcutaneous fat area. Gene ontology analysis was performed to examine the impact of visceral fat area (VFA) and subcutaneous fat area (SFA) on the percentage of obesity-associated genes (relative to total number of genes), such as inflammation (GO:0006954), oxidative stress (GO:0006979), immune response (GO:0006955), lipid metabolism (GO:0006629), and glucose metabolism (GO:0006006)
Discussion
The major finding of the present study was that visceral fat, but not subcutaneous fat, in obese individuals had a significant impact on peripheral blood cells gene expression profile. While similar results were reported previously by our group [8, 9], these studies had several limitations: (1) VFA was estimated by abdominal bioelectrical impedance analysis (BIA), rather than by CT. The latter is recognized as the gold standard method for fat area measurement [12, 15, 16]. (2) The majority of the subjects enrolled in the above previous studies were diabetics (75%) with a mean HbA1c of 8.1%. The inclusion of such patients could have influenced the results. (3) Impact of SFA on gene expression level in peripheral blood cells could not be determined under abdominal BIA procedure. The present study is clinically more significant as it included precise measurement of VFA and SFA by CT scan and negligible diabetic conditions.
The biological differences between visceral and subcutaneous fat have been investigated. The rate of lipolysis and lipogenesis activities are higher in adipocytes of visceral fat tissue than those of subcutaneous fat tissue [17, 18], suggesting that visceral fat accumulation increases free fatty acids (FFA) in the portal vein, accelerates hepatic lipogenesis, and results in dyslipidemia involving high FFA level in the bloodstream. Visceral fat accumulation also enhances inflow of glycerol into the liver and hepatic glucose production through adipose and hepatic glycerol channels; aquaporin 7 and 9, respectively [19]. Furthermore, adipose mRNA levels dynamically change in visceral fat compared to subcutaneous fat, especially in obese subjects. As BMI increases, the mRNA levels of adiponectin and peroxisome proliferator-activated receptor gamma (PPARγ) are reduced, while mRNA level of NADPH oxidase subunit p22, promoting reactive oxygen species (ROS), is augmented, in visceral fat, but not in subcutaneous fat [20]. Visceral fat accumulation is also a major risk for the reduction of circulating adiponectin (hypoadiponectinemia) [1]. Collectively, compared to subcutaneous fat, visceral fat accumulation largely and pathologically alters not only its own fat tissue, but also circulating substances and metabolic outcome. It is therefore conceivable that these visceral fat-mediated changes can also alter the gene expression profile in peripheral blood cells.
Increasing evidence indicates that chronic low-grade inflammation in the adipose tissue, especially in visceral fat, is located upstream of the metabolic syndrome [21, 22]. Gut microbiota also accelerates inflammatory changes in visceral fat [7]. Various immune cells infiltrate adipose tissue and cause inflammatory changes through direct cell–cell interaction and/or indirect cytokine-mediated intercellular communication. It is not hard to imagine that such interactions among immune cells and adipocytes influence peripheral blood cells, but such processes have not been confirmed yet. The present study also suggests that gene expression profile of peripheral blood cells reflects local inflammatory changes in visceral fat.
Interestingly, KLF10, a member of the Krüppel-like family of transcription factors, showed the most significant and negative correlation with VFA (Table 3). KLF10 is augmented through the transforming growth factor-β (TGF-β)-Smad signaling pathway [23]. It plays a crucial role in TGF-β-mediated induction of regulatory T-cells (Treg) from naive T-cells [24]. In mice lacking KLF10, Treg activity was reduced and proinflammatory changes were accelerated. Transfer of KLF10-deficient T-cells failed to suppress the development of atherosclerosis in apolipoprotein E knockout mice with high-fat diet [25]. KLF10-deficient mice also showed hyperglycemia in males and hypertriglyceridemia in females [26]. KLF10 has been shown to regulate 20–30% of hepatic genes related to glucose and lipid metabolism [26]. Genetic variants of KLF10 are associated with susceptibility to type 2 diabetes [27]. However, KLF10 mRNA expressions were not significantly correlated with diabetes or dyslipidemia in present study. To confirm the association between KLF10 expressions in peripheral blood cells and diabetes or dyslipidemia, further investigations would be desired in some other populations, different from present clinical profiles, such as non-obese or non-diabetic subjects. Present data provides a possibility that visceral fat adiposity-associated reduction in peripheral blood KLF10 mRNA level is related to the pathogenesis of the metabolic syndrome, although further clinical studies would be needed in future.
The present study has several limitations. The study population was small and the proportion of female was low. Several participants received medications such as statins and ACE-I/ARBs. Importantly, the majority of subjects were obese and showed abundant accumulation of visceral fat according to the Japanese criteria; the study included only one subject with VFA below 100 cm2. The full impact of VFA on the gene expression profile of peripheral blood cells has not been determined previously and should be examined also in non-obese individuals. In the present study, among the top 30 genes that correlated positively with VFA (Table 3), 14 (46.7%) genes were up-regulated in obesity, and among the top 30 genes that correlated negatively with VFA (Table 3), 17 (56.7%) genes were down-regulated in obesity. Unfortunately, a control group of non-obese subjects could not be included in the present study for ethical reasons (exposure of such subjects to CT scanning). For this reason, no data are available for the correlation of VFA and SFA to the gene expression profile of peripheral blood cells in non-obese subjects. Therefore, our results can only be applied to obese individuals.
Conclusions
The present study demonstrated that accumulation of visceral fat, but not that of subcutaneous fat, alters the gene expression profile of peripheral blood cells in obese Japanese subjects. The results should enhance our understanding of the pathogenesis of the metabolic syndrome.
Authors’ contributions
NM, YY, KY, SN, MY, TF, KM, and YM designed the study. YO, YY, KY, and YM contributed blood samples. YO, NM, and SN performed the statistical analysis. MY, YT, SM, HN, SF, YF, SK, and HN contributed to the discussion. YO, NM, and SN drafted the manuscript, and YY, KY, TF, KM, YM, and IS edited the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Kayoko Ohashi from the Department of Metabolic Medicine, Graduate School of Medicine, Osaka University, for the excellent technical assistance. We also thank all members of the IIIrd Laboratory (Adiposcience Laboratory), Department of Metabolic Medicine, Graduate School of Medicine, Osaka University, and Department of Endocrinology and Metabolism, Sumitomo Hospital, for the helpful discussion on the project.
Competing interests
The authors declare that they have no competing interests.
Availability of data and material
The raw microarray data are deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO Series GSE85226).
Ethics approval and consent to participate
Written informed consent was obtained from each patient after explaining the purpose of study. The study protocol was approved by the human ethics committees of Sumitomo Hospital and Osaka University. This study was also registered with the University hospital Medical Information Network (UMIN #000001663).
Funding
This work was supported in part by the Japan Society for the Promotion of Science (JSPS)-Grants-in-Aid for Scientific Research (KAKENHI) #JP25461386 (to NM), #JP16K09782 (to MY) and #JP26293221 (to TF), Takeda Science Foundation (to NM), and Japan Foundation for Applied Enzymology (to YF). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Yoshinari Obata, Email: obata-yoshinari@endmet.med.osaka-u.ac.jp.
Norikazu Maeda, Phone: +81-6-6879-3732, Email: norikazu_maeda@endmet.med.osaka-u.ac.jp.
Yuya Yamada, Email: yamada-yuuya@sumitomo-hp.or.jp.
Koji Yamamoto, Email: yamamoto-kouji@sumitomo-hp.or.jp.
Seiji Nakamura, Email: nakamura@dna-chip.co.jp.
Masaya Yamaoka, Email: myamaoka@imed2.med.osaka-u.ac.jp.
Yoshimitsu Tanaka, Email: ytanaka0325@endmet.med.osaka-u.ac.jp.
Shigeki Masuda, Email: masuda@endmet.med.osaka-u.ac.jp.
Hirofumi Nagao, Email: nagao@endmet.med.osaka-u.ac.jp.
Shiro Fukuda, Email: s.fukuda@endmet.med.osaka-u.ac.jp.
Yuya Fujishima, Email: y.fujishima@endmet.med.osaka-u.ac.jp.
Shunbun Kita, Email: shunkita@endmet.med.osaka-u.ac.jp.
Hitoshi Nishizawa, Email: hitoshin1127@endmet.med.osaka-u.ac.jp.
Tohru Funahashi, Email: momo10271122@gmail.com.
Ken-ichi Matsubara, Email: matsubara@dna-chip.co.jp.
Yuji Matsuzawa, Email: matsuzawa-yuji@sumitomo-hp.or.jp.
Iichiro Shimomura, Email: ichi@endmet.med.osaka-u.ac.jp.
References
- 1.Matsuzawa Y. Therapy insight: adipocytokines in metabolic syndrome and related cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2006;3:35–42. doi: 10.1038/ncpcardio0380. [DOI] [PubMed] [Google Scholar]
- 2.Okauchi Y, Kishida K, Funahashi T, Noguchi M, Morita S, Ogawa T, et al. 4-year follow-up of cardiovascular events and changes in visceral fat accumulation after health promotion program in the Amagasaki visceral fat study. Atherosclerosis. 2010;212:698–700. doi: 10.1016/j.atherosclerosis.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Hiuge-Shimizu A, Kishida K, Funahashi T, Ishizaka Y, Oka R, Okada M, et al. Absolute value of visceral fat area measured on computed tomography scans and obesity-related cardiovascular risk factors in large-scale Japanese general population (the VACATION-J study) Ann Med. 2012;44:82–92. doi: 10.3109/07853890.2010.526138. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzawa Y, Funahashi T, Kihara S, Shimomura I. Adiponectin and metabolic syndrome. Arterioscler Thromb Vasc Biol. 2004;24:29–33. doi: 10.1161/01.ATV.0000099786.99623.EF. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzawa Y, Funahashi T, Nakamura T. The concept of metabolic syndrome: contribution of visceral fat accumulation and its molecular mechanism. J Atheroscler Thromb. 2011;18:629–639. doi: 10.5551/jat.7922. [DOI] [PubMed] [Google Scholar]
- 6.Hotamisligil GS, Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat Rev Immunol. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 8.Yamaoka M, Maeda N, Nakamura S, Kashine S, Nakagawa Y, Hiuge-Shimizu A, et al. A pilot investigation of visceral fat adiposity and gene expression profile in peripheral blood cells. PLoS ONE. 2012;7:e47377. doi: 10.1371/journal.pone.0047377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaoka M, Maeda N, Nakamura S, Mori T, Inoue K, Matsuda K, et al. Gene expression levels of S100 protein family in blood cells are associated with insulin resistance and inflammation. Biochem Biophys Res Commun. 2013;433:450–455. doi: 10.1016/j.bbrc.2013.02.096. [DOI] [PubMed] [Google Scholar]
- 10.Lee H, Chu SH, Park JY, Park HK, Im JA, Lee JW. Visceral adiposity is associated with SIRT1 expression in peripheral blood mononuclear cells: a pilot study. Endocr J. 2013;60:1269–1273. doi: 10.1507/endocrj.EJ13-0207. [DOI] [PubMed] [Google Scholar]
- 11.Examination Committee of Criteria for ‘Obesity Disease’ in Japan. Japan Society for the Study of Obesity New criteria for ‘obesity disease’ in Japan. Circ J. 2002;66:987–992. doi: 10.1253/circj.66.987. [DOI] [PubMed] [Google Scholar]
- 12.Yoshizumi T, Nakamura T, Yamane M, Islam AH, Menju M, Yamasaki K, et al. Abdominal fat: standardized technique for measurement at CT. Radiology. 1999;211:283–286. doi: 10.1148/radiology.211.1.r99ap15283. [DOI] [PubMed] [Google Scholar]
- 13.Committee of the Japan Diabetes Society on the Diagnostic Criteria of Diabetes Mellitus. Seino Y, Nanjo K, Tajima N, Kadowaki T, Kashiwagi A, Araki E, et al. Report of the committee on the classification and diagnostic criteria of diabetes mellitus. J Diabetes Investig. 2010;1:212–228. doi: 10.1111/j.2040-1124.2010.00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Japanese Society of Nephrology Evidence-based practice guideline for the treatment of CKD. Clin Exp Nephrol. 2009;13:537–566. doi: 10.1007/s10157-009-0237-8. [DOI] [PubMed] [Google Scholar]
- 15.Tokunaga K, Matsuzawa Y, Ishikawa K, Tarui S. A novel technique for the determination of body fat by computed tomography. Int J Obes. 1983;7(5):437–445. [PubMed] [Google Scholar]
- 16.Matsuzawa Y, Shimomura I, Nakamura T, Keno Y, Tokunaga K. Pathophysiology and pathogenesis of visceral fat obesity. Ann N Y Acad Sci. 1995;748:399–406. doi: 10.1111/j.1749-6632.1994.tb17336.x. [DOI] [PubMed] [Google Scholar]
- 17.Smith U, Hammersten J, Björntorp P, Kral JG. Regional differences and effect of weight reduction on human fat cell metabolism. Eur J Clin Invest. 1979;9:327–332. doi: 10.1111/j.1365-2362.1979.tb00892.x. [DOI] [PubMed] [Google Scholar]
- 18.Mittelman SD, Van Citters GW, Kirkman EL, Bergman RN. Extreme insulin resistance of the central adipose depot in vivo. Diabetes. 2002;51:755–761. doi: 10.2337/diabetes.51.3.755. [DOI] [PubMed] [Google Scholar]
- 19.Maeda N, Funahashi T, Shimomura I. Metabolic impact of adipose and hepatic glycerol channels aquaporin 7 and aquaporin 9. Nat Clin Pract Endocrinol Metab. 2008;4:627–634. doi: 10.1038/ncpendmet0980. [DOI] [PubMed] [Google Scholar]
- 20.Hirata A, Maeda N, Nakatsuji H, Hiuge-Shimizu A, Okada T, Funahashi T, et al. Contribution of glucocorticoid-mineralocorticoid receptor pathway on the obesity-related adipocyte dysfunction. Biochem Biophys Res Commun. 2012;419:182–187. doi: 10.1016/j.bbrc.2012.01.139. [DOI] [PubMed] [Google Scholar]
- 21.Badman MK, Flier JS. The gut and energy balance: visceral allies in the obesity wars. Science. 2005;307:1909–1914. doi: 10.1126/science.1109951. [DOI] [PubMed] [Google Scholar]
- 22.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI200525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC. Functional role of KLF10 in multiple disease processes. BioFactors. 2010;36:8–18. doi: 10.1002/biof.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–253. doi: 10.1038/ni1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Z, Wara AK, Icli B, Sun X, Packard RR, Esen F, et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(–) T cells and T regulatory cells. J Biol Chem. 2009;284:24914–24924. doi: 10.1074/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillaumond F, Gréchez-Cassiau A, Subramaniam M, Brangolo S, Peteri-Brünback B, Staels B, et al. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol. 2010;30:3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutierrez-Aguilar R, Benmezroua Y, Balkau B, Marre M, Helbecque N, Charpentier G, et al. Minor contribution of SMAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab. 2007;33:372–378. doi: 10.1016/j.diabet.2007.06.002. [DOI] [PubMed] [Google Scholar]


