Abstract
The reproductive modes of anurans (frogs and toads) are the most diverse of terrestrial vertebrates, and a major challenge is identifying selective factors that promote the evolution or retention of reproductive modes across clades. Terrestrialized anuran breeding strategies have evolved repeatedly from the plesiomorphic fully aquatic reproductive mode, a process thought to occur through intermediate reproductive stages. Several selective forces have been proposed for the evolution of terrestrialized reproductive traits, but factors such as water systems and co‐evolution with ecomorphologies have not been investigated. We examined these topics in a comparative phylogenetic framework using Afrobatrachian frogs, an ecologically and reproductively diverse clade representing more than half of the total frog diversity found in Africa (∼400 species). We infer direct development has evolved twice independently from terrestrialized reproductive modes involving subterranean or terrestrial oviposition, supporting evolution through intermediate stages. We also detect associations between specific ecomorphologies and oviposition sites, and demonstrate arboreal species exhibit an overall shift toward using lentic water systems for breeding. These results indicate that changes in microhabitat use associated with ecomorphology, which allow access to novel sites for reproductive behavior, oviposition, or larval development, may also promote reproductive mode diversity in anurans.
Keywords: Africa, Afrobatrachia, anurans, direct development, reproductive mode
Across terrestrial vertebrates, amphibians have the greatest diversity of reproductive modes, which are often accompanied by a wide range of ecologies (Duellman and Trueb 1986). In particular, the order Anura (frogs and toads) exhibits incredible variation with at least 40 distinct reproductive modes, and further fine‐scale divisions resulting in nearly 60 possible modes (Boulenger 1886; Salthe and Duellman 1973; Duellman 1985; Duellman and Trueb 1986; Haddad and Prado 2005; Altig and Crother 2006; Altig and McDiarmid 2007; Duellman 2007; Wells 2007; Iskandar et al. 2014; Crump 2015). These modes are defined on the basis of oviposition site, egg and clutch characteristics, larval environment and development, and the degree of parental care. The proposed plesiomorphic reproductive condition of anurans is characterized by aquatic oviposition with a free‐swimming exotrophic larval stage (Duellman 1985), which is the most widespread mode among extant anurans. This mode is recovered as the ancestral state of anurans using phylogenetic comparative methods, and also appears to be the basal condition for a majority of frog families (Gomez‐Mestre et al. 2012). Much of the variation in reproductive mode seems to represent steps toward an independence from water, in which many anurans rely on for oviposition, larval development, or both (Lutz 1947, 1948; Salthe and Duellman 1973; Duellman and Trueb 1986; Wells 2007). The conventional view posits that many of these modes represent a long sequence of changes leading to greater terrestriality, with the terminal stage being direct development in which endotrophic larvae develop in terrestrial eggs and hatch as miniature versions of the adults. However, the evolution of direct development and other highly terrestrialized modes through intermediate stages has recently been challenged, and the switch to direct development may also occur from aquatic breeders, thereby circumventing intermediate stages (Gomez‐Mestre et al. 2012).
There are a number of selective pressures that can promote terrestrialized breeding strategies, including avoidance of aquatic predation (Lutz 1948; Tihen 1960; Crump 1974; Prado et al. 2002; Haddad and Prado 2005) or parasites (Todd 2007), precipitation and humidity levels (da Silva et al. 2012; Gomez‐Mestre et al. 2012) especially in forested regions (Poynton 1964; Gomez‐Mestre et al. 2012; Müller et al. 2013), and habitat instability (Crump 1974; Magnusson and Hero 1991). The physical environment may also play a role, as fast‐flowing water may not be conducive to aquatic oviposition and fertilization (Goin and Goin 1962), and the seasonal lack of water in some montane systems may actually promote complete terrestrial development (Müller et al. 2013). Ecomorphological differentiation among species may also drive reproductive mode diversity, particularly if morphological adaptations allow access to novel microhabitats or spatial resources. In these cases, changes in reproductive traits such as calling site, oviposition site, or clutch characteristics may occur with shifts in adult habitat use, thereby facilitating reproductive spatial partitioning. Though the repeated independent evolution of particular frog ecomorphologies is well characterized, explicit tests for associations between adult ecologies and key reproductive traits have not been conducted.
There are over 800 frog species in 17 families occurring in sub‐Saharan Africa, and although a majority of species exhibit the fully aquatic reproductive mode, more than 15 terrestrialized reproductive modes have been documented. Remarkably, 11 of these terrestrialized reproductive modes occur in a single African‐endemic clade termed Afrobatrachia (sensu Frost et al. 2006). This radiation contains ∼400 species in four families (Arthroleptidae, Brevicipitidae, Hemisotidae, Hyperoliidae) that together represent over half of the total frog species diversity in sub‐Saharan Africa (AmphibiaWeb 2015; Frost 2015). The species in this clade are diverse in terms of their ecomorphologies, behaviors, and general life histories, and include a number of charismatic taxa such as the fossorial “rain frogs” (genus Breviceps; Fig. 1B), the “hairy frog” (genus Trichobatrachus; Fig. 1G), and the colorful hyperoliid reed frogs (Fig. 1S–W). The main diversity of Afrobatrachians is found primarily in lowland and montane forests in which they display a number of adult ecologies including arboreality, terrestriality, and fossoriality. The dozen different reproductive modes present in Afrobatrachians mainly involve variations in the site of oviposition (in water, subterranean nests, terrestrial sites, or arboreal locations), water system (lentic or lotic), and location of larval development (exotrophic aquatic tadpole vs. endotrophic terrestrial tadpole). Direct development, the most terrestrialized reproductive mode, occurs in Brevicipitidae (FitzSimons and Van Dam 1929; Wager 1965; Channing 2001) and the arthroleptid genus Arthroleptis (Guibé and Lamotte 1958; Wager 1986).
Figure 1.
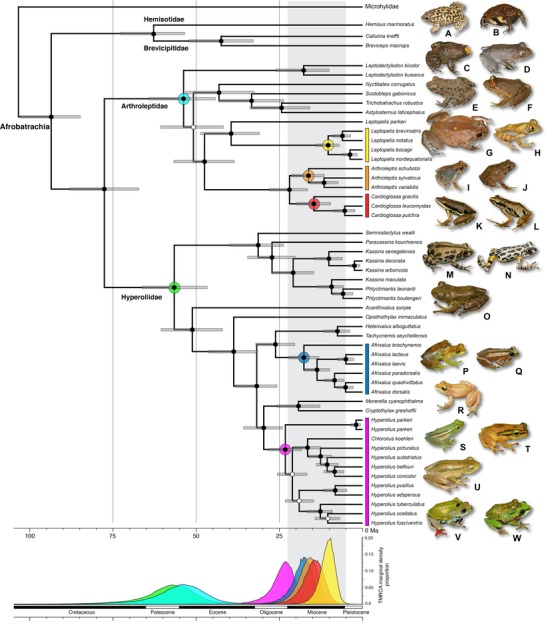
Bayesian maximum clade credibility chronogram of the Afrobatrachia obtained using BEAST, illustrating divergence date estimates and the 95% highest posterior density region of dates (indicated by grey bars). Filled circles on nodes represent high support (PPB > 0.95); open circles indicate PPB < 0.95. Nodes of interest are color coded: purple, Afrobatrachia; light blue, Arthroleptidae; green, Hyperoliidae; yellow, Leptopelis; orange, Arthroleptis; red, Cardioglossa; dark blue, Afrixalus; magenta, Hyperolius. The TMRCA marginal density proportion for each of these specific nodes, based on the combined results of all calibrations strategies, is illustrated below the chronogram, with distributions matching the same color scheme. Families are represented by the following: Hemisotidae (A); Brevicipitidae (B); Arthroleptidae (C: Leptodactylodon, D: Nyctibates, E: Scotobleps, F: Astylosternus, G: Trichobatrachus, H: Leptopelis, I–J: Arthroleptis, K–L: Cardioglossa); Hyperoliidae (M–N: Kassina, O: Phlyctimantis, P–Q: Afrixalus, R: Cryptothylax, S–W: Hyperolius). Photos A–Q, S–W by D. M. Portik, photo R by G. Jongsma.
The high ecological and reproductive mode diversity in Afrobatrachians allows testing of hypotheses regarding the evolution of direct development, terrestrialized oviposition, and the co‐evolution of ecomorphology and reproductive traits. The presence of direct development in multiple genera of Afrobatrachians could be explained by multiple independent origins, or by the retention of this state in a common ancestor accompanied by repeated losses in closely related taxa. In either scenario, identifying if the fully aquatic reproductive mode or a more terrestrialized intermediate mode precedes direct development will determine if this complex trait evolves through intermediate stages. The variation in Afrobatrachian reproductive modes may be partially driven by specific adult ecologies if functional links between ecology and reproductive traits involving spatial resources (oviposition site, water system) exist. Patterns such as the long‐term retention of both character states, or the repeated independent evolution of the linked character states, may point to such associations.
In this study, we use a comparative phylogenetic approach to investigate the evolution of reproductive mode diversity in Afrobatrachian frogs. Though previous molecular phylogenetic studies provide a consensus on the family‐level relationships within the Afrobatrachia, the genus‐ and species‐level relationships are not strongly resolved (Bossuyt et al. 2006; Frost et al. 2006; Roelants et al. 2007; Pyron and Wiens 2011). Here, we conduct the most comprehensive analysis of phylogenetic relationships within the Afrobatrachia by using a combination of newly generated and published sequence data. We explore the evolution of direct development, including the number of origins and whether the preceding reproductive mode is fully aquatic or involves terrestrialized oviposition. We test for associations among adult ecology, oviposition site, and water system to examine the influence of ecomorphology on reproductive mode diversity. Finally, we examine if terrestrialized reproductive modes are more strongly associated with breeding in lotic systems.
Methods
SAMPLING
To resolve higher‐level relationships within the Afrobatrachia, we generated new multilocus sequence data for as many genera as possible within the families Hemisotidae, Brevicipitidae, Arthroleptidae, and Hyperoliidae. This resulted in a total of 51 ingroup species with multiple representatives from each genus, when possible. We sampled all eight arthroleptid genera, 13 of 18 hyperoliid genera, two of five brevicipitid genera, and the single hemisotid genus, for a total of 25 genera. Regarding the genera not included, samples were unavailable for five hyperoliid genera (Alexteroon, Arlequinus, Callixalus, Chrysobatrachus, Kassinula; totaling seven species), and we did not densely sample the Brevicipitidae because relationships among these genera were recently published (Loader et al. 2014). Within species‐rich genera, we chose species that represent major clades and divergent taxa based on published or unpublished mtDNA barcode data (Blackburn 2008; D. C. Blackburn and D. M. Portik, unpubl. data). In addition, to test the monophyly of the genus Leptopelis, we sampled each of the subgenera proposed by Laurent (1941). Because of complications related to polyploidy in the genus Astylosternus (Bogart and Tandy 1981), we included only one representative of a diploid species. We included one species (Phrynomantis microps) of the family Microhylidae as an outgroup because previous studies demonstrate this family to be the sister clade to the Afrobatrachia (e.g., Bossuyt et al. 2006; Frost et al. 2006; Roelants et al. 2007; Pyron and Wiens 2011). The museum information for all 53 specimens is provided in the Appendix.
MOLECULAR DATA
Whole genomic DNA was extracted from ethanol‐ or RNALater‐preserved liver, muscle, or toe clip samples using a high‐salt DNA extraction (Aljanabi and Martinez 1997). We obtained sequence data for five nuclear loci and one mtDNA locus using primers listed in Table 1: POMC (624 bp), RAG‐1 (777 bp), TYR (573 bp), FICD (524 bp), KIAA2013 (540 bp), and 16S (578 bp). Polymerase chain reactions (PCRs) were carried out in 12.5 μl volumes consisting of the following: 1.4 μl Roche 10× (500 mM Tris/HCl, 100 mM KCl, 50 mM (NH4)2 SO4, 20 mM MgCl2, pH 8.3), 1.1 μl 25 mM MgCl2, 0.22 μl 2 mM DNTPs, 0.22 μl 10.0 μM forward primer, 0.22 μl 10.0 μM reverse primer, 7.25 μl H2O, 1.1 μl betaine, 1.1 μl BSA, 0.08 μl Taq, and 1.0 μl DNA. Thermocycling schemes and amplifications of FICD and KIAA2013 followed the specific nested PCR protocol of Shen et al. (2013). Remaining genes were amplified using the following thermocycling protocol: an initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 sec, a gene‐specific annealing temp (55°C: RAG‐1, TYR; 51°C: POMC) for 30 sec, 72°C for 60 sec, and a final extension at 72°C for 10 min. The PCR amplifications were visualized on an agarose gel and cleaned using ExoSAP‐IT (USB). Gene products were sequenced using BigDye version 3.1 on an ABI3730 (Applied Biosystems). All forward and reverse sequences were assembled using Geneious (Kearse et al. 2012) and aligned using MUSCLE (Edgar 2004), and subsequently translated to ensure conservation of reading frame. The final concatenated alignment of the five nuclear loci totals 3038 bp, with 1150 parsimony informative characters (37.9%) and 135 autapomorphic characters (4.4%), with only 10% missing data. The percentage of parsimony informative characters to total base pairs per gene was relatively consistent and is as follows: RAG‐1, 30.1%; FICD, 34.9%; TYR, 36.3%; KIAA2013, 37.5%; POMC, 39.7%. All sequences are deposited in GenBank: KX492593–KX492885.
Table 1.
A list of primers, primer sequences, and sources for nuclear genes sequenced in this study
| Gene | Primer name | Sequence (5′–3′) | Source |
|---|---|---|---|
| POMC | POMC‐1 | GAATGTATYAAAGMMTGCAAGATGGWCCT | Wiens et al. (2005) |
| POMC‐7 | TGGCATTTTTGAAAAGAGTCAT | Smith et al. (2007) | |
| RAG‐1 | RAG1 DCB1Fi | CTTCCGTGGAACAGGATATGA | Present study |
| RAG1 DCB1R | CCAGATTCGTTGCCTTCACT | Present study | |
| TYR | TyrC | GGCAGAGGAWCRTGCCAAGATGT | Bossuyt and Milinkovitch (2000) |
| TyrG | TGCTGGCRTCTCTCCARTCCCA | Bossuyt and Milinkovitch (2000) | |
| FICD | FICD F1 | CCKCTNGTNGARGARATHGAYCA | Shen et al. (2013) |
| FICD R1 | TYTCNGTRCAYTTNGCDATRAA | Shen et al. (2013) | |
| FICD F2 | AGGGTTTTCCCAGTCACGACTACTAYCAYCAYATHTAYCAYAC | Shen et al. (2013) | |
| FICD R2 | AGATAACAATTTCACACAGGAARGGCCKVACRTCNCCYTCRTT | Shen et al. (2013) | |
| KIAA2013 | KIAA2013 F1 | CTSAANTAYGCNGAYCAYTGYTT | Shen et al. (2013) |
| KIAA2013 R1 | CCNGGNCCRCARTAYTCRTTRTA | Shen et al. (2013) | |
| KIAA2013 F2 | AGGGTTTTCCCAGTCACGACACYATGCAYGCNGAGAAYYTGTGG | Shen et al. (2013) | |
| KIAA2013 R2 | AGATAACAATTTCACACAGGGANGCCACNCTRAACCARAA | Shen et al. (2013) |
GENBANK SAMPLING
In addition to generating new multilocus sequence data for 51 species, we included GenBank sequences for the purpose of expanding our taxon sampling for character analyses. An additional 118 additional species were included with various combinations of RAG‐1 (n = 16), TYR (n = 34), and 16S (n = 112), though most species are represented only by 16S data (Table S1). With the addition of these samples from GenBank, we included 169 ingroup species for phylogenetic analyses. Though this sampling represents ∼43% of the total species of Afrobatrachians, 27 of 32 Afrobatrachian genera are represented and species rich genera are sampled proportional to their diversity. The full alignment of five nuclear markers and 16S sequence data is available on the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.sh0h0.
PHYLOGENETIC ANALYSES
To explore higher‐level phylogenetic relationships of Afrobatrachians, we chose to analyze the nuclear‐only dataset, using both maximum‐likelihood (ML) and Bayesian analyses. We used PartitionFinder (Lanfear et al. 2012) to simultaneously determine our best partitioning strategy and models for each partition subset. The greedy search algorithm was employed, and model selection was conducted using the BIC. The optimal partitioning strategy includes five partitions and models are as follows: partition one (FICD codon position (cp) 1, KIAA2013 cp1, RAG‐1 cp1): GTR+G; partition two (FICD cp2, KIAA2013 cp2, RAG‐1 cp2): HKY+G; partition three (FICD cp3, KIAA2013 cp3, RAG‐1 cp3): HKY+G; partition four (KIAA2013 cp3, TYR cp3): GTR+G; and partition five (POMC cp1, POMC cp2, TYR cp1, TYR cp2): HKY+G+I. We used MrBayes version 3.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) to execute two parallel runs with four MCMC chains, and analyses were run for 20,000,000 generations with sampling every 1000 generations, resulting in 20,000 trees. Runs were assessed using Tracer version 1.5.0 (Rambaut and Drummond 2009) to ensure key parameters had reached stationarity (ESS values >150). The first 25% of the total number of generations were discarded as burn‐in and a 50% majority rule consensus tree was calculated from the combined remaining trees of both parallel runs. We performed ML analyses of the partitioned dataset using GARLI version 2.0 (Zwickl 2006) to execute 1000 nonparametric bootstrap replicates using the default stopping criteria. A 50% majority rule consensus tree was generated from the 1000 replicates.
DIVERGENCE DATING ANALYSES
We conducted two sets of divergence dating analyses for the purpose of (1) estimating divergence times for the nuclear‐only dataset, and (2) to create sets of ultrametric trees with the expanded sampling data set for character analyses. Dating analyses were carried out using BEAST version 1.8 (Drummond et al. 2012) using multiple calibration strategies based on results from Roelants et al. (2007), Kurabayashi and Masayuki (2013), and Loader et al. (2014). We used four secondary calibration points constraining the most recent common ancestors (MRCAs) of (Arthroleptidae + Hyperoliidae + Hemisotidae + Brevicipitidae) to 92.8 Ma ± 5.0 SD, (Hemisotidae + Brevicipitidae) to 65.9 Ma ± 6.5 SD, and Brevicipitidae to 45.4 Ma ± 8.0 SD. We applied normal distributions to these calibrated nodes, covering the range of the confidence intervals reported in previous studies, as there is no justification for weighting these secondary calibrations toward a minimum or maximum bound. To explore the effects of calibration strategy, we used all permutations of the three calibrations, including single‐calibration analyses, for a total of seven unique calibrated analyses for the nuclear‐only data set. Based on these results, the expanded sampling dataset was analyzed using the full calibration set. Each analysis was run 20,000,000 generations with sampling every 2000 generations. For all analyses, we used the Yule model of speciation as the tree prior, applied an uncorrelated relaxed lognormal clock, and unlinked clock and substitution models. Substitution models and partitioning schemes were identical to those used in phylogenetic analyses. Runs were assessed using Tracer version 1.5.0 (Rambaut and Drummond 2009) to examine convergence. A burn‐in of 25% was discarded and maximum clade credibility trees were created from the remaining 7500 trees. For nodes of interest, divergence date means, confidence intervals, and marginal density proportions were obtained from each of the seven calibrated analyses.
ANCESTRAL CHARACTER ESTIMATIONS AND TRAIT ASSOCIATIONS
We collected data for several character sets focused on ecological and reproductive traits, which were coded as multistate discrete characters. These character sets include adult ecology (fossorial, terrestrial, arboreal), oviposition site (aquatic, subterranean, terrestrial, arboreal), tadpole water system (lentic, lotic, or none in the case of direct development), and reproductive mode. There are many ways to categorize the reproductive modes of amphibians (reviewed in Crump 2015), but here we focus on the degree of terrestrialization of breeding strategy, which examines the combination of oviposition site and the site of tadpole development. We categorize reproductive mode to include the following four states: (1) aquatic eggs and aquatic larvae, (2) terrestrial eggs and aquatic larvae, (3) arboreal eggs and aquatic larvae, and (4) terrestrial eggs and terrestrial larvae (including direct development). We were able to score characters for 151 taxa based on an exhaustive literature search and/or personal observations (FitzSimons and Van Dam 1929; Guibé and Lamotte 1958; Wager 1965; Amiet 1972; Wager 1986; Schiøtz 1999; Channing 2001; Channing and Howell 2006; Amiet 2012; AmphibiaWeb 2015). Taxa with incomplete or absent data were excluded from analyses (18 species). Information for many species is based on direct accounts, but reproductive traits are sometimes assumed or extrapolated across certain genera (Arthroleptis, Breviceps, Callulina, Cardioglossa, Leptodactylodon) in the literature or databases. A summary of the character data is provided in Table S2. Although our species sampling is incomplete (∼40%), our dataset captures the full diversity of reproductive modes of Afrobatrachians, including variation across and within genera.
The ancestral states of adult ecology and reproductive mode were examined using three approaches, including ML, stochastic character mapping, and Bayesian inference (BI). Using the ACE function of the APE package in R (Paradis et al. 2004), the likelihoods of three transition models (equal rates [ER], symmetrical rates [SYM], or all rates different [ARD]) were determined by ML, and subsequent likelihood ratio tests were conducted to determine the best‐fitting model for each character set. The best fitting models were used for all subsequent analyses. ML estimation of ancestral states was carried out using ACE to determine the marginal ancestral state reconstruction of the root and the conditional scaled likelihoods of all remaining nodes. To estimate the marginal probabilities for all nodes based on joint sampling, stochastic mapping was implemented using the make.simmap function of the phytools package in R (Revell 2012). To account for branch length and topological uncertainty, we performed mapping using 100 replicates on 100 randomly selected trees from the posterior distribution of trees resulting from BEAST, resulting in a total of 10,000 mapped trees. The number of transitions between character states and the proportion of time spent in each state were summarized using phytools. Finally, the root state was estimated in a Bayesian framework using BayesTraits version 2 (Pagel et al. 2004). Alternative root states were compared by fixing the root state using the “fossil node” implementation for model testing, and an unconstrained analysis was performed to obtain the posterior probability of the root state. Under the multistate model of evolution, analyses were run using reversible jump MCMC for 50,000,000 post‐burn‐in generations, sampling every 5000 generations. Marginal likelihoods were calculated using stepping‐stone sampling with 100 samples and 10,000 iterations per sample. Alternative hypotheses were compared using log Bayes Factors (logBF = 2(log marginal likelihood[model 1] – log marginal likelihood[model 2]), with logBF > 5 considered as support. Analyses incorporated the topologies of 100 randomly selected trees from the posterior distribution obtained from BEAST to account for topological and branch length uncertainty. All scripts and data used for these analyses are deposited on Dryad: doi:10.5061/dryad.sh0h0.
To investigate potential associations among adult ecology, oviposition site, and water systems, analyses were conducted using BayesTraits version 2 (Pagel et al. 2004). The discrete dependent and discrete independent models of evolution were used to examine correlations between character states across different character sets. Correlations were investigated between states of adult ecology and oviposition site, adult ecology and tadpole water system, and oviposition site and tadpole water system. Analyses were performed using ML and BI. ML analyses were conducted using 1000 attempts per tree, across 100 randomly selected trees from the posterior distribution obtained from BEAST. Bayesian analyses were run using reversible jump MCMC for 50,000,000 post‐burn‐in generations, sampling every 5000 generations. Marginal likelihoods were calculated using stepping‐stone sampling with 100 samples and 10,000 iterations per sample. Because these are nested models, the alternative hypotheses of dependent versus independent models of evolution were compared using a likelihood ratio test, or using logBF, where logBF = 2(log marginal likelihood[dependent model] – log marginal likelihood[independent model]). Analyses incorporated the topologies of 100 randomly selected trees from the posterior distribution of trees resulting from BEAST.
For character states exhibiting dependent evolution, the z‐scores of character transitions were estimated (Pagel and Meade 2006). The z‐scores were calculated from the proportion of the number of occurrences of a value of zero estimated for relevant transition parameters, and are expressed as a percentage. The z‐scores allow examination of the contingency of evolutionary changes, with high z‐scores signifying unlikely paths for evolutionary transformations of character pairs. In this framework, it can be evaluated if state changes of a particular character are contingent upon the background state of the other character.
We performed simulations to assess the proportion of significant character associations that might be recovered by chance (type I error) based on the number of character states and tips in our tree. Using the q‐matrices obtained using SIMMAP, we conducted 500 independent simulations of the multistate discrete traits with the sim.history function in phytools (Revell 2012). These traits included adult ecology (three states), water system (two states), and oviposition site (four states). For each simulation, a single state was chosen randomly for each of the three traits and correlations between these states were examined (adult ecology and water system, adult ecology and oviposition site, and oviposition site and water system). This resulted in three correlation analyses per simulation, for a total of 1500 correlation analyses. Analyses were performed on the single best tree with ML and BI using BayesTraits version 2 (Pagel et al. 2004). Analyses were conducted using 1000 searches or reversible jump MCMC for 1,000,000 post‐burn‐in generations. Significance was calculated using a likelihood ratio test or logBF as described above. We summarized the overall number of significant associations detected for these nonassociated traits using either ML (P < 0.01) or BI (logBF > 10), but also counted how many comparisons produced significant associations using both methods (P < 0.01 and logBF > 10). The scripts for simulating characters and performing correlation analyses, along with the resulting simulation data, are deposited on Dryad: doi:10.5061/dryad.sh0h0.
Results
PHYLOGENETIC RELATIONSHIPS
The phylogenetic relationships within the Afrobatrachia are concordant across analyses, and both the monophyly of and relationships among the four Afrobatrachian families are resolved with high support (Figs. 1, 2). A sister relationship is recovered with high support for Hemisotidae and Brevicipitidae, as well as for Arthroleptidae and Hyperoliidae. Within the family Arthroleptidae, four major clades are consistently recovered: the genus Leptodactylodon; the genus Leptopelis; a clade containing Nyctibates, Scotobleps, Astylosternus, and Trichobatrachus; and a clade containing Cardioglossa and Arthroleptis (Fig. 1). The relationships among these four clades are not fully resolved, though some analyses recover the genus Leptodactylodon as sister to the other two major clades (Fig. 1). The sister relationship between the genus Leptopelis and the clade consisting of Cardioglossa and Arthroleptis is recovered in most analyses. The species rich genera (Arthroleptis, Cardioglossa, and Leptopelis) are each recovered as monophyletic in all analyses. The species Leptopelis parkeri is found to be sister to and highly divergent from a clade containing all other Leptopelis (Fig. 1).
Figure 2.
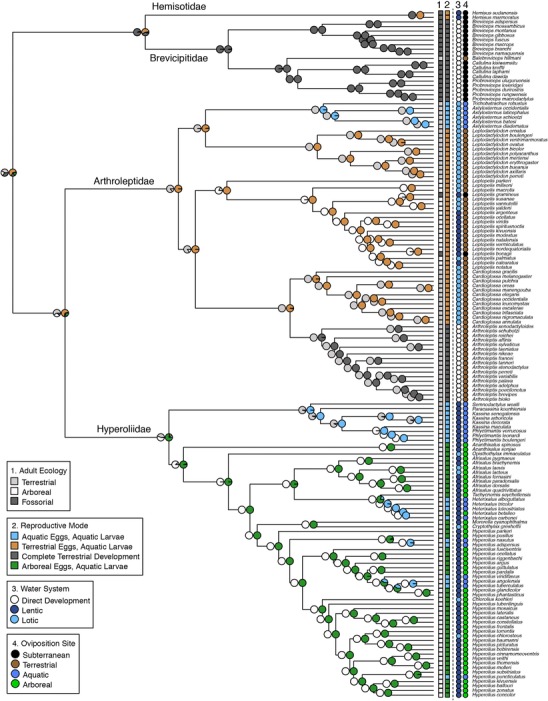
Mapping of ecological, habitat, and reproductive character states onto the expanded sampling time‐calibrated phylogeny of Afrobatrachians. Boxes represent characters used for ancestral state reconstructions, with pie charts at nodes representing posterior probabilities of character states. Circles represent characters used for correlated evolution analyses. Numbers above boxes or circles match to character legends.
Within the Hyperoliidae, there is strong support for two subclades comprising, respectively, the “kassinoids” and all other hyperoliids (Figs. 1, 2). The kassinoid genera represented in our dataset include Semnodactylus, Paracassina, Phlyctimantis, and Kassina. Within the kassinoids, we find that Kassina maculata is more closely related to Phylctimantis than to other species of Kassina, and we recommend that this species be transferred to Phlyctimantis (see Supporting Information). The genus Acanthixalus and Opisthothylax form the earliest diverging lineages of the remaining hyperoliids. Heterixalus and Tachycnemis, endemic to the islands of Madagascar and the Seychelles, respectively, are recovered as sister taxa, and are in turn sister to the species rich genus Afrixalus, which is recovered as monophyletic. The genera Cryptothylax and Morerella form a strongly supported clade that is sister to a clade comprising the genus Hyperolius and the monotypic genus Chlorolius, which is embedded well within Hyperolius. Relationships within the species‐rich genus Hyperolius are not strongly resolved. We discuss the systematics and taxonomic implications of these results in Supporting Information.
DIVERGENCE DATING ESTIMATES
The dating analyses resulting from the seven calibration strategies produced identical topologies and similar mean estimates for dates (Fig. 1, Table 2). In general, the analyses incorporating fewer calibration points resulted in dating estimates with larger 95% highest posterior density regions (95% HPD), with little difference in means (Table 2). We therefore present results from the analysis incorporating all calibrations (Afr, [H+B], B; Table 2; Fig. 1; Supporting Information). Results from the expanded sampling data set, including the nuclear‐only data set and the combined mtDNA and nuclear data sets, are provided in Supporting Information.
Table 2.
Divergence dating estimates and 95% HPD intervals obtained for groups of interest
| Analysis | Afrobatrachia | Hyperoliidae | Arthroleptidae | Arthroleptis | Cardioglossa | Leptopelis | Hyperolius | Afrixalus |
|---|---|---|---|---|---|---|---|---|
| Afr, (H+B), B | 93.6 (84.8–102.8) | 56.6 (47.0–66.6) | 53.7 (43.9–63.9) | 16.2 (11.6–21.2) | 14.6 (9.6–19.9) | 10.3 (7.0–14.0) | 23.3 (18.3–28.3) | 17.6 (13.2–22.4) |
| (H+B), B | 103.8 (73.9–138.0) | 62.8 (43.6–83.8) | 59.0 (40.4–79.5) | 17.7 (11.3–25.6) | 15.8 (9.1–23.3) | 11.3 (6.5–16.1) | 25.8 (17.5–34.5) | 19.5 (12.5–26.8) |
| (H+B) | 100.9 (71.0–133.2) | 60.6 (40.5–81.0) | 57.5 (39.5–77.6) | 17.2 (10.7–24.0) | 15.4 (9.0–22.5) | 11.1 (6.4–15.9) | 24.9 (16.5–33.7) | 18.8 (12.1–26.1) |
| B | 96.1 (47.1–151.6) | 58.0 (26.4–91.6) | 55.0 (25.2–87.0) | 16.3 (7.4–26.7) | 14.7 (5.8–24.5) | 10.5 (4.4–17.3) | 23.8 (10.8–37.9) | 18.1 (7.8–29.0) |
| Afr, B | 92.7 (82.9–101.9) | 56.3 (46.2–66.2) | 53.3 (43.4–64.2) | 16.0 (11.6–20.9) | 14.4 (9.0–19.5) | 10.2 (6.8–13.8) | 23.0 (18.3–27.8) | 17.5 (12.9–22.0) |
| Afr, (H+B) | 93.3 (84.0–102.6) | 56.5 (46.6–66.7) | 53.2 (43.0–63.2) | 16.0 (11.4–20.7) | 14.3 (9.4–19.5) | 10.2 (6.8–13.8) | 23.2 (18.6–28.3) | 17.6 (13.0–22.2) |
| Afr | 91.9 (82.2–101.8) | 55.1 (44.8–65.2) | 52.5 (42.3–62.9) | 15.7 (11.3–20.5) | 14.0 (9.0–19.0) | 10.1 (6.8–13.7) | 22.7 (17.8–27.8) | 17.1 (12.5–21.8) |
Afr, tmrca of Afrobatrachia (Hemisotidae, Brevicipitidae, Arthroleptidae, Hyperoliidae); (H+B), tmrca of Hemisotidae and Brevicipitidae; B, tmrca of Brevicipitidae.
The estimates for the TMRCA of the families Hyperoliidae and Arthroleptidae are similar, being approximately 56.6 Ma (95% HPD: 47.0–66.6 Ma) and 53.7 Ma (95% HPD: 43.9–63.9 Ma), respectively (Fig. 1, Table 2). Early diversification events in the species‐rich genera appear to be concentrated in the Late Oligocene and Early Miocene (Fig. 2). Diversification began to occur in the genus Arthroleptis approximately 16.2 Ma (95% HPD: 11.6–21.2 Ma) and in Cardioglossa approximately 14.6 Ma (95% HPD: 9.6–19.9 Ma). Leptopelis parkeri, endemic to the Eastern Arc Mountains, is estimated to have diverged from all other Leptopelis approximately 39.5 Ma (95% HPD: 31.0–48.7 Ma), with diversification of the remaining species beginning around 10.3 Ma (95% HPD: 7.0–14.0 Ma). Within hyperoliids, the TMRCA for the genus Afrixalus is estimated to be 17.6 Ma (95% HPD: 13.2–22.4 Ma), and diversification in Hyperolius, the largest Afrobatrachian genus, appears to have begun approximately 23.3 Ma (95% HPD: 18.3–28.3 Ma).
The Malagasy and Seychelles Island hyperoliids, comprised of the genera Heterixalus and Tachycnemis, appear to have diverged from their closest mainland African relatives (Afrixalus) approximately 26.1 Ma (95% HPD: 20.4–32.1 Ma). This divergence represents a major overseas dispersal event, which is supported by favorable oceanic currents in the Mozambique Channel prior to the Miocene (Ali and Huber 2010; Samonds et al. 2012; Tolley et al. 2013). Pyron (2014) estimated this divergence to be 51 Ma, and although our results are much younger, both fall in the expected oceanic current time interval predicted (>24 Ma). Heterixalus and Tachycnemis are estimated to have diverged from one another 7.6 Ma (95% HPD: 3.8–11.6 Ma).
EVOLUTION OF ADULT ECOLOGY AND REPRODUCTIVE MODE
Comparisons of the transition rate models revealed that more complex models (SYM, ARD) were not significantly better fits than a simple one‐rate model (ER); therefore the ER transition model was used for subsequent analyses. The ML ancestral character estimations of adult ecology provide ambiguous results for the state of the common ancestor (Table 3). Bayesian hypothesis testing performed by fixing root states revealed no significant differences in marginal likelihoods across models (fossorial: –48.87, arboreal: –49.72, terrestrial: –49.05; logBF < 5). Transition matrices reveal fossoriality has arisen from terrestriality (common ancestor of hemisotids and brevicipitids), as well as arboreality (within the genus Leptopelis, Fig. 3A). The most common ecological transition is from terrestriality to arboreality, and it has occurred independently at least four times (Fig. 3A). The transitions have taken place in the families Arthroleptidae (Leptopelis) and Hyperoliidae (Phlyctimantis, Kassina arboricola, mrca of non‐kassinoid hyperoliids; Fig. 2). There are no transitions from fossoriality to arboreality across Afrobatrachians.
Table 3.
The marginal probability of the root state for adult ecology and reproductive mode
| Character set | Character states | Scaled root likelihood |
|---|---|---|
| Adult ecology | Terrestrial | 0.43 |
| Arboreal | 0.22 | |
| Fossorial | 0.35 | |
| Reproductive mode | Aquatic/aquatic | 0.04 |
| Terrestrial/aquatic | 0.65 | |
| Arboreal/aquatic | 0.15 | |
| Terrestrial/terrestrial | 0.16 |
The probabilities are based on maximum‐likelihood analyses using ACE. The highest probability state is bolded.
Figure 3.
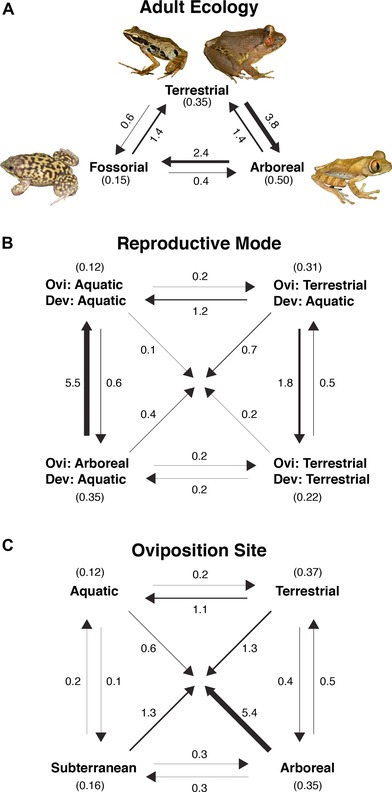
Estimated number of evolutionary changes among (A) adult ecology, (B) reproductive modes, or (C) oviposition site inferred from stochastic character mapping using 100 replicates on each of 100 randomly selected ultrametric trees (10,000 mapped trees). Width of arrows is proportional to estimated number of changes.
Ancestral character estimates of reproductive mode provide support for a terrestrialized breeding strategy present in the common ancestor of the Afrobatrachia, specifically terrestrial oviposition with aquatic larvae (Fig. 2). Comparisons of the marginal likelihoods of fixed root states inferred by Bayesian analysis support a terrestrial oviposition/aquatic larvae root state (terrestrial/aquatic: –62.75, aquatic/aquatic: –65.73, terrestrial/terrestrial: –65.43, arboreal/aquatic: –64.81; logBF: 5.96, 5.36, 4.12, respectively), and ML analyses estimate this root state with a marginal probability of 0.65 (compared to 0.15, 0.16, and 0.04 of other states, Table 3). The number of transitions between these four reproductive modes varies (Fig. 3B), but few transitional paths occur frequently. The complete terrestrial reproductive mode only arises from the terrestrial/aquatic reproductive mode, rather than from the aquatic/aquatic or arboreal/aquatic states (Fig. 3B), and is confirmed as originating independently in the family Brevicipitidae and the arthroleptid genus Arthroleptis (Fig. 2). Transitions to the arboreal/aquatic reproductive mode and aquatic/aquatic mode occur disproportionately from the terrestrial/aquatic reproductive mode (1.8 transitions and 1.2 transitions, respectively), which is the inferred ancestral state. Transitions to terrestrial oviposition and aquatic larvae from the other three reproductive modes are estimated in very low frequency, suggesting they are not likely (Fig. 3B). There are at least five reversals from arboreal oviposition to aquatic oviposition, all of which occur in hyperoliid frogs (Fig. 2). These reversals are also reflected in the transition matrix of oviposition sites (Fig. 3C), in which the transition from arboreal to aquatic oviposition has occurred at least five times.
TRAIT ASSOCIATIONS
The simulations of nonassociated character states indicate the level of type I error is 4.6% using ML (1433 nonsignificant, 67 significant associations), and 26.9% using BI (1182 non‐significant, 318 significant associations). Though these methods do recover false‐positive associations between random character states, of the 1500 total comparisons only seven were detected as significantly associated using both methods (<1%). To be conservative, we therefore only emphasize results that show significant associations based on both analysis types.
The exploration of correlations between adult ecology, oviposition site, and larval water system revealed several sets of significantly associated characters (Table 4). There are significant associations between arboreal ecology and arboreal oviposition (likelihood ratio = 19.52, P < 0.001; logBF = 10.70), between fossorial ecology and subterranean oviposition (likelihood ratio = 31.00, P < 0.001; logBF = 25.68), and between arboreal ecology and lentic water systems (likelihood ratio = 14.24, P = 0.007; logBF = 7.36). No significant associations between lotic water systems and oviposition were found. The z‐scores for fossorial ecology and subterranean oviposition reveal that the most likely pathway was a gain of fossoriality, followed by a gain of subterranean oviposition (Fig. S1). The z‐scores for arboreality and arboreal oviposition suggest a reversal from arboreal ecology accompanied by the retention arboreal oviposition is not likely, but that the loss of arboreal oviposition in arboreal species occurs (Fig. S1). These patterns are observed in the derived hyperoliids, with several species exhibiting a transition from arboreal oviposition to aquatic oviposition (Fig. 2).
Table 4.
Results of character correlations based on maximum‐likelihood and Bayesian analyses
| Maximum likelihood | Bayesian Inference | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Character 1 | Character 2 | Dependent | Independent | Likelihood ratio | df | P‐value | Dependent | Independent | logBF |
| Arboreal ecology | Aquatic oviposition | −59.21 | −65.19 | 11.96 | 4 | 0.018 | −75.96 | −77.68 | 3.44 |
| Arboreal oviposition | −55.74 | −65.50 | 19.52 | 4 | < 0.001 | −68.08 | −73.43 | 10.70 | |
| Terrestrial oviposition | −44.94 | −49.67 | 9.46 | 4 | 0.051 | −55.64 | −57.64 | 4.00 | |
| Terrestrial ecology | Aquatic oviposition | −53.09 | −60.37 | 14.56 | 4 | 0.006 | −71.05 | −71.66 | 1.22 |
| Arboreal oviposition | −55.18 | −59.05 | 7.74 | 4 | 0.102 | −67.46 | −67.10 | −0.72 | |
| Terrestrial oviposition | −37.39 | −43.22 | 11.66 | 4 | 0.020 | −50.93 | −51.92 | 1.98 | |
| Fossorial ecology | Subterranean oviposition | −18.35 | −33.85 | 31.00 | 4 | < 0.001 | −32.40 | −45.24 | 25.68 |
| Aquatic oviposition | Lentic system | −70.37 | −75.69 | 10.64 | 4 | 0.031 | −84.09 | −84.99 | 1.80 |
| Lotic system | −76.54 | −79.75 | 6.42 | 4 | 0.170 | −87.92 | −87.82 | −0.20 | |
| Arboreal oviposition | Lentic system | −69.95 | −73.51 | 7.12 | 4 | 0.130 | −80.11 | −80.02 | −0.18 |
| Lotic system | −75.20 | −77.02 | 3.64 | 4 | 0.457 | −85.33 | −86.96 | 3.26 | |
| Terrestrial oviposition | Lentic system | −52.62 | −57.69 | 10.14 | 4 | 0.038 | −70.33 | −71.19 | 1.72 |
| Lotic system | −59.05 | −61.23 | 4.36 | 4 | 0.359 | −75.97 | −77.31 | 2.68 | |
| Arboreal ecology | Lentic system | −63.91 | −71.03 | 14.24 | 4 | 0.007 | −76.49 | −80.17 | 7.36 |
| Lotic system | −70.15 | −73.89 | .48 | 4 | 0.113 | −81.80 | −82.02 | 0.44 | |
| Terrestrial ecology | Lentic system | −63.71 | −68.13 | 8.84 | 4 | 0.065 | −70.91 | −71.23 | 0.64 |
| Lotic system | −57.40 | −64.58 | 14.36 | 4 | 0.006 | −76.92 | −77.17 | 0.50 | |
Model values are log‐likelihood values for maximum‐likelihood analyses, whereas Bayesian models are marginal likelihoods calculated by stepping stone sampling. Significant values, for which the dependent model of evolution is significantly better than the independent model, are expressed in bold (P‐value < 0.01, logBF > 5).
Discussion
EVOLUTION OF DIRECT DEVELOPMENT
Among the 40 described reproductive modes for anurans, the most commonly observed mode is the plesiomorphic state involving aquatic oviposition followed by an exotrophic free‐swimming tadpole stage (Duellman 1985), a condition recovered as the basal state for a majority of frog families (Gomez‐Mestre et al. 2012). Our phylogenetic model testing suggests the common ancestor of the 400 species of Afrobatrachians did not possess this condition, but rather had a reproductive mode of terrestrial eggs and a free‐swimming exotrophic tadpole (Fig. 2). We find the presence of direct development in Brevicipitidae and Arthroleptis is therefore best explained by two independent origins. Conversely, the terrestrialized reproductive mode present in hemisotids and a majority of arthroleptids can be explained by the retention of the ancestral state of terrestrial eggs with aquatic larvae, rather than convergence (Fig. 2). Though we had no temporal predictions for the evolution of direct development, this trait appeared much earlier in brevicipitids (31.9–48.4 Ma) than in the genus Arthroleptis (13.2–22.0 Ma). In both groups there is long‐term retention of direct development, with no evidence for subsequent transitions back to a more aquatic reproductive mode. We also find that direct development has evolved from modes involving subterranean or terrestrial oviposition, but not from arboreal oviposition. Gomez‐Mestre et al. (2012) recover an ancestral state of the fully aquatic reproductive mode for Afrobatrachians, which can be attributed to differences in phylogenetic topology, taxon sampling, outgroup rooting, and characters analyzed. However, their result also supports our conclusions, as direct development is not inferred as the ancestral state for this group.
The “intermediate stages” hypothesis for anuran reproductive modes posits that the evolution of direct development should proceed in a sequence through prior stages involving progressively more terrestrialized characteristics, though not requiring a linear trajectory through all possible reproductive modes (Salthe and Duellman 1973; Duellman and Trueb 1986; Wells 2007; Crump 2015). In their study, Gomez‐Mestre et al. (2012) found evidence that direct development evolved from the fully aquatic mode nearly as many times as modes involving terrestrial eggs, challenging this hypothesis. Here, we find the independent evolution of direct development in Brevicipitidae and the genus Arthroleptis supports the “intermediate stages” hypothesis, as the sister taxa of these two groups (Hemisotidae and Cardioglossa, respectively) each exhibit a terrestrialized reproductive mode (Fig. 2). In addition to our work, the evolution of complete terrestrial development has recently been explored with improved sampling across other anuran clades. Meegaskumbura et al. (2015) convincingly demonstrate that direct development has evolved at least twice in rhacophorid tree frogs, and in both cases from terrestrialized reproductive modes involving arboreal eggs and aquatic larvae. Pereira et al. (2015) examined the evolution of reproductive modes in leptodactyline frogs, and concluded this group does not exhibit gradual evolution toward a fully terrestrial reproductive mode. The authors determined complete terrestrial development arises from an aquatic state; however, the aquatic state referred to involves the creation of a floating foam nest that removes the eggs from the water. This represents a terrestrialized reproductive mode, and because this mode gave rise to complete terrestrial development rather than the fully aquatic reproductive mode, we conclude their results are consistent with the “intermediate stages” hypothesis.
The detection of transitions from the fully aquatic reproductive mode to direct development may be an artifact resulting from the failure to observe intermediate states. The lack of intermediate states can stem from coarse phylogenetic or phenotypic resolution, as previously highlighted by Meegaskumbura et al. (2015) and noted by Gomez‐Mestre et al. (2012). This issue can be ameliorated through fine‐scale phylogenetic and phenotypic sampling of a clade, which decreases the potential for errors in ancestral character estimation. Cases involving transitions from fully aquatic breeding to direct development in rhacophorids (Philautus; Meegaskumbura et al. 2015) and in Afrobatrachians (Arthroleptis; this study) appear to be artifacts, which have been resolved through improved phylogenetic reconstructions and accurate character assignments. As the availability of species sampling and reproductive mode information improves for other clades containing direct developers, we expect support for the intermediate stages hypothesis will continue to grow.
PATTERNS OF WATER SYSTEM USE
The terrestrialization of anuran breeding strategies may be a result of selective pressures from flowing water associated with lotic ecosystems (Alcala 1962; Goin and Goin 1962). This is an alternative to the hypothesis that aquatic predation in the egg and larval stage is the dominant selective pressure (Tihen 1960; Poynton 1964; Magnusson and Hero 1991; Haddad and Prado 2005). Although initially characterizing montane systems (where flow is typically faster), there are a variety of lotic habitats in lowlands that present similar challenges for egg deposition, fertilization, and tadpole development. If the use of lotic habitats requires some degree of reproductive terrestrialization, these patterns should emerge through broad scale comparisons of the evolution of reproductive mode and water system preference, regardless of elevation. Based on our analyses of the coevolution of reproductive traits and water systems, we did not find support for associations between terrestrialized oviposition sites and lotic systems (Table 4, Fig. 2). However, we caution against a strict interpretation of our results because Afrobatrachians disproportionately exhibit terrestrialized reproductive modes. To more appropriately test this hypothesis, all African anurans should be included, many of which exhibit the fully aquatic reproductive mode that is proposed as problematic for breeding in lotic habitats. Such a test would determine the relationship between lotic systems and oviposition type, including whether species associated with lotic systems disproportionately possess non‐aquatic oviposition.
We detected a strong association between the evolution of arboreality and use of lentic water systems for breeding (Table 4). This evolves convergently and is largely consistent across Afrobatrachians regardless of oviposition sites (Fig. 2). The availability of suitable habitat for breeding behavior such as calling and oviposition may provide an explanation for this pattern. Many arboreal species call from emergent vegetation in ponds, which for hyperoliids are often the same sites used to deposit eggs overhanging the water (Schiøtz 1999; Channing 2001; Channing and Howell 2006; Amiet 2012). This emergent vegetation is typically more prevalent in lentic systems and absent in the waters of swiftly moving streams. Another potential explanation for this relationship is the high proportion of terrestrialized oviposition sites used by arboreal hyperoliid species, which may be more advantageous in lentic systems. This indirect relationship could be driven by selective pressures for nonaquatic oviposition, such as the high level of aquatic predation of the eggs or young larvae occurring in pond systems (Lutz 1948; Crump 1974; Magnusson and Hero 1991; Haddad and Prado 2005). In addition to reducing predation on egg masses, the delay in hatching from non‐aquatic oviposition sites often produces larger tadpoles (Salthe and Duellman 1973; Duellman and Trueb 1986), which may further reduce predation pressure. Together, the combination of habitat structure for reproductive behaviors and larval predation pressures may drive the shift to lentic breeding in arboreal Afrobatrachian species. This association has not been previously demonstrated in frogs. Further work incorporating other arboreal anuran lineages can clarify whether there is a more general adaptive relationship between arboreality and breeding in lentic water systems, including identifying specific selective factors.
Beyond ecology and reproductive traits, we also observed family level differences in water system use. Our results indicate arthroleptids are predominately using lotic systems (with the exception of Leptopelis), whereas hyperoliids are primarily using lentic systems (Fig. 2). These preferences, in combination with additional divergences in oviposition site, essentially limit the competition for spatially distributed reproductive resources at the clade level (Fig. 2). This spatial reproductive partitioning may partially explain the high numbers of sympatric arthroleptid and hyperoliid species found across the Afrotropics.
ECOMORPHOLOGY AND REPRODUCTIVE TRAITS
Ecomorphological differentiation can increase reproductive mode diversity if morphological adaptations allowing shifts to novel microhabitats are accompanied by changes in reproductive traits involving spatial resources. Frog reproductive mode diversity is largely driven by the location of egg deposition, which is a combination of oviposition site and water source (Duellman and Trueb 1986; Haddad and Prado 2005; Crump 2015). Access to oviposition sites located in particular microhabitats may be a direct consequence of ecomorphology, and here we find strong evidence for associations between arboreality and arboreal oviposition and between fossoriality and subterranean oviposition (Fig. 2, Table 4). We also find support for a link between arboreality and the use of lentic water systems for breeding, but detect no relationship between ecomorphology or terrestrialized oviposition and lotic water systems (Table 4).
The reproductive diversity of Afrobatrachians is enhanced by the presence of fossorial and arboreal lineages, which are exploiting novel microhabitats for oviposition. These arboreal or subterranean sites are not used by any terrestrial Afrobatrachian lineages, suggesting site access is ecomorph‐dependent. All fossorial Afrobatrachian lineages exhibit subterranean oviposition, which produces a pattern of replicated evolution of these states across lineages (Fig. 2). The arboreal Afrobatrachian lineages exhibit greater variation in oviposition site, including terrestrial, aquatic, and arboreal sites. Here, the trait association appears to be driven by the origin and retention of arboreal oviposition in the derived hyperoliids (Fig. 2). Other arboreal groups retain the plesiomorphic oviposition site of their common ancestor, including the two transitions to arboreality in the kassinoids (genus Phlyctimantis, Kassina arboricola) and the arthroleptid genus Leptopelis. Arboreality is not always accompanied by the evolution of arboreal oviposition, but there are no examples of terrestrial or fossorial Afrobatrachian lineages with arboreal oviposition. In this manner, arboreality appears to be a precursor to arboreal oviposition in this group, and access to arboreal oviposition sites is limited to this ecomorph.
Our work demonstrates that the evolution of distinct Afrobatrachian adult ecomorphs allowed transitions to novel oviposition sites. However, the role of ecomorphology in the overall evolution of reproductive traits across all frogs requires further study. Exploring the relationship between morphological diversity and reproductive mode diversity may help to explain the repeated evolution of particular terrestrialized reproductive traits. Key topics for future study include whether increased diversity of adult ecomorphs in a clade is positively related to increased reproductive mode diversity, as well as whether specific ecomorphologies might promote or constrain the evolution of particular reproductive traits, especially those that are spatially distributed in habitats.
Associate Editor: M. Alfaro
Handling Editor: M. Servedio
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Figure S1. A summary of z‐scores for transitions between character sets.
Table S1. GenBank numbers of published sequences used in molecular analyses.
Table S2. List of ecological and reproductive characters associated with taxa.
Supporting File 3. Summary of taxonomic recommendations.
Supporting File 4. Time‐calibrated tree of nucDNA‐only data set.
Supporting FIle 5. Time‐calibrated tree of nucDNA and mtDNA data set.
ACKNOWLEDGMENTS
Fieldwork and lab work were supported by National Science Foundation grant DEB‐1202609 to DCB, and under the approval of the Institutional Animal Care and Use Committee (2014‐2) at the California Academy of Sciences. The Cameroon Ministry of Forests and Wildlife (MINFOF) and Ministry of Scientific Research and Innovation (MINRESI) provided necessary permits for conducting research and exportation. Additional tissue samples were provided by M.‐O. Rödel, M. Hirschfeld, and T. Colston. This article benefitted greatly from discussions with J. A. McGuire, A. Corl, R. C. Bell, G. Jongsma, H. C. Liedtke, and the McGuire lab group. We thank M. Alfaro, R. A. Pyron, and F. T. Burbrink for comments that greatly improved the quality of this article.
DATA ARCHIVING
We have archived several scripts and data sets on the Dryad Digital Repository: doi 10.5061/dryad.sh0h0.
Table A1.
List of species and associated catalog or field numbers included for multilocus molecular data collection
| Family | Genus | Species | Museum number |
|---|---|---|---|
| Arthroleptidae | Arthroleptis | schubotzi | CAS 250728 |
| sylvaticus | MH0314 | ||
| variabilis | CAS 207824 | ||
| Astylosternus | laticephalus | MVZ 244909 | |
| Cardioglossa | gracilis | NCSM 78888 | |
| pulchra | 0951N | ||
| leucomystax | MK067 | ||
| Leptodactylodon | bicolor | MCZ A 139599 | |
| bueanus | MCZ A 137970 | ||
| Leptopelis | bocagii | CAS 250770 | |
| brevirostris | CAS 207830 | ||
| nordequatorialis | MCZ A 139611 | ||
| notatus | CAS 253554 | ||
| parkeri | CAS 168787 | ||
| Nyctibates | corrugatus | MCZ A 136788 | |
| Scotobleps | gabonicus | MH0406 | |
| Trichobatrachus | robustus | AMCC 117634 | |
| Brevicipitidae | Breviceps | macrops | CAS 193965 |
| Callulina | kreffti | CAS 168715 | |
| Hemisotidae | Hemisus | marmoratus | MVZ 244947 |
| Hyperoliidae | Acanthixalus | sonjae | MORAS2 |
| Afrixalus | brachynemis | MVZ 265821 | |
| dorsalis | CAS 253854 | ||
| lacteus | CAS 253843 | ||
| laevis | CAS 253803 | ||
| paradorsalis | CAS 249943 | ||
| quadrivittatus | MCZ A 138087 | ||
| Chlorolius | koehleri | MHO 536 | |
| Cryptothylax | greshoffii | MVZ 234714 | |
| Heterixalus | alboguttatus | MVZ 241451 | |
| Hyperolius | adspersus | CAS 254256 | |
| balfouri | CAS 253643 | ||
| concolor | CAS 253869 | ||
| fusciventris | CAS 254006 | ||
| ocellatus | CAS 254075 | ||
| parkeri | FMNH 274372 | ||
| parkeri | MVZ 233914 | ||
| picturatus | UWBM 5723 | ||
| pusillus | MVZ 233894 | ||
| substriatus | MVZ 265976 | ||
| tuberculatus | CAS 207717 | ||
| Kassina | arboricola | UWBM 5746 | |
| decorata | MCZ FS 34409 | ||
| maculata | SAIAB 88587 | ||
| senegalensis | MVZ 234142 | ||
| Morerella | cyanophthalma | Ba04.3 | |
| Opisthothylax | immaculatus | MVZ 234815 | |
| Paracassina | kounhiensis | TJC 869 | |
| Phlyctimantis | boulengeri | ADL 3890 | |
| leonardi | CAS 253978 | ||
| Semnodactylus | wealli | SANBI 4627 | |
| Tachycnemis | seychellensis | ||
| Microhylidae | Phrynomantis | microps | MVZ 249480 |
LITERATURE CITED
- Alcala, A. C. 1962. Breeding behavior and early development of frogs of Negros, Phillipine Islands. Copeia 1962:679–726. [Google Scholar]
- Ali J. R., and Huber M.. 2010. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 463:653–680. [DOI] [PubMed] [Google Scholar]
- Aljanabi, S. , and Martinez I.. 1997. Universal and rapid salt‐extraction of high quality genomic DNA for PCR‐based techniques. Nucl. Acids Res. 25:4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altig, R. , and Crother B. I.. 2006. The evolution of three deviations from the biphasic anuran life cycle: alternatives to selection. Herpetol. Rev. 37:321–325. [Google Scholar]
- Altig, R. , and McDiarmid R. W.. 2007. Morphological diversity and evolution of egg and clutch structure in amphibians. Herpetol. Mono. 27:1–32. [Google Scholar]
- Amiet, J.‐L. 1972. Le Cardioglossa camerounaises.‐Science et Nature, Paris 114:11–24. [Google Scholar]
- Amiet, J.‐L. 2012. Les Rainettes du Cameroun (Amphibiens Anoures). La Nef des Livres, Saint‐Nazaire, France. [Google Scholar]
- AmphibiaWeb . 2015. Information on amphibian biology and conservation. Berkeley, CA: Available at http://www.amphibiaweb.org/. Accessed September 2015. [Google Scholar]
- Boulenger, G. A. 1886. Remarks in connection with the preceding note. Ann. Mag. Nat. Hist. 17:463–464. [Google Scholar]
- Blackburn, D. C. 2008. Biogeography and evolution of body size and life history of African frogs: phylogeny of squeakers (Arthroleptis) and long‐fingered frogs (Cardioglossa) estimated from mitochondrial data. Mol. Phylogenet. Evol. 49:806–826. [DOI] [PubMed] [Google Scholar]
- Bogart, J. P. , and Tandy M.. 1981. Chromosome lineages in African ranoid frogs. Monitore Zoologico Italiano. Nuova Serie, Supplemento. Firenze 15:55–91. [Google Scholar]
- Bossuyt, F. , and Milankovitch M. C.. Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. P. Natl. Acad. Sci. USA 97:6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt, F. , Brown R. F., Hillis D. M., Cannatella D. C., and Milinkovitch M. C.. 2006. Phylogeny and biogeography of a cosmopolitan frog radiation: Late Cretaceous diversification resulted in continent‐scale endemism in the family Ranidae. Syst. Biol. 55:579–594. [DOI] [PubMed] [Google Scholar]
- Channing, A. 2001. Amphibians of Central and Southern Africa. Cornell University Press, Ithaca, NY. [Google Scholar]
- Channing, A. , and Howell K. M.. 2006. Amphibians of East Africa. Edition Chimaira, Frankfurt, Germany. [Google Scholar]
- Crump, M. L. 1974. Reproductive strategies in a tropical anuran community. Univ. Kans. Mus. Nat. Hist. Misc. Publ. 61:1–68. [Google Scholar]
- Crump, M. L. 2015. Anuran reproductive modes: evolving perspectives. J. Herpetol. 49:1–16. [Google Scholar]
- da Silva, F. R. , Almeida‐Neto M., do Prado V. H. M., Haddad C. F. B., and Rossa‐Feres D. C.. 2012. Humidity levels drive reproductive modes and phylogenetic diversity of amphibians in the Brazilian Atlantic forest. J. Biogeogr. 39:1720–1732. [Google Scholar]
- Drummond, A. J. , Suchard M. A., Xie D., and Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duellman, W. E. 1985. Reproductive modes in anuran amphibians: phylogenetic significance of adaptive strategies. S. Afr. J. Sci. 81:174–178. [Google Scholar]
- Duellman, W. E. 2007. Amphibian life histories: their utilization in phylogeny and classification Pp. 2843–2892 in Heatwole H. and Tyler M. J., eds. Amphibian biology. Vol. 7, systematics. Surrey Beatty and Sons, NSW, Australia. [Google Scholar]
- Duellman, W. E. , and Trueb L.. 1986. Biology of amphibians. McGraw‐Hill. New York, USA. [Google Scholar]
- Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzSimons, V. , and Van Dam G.. 1929. Some observations on the breeding habitats of Breviceps . Ann. Transvaal Mus. 13:152–153. [Google Scholar]
- Frost, D. R. , Grant T., Faivovich J., Bain R. H., Haas A., Haddad C. F. B., de Sa R. O., Channing A., Wilkinson M., Donnellan S. C., et al. 2006. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 297:1–370. [Google Scholar]
- Frost, D. R. 2015. Amphibian Species of the World: an Online Reference. Version 6.0. American Museum of Natural History, New York. Available from: http://research.amnh.org/vz/herpetology/amphibia. Accessed September 2015.
- Goin, O. B. , and Goin C. J.. 1962. Amphibian eggs and the montane environment. Evolution 16:364–371. [Google Scholar]
- Gomez‐Mestre, I. , Pyron R. A., and Wiens J. J.. 2012. Phylogenetic analyses reveal unexpected patterns in the evolution of reproductive modes in frogs. Evolution 66:3687–3700. [DOI] [PubMed] [Google Scholar]
- Guibé, P. J. and Lamotte M.. 1958. Morphologie et reproduction par développement direct d'un anoure du Mont Nimba, Arthroleptis crusculum Angel. Bull. Mus. Natl. Hist. Nat. 2:125–133. [Google Scholar]
- Haddad, C. F. B. , and Prado C. P. A.. 2005. Reproductive modes in frogs and their unexpected diversity in the Atlantic forest of Brazil. BioScience 55:207–217. [Google Scholar]
- Huelsenbeck, J. P. , and Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. [DOI] [PubMed] [Google Scholar]
- Iskandar, D. T. , Evans B. J., and McGuire J. A.. 2014. A novel reproductive mode in frogs: a new species of fanged frog with internal fertilization and birth of tadpoles. PLoS One 10(3):e0119988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir R., Wilson A., Stones‐Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi, A. , and Sumida M.. 2013. Afrobatrachian mitochondrial genomes: genome reorganization, gene rearrangement mechanisms, and evolutionary trends of duplicated and rearranged genes. BMC Genomics 14:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R. , Calcott B., Ho S. Y. W., and Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29:1695–701. [DOI] [PubMed] [Google Scholar]
- Laurent, R. F. 1941. Contribution à l'ostéologie et à la systématique des rhacophorides africains. Revue de Zoologie et de Botanique Africaines. Tervuren 35:85–111. [Google Scholar]
- Lawson, L. P. , and Collett L.. 2011. Herpetofauna of montane areas of Tanzania. 1. Results from two amphibian surveys of Malundwe Mountain, Mikumi National Park. Fieldiana Life Earth Sci. 4:74–80. [Google Scholar]
- Loader, S. P. , Ceccarelli F. S., Menegon M., Howell K. M., Kassahun R., Mengistu A. A., Saber S. A., Gebresenbet F., de Sa R., Davenport T. R. B., et al. 2014. Persistence and stability of Eastern Afromontane forests: evidence from brevicipitid frogs. J. Biogeogr. 41:1781–1792. [Google Scholar]
- Lutz, B. 1947. Trends towards non‐aquatic and direct development in frogs. Copeia 1947:242–252. [Google Scholar]
- Lutz, B. 1948. Ontogenetic evolution in frogs. Evolution 2:29–39. [DOI] [PubMed] [Google Scholar]
- Magnusson, W. E. , and Hero J.‐M.. 1991. Predation and the evolution of complex oviposition behavior in Amazonian rainforest frogs. Oecologica 86:10–18. [DOI] [PubMed] [Google Scholar]
- Meegaskumbura, M. , Senevirathne G., Biju S. D., Garg S., Meegaskumbura S., Pethiyagoda R., Hanken J., and Schneider C. J.. 2015. Patterns of reproductive‐mode evolution in Old World tree frogs (Anura, Rhacophoridae). Zool. Scr. 44:509–522. [Google Scholar]
- Müller, H. , Liedtke H. C., Menegon M., Beck J., Ballesteros‐Mejia L., Nagel P., and Loader S. P.. 2013. Forests as promoters of terrestrial life‐history strategies in East African amphibians. Biol. Lett. 9:20121146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel, M. , and Meade A.. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible‐jump Markov chain Monte Carlo. Am. Nat. 167:808–825. [DOI] [PubMed] [Google Scholar]
- Pagel, M. , Meade A., and Barker D.. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53:673–684. [DOI] [PubMed] [Google Scholar]
- Paradis, E. , Claude J., and Strimmer K.. 2004. APE: analysis of phylogenetics and evolution in R language. Bioinformatics 20:289–290. [DOI] [PubMed] [Google Scholar]
- Pereira, E. B. , Collevatti R. G., Kokubum M. N. C., Miranda N. E. O., and Maciel N. M.. 2015. Ancestral reconstruction of reproductive traits shows no tendency toward terrestriality in leptodactyline frogs. BMC Evol. Biol. 15:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynton, J. C. 1964. Relationships between habitat and terrestrial breeding in amphibians. Evolution 18:131. [Google Scholar]
- Portik, D.M. , and Blackburn D.C.. 2016. Data from: The evolution of reproductive diversity in Afrobatrachia: A phylogenetic comparative analysis of an extensive radiation of African frogs Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.sh0h0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, C. P. A. , Uetanabaro M., and Haddad C. F. B.. 2002. Description of a new reproductive mode in Leptodactylus (Anura, Leptodactylidae), with a review of the reproductive specialization towards terrestriality in the genus. Copeia 2002:1128–1133. [Google Scholar]
- Pyron, R. A. 2014. Biogeographic analysis reveals ancient continental vicariance and recent oceanic dispersal in amphibians. Syst. Biol. 63:779–797. [DOI] [PubMed] [Google Scholar]
- Pyron, R. A. , and Wiens J. J.. 2011. A large‐scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61:543–583. [DOI] [PubMed] [Google Scholar]
- Rambaut, A. , and Drummond A. J.. 2009. Tracer v1.5.0 and higher. Available at http://beast.bio.ed.ac.uk.
- Revell, L. J. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3:217–223. [Google Scholar]
- Roelants, K. , Gower D. J., Wilkinson M., Loader S. P., Biju S. D., Guillaume K., Moriau L., and Bossuyt F.. 2007. Global patterns of diversification in the history of modern amphibians. Proc. Natl. Acad. Sci. USA 104: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F. , and Huelsenbeck J. P.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Salthe, S. N. , and Duellman W. E.. 1973. Quantitative constraints associated and reproductive mode in anurans Pp. 229–249 in Vial J. L., ed., Evolutionary biology of the anurans. Univ. of Missouri Press, Missouri, USA. [Google Scholar]
- Samonds, K. E. , Godfrey L. R., Ali J. R., Goodman S. M., Vences M., Sutherland M. R., Irwin M. T., and Krause D. W.. 2012. Spatial and temporal arrival patterns of Madagascar's vertebrate fauna explained by distance, ocean currents, and ancestor type. Proc. Natl. Acad. Sci. USA 109:5352–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. X. , Liang D., Feng Y. J., Chen M. Y., and Zhang P.. 2013. A versatile and highly efficient toolkit including 102 nuclear markers for vertebrate phylogenomics, tested by resolving the higher level relationships of the Caudata. Mol. Biol. Evol. 30:2235–2248. [DOI] [PubMed] [Google Scholar]
- Schiøtz, A. 1999. Treefrogs of Africa. Edition Chimaira, Frankfurt, Germany. [Google Scholar]
- Smith, S. A. , Stephens P. R., and Wiens J. J.. 2005. Replicate patterns of species richness, historical biogeography, and phylogeny in Holarctic treefrogs. Evolution 59:2433–2450. [PubMed] [Google Scholar]
- Tihen, J. A. 1960. Comments on the origin of the amniote egg. Evolution 14:528–531. [Google Scholar]
- Todd, B. D. 2007. Parasites lost? An overlooked hypothesis for the evolution of alternative reproductive strategies in amphibians. Am. Nat. 170:7793–799. [DOI] [PubMed] [Google Scholar]
- Tolley, K. A. , Townsend T. M., and Vences M.. 2013. Large‐scale phylogeny of chameleons suggests African origins and Eocene diversification. Proc. Biol. Sci. 280:20130184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager, V. A. 1965. The frogs of South Africa. Purnell and Sons, Cape Town, South Africa. [Google Scholar]
- Wager, V. A. 1986. Frogs of South Africa, their fascinating life stories. Delta Books, Johannesburg, South Africa. [Google Scholar]
- Wiens, J. J. , Fetzner J. W. Jr., Parkinson C. L., and Reeder T. W.. 2005. Hylid frog phylogeny and sampling strategies for speciose clades. Systematic Biology 54:719–748. [DOI] [PubMed] [Google Scholar]
- Wells, K. D. 2007. The ecology and behavior of amphibians. Univ. of Chicago Press, Illinois, USA. [Google Scholar]
- Zwickl, D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. thesis, The University of Texas at Austin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Figure S1. A summary of z‐scores for transitions between character sets.
Table S1. GenBank numbers of published sequences used in molecular analyses.
Table S2. List of ecological and reproductive characters associated with taxa.
Supporting File 3. Summary of taxonomic recommendations.
Supporting File 4. Time‐calibrated tree of nucDNA‐only data set.
Supporting FIle 5. Time‐calibrated tree of nucDNA and mtDNA data set.


