Abstract
We propose a joint model to analyze the structure and intensity of the association between longitudinal measurements of an ordinal marker and time to a relevant event. The longitudinal process is defined in terms of a proportional‐odds cumulative logit model. Time‐to‐event is modeled through a left‐truncated proportional‐hazards model, which incorporates information of the longitudinal marker as well as baseline covariates. Both longitudinal and survival processes are connected by means of a common vector of random effects.
General inferences are discussed under the Bayesian approach and include the posterior distribution of the probabilities associated to each longitudinal category and the assessment of the impact of the baseline covariates and the longitudinal marker on the hazard function. The flexibility provided by the joint model makes possible to dynamically estimate individual event‐free probabilities and predict future longitudinal marker values.
The model is applied to the assessment of breast cancer risk in women attending a population‐based screening program. The longitudinal ordinal marker is mammographic breast density measured with the Breast Imaging Reporting and Data System (BI‐RADS) scale in biennial screening exams. © 2016 The Authors. Statistics in Medicine Published by John Wiley & Sons Ltd.
Keywords: BI‐RADS scale, Latent process, left‐truncated proportional‐hazards model, Proportional‐odds cumulative logit model
1. Introduction
The current evidence on benefits and harms supports the personalization of screening as a crucial step to improve early detection of breast cancer 1, 2. A number of risk models were designed to measure the individual probability of developing breast cancer 3, 4, 5. In the context of individualized breast cancer screening, the utility of these risk models has been questioned because of their low discrimination power 6. The inclusion of a baseline measure of breast density – a characteristic of the breast tissue – in the risk models improved the accuracy of the breast cancer risk estimate 7, 8, 9, 10.
Several studies have shown that high breast density is associated with increased breast cancer risk 7, 11, 12, 13, 14, with risk estimates in the range four‐fold to six‐fold for women with very high breast density compared with women with low breast density 11, 12. Other studies have examined whether changes in breast density over time are associated to changes in breast cancer risk 15, 16, 17, 18, 19, 20, 21 and have suggested that monitoring changes in breast density could help to identify women at greater risk of disease. In most of the cases, the statistical methods used did not account for relevant characteristics of prospective studies like non‐ignorable dropout mechanisms or internal time‐dependent covariates 22.
Joint modeling of longitudinal and time‐to‐event data is an increasingly productive area of statistical research that assesses the association between longitudinal and survival processes. It enhances longitudinal modeling by allowing for the inclusion of non‐ignorable dropout mechanisms and survival modeling by the inclusion of internal time‐dependent covariates 22. Joint models were introduced during the 90s 23, 24, 25 and since then, have been applied to a great variety of studies in epidemiological and biomedical areas. Shared‐parameter models are a type of joint models where the longitudinal and time‐to‐event processes are connected by means of a common set of subject‐specific random effects. These models make possible to quantify both the population and individual effects of the underlying longitudinal outcome on the risk of an event and obtain individualized time‐dynamic predictions. Recently, Rizopoulos proposed an overview of the theory and applications of joint modeling 26 and developed the JM 27 and JMbayes 28 R packages for the frequentist and Bayesian shared‐effects' approaches, respectively. Serrat et al. illustrate the application of both statistical approaches to joint modeling longitudinal measures of prostate specific antigen and prostate cancer detection in men participating in a screening trial 29.
When longitudinal outcomes are ordinal, joint models become more complex. Different approaches, that use constraints in the probabilities of the categorical outcomes or the discretization of a continuous latent variable, have been proposed 30, 31, 32, 33. The non‐linear and longitudinal nature of the data produce a complex likelihood function, difficult to maximize under the frequentist paradigm. This could be a reason why the standard software for joint models does not include longitudinal ordinal variables yet. Some relevant works on the subject use the frequentist 34, 35, 36 and Bayesian 33, 37, 38 approaches, respectively.
The objective of this paper is to propose a Bayesian joint model for assessing the structure and intensity of the association between longitudinal measures of an ordinal marker and a time‐to‐event outcome. In particular, we use a proportional‐odds cumulative logit model 30 for the ordinal measurements and a proportional hazard model with left‐truncation for the time to an event of interest. We have applied the model to analyze the risk of breast cancer in women attending a population‐based screening program with regard to repeated measurements of mammographic breast density.
Section 2 presents a description of the motivating dataset. Section 3 formulates the joint model and discusses general inferences for (1) dynamic probabilities associated to the different ordinal categories, (2) the impact of baseline covariates and the longitudinal marker on the hazard function, (3) dynamic estimation of survival probabilities, and (4) prediction of future longitudinal outcomes. Section 4 applies the model developed in Section 3 to study age at diagnosis of breast cancer in women who participate in a population‐based screening program. Finally, Section 5 contains a discussion and some conclusions.
2. Motivating data
2.1. Study design and study population
This is an observational prospective study including 13,760 women that participated for the first time in the breast cancer early‐detection program in the Vallès Occidental Est (BCEDP‐VOE) area in Catalonia (Spain), between October 1995 and June 1998. The BCEDP‐VOE invites women aged 50–69 years for biennial mammographic exams. At study entry, the participants were 50–70 years old and did not have a personal history of breast cancer. They were followed for vital status or possible diagnosis of breast cancer until December 2013 39, 40, 41.
Of the initial 13,760 women, we excluded seven without follow‐up data, as well as 38 women who were diagnosed with breast cancer and nine who died within six months of baseline. Twenty‐one women were also excluded for not having breast density measurements within the 50–70 age interval. We analyzed invasive breast cancer and ductal carcinoma in situ diagnosed during follow‐up. The final sample included 13,685 women, with 431 diagnosed with breast cancer.
2.2. Variables and data description
At the first mammographic exam, the study participants answered a questionnaire that included information on family history of breast cancer, prior breast procedures, age at menarche, age at first birth, and menopausal status. Family history refers to absence/presence of first‐degree relatives with breast cancer. Prior breast procedures included breast biopsy, fine needle aspiration, cyst aspiration, breast reconstruction, lumpectomy, and surgical treatment.
Breast density is a characteristic of the breast tissue that is reflected in mammograms. Breasts are considered dense if the connective and epithelial tissues predominate over the fatty tissue. At all mammographic exams, breast density was rated and recorded according to the BI‐RADS system 40, 42 that categorizes breast density in four groups: a, almost entirely fatty (low density); b, scattered fibroglandular densities (medium density); c, heterogeneously dense (high density); and d, extremely dense (very high density). This longitudinal breast density data is a remarkable and unique characteristic of the BCEDP‐VOE among the breast cancer screening programs in Spain. Breast density measures within 6 months before breast cancer diagnosis could be affected by the presence of preclinical breast cancer; therefore, they were excluded. The mammographic exams performed before age 50 or after age 70 were excluded in order to avoid sample biases. A total of 81,621 measures of breast density were included in the longitudinal analysis, with median [range] 4 [1 to 9] and 6 [1 to 15] measures for women with or without breast cancer diagnosis, respectively.
We considered that the time origin for the event of interest (diagnosis of breast cancer) was age 50 years, the lower limit of the screening age interval. We defined the time‐to‐event as the time elapsed from the origin to diagnosis of breast cancer. For women without a breast cancer diagnosis at the study end, the censoring time was obtained as the minimum of time to death and time to the last screening exam plus 2.5 years that correspond to the active follow‐up for cancer identification. It is important to remark that women who entered the program over 50 years had delayed entry times that may induce length biased sampling or left truncation.
Among 13,685 women aged 50 years and older, 431 developed breast cancer – 336 invasive cancers and 95 ductal carcinoma in situ –, and 513 died within 2.5 years of the last mammogram. Median follow‐up was 12.7 years for women without breast cancer and 8.2 years for women with breast cancer. Table 1 shows the baseline characteristics of women and the breast density measurements at first and last examination according to their breast cancer diagnosis status at the end of follow‐up. High breast density categories were more prevalent among women with breast cancer, in both the first and the last mammogram. Furthermore, between the first and last exams, the prevalence of low density categories increased, as described in the literature.
Table 1.
Baseline risk factors according to breast cancer diagnosis status at the end of follow‐up.
| No breast cancer | Breast cancer | |||
|---|---|---|---|---|
| N = 13,254 (%) | N = 431 (%) | |||
| Family history of breast cancer | ||||
| No | 12,539 | (94.8) | 388 | (90.2) |
| Yes | 686 | (5.2) | 42 | (9.8) |
| Prior breast procedures | ||||
| No | 12,318 | (92.9) | 374 | (86.8) |
| Yes | 936 | (7.1) | 57 | (13.2) |
| Breast density at first examination | ||||
| (baseline breast density) | ||||
| a: Almost entirely fatty | 2959 | (23.4) | 56 | (13.9) |
| b: Scattered fibroglandular densities | 5353 | (42.3) | 138 | (34.2) |
| c: Heterogeneously dense | 2301 | (18.2) | 103 | (25.6) |
| d: Extremely dense | 2037 | (16.1) | 106 | (26.3) |
| Breast density at last examinationa | ||||
| (women with at least two examinations) | ||||
| a: Almost entirely fatty | 2284 | (18.1) | 35 | (9.4) |
| b: Scattered fibroglandular densities | 7957 | (63.0) | 201 | (54.0) |
| c: Heterogeneously dense | 1475 | (11.7) | 71 | (19.1) |
| d: Extremely dense | 919 | (7.3) | 65 | (17.5) |
If breast cancer was diagnosed within 6 months following the last mammography, the last breast density considered was the previous one, whenever it was not coincident with the baseline measure.
To illustrate the longitudinal breast density measurements, we randomly selected eight women without and eight women with cancer (Figure 1). A high variability of the breast density trajectories can be observed: some women experience an increase of breast density, while others remain stable, fluctuate, or experience a decrease. The plots show the biennial periodicity of the screening exams, as well as the unbalanced number of measures between women, because of different reasons. Not all women entered the study at the same age, not all the scheduled screening exams were taken, or not always breast density was rated.
Figure 1.
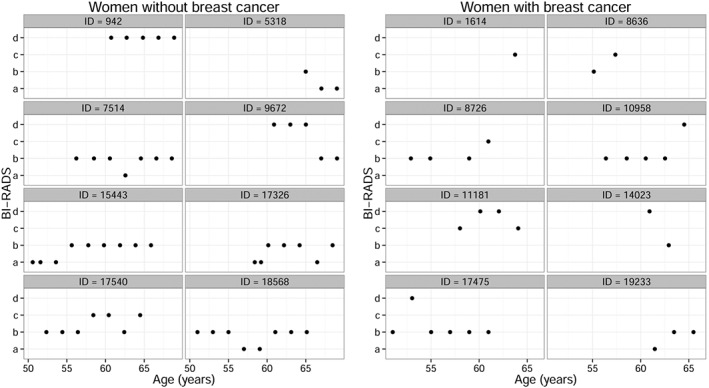
Subject‐specific profiles of BI‐RADS measures for 16 randomly selected women. The left panel corresponds to eight women without breast cancer, and the right panel corresponds to eight women with breast cancer.
3. The joint model
We propose a model with two submodels: (1) a proportional‐odds cumulative logit model for the longitudinal ordinal measurements based on the idea of a continuous latent variable 30, 33 and (2) a left‐truncated proportional hazard model for the time‐to‐event, which incorporates information from the longitudinal process. Both processes are connected through a shared vector of random effects, which, in the presence of covariates and parameters, endows them with conditional independence 26.
Let {D 1,D 2,…,D K} denote the set of ordinal categories and y ij the longitudinal category of individual i, i = 1,…,n, at time t ij, j = 1,…,n i. We assumed an underlying continuous latent variable that determines the ordinal category of individual i at time t ij. This latent variable has no interest per se, but it is useful for motivating and interpreting the cumulative logit model. The relationship between y ij and is stated as
where −∞ = γ 0<γ 1<⋯<γ K − 1<γ K=∞ are unknown cut‐points. We assumed a logistic distribution for , Lo(m ij,s = 1), with location parameter m ij (mean) and a common scale parameter s = 1 for achieving identifiability. The choice of that distribution implies a logit link for the cumulative probabilities
| (1) |
and therefore,
Despite the fact that s = 1 in the logistic distribution of the latent variable, the model is overparameterized (any set of k probabilities can be obtained increasing the cut‐points in the same quantity). To obtain an identifiable model, we arbitrarily introduced a reference point on the latent scale, in particular γ K/2=0 if K is even and γ (K − 1)/2=0 or γ (K + 1)/2=0 if K is odd.
We considered a mixed‐effects model to describe the subject‐specific time trajectories of the latent variable
| (2) |
where is a P dimensional vector of covariates relevant to the longitudinal process, as indicated by superscript (l), for individual i at time t ij with regression coefficients (populational) β; z ij the vector of explanatory variables attached to the vector of random effects b i for the ith individual at time t ij; and ε ij an error term for the ith individual at time t ij, modeled in terms of a logistic distribution, Lo(0,1). The random effects b = (b 1,…,b n)T are conditionally i.i.d. (given the hyperparameter vector ϕ) with (b i∣ϕ) ∼ f(b i∣ϕ).
Let T i, i = 1,…,n, be the observed event time for the ith subject, obtained as the minimum between the true failure time, , and the right‐censoring time, C i, . The event indicator takes the value 1 if the observed time corresponds to a true event time, and 0 otherwise. In addition, event times corresponding to individuals who enter the study at delayed entry times introduce left‐truncation, thus defining the subsequent hazard function as zero in the period before the entrance of the individual to the system 43. In particular, we consider the hazard function of in terms of a left‐truncated relative risk regression model
| (3) |
and zero otherwise, where h 0(t) is the baseline risk function; represents the vector of baseline covariates relevant to the survival process, as indicated by superscript (s), with associated coefficients η; α assesses the effect of the longitudinal marker of subject i on the event of interest in terms of the latent variable mean; a i is the delayed entry time of individual i. It is important to comment that left‐truncated data will add computational complexity to the modeling because the likelihood function corresponding to this type of data will incorporate conditional probabilities that contain the information that the individual is alive in the period between their theoretical entrance at time zero and their real entrance to the system.
To complete the Bayesian model, it is necessary to elicit a prior distribution, π(θ) for all the unknown parameters and hyperparameters of the joint model θ = (β,η,α,γ,ϕ)T. Our joint model contains parameters and hyperparameters, θ, and random effects b. From a Bayesian perspective, , where represents all the data collected from the longitudinal and the survival processes, is the joint posterior distribution of the parameters, hyperparameters, and random effects, which can be obtained by the hierarchical modeling
| (4) |
is the likelihood function of θ and b for data , f(b∣ϕ) the distribution of the random effects b given ϕ introduced before, and π(θ) the prior distribution for θ. Markov Chain Monte Carlo (MCMC) simulation methods allow to obtain an approximated random sample from the posterior , which is the key element and the starting point of all relevant inferences.
Finally, it is worth noting that when inference is carried out under the Bayesian formulation, the shared joint model will induce conditional independence between the longitudinal and the survival processes given not only the random effects and covariates but also given all the parameters and hyperparameters in the model, as a result of its stochastic role in Bayesian inference.
3.1. Dynamic probabilities associated to ordinal categories
From expression ((1)), the probability distribution of the ordinal marker y it for individual i at time t can be computed in terms of the logistic distribution of their latent variable as
| (5) |
These probabilities depend on θ, b i, and the relevant covariates associated to that individual. Consequently, we could use the posterior marginal distribution for computing the posterior distribution, , of all the relevant dynamic probabilities for each individual in the study.
A complementary and overall perspective of the temporal evolution of the different categories of the ordinal marker is based on the marginal distribution
| (6) |
which is computed by integrating out the random effects of the conditional distribution ((5)). This distribution only depends on θ. It can be interpreted as the time‐specific population distribution of the longitudinal marker for a generic individual of the population with covariate values . Consequently, we can use our current information about θ expressed through and compute the posterior distribution for each ordinal category D k. This posterior distribution provides point estimates of these relevant probabilities such as posterior expectations
| (7) |
as well as credible intervals for measuring the uncertainty of the estimation.
Our model also allows to explore the estimated relationship between the ordinal and latent variables. We could address the posterior distribution of the latent variable for each time with regard to each individual in the study or a generic one. The logistic distribution, Lo(m it,s = 1), for the latent variable is a conditional distribution with an unknown mean that depends on θ and b i. The subsequent marginal distribution can be obtained as in ((6)) integrating out the random effects and can be interpreted as a time‐specific population distribution of the latent variable for a generic individual. Again, this marginal distribution is also a conditional distribution that depends on the population parameters θ and the posterior distribution of the latent variable can be estimated as
| (8) |
3.2. Impact of the covariates on the risk of the event
The hazard ratio (HR) of an individual with covariates x having the event as compared with an individual with covariates x ∗ is exp , where P is the number of covariates. This hazard ratio only depends on η, the vector of regression coefficients in ((3)). Consequently, its posterior distribution,
| (9) |
computed from the approximate MCMC sample from the posterior marginal , provides all the relevant information about that HR.
The association parameter α allows to assess the relationship of the mean of the latent density with the hazard function but does not provide a direct link with the ordinal longitudinal variable. To facilitate an interpretation of the association parameter in terms of the ordinal measurements, we propose the following ad‐hoc procedure:
Compute the posterior mean, E , of the cut‐points γ k and construct the posterior intervals , k = 1,…,K.
- Define for a given time t a representative value , k = 1,…,K of the mean of each ordinal category in the latent scale as follows
- Compute the median of the posterior distribution ((8)) in each interval , and consider them, , as the representative value of the latent variable y ∗ in each ordinal category.
- For each , generate a value of the latent mean according to the general formulation ((2)) y ∗=m + ε, or equivalently m = y ∗−ε, where ε is a random error with logistic distribution Lo(0,1).
Approximate the conditional HR, given α, of an individual in the ordinal category k having the event versus an individual in category k ′ at time t as e .
Compute the posterior distribution of the approximate HR in step 3 from the marginal posterior distribution of α as .
3.3. Prediction
Bayesian reasoning approaches the estimation of the conditional survival probability of an individual i with a given history provided by their baseline covariates and longitudinal follow‐up (which guarantees that their survival time is greater than the time, , of their last longitudinal measurement) through the posterior distribution , . This posterior contains all relevant information about the location and variability of this conditional survival probability over time. In particular, its posterior mean can be more easily computed as
| (10) |
where the conditional probability does not depend on the particular longitudinal trajectory, , as a result of the induced independence of the shared effects joint model, and is the marginal posterior distribution of the common parameters' and random effects' vector for individual i, given and .
We could also approach prediction of a future longitudinal measurement of an individual in the study 44. The posterior predictive distribution of a new longitudinal measurement at the time of a future scheduled appointment for individual i with covariates x i and longitudinal ordinal history is given by
| (11) |
where
is obtained from ((1)), with . The conditional probability is independent on the survival history, , as a consequence of the shared random effects joint model. Note also that the different longitudinal measurements of the same individual are independent given (θ,b i).
The previous two posteriors both apply to individuals in the study and to individuals of the population that could be involved in the study in the future. In this framework, some discussion about the posterior distribution becomes necessary. If the interest concentrates on a specific individual in the study, such as individual i, for whom we want to estimate the conditional probability ((10)) or the predictive distribution ((11)) from the current data , the information provided by and is already included in . Consequently, . If the interest focuses on sequentially estimate ((10)) or/and predict ((11)) as a result of their follow‐up, we would need to sequentially update the current posterior distribution with all that new relevant follow‐up information, in particular new longitudinal measurements and the updated survival time.
Dynamic posterior estimation and prediction for individuals of the population who have not participated in the study are also possible. Let us consider now a new subject i ′ who initially has not participated in the inferential process and enters the study at time with given values of the baseline covariates. The posterior distribution of their unconditional survival probability is with posterior mean
| (12) |
Prediction of their first longitudinal measurement planned at a fixed time is
| (13) |
If as a consequence of the follow‐up of this individual, we would like to compute posterior probabilities of the type ((10)) and/or ((11)), the subsequent marginal posterior distribution will come from the joint posterior distribution , which includes the common parameters and hyperparameters θ, the vector of random effects b associated to the original individuals in the study and those of that new individual considered, .
From a Bayesian point of view, the incorporation of sequential information from an individual who is already involved in the study or from the follow‐up of a future subject implies the need of sequentially update the posterior distribution . In the case of studies based on samples with large sample size, we would expect a minimal change in the estimation of the common parameters but possibly not in the subject specific random effects. The process of updating a posterior distribution for which we only have an approximate random sample and not an analytical distribution is conceptually easy but not so in practice. The main tools to carry out this computational process are based on sequential MCMC methods 45, 46 and although are beyond the scope of this paper are a current aim of our research team. Rizopoulos 47 proposes, as an approximation of the subsequent posterior distribution, updating the specific random effects associated with individuals. In particular, the author uses empirical Bayesian estimation for the random effects and an asymptotic normal distribution, based on maximum likelihood estimation, for the common population parameters. Taylor et al. 48 also separately update the vector of random effects by using a quick MCMC based on a prior distribution for the population parameters coming from the marginal posterior .
4. Joining longitudinal breast density and age at breast cancer detection
Let {D 1,D 2,D 3,D 4} denote the set of BI‐RADS breast density categories {a,b,c,d}, which represent low, medium, high, and very high density, respectively, y ij the breast density category of woman i, i = 1,…,n, at time t ij(age 50 + t ij), j = 1,…,n i and her subsequent underlying continuous latent value.
Following (3), the connection between both processes is
where −∞ = γ 0<γ 1<γ 2<γ 3<γ 4=∞ are unknown cut‐points, with γ 2=0, and Lo(m ij,s = 1) represents the corresponding logistic distribution for .
Considering the evidence of a decreasing trend of breast density with age and a linear trajectory for the longitudinal latent breast density of woman i
| (14) |
where (β 0,β 1)T and (b i0,b i1)T are the fixed (population) and random effects (individual) for the intercept and the slope term, respectively, and ε it the error term. Random effects (b 0,b 1)T, where b 0=(b 01,…,b 0n)T and b 1=(b 11,…,b 1n)T, are assumed conditionally i.i.d. with (b i0∣σ 0) ∼ N(0,σ 0) and (b i1∣σ 1) ∼ N(0,σ 1).
The hazard function of age at breast cancer diagnosis is defined in terms of the left truncated relative risk regression model
| (15) |
where is the baseline risk function of a Weibull distribution, We ; family history of breast cancer (Famhist) and prior breast procedures (Brstproc) are dychotomous baseline covariates with associated coefficients η 1 and η 2, respectively; α assesses the effect of the individual trajectory of breast density on breast cancer risk in terms of the latent breast density mean; and a i is the age over 50 at which woman i enters the screening program, thus providing the left truncated time 43.
We assumed prior independence among all the parameters and hyperparameters as a default specification and, with the aim of giving all inferential prominence to the data, we elicited wide proper prior distributions. For the parameters in the longitudinal submodel, we followed Lunn et al. 30 except for the standard deviations. In particular, we selected N(0,100) for the β's regression coefficients. The ordinal constraint for the cutpoints of the latent scale, −∞ < γ 1<γ 2=0 < γ 3<γ 4=∞, was expressed by truncating the subsequent prior distributions in the appropriate parametric subspace
where is the indicator function. Prior means for γ 1 and γ 3, respectively, correspond to the first and third quartiles of a logistic distribution Lo(0,1) in order to provide the same prior probability to each response category. For the standard deviations σ 0 and σ 1, we choose a uniform distribution, Un(0,10). In the case of the survival submodel, we selected N(0,100) for the η's regression coefficients as well as for the association coefficient α, and a gamma distribution Ga(1,1) for the parameter λ of the baseline hazard function because it mimics a constant baseline hazard function 49.
4.1. Posterior distribution
The posterior distribution was computed in terms of the hierarchical modeling ((4)) and approximated using MCMC simulation methods through the JAGS software 50. In particular, we run three MCMC chains with 100,000 iterations, 10,000 of which were used for the burn‐in period. The chains were thinned by only storing every 270th iteration in order to reduce autocorrelation in the saved sample. Trace plots of the simulated values of the three chains appear overlapping one another indicating stabilization. Convergence of the chains to the posterior distribution was assessed through the potential scale reduction factor, , and the effective number of independent simulation draws, n eff, 51 and 52, respectively. compares the within‐chain variance to the estimated variance of the posterior distribution in such a way that values near 1 indicate that the simulated process has reached the posterior distribution. n eff deals with the level of autocorrelation of the chains simulated values, so that n eff>100 indicates that sufficient MCMC samples have been obtained.
Table 2 summarizes the approximate MCMC random sample from the marginal posterior distribution through the mean, median, standard deviation, 2.5% and 97.5% percentiles. The last column of Table 2 contains the probability that the corresponding parameter is positive: a 0.5 probability would indicate that a positive value of the parameter is equally likely that a negative one, hence indicating little relevance of the corresponding variable (given the remaining covariates). This is not the case for the parameters of our model with probabilities that show a clear preference for being above or under zero.
Table 2.
Posterior summaries of the parameters and hyperparameters of the breast cancer joint model.
| Mean | SD | 2.5% | Median | 97.5% | P(·>0|D) | |
|---|---|---|---|---|---|---|
| β 0 | −1.4262 | 0.0346 | −1.4964 | −1.4251 | −1.3608 | 0.0000 |
| β 1 | −0.0524 | 0.0018 | −0.0560 | −0.0524 | −0.0489 | 0.0000 |
| σ 0 | 2.6067 | 0.0227 | 2.5643 | 2.6059 | 2.6534 | |
| σ 1 | 0.0053 | 0.0018 | 0.0015 | 0.0053 | 0.0087 | |
| γ 1 | −4.6994 | 0.0269 | −4.7521 | −4.6998 | −4.6489 | |
| γ 3 | 1.7362 | 0.0156 | 1.7060 | 1.7364 | 1.7675 | |
| λ | 1.5366 | 0.1044 | 1.3287 | 1.5387 | 1.7386 | |
| η 0 | −7.6066 | 0.3369 | −8.2476 | −7.6011 | −6.9337 | 0.0000 |
| η 1 | 0.6227 | 0.1716 | 0.2747 | 0.6308 | 0.9517 | 0.9984 |
| η 2 | 0.4535 | 0.1440 | 0.1644 | 0.4600 | 0.7210 | 1.0000 |
| α | 0.1490 | 0.0207 | 0.1089 | 0.1496 | 0.1887 | 1.0000 |
The marginal posterior distribution associated to the population intercept β 0 and slope β 1 of the mean of the latent breast density clearly states that both are negative, , indicating decreasing values over time of the true latent breast density and therefore a higher probability of being in the lower breast density categories with age. The variability associated with the random intercept is important, E , as a sign of high population heterogeneity with regard to initial breast density. In contrast, there is small variability in the subject‐specific slopes, E , which denotes that subject‐specific trajectories of the true latent breast density do not differ much from the population trend. The estimation of the cut‐points γ 1 and γ 3 is very stable and accurate. The posterior means of the coefficients associated to the baseline covariates, family history of breast cancer, and prior breast procedures, 0.6227 and 0.4535, respectively, indicate an increase of risk of breast cancer detection when women have one or both of these risk factors. These values are consistent with the ones reported in the literature. The strength of the association between the breast density and age at breast cancer diagnosis is assessed through their posterior expectation =0.149 and 95% credible interval (0.1089, 0.1887). In addition, the posterior probability 1 for that coefficient being positive provides strong support on the connection between breast density and breast cancer risk.
4.2. Probabilities associated to breast density BI‐RADS categories
Figure 2 (top) shows the posterior mean and 95% credible interval of the posterior distribution associated to each BI‐RADS category for a generic woman in the study. Probabilities associated to category BI‐RADS b are always higher than 0.5, and grow slightly with age. Probabilities for categories a, c, and d are initially very similar, but categories c and d decrease with age following a similar pattern while category a increases (see Table S1 in Appendix). The information provided by credible intervals is very valuable, thus indicating high precision in the estimated means.
Figure 2.
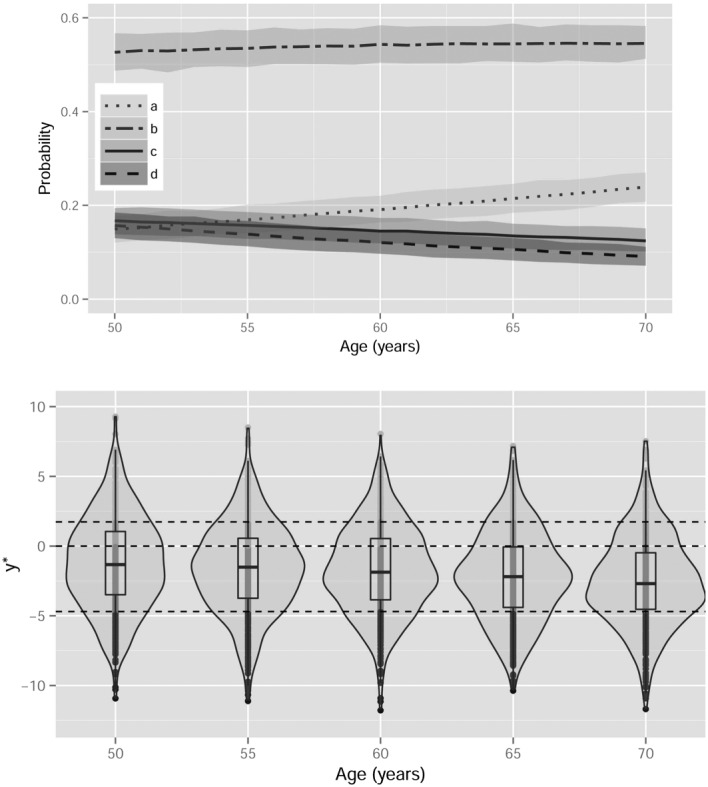
Age‐specific population distribution of breast density. Posterior mean and 95% credible interval of the probability associated to each BI‐RADS category with respect to age (top) and violin plot of the posterior marginal distribution of the latent breast density of an average woman at ages 50, 55, 60, 65, and 70 (bottom). Horizontal dotted lines represent the posterior mean of the cut‐points, thus approximately indicating the region of the latent density corresponding to ordinal BD categories a,b,c,andd (from bottom to top).
Figure 2 (bottom) shows a violin plot (a combination of a kernel density plot and a boxplot) of the posterior marginal distribution of the latent breast density at ages 50, 55, 60, 65 and 70. The four categories of the ordinal breast density are marked with regard to the posterior mean of the cut points γ 1 and γ 3, and γ 2=0. The visual comparison between real and latent results is very interesting. We clearly appreciate that the posterior marginal distribution of the latent breast density tends towards lower values with age. In addition, the bottom tail of the distributions increase with age in detriment of the top tail, thus indicating the general decreasing of breast density with age.
4.3. Assessment of the impact of the study variables on breast cancer risk
Relevant HRs arise from the combination of baseline covariate categories. Figure 3 shows the posterior distribution of the HRs of a breast cancer diagnosis for family history of breast cancer, prior breast procedures, and both risk factors, with posterior means 1.864, 1.574, and 2.934, respectively. The marginal effects of each covariate are relevant, with posterior probabilities 0.998 and 1.000 associated to HR values greater than 1 for family history of breast cancer and prior breast procedures, respectively.
Figure 3.
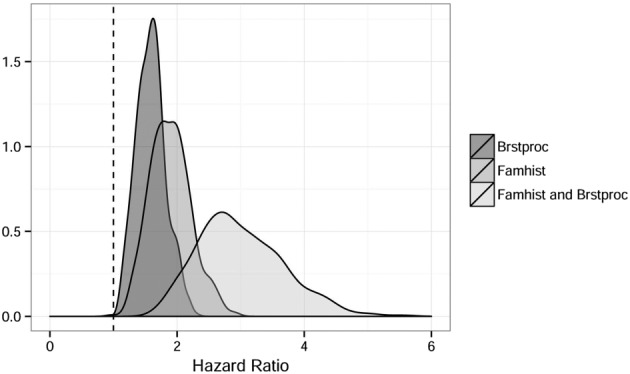
Posterior distribution of the hazard ratios associated to family history of breast cancer and prior breast procedures.
Following the ad‐hoc procedure presented in Section 3.2, Figure 4 shows the posterior mean and 95% credible intervals with regard to age of the approximate HR of a breast cancer diagnosis for women with breast densities b, c, and d compared with women with the same covariate values and breast density a. Changes in breast density from category a towards more dense categories have a strong effect on breast cancer risk. We observe posterior means of the HR around four for category d versus a, and HRs greater than 1 (around 1.7 and 2.6) when comparing categories b and c versus a, respectively. A gently wavy behavior for the posterior distributions and credible intervals of the HRs with respect to age can be appreciated, as a consequence of the simulation of the logistic error in the procedure.
Figure 4.
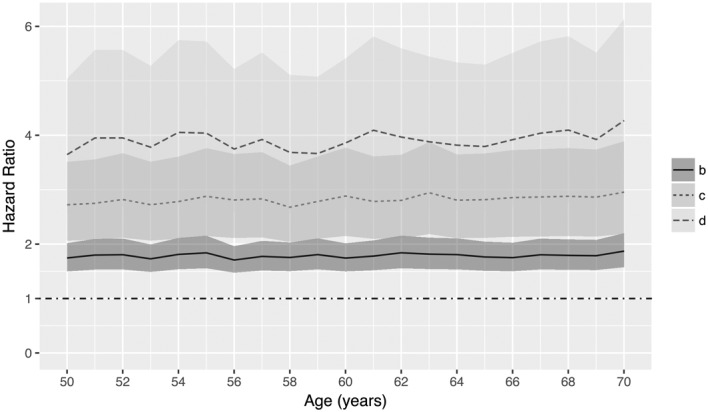
Approximate posterior mean and 95% credible interval with regard to age of the HRs of a BC diagnosis for women withbreast density b,c, andd as compared with women with the same covariates and BD measurement a.
4.4. Prediction
Figure 5 shows the posterior mean ((10)) and 95% credible band for four of the women without cancer in Figure 1, at the end of follow‐up. As expected, breast cancer‐free probabilities are very high and decrease with age. It is worth noting the narrowness of the bands corresponding to women with a higher number of density measurements, possibly because of the precision of the random effects estimates. Women with high breast density values seem to have lower breast cancer‐free survival probabilities. However, we must also consider the baseline risk factors. Thus, disease‐free survival is higher for woman 942, who has a stable very high breast density, than for woman 9672, who experiences a decrease in density. This result can be attributed to presence of prior breast procedures in woman 9672 and absence of them in woman 942. Both women do not have family history of breast cancer.
Figure 5.
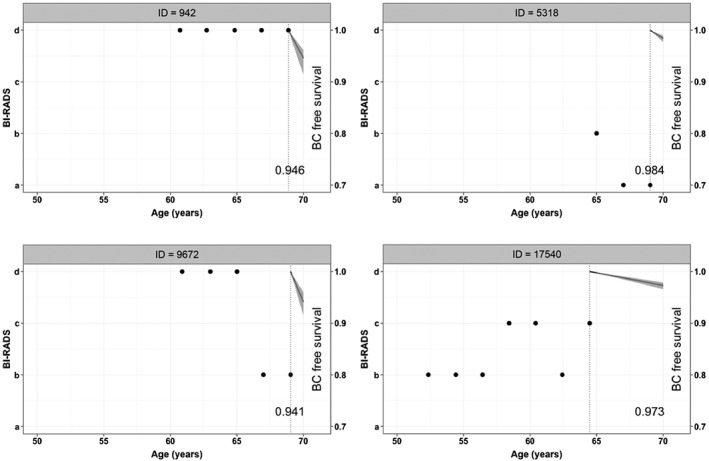
Posterior mean and 95% credible band of the probability of a breast cancer‐free diagnosis for women with IDs 942; 5318; 9672; and 17540 without breast cancer at the end of the follow‐up. The value of the probability at the lower right of each graphic is the subsequent posterior mean at 70 years.
Figure 6 shows the posterior predictive distribution of the ordinal breast density categories for the women in Figure 5. We appreciate a great variability among the predicted BI‐RADS trajectories, and for most of the selected women, category b has the highest probabilities over age. But, for woman 5318 category a is always the most prevalent with an increasing trend over age, followed by categories b, c and d. These results are in line with the observed breast density trajectory for this woman: three breast density measurements with values b, a and a at late ages 65, 67, and 69, respectively. In contrast, for woman 942 category d is always the most prevalent with a decreasing trend over age. These results are also consistent with the observed breast density trajectory for this woman, stable with very high breast density at relatively late ages 61, 63, 65, 67, and 69.
Figure 6.
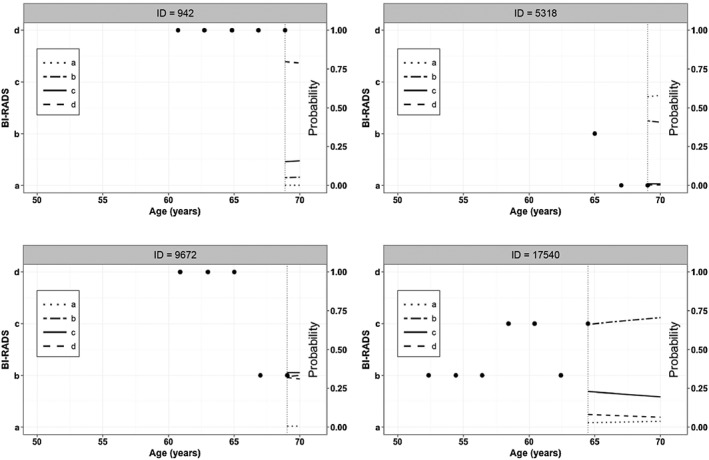
Posterior predicted mean of the breast density in the BI‐RADS scale over age for women with IDs with IDs 942; 5318; 9672; and 17540 without breast cancer at the end of the follow‐up.
5. Discussion
We propose a Bayesian joint model that combines the information provided by a longitudinal ordinal process and a left‐truncated time‐to‐event outcome. The joint density of both processes is approached through a shared‐parameter model which generates a structure of association and conditional independence between both outcomes by means of a vector of common random effects.
We chose a latent variable formulation for the longitudinal process which translated the ordinal scale to the framework of linear mixed models, with a logistic distribution for the measurement error. The latent variable approach facilitates the computational implementation of the model but introduces complexity in the interpretation of results. We assume a logistic distribution for the latent variable that implies the logistic link for cumulative probabilities. Other models might be also appropriate. The most usual alternatives are the normal and the extreme value distributions, which result in the probit and complementary log‐log regression links, respectively. It is widely accepted that probit and logit links produce similar results. This also occurs in our study (results not shown), where we have implemented the probit link to assess the robustness of the model. This is not the case for the extreme value distribution that, unlike the logistic and normal ones, is not symmetrical.
We consider that the cut‐points that relate the ordinal and latent variables are common for all individuals and time. This assumption may produce some stiffness in the longitudinal model. Thus, individual or time‐specific cut‐points might endow of more flexibility to the longitudinal latent variable at the expense of a more complex model. Dealing with more than four categories in the ordinal variable is not straightforward. One of the reasons for this is that one or both of the endpoints of the truncated intervals in which the marginal prior distribution of each unknown cut‐point is defined can be also unknown 53. The estimation of these models involves important computational issues in the MCMC sampling that have provided many discussion and proposals, such as hybrid Metropolis–Hasting algorithms to sample from the subsequent posterior distribution 54, 55.
Robustness is a major statistical concern in Hierarchical Bayesian models because it can be affected by an inappropriate choice of hyperprior distributions for hyperparameters. We have tested the sensitivity of the model using other prior specifications for the hyperprior distribution of the random effects scale parameter. In particular, we have considered a wide uniform distribution, Un(0, 100), as an alternative to the elicited Un(0,10) in the paper and inverse‐gamma hyperdistributions, IGa(0.01, 0.01) and IGa(0.001, 0.001), because of their common use in Bayesian applications. All of them provided almost identical results (not shown in the paper), possibly because of the large sample size.
Our proposal could be applied to a variety of real problems devoted to analyze time‐to‐event outcomes with temporal ordinal endogeneous covariates. We explored the role of death prior to breast cancer diagnosis as a competing risk. The cumulative incidences estimated with the Kaplan–Meier method or the competing risks with cause‐specific hazards approach are very similar, even at older ages (See Figure A1 in the Supplementary file). Therefore, even though the censoring due to the competing risk “death” was informative, it would hardly affect our estimates. Event times have been modeled in terms of relative risk models with left‐truncation as a corrective mechanism for the delayed entry bias. Left truncation is common in observational studies of risk factors, where not all the participants enter the study at time zero. We select the Weibull distribution as baseline risk function because it is a traditional model for survival data with a great flexibility in representing different types of risk. The exploration of more sophisticated baseline risk functions, which include multimodality and heavy tailed distributions, in the area of Bayesian joint modeling is a relevant subject with strong connections with the specification of prior distributions. See 56 for a detailed explanation of piecewise constant hazard models and Gamma processes. In addition, the latent linear mixed model is a flexible model that can accommodate heterogeneous trajectories, from linear to complex functions. This is the case of linear models expressed in terms of spline bases to accommodate non‐linear profiles 57.
We have used our joint model for analyzing the relationship between mammographic breast density and breast cancer risk in women attending a public screening program. A linear subject‐specific trajectory of the latent variable is included in a relative risks survival model together with two of the most known breast cancer risk factors, family history of breast cancer and previous breast procedures. Our joint model for breast cancer and breast density is a good starting point that provides results consistent with the literature 16, 18, 19, 21. They are the basis for a rationale for extending the model and assessing its adequacy and accuracy. Evaluating the ability of breast density to predict time to breast cancer diagnosis, under our joint model, by means of calibration and discrimination measures 58 is a major concern of our research. The discriminative power of our model should offset its complexity. Discrimination measures based on receiver operating characteristic curves are commonly used for assessing predictive accuracy. We are currently exploring a general latent variable approach that could be appropriate.
In contrast to studies published to date, our study is the first to have used the complete longitudinal history of breast density for assessing breast cancer risk over time, at population and individual level. Potential benefits of the proposed joint model include obtaining individual predictions of time‐free of breast cancer at age u > t, given the observed responses up to age t, and also individual longitudinal predictions of future breast density values. Thus, a joint model similar to that shown here could be used for surveillance of breast cancer risk over time, for scheduling screening exams based on individual dynamic predictions, and also in discussing prevention strategies for those at high risk 16.
Supporting information
Supporting info item
Acknowledgements
This paper was partially supported by the research grants Combination and Propagation of Uncertainties (ComPro_UN, MTM2013‐42323‐P), Statistical methods for clinical trials, complex censoring schemes and integrative omics data analysis (MTM2015‐64465‐C2‐1‐R), and Women participation in decisions and strategies on early detection of breast cancer (PI14/00113) from the Spanish Ministry of Economy and Competitiveness, ACOMP/2015/202 from the Generalitat Valenciana, and GRBIO‐2014‐SGR464 and GRAES‐2014‐SGR978 from the Generalitat de Catalunya.
We thank Dimitris Rizopoulos and Arindom Chakraborty for their advice on joint modeling. We are also indebted to the research group Grup de Recerca en Anàlisi Estadística de la Supervivència (GRASS), specially to Carles Serrat, for their support and fruitful discussions. We also thank Núria Torà and Marina Pont for their work on data management, and JP Glutting for review and editing. We wish to acknowledge three anonymous referees and the Associate Editor for their useful comments that improved very much the original version of the paper.
We also would like to thank the staff of the Cancer Screening Office and the radiologists of the BCEDP‐VOE, for their valuable help in obtaining the data we analyzed. And, last but not least, we appreciate all women that kindly answered the questionnaire.
Armero, C. , Forné, C. , Rué, M. , Forte, A. , Perpiñán, H. , Gómez, G. , and Baré, M. (2016) Bayesian joint ordinal and survival modeling for breast cancer risk assessment. Statist. Med., 35: 5267–5282. doi: 10.1002/sim.7065.
The copyright line for this article was changed on 26 September 2016 after original online publication.
References
- 1. Onega T, Beaber EF, Sprague BL, Barlow WE, Haas JS, Tosteson AN, Schnall MD, Armstrong K, Schapira MM, Geller B, Weaver DL, Conant EF. Breast cancer screening in an era of personalized regimens: a conceptual model and National Cancer Institute initiative for risk‐based and preference‐based approaches at a population level. Cancer 2014; 19:2955–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vilaprinyo E, Forné C, Carles M, Sala M, Pla R, Castells X, Domingo L, Rue M, Interval Cancer Study Group (INCA). Cost‐effectiveness and harm‐benefit analyses of risk‐based screening strategies for breast cancer. PLoS One 2014; 9:e86858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, Mulvihill JJ. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. Journal of the National Cancer Institute 1989; 81:1879–1886. [DOI] [PubMed] [Google Scholar]
- 4. Gail MH. Personalized estimates of breast cancer risk in clinical practice and public health. Statistics in Medicine 2011; 30(10):1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Statistics in Medicine 2004; 23(7):1111–1130. [DOI] [PubMed] [Google Scholar]
- 6. Anothaisintawee T, Teerawattananon Y, Wiratkapun C, Kasamesup V, Thakkinstian A. Risk prediction models of breast cancer: a systematic review of model performances. Breast Cancer Research and Treatment 2012; 133(1):1–10. [DOI] [PubMed] [Google Scholar]
- 7. Tice JA, Cummings SR, Ziv E, Kerlikowske K. Mammographic breast density and the Gail model for breast cancer prediction in a screening population. Breast Cancer Research and Treatment 2005; 94:115–122. [DOI] [PubMed] [Google Scholar]
- 8. Chen J, Pee D, Ayyagari R, Graubard B, Schairer C, Byrne C, Benichou J, Gail MH. Projecting absolute invasive breast cancer risk in white women with a model that includes mammographic density. Journal of the National Cancer Institute 2006; 98:1215–1226. [DOI] [PubMed] [Google Scholar]
- 9. Barlow WE, White E, Ballard‐Barbash R, Vacek PM, Titus‐Ernstoff L, Carney PA, Tice JA, Buist DS, Geller BM, Rosenberg R, Yankaskas BC, Kerlikowske K. Prospective breast cancer risk prediction model for women undergoing screening mammography. Journal of the National Cancer Institute 2006; 98:1204–1214. [DOI] [PubMed] [Google Scholar]
- 10. Tice JA, Cummings SR, Smith‐Bindman R, Ichikawa L, Barlow WE, Kerlikowske K. Using clinical factors and mammographic breast density to estimate breast cancer risk: development and validation of a new predictive model. Annals of Internal Medicine 2008; 148:337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Byrne C, Schairer C, Wolfe JN, Parekh N, Salane M, Brinton LA, Hoover RN, Haile R. Mammographic features and breast cancer risk: effects with time, age, and menopause status. Journal of the National Cancer Institute 1995; 87:1622–1629. [DOI] [PubMed] [Google Scholar]
- 12. Boyd NF, Lockwood GA, Byng JW, Tritchler DL, Yaffe MJ. Mammographic densities and breast cancer risk. Cancer Epidemiology, Biomarkers & Prevention 1998; 7:1133–44. [PubMed] [Google Scholar]
- 13. Vacek PM, Geller BM. A prospective study of breast cancer risk using routine mammographic breast density measurements. Cancer Epidemiology, Biomarkers & Prevention 2004; 13:715–722. [PubMed] [Google Scholar]
- 14. Vachon CM, van Gils CH, Sellers TA, Ghosh K, Pruthi S, Brandt KR, Pankratz VS. Mammographic density, breast cancer risk and risk prediction. Breast Cancer Research 2007; 9:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Gils CH, Hendriks JHCL, Holland R, Karssemeijer N, Otten JDM, Straatman H, Verbeek ALM. Changes in mammographic breast density and concomitant changes in breast cancer risk. European Journal of Cancer Prevention 1999; 8:509–515. [DOI] [PubMed] [Google Scholar]
- 16. Kerlikowske K, Ichikawa L, Miglioretti DL, Buist DS, Vacek PM, Smith‐Bindman R, Yankaskas B, Carney PA, Ballard‐Barbash R, Consortium NIoHBCS . Longitudinal measurement of clinical mammographic breast density to improve estimation of breast cancer risk. Journal of the National Cancer Institute 2007; 99:386–395. [DOI] [PubMed] [Google Scholar]
- 17. Vachon CM, Pankratz VS, Scott CG, Maloney SD, Ghosh K, Brandt KR, Milanese T, Carston MJ, Sellers TA. Longitudinal trends in mammographic percent density and breast cancer risk. Cancer Epidemiology, Biomarkers & Prevention 2007; 16:921–928. [DOI] [PubMed] [Google Scholar]
- 18. Lokate M, Stellato RK, Veldhuis WB, Peeters PHM, van Gils CH. Age‐related changes in mammographic density and breast cancer risk. American Journal of Epidemiology 2013; 178:101–109. [DOI] [PubMed] [Google Scholar]
- 19. Work ME, Reimers LL, Quante AS, Crew KD, Whiffen A, Beth Terry M. Changes in mammographic density over time in breast cancer cases and women at high risk for breast cancer. International Journal of Cancer 2014; 135:1740–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huo CW, Chew GL, Britt KL, Ingman WV, Henderson MA, Hopper JL, Thompson EW. Mammographic density ‐ a review on the current understanding of its association with breast cancer. Breast Cancer Research and Treatment 2014; 144:479–502. [DOI] [PubMed] [Google Scholar]
- 21. Kerlikowske K, Gard CC, Sprague BL, Tice JA, Miglioretti DL. One versus two breast density measures to predict 5‐ and 10‐year breast cancer risk. Cancer Epidemiology, Biomarkers & Prevention 2015; 24(6):889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data 2nd ed. Wiley: Hoboken, 2002. [Google Scholar]
- 23. Tsiatis AA, DeGruttola V, Wulfsohn MS. Modeling the relationship of survival to longitudinal data measured with error. Applications to survival and CD4 counts in patients with AIDS. Journals ‐ American Statistical Association 1995; 90:27–37. [Google Scholar]
- 24. Faucett CL, Thomas DC. Simultaneously modelling censored survival data and repeatedly measured covariates: a Gibbs sampling approach. Statistics in Medicine 1996; 15:1663–1685. [DOI] [PubMed] [Google Scholar]
- 25. Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics 1997; 53:330–339. [PubMed] [Google Scholar]
- 26. Rizopoulos D. Joint Models For Longitudinal and Time‐to‐Event Data with Applications in R. CRC Press, Biostatistics Series: Boca Raton, FL, 2012. [Google Scholar]
- 27. Rizopoulos D. JM: An R package for the joint modelling of longitudinal and time‐to‐event data. Journal of Statistical Software 2010; 35:1–33.21603108 [Google Scholar]
- 28. Rizopoulos D. JMbayes: Joint modeling of longitudinal and time‐to‐event data under a bayesian approach, 2015. Available from: http://CRAN.R-project.org/package=JMbayes, r package version 0.7‐2, [last access date was December 28, 2015].
- 29. Serrat C, Rué M, Armero C, Piulachs X, Perpiñán H, Forte A, Páez A, Gómez G. Frequentist and bayesian approaches for a joint model for prostate cancer risk and longitudinal prostate‐specific antigen data. Journal of Applied Statistics 2015; 42(6):1223–1239. [Google Scholar]
- 30. Lunn DJ, Wakefield J, Racine‐Poon A. Cumulative logit models for ordinal data: a case study involving allergic rhinitis severity scores. Statistics in Medicine 2001; 20:2261–2285. [DOI] [PubMed] [Google Scholar]
- 31. Chakraborty A, Das K. Inferences for joint modelling of repeated ordinal scores and time to event data. Computational and Mathematical Methods in Medicine 2010; 11:281–295. [DOI] [PubMed] [Google Scholar]
- 32. Li N, Elashoff RM, Li G, Saver J. Joint modeling of longitudinal ordinal data and competing risks survival times and analysis of the NINDS rt‐PA stroke trial. Statistics in Medicine 2010; 29:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo S. A Bayesian approach to joint analysis of multivariate longitudinal data and parametric accelerated failure time. Statistics in Medicine 2014; 33:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hogan JW, Laird N. Mixture models for the joint distribution of repeated measures and event times. Statistics in Medicine 1997; 28:239–257. [DOI] [PubMed] [Google Scholar]
- 35. Chakraborty A. Bounded influence function based inference in joint modelling of ordinal partial linear model and accelerated failure time model. Statistical Methods in Medical Research 2014; 11. DOI 0962280214531570, [first published on April 25, 2014]. [DOI] [PubMed] [Google Scholar]
- 36. Proust‐Lima C, Dartigues JF, Jacqmin‐Gadda H. Joint modeling of repeated multivariate cognitive measures and competing risks of dementia and death: a latent process and latent class approach. Statistics in Medicine 2016; 35:382–398. [DOI] [PubMed] [Google Scholar]
- 37. He B, Luo S. Joint modeling of multivariate longitudinal measurements and survival data with applications to Parkinson disease. Statistical Methods in Medical Research 2016; 25(4):1346–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu F, Li Q. A Bayesian model for joint analysis of multivariate repeated measures and time to event data in crossover trials. Statistical Methods in Medical Research 2016; 25(5):2180–2192. [DOI] [PubMed] [Google Scholar]
- 39. Baré ML, Montes J, Florensa R, Sentís M, Donoso L. Factors related to non‐participation in a population‐based breast cancer screening programme. European Journal of Cancer Prevention 2003; 12:487–94. [DOI] [PubMed] [Google Scholar]
- 40. Baré M, Bonfill X, Andreu X. Relationship between the method of detection and prognostic factors for breast cancer in a community with a screening programme. Journal of Medical Screening 2006; 13:183–91. [DOI] [PubMed] [Google Scholar]
- 41. Baré M, Sentís M, Galceran J, Ameijide A, Andreu X, Ganau S, Tortajada L, Planas J. Interval breast cancers in a community screening programme: frequency, radiological classification and prognostic factors. European Journal of Cancer Prevention 2008; 17:414–21. [DOI] [PubMed] [Google Scholar]
- 42. American College of Radiology. Breast Imaging Reporting and Data Systemő(BI‐RADSő) 2013.
- 43. Uzunoḡullari a, Wang J‐L. Comparison of hazard rate estimators for left truncated and right censored data. Biometrika 1992; 79:297–310. [Google Scholar]
- 44. Armero C, Forte A, Perpiñán H, Sanahuja MJ, Agustí S. Bayesian joint modeling for assessing the progression of Chronic Kidney Disease in children. Statistical Methods in Medical Research 2016. DOI: 10.1177/0962280216628560. [DOI] [PubMed] [Google Scholar]
- 45. Del Moral P, Doucet A, Jasra A. Sequential Monte Carlo samplers. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2006; 68:411–436. [Google Scholar]
- 46. Andrieu C, Doucet A, Holenstein R. Particle Markov Chain Monte Carlo methods. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2010; 72:269–342. [Google Scholar]
- 47. Rizopoulos D. Dynamic predictions and prospective accuracy in joint models for longitudinal and time‐to‐event data. Biometrics 2011; 67:819–829. [DOI] [PubMed] [Google Scholar]
- 48. Taylor JMG, Park Y, Ankerst DP, Proust‐Lima C, Williams S, Kestin L, Bae K, Pickles T, Sandler H. Real‐time individual predictions of prostate cancer recurrence using joint models. Biometrics 2013; 69(1):206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo X, Carlin BP. Separate and joint modeling of longitudinal and event time data using standard computer packages. The American Statistician 2004; 58:1–9. [Google Scholar]
- 50. Plummer M. JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling. Proceedings of the 3rd International Workshop on Distributed Statistical Computing (DSC 2003), Vienna, Austria, 2003. [Google Scholar]
- 51. Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical Science 1992; 7:457–511. [Google Scholar]
- 52. Kass RE, Carlin BP, Gelman A, Neal R. Markov Chain Monte Carlo in Practice: A Roundtable Discussion. The American Statistician 1998; 52:93–100. [Google Scholar]
- 53. Albert JH, Siddhartha C. Bayesian Analysis of Binary and Polychotomous Response Data. Journal of the American Statistical Association 1993; 88:669–679. [Google Scholar]
- 54. Cowles MK. Accelerating Monte Carlo Markov chain convergence for cumulative‐link generalizad linear models. Statistics and Computing 1996; 6:101–111. [Google Scholar]
- 55. Nandram B, Chen MH. Reparameterizing the generalized linear model to accelerate Gibbs sampler convergence. Statistics in Medicine 1996; 54:129–144. [Google Scholar]
- 56. Ibrahim JG, Chen M‐H, Sinha D. Bayesian Survival Analysis. Springer: New York, 2001. [Google Scholar]
- 57. Andrinopoulou E‐R, Rizopoulos D, Takkenberg JJM, Lesaffre E. Joint modeling of two longitudinal outcomes and competing risk data. Statistics in Medicine 2014; 33(18):3167–3178. [DOI] [PubMed] [Google Scholar]
- 58. Rizopoulos D, Verbeke G, Molenberghs G. Multiple‐imputation‐based residuals and diagnostic plots for joint models of longitudinal and survival outcomes. Biometrics 2010; 66:20–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting info item


