Suppressing chloroplastic HCF106 and THF1 expression enhances drought resistance in Arabidopsis.
Abstract
Chloroplast as the site for photosynthesis is an essential organelle in plants, but little is known about its role in stomatal regulation and drought resistance. In this study, we show that two chloroplastic proteins essential for thylakoid formation negatively regulate drought resistance in Arabidopsis (Arabidopsis thaliana). By screening a mutant pool with T-DNA insertions in nuclear genes encoding chloroplastic proteins, we identified an HCF106 knockdown mutant exhibiting increased resistance to drought stress. The hcf106 mutant displayed elevated levels of reactive oxygen species (ROS) in guard cells, improved stomatal closure, and reduced water loss under drought conditions. The HCF106 protein was found to physically interact with THF1, a previously identified chloroplastic protein crucial for thylakoid formation. The thf1 mutant phenotypically resembled the hcf106 mutant and displayed more ROS accumulation in guard cells, increased stomatal closure, reduced water loss, and drought resistant phenotypes compared to the wild type. The hcf106thf1 double mutant behaved similarly as the thf1 single mutant. These results suggest that HCF106 and THF1 form a complex to modulate chloroplast function and that the complex is important for ROS production in guard cells and stomatal control in response to environmental stresses. Our results also suggest that modulating chloroplastic proteins could be a way for improving drought resistance in crops.
Drought is a frequently occurring environmental condition that causes enormous economic losses in agriculture. Under drought conditions, plants as sessile organisms have to deal with water deficit by increasing water uptake and reducing water loss. Building a deeper and larger root system helps ensure water absorption from the soil. Plants can also adjust physiologically to increase water use efficiency in order to maintain growth under drought conditions. Minimizing water loss is often a common strategy for plants to cope with water deficit, which is largely executed through stomatal closure.
Stomatal closure under drought and other stress conditions is mediated by signaling molecules such as abscisic acid (ABA), reactive oxygen species (ROS), Ca2+, etc. (Murata et al., 2015). Drought stress promotes ABA accumulation, and ABA triggers an increase in cytosolic Ca2+. The increase in [Ca2+]cyt helps to activate plasma membrane anion channels, causing the efflux of anions such as Cl− and NO3−, which depolarizes the plasma membrane of guard cells. This process in turn enhances K+ efflux through the outward rectifying K+ channel (Kim et al., 2010). Consequently, the turgor pressure of guard cells decreases due to solutes and water efflux and the stoma closes. Early signaling events in ABA-triggered stomatal closure include binding of ABA to the RCAR/PYR1/PYL receptors; the ABA-bound receptor proteins then interact with clade A protein phosphatase 2Cs (PP2Cs), resulting in the disruption of PP2C-SnRK2 protein complexes and activation of the SNF1-related protein kinase SnRK2s. The active SnRK2s phosphorylate downstream effector proteins, leading to stomatal closure and other ABA responses (Munemasa et al., 2015).
The OPEN STOMATAL1 (OST1) is one of the SnRK2s activated by ABA (Mustilli et al., 2002). OST1 can directly phosphorylate the anion channel SLAC1, thus promoting anion efflux and stomatal closure (Vahisalu et al., 2008). Moreover, the plasma membrane-bound NADPH oxidase RbohF is also a direct target of OST1. Phosphorylation of RbohF activates its production of O2.− and subsequent formation of H2O2 in the apoplastic compartment (Sirichandra et al., 2009; Acharya et al., 2013). ROS are thought to be key second messengers that integrate stress and ABA signaling in stomatal movement. ROS could be sensed by AtGPX3 and the PP2C proteins ABI1 and ABI2. AtGPX3 was proposed to resemble the yeast GPX-like enzyme ORP1 and to function as a ROS transducer and scavenger in guard cells. AtGPX3 physically interacts with ABI2 and ABI1, and oxidized AtGPX3 significantly reduces the phosphatase activity of ABI2 in vitro (Miao et al., 2006). Downstream ROS targets include the components of the MAP kinase cascade MPK3, MPK9, and MPK12. These MAPKs are activated by ABA and H2O2 and are involved in ROS-mediated ABA signaling in stomatal closure (Zhang et al., 2014). In addition, GHR1, a plasma membrane receptor-like kinase, also plays an important role in ABA- and H2O2-mediated stomatal closure. GHR1 physically interacts with, phosphorylates, and activates SLAC1, while ABI2 antagonizes the activation of SLAC1 by GHR1 (Hua et al., 2012).
The apoplastic ROS burst generated by the plasma membrane NADPH oxidases is believed to be the major source of ROS for ABA signaling in stomatal closure. However, other sources of ROS produced through cell wall-associated peroxidases and enzymes in the peroxisome also contribute to ROS-mediated stomatal closure (Murata et al., 2015). Interestingly, chloroplast as a major ROS producer under both normal and stress conditions has not been considered as an important source of ROS-mediating stomatal closure. Chloroplast has been implicated in Ca2+-regulated stomatal movement (Weinl et al., 2008), but whether chloroplast-originated ROS are involved in stomatal regulation in response to stress conditions is unclear.
HCF106 is a component of the thylakoid twin-Arg translocation (cpTat) system, and cpTat is one of the two thylakoid protein translocation systems responsible for translocating stromal proteins into the lumen (Robinson and Bolhuis, 2004). The cpTat translocase includes Tha4, HCF106, and cpTatC, which correspond to the bacterial counterparts TatA, TatB, and TatC, respectively. The cpTat transports photosynthetic components, including proteins for both PSII and PSI, thus making this translocation system essential, and null mutants of each of the complex proteins are chlorotic and unviable at seedling stage (Voelker and Barkan, 1995; Motohashi et al., 2001). Thylakoid formation 1 (THF1) is a chloroplastic protein located in the outer membrane and the stroma. Mutation in the THF1 gene results in variegated leaves (Wang et al., 2004; Huang et al., 2006). THF1 interacts with the G protein α-subunit GPA1, regulates the FtsH protease activity and is vital for chloroplast development (Huang et al., 2006; Zhang et al., 2009).
In this study, we screened a T-DNA insertional mutant pool for mutations conferring drought resistance in Arabidopsis (Arabidopsis thaliana). Several mutant lines showing an increased drought resistance were isolated, and one of the mutant lines was identified as a weak allele of the hcf106 mutant. HCF106 physically interacts with THF1, and mutations in both genes resulted in drought resistance. Both hcf106 and thf1-1 mutants displayed reduced stomatal aperture and reduced water loss. We deduce that mutations in HCF106 and THF1 resulted in increased ROS accumulation in guard cells, thus promoting stomatal closure, preventing water loss, and conferring drought resistance.
RESULTS
Identification of the Drought-Resistant hcf106 Mutant
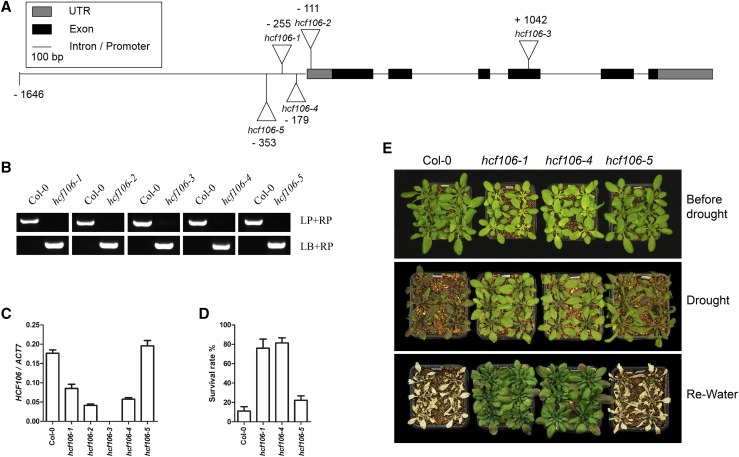
To establish the role of chloroplastic proteins in drought resistance in plants, we collected more than 1000 mutant lines with T-DNA insertions in the nuclear genes encoding chloroplastic proteins from the ABRC and performed a screening for mutants showing altered response to drought stress. Several mutants exhibiting drought resistance were identified, and one of the drought-resistant mutants, SALK_067017C, showed a reduced wilting phenotype compared with the wild-type (Columbia-0) plants under drought stress (Supplemental Fig. S1). The SALK_067017C line has a T-DNA insertion in the promoter region of the nuclear gene AT5G52440 previously named as High Chlorophyll Fluorescence 106 (HCF106); thus, this mutant was designated as hcf106-1. To confirm that the hcf106-1 mutation is responsible for the drought-resistant phenotype, four additional T-DNA insertion mutant alleles of the HCF106 gene, SALK_044421C, SALK_020680, SAIL_760_H06, and SAIL_831_E01 were obtained from the ABRC, and the homozygous lines were renamed as hcf106-2, hcf106-3, hcf106-4, and hcf106-5, respectively (Fig. 1B). The hcf106-2 and hcf106-3 mutants have T-DNA insertions in the 5′UTR and the fourth exon of the HCF106 gene, respectively, and the hcf106-4 and hcf106-5 have T-DNA insertions in the promoter region of the HCF106 gene (Fig. 1A). Quantitative real-time (qRT)-PCR analysis revealed that the transcript level of HCF106 was markedly reduced in hcf106-1 to -4 mutants, but was not significantly altered in the hcf106-5 mutant (Fig. 1C). The hcf106-2 and hcf106-3 mutants exhibited albino lethal phenotype under normal growth conditions (Supplemental Figs. S2A and S6A), indicating that HCF106 is an essential gene for Arabidopsis. The hcf106-1 mutant exhibited a more severe growth phenotype than the hcf106-4 mutant at the etiolated seedling stage, whereas both hcf106-1 and hcf106-4 performed similarly during mature plant stage (Supplemental Fig. S2). Under the drought stress condition tested, both hcf106-1 and hcf106-4 mutants showed a clear resistant phenotype as indicated by both survival rate and morphological appearance, while hcf106-5, in which the HCF106 gene expression was not affected, was as sensitive as the wild type (Fig. 1, D and E). These results confirm that the drought-resistant phenotype is indeed attributed to the mutations in the HCF106 gene.
Figure 1.
HCF106 mutants exhibit drought resistance phenotype. A, The scheme of T-DNA insertions of the hcf106 mutant alleles. B. PCR verification of the homozygous T-DNA insertion alleles of hcf106. LP, left primer. RP, right primer. LB, primer of T-DNA left border. C, Quantitative measurement of the relative expression level of HCF106 in hcf106-1, hcf106-2, hcf106-3, hcf106-4, hcf106-5, and Col-0. Values are means ± sd (n = 3). D, The survival rate of 21-d-old plants of hcf106-1, hcf106-4, hcf106-5, and Col-0 grown under drought stress in soil for 12 d and recovered for 5 d. Values are means ± sd (n = 3). E, Drought resistance assay of hcf106-1, hcf106-4, hcf106-5, and Col-0. Twenty-one-day-old plants were drought stressed for 10 d and rewatered for 5 d.
Molecular Complementation of the hcf106 Mutant
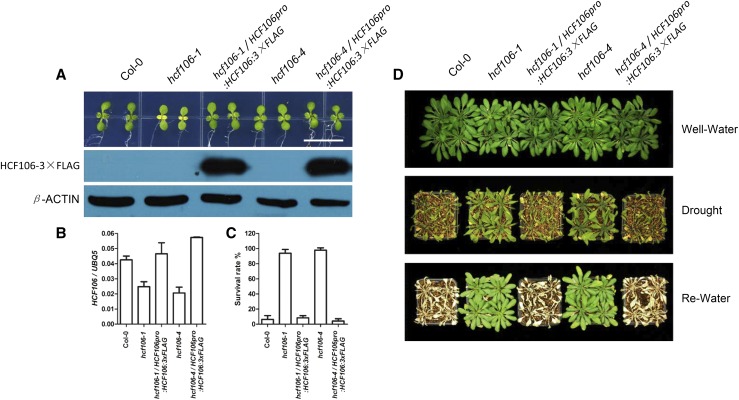
The genomic sequence of the HCF106 gene with its native promoter was fused with 3×FLAG tag to create HCF106pro:HCF106-3×FLAG construct. This construct was introduced into hcf106-1 and hcf106-4 mutants by Agrobacterium tumefaciens-mediated transformation. More than 20 transgenic lines expressing HCF106 were obtained, and these transgenic lines displayed similar growth and developmental phenotypes with the wild type under normal growth conditions. Two transgenic lines, hcf106-1/HCF106pro:HCF106:3×FLAG and hcf106-4/HCF106pro:HCF106:3×FLAG, were selected for further analysis (Fig. 2A). qRT-PCR analysis indicated that the HCF106 transcript levels in these two lines were recovered to the wild type level (Fig. 2B), and immunoblotting assay showed that HCF106-3×FLAG fusion protein was expressed in the transgenic plants (Fig. 2A). Twenty-one-day-old wild-type (Col-0), hcf106-1, hcf106-4, and the complementation transgenic plants grown in soil were subjected to drought treatment for 10 d. After rewatering for 5 d, nearly 90% of the hcf106-1 and hcf106-4 mutant plants survived, while the survival rates of hcf106-1/HCF106pro:HCF106:3×FLAG (∼8%) and hcf106-4/HCF106pro:HCF106:3×FLAG (∼4%) lines were comparable to that of the wild type (∼6%; Fig. 2, C and D). These results indicated that the HCF106 gene complemented the hcf106 mutant phenotype and is thus a negative regulator of plant drought resistance.
Figure 2.
Molecular complementation of the drought-tolerant phenotype of hcf106 mutants. A, The phenotype of hcf106 and complementation plants at early seedling stage. Top, 7-d-old seedlings; bottom, immunoblot analysis of HCF106-3×FLAG; β-ACTIN was used as a loading control; bar = 1 cm. B, HCF106 expression level in Col-0, hcf106-1, hcf106-4, hcf106-1 complementation line, and hcf106-4 complementation line. Values are means ± sd (n = 3). C, Survival rate of the complementation plants after drought and rewatering treatment (n = 4 biological replicates, 16 plants of each replicate). Data represent means ± sd. D, Col-0, hcf106-1, hcf106-4, hcf106-1 complementation line, and hcf106-4 complementation line grown under normal growth conditions for 21 d, drought treated for 10 d, and then recovered for 5 d.
Subcellular Localization and Tissue-Specific Expression of HCF106
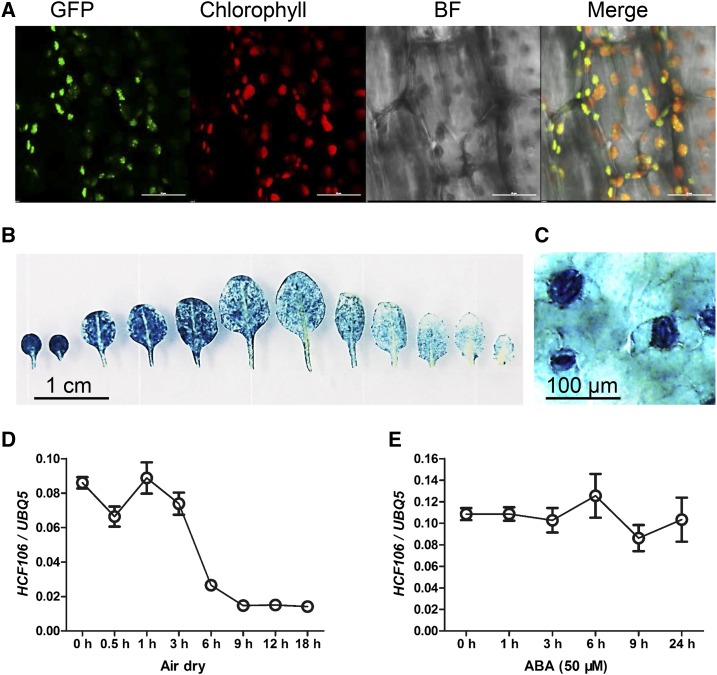
The maize HCF106 is a homolog of the E. coli tatB and is located in the thylakoids with its C-terminal region and amphipathic helix in the stroma (Settles et al., 1997). In order to assess the subcellular localization of the Arabidopsis HCF106, transgenic plants expressing genomic HCF106 fused with the green fluorescent protein (HCF106-GFP) driven by the HCF106 native promoter or CaMV35S promoter were generated. Confocal microscopy revealed that the HCF106-GFP fusion protein was specifically localized in the chloroplasts with some discrete bright spots (Fig. 3A; Supplemental Fig. S3, A and B). This observation indicated that Arabidopsis HCF106 protein is also a chloroplast protein. To determine the tissue expression pattern of HCF106, transgenic Arabidopsis plants containing a GUS reporter gene driven by the HCF106 promoter were analyzed. GUS activity was observed in most of the tissues in a seedling, with strong GUS staining in leaf cells and the vascular tissues of the root (Fig. 3B; Supplemental Fig. S3, C and D). In the leaf, GUS activity was mainly detected in guard cells (Fig. 3C). Additionally, we examined whether the expression of HCF106 is responsive to drought stress and exogenous ABA. qRT-PCR analysis showed that the HCF106 transcript was down-regulated by air dry for 3 to 18 h but that the expression was not significantly altered by ABA treatment for 1 to 24 h (Fig. 3, D and E).
Figure 3.
Subcellular localization and tissue-specific expression of HCF106. A, Subcellular localization of HCF106-GFP in hypocotyl of transgenic Arabidopsis seedlings expressing genomic HCF106-GFP driven by the 35S promoter. Chlorophyll, autofluorescence of chlorophyll. BF, bright field. Bars = 20 μm. B and C, GUS activity in the rosette leaves (B) and leaf epidermis (C) of 21-d-old transgenic Arabidopsis containing the fusion HCF106pro-GUS. D and E, HCF106 expression level in 21-d-old plant leaves treated with air drying (D) or 50 μm ABA (E). Values are means ± sd (n = 3).
To assess whether overexpression of HCF106 in Arabidopsis could affect drought resistance, we examined two independent 35S:HCF106-GFP transgenic lines with two independent 35S:GFP transgenic lines as controls. qRT-PCR assay confirmed that HCF106 expression was increased more than 10 times in the two overexpression lines relative to the GFP-only control plants (Supplemental Fig. S4B). Drought assay (Supplemental Fig. S4, A and C) and water loss analysis (Supplemental Fig. S4D) indicated that drought resistance remained unaffected in the HCF106 overexpression transgenic lines compared with the GFP-only controls.
HCF106 Is Important for Stomatal Control and Affects H2O2 Accumulation in Guard Cells
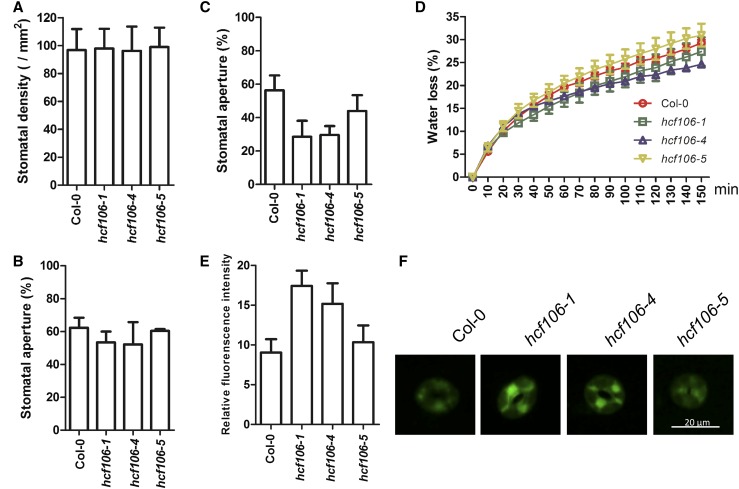
To elucidate the physiological mechanism of drought resistance conferred by the hcf106 mutations, we first examined the rate of water loss in Col-0, hcf106-1, hcf106-4, and hcf106-5. The detached leaves of hcf106-1 and hcf106-4 showed less water loss than those of Col-0 and hcf106-5 plants (Fig. 4D). This indicates that drought resistance in the hcf106 mutants is probably due to a decrease in water loss. Since stomata are the major sites in leaves that regulate water loss in response to drought conditions, we then examined the stomatal aperture (ratio of the width to length of the aperture) of Col-0 and hcf106 mutant leaves. Under normal growth conditions, the stomatal apertures of hcf106-1, hcf106-4, and hcf106-5 mutants were 53.4%, 52.2%, and 60.4%, respectively, compared with 62.4% in Col-0 (Fig. 4B). After drought treatment, the stomatal apertures of hcf106-1, hcf106-4, and hcf106-5 mutants were 28.5%, 29.6%, and 44.0%, respectively, compared with 56.3% in Col-0 (Fig. 4C). The stomatal density and guard cell size were also measured. The hcf106 mutants had no significant difference in stomatal density (Fig. 4A) and guard cell size (Supplemental Fig. S9C) when compared with the wild-type Col-0. ABA is a well-known hormonal regulator of stomatal movement and promotes stomatal closure in response to drought stress. We therefore measured the endogenous ABA contents, and the data revealed no significant difference between wild type and the hcf106 mutants under drought stress conditions (Supplemental Fig. S5A). Seed germination of Col-0, hcf106-1, and hcf106-4 also did not show difference in response to exogenous ABA (Supplemental Fig. S5B).
Figure 4.
Knockdown HCF106 enhances stomatal closure, reduces water loss, and increases H2O2 accumulation. A, Stomatal density and B, stomatal aperture of the middle leaves of 4-week-old Col-0, hcf106-1, hcf106-4, and hcf106-5 under normal growth conditions. Data represent means ± sd (n = 3). C, Stomatal aperture of the middle leaves of 3-week-old Col-0, hcf106-1, hcf106-4, and hcf106-5 after drought stress for 7 d. D, Water loss in Col-0, hcf106-1, hcf106-4, and hcf106-5 leaves (n = 3, each containing five fully expanded leaves from 4-week-old plants). Data represent means ± sd (n = 3). E, Quantitative analysis of H2O2 levels in guard cells of Col-0, hcf106-1, hcf106-4, and hcf106-5 (n = 4 leaves, 12 stomata per leaf from 21-d-old plants after drought stress for 7 d). Data represent means ± sd. F, H2O2 accumulation in guard cells of Col-0, hcf106-1, hcf106-4, and hcf106-5 labeled with CM-H2DCFDA (n = 3 leaves, 15 stomata per leaf from 21-d-old plants following drought stress for 7 d).
ROS were implicated in stomatal closure; thus, the levels of H2O2 and superoxide accumulation in wild type and mutant leaves were assayed by using histochemical staining. The levels of H2O2 and superoxide in the mutant leaves were higher than those in the wild-type leaves after drought stress treatment (Supplemental Fig. S9, A and B). Furthermore, H2O2 accumulation in guard cells was measured by using the fluorescent probe CM-H2DCFDA. The relative fluorescence intensities in the guard cells of hcf106-1 and hcf106-4 mutants were significantly higher than that in the wild type (Fig. 4, E and F), indicating that the guard cells of the mutants accumulated more H2O2 than those of the wild type under drought condition.
HCF106 Physically Interacts with THF1
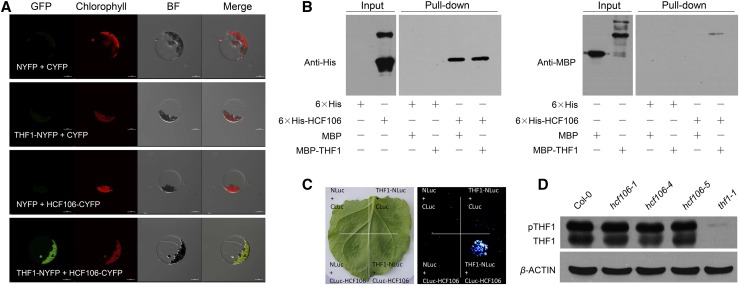
The essential role of HCF106 in chloroplast development was investigated by using ultrastructural analysis. The chloroplasts in the leaves of 6-d-old hcf106-2 and hcf106-3 seedlings were filled with numerous vesicles and failed to accumulate stromal lamellae, which resembles the structural defects of chloroplasts in the previously reported thf1 mutant (Supplemental Fig. S6B). The role of HCF106 in thylakoids formation was further studied by identifying HCF106-intercting proteins using coimmunoprecipitation (co-IP) of chloroplastic proteins followed by mass spectrometry analysis. The immunoprecipitation-mass spectrometry analysis showed that most of the potential HCF106-associated proteins are photosynthesis system proteins, including PSI components, PSII subunits, ATPase, and oxidase (Supplemental Table S1). Interestingly, THF1 was also coimmunoprecipitated with HCF106 protein, suggesting an interaction between these two proteins. HCF106-THF1 interaction was further verified by using bimolecular fluorescence complementation (BiFC) and firefly luciferase complementation imaging assay (LCI; Chen et al., 2008). Transient expression of both HCF106-CYFP and THF1-NYFP fusion genes in Arabidopsis mesophyll protoplasts of Col-0 resulted in yellow fluorescence in the chloroplasts (Fig. 5A), which indicates an interaction between HCF106 and THF1. The LCI assay in Nicotiana benthamiana leaf further supported the interaction between HCF106 and THF1 (Fig. 5C). To further test whether HCF106 directly interacts with THF1, we used recombinant 6×His-fused HCF106 and MBP-tagged THF1 proteins purified from Escherichia coli and performed a His pull-down assay. This assay indicated that HCF106 physically interacts with THF1 in vitro (Fig. 5B). The transcript level of THF1 was not affected by the hcf106 mutations (Supplemental Fig. S7D), but the THF1 protein level (the nonphosphorylated form) appeared slightly decreased in the hcf106-1 and hcf106-4 mutants (Fig. 5D). This suggests that, by forming a complex, HCF106 may help stabilize the THF1 protein in the chloroplasts.
Figure 5.
HCF106 physically interacts with THF1. A, BiFC assay in Arabidopsis protoplasts showing the interaction between HCF106 and THF1. B, In vitro pull-down of MBP-tagged THF1 using 6×His-tagged HCF106 as determined by immunoblotting with anti-His antibody and anti-MBP antibody. C, Luciferase imaging of N. benthamiana leaf coinfiltrated with the Agrobacteria strains containing THF1-NLuc and/or CLuc-HCF106. D, THF1 protein levels in leaves of 4-week-old Col-0, hcf106-1, hcf106-4, hcf106-5, and thf1-1 determined with immunoblots. pTHF1, phosphorylated form of THF1; β-ACTIN was used as a loading control.
Knockout of THF1 Gene Enhances Drought Resistance
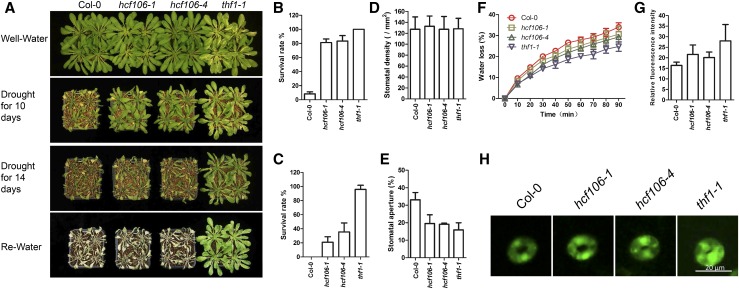
The tissue-specific expression of THF1 gene was investigated by using promoter-GUS analysis. GUS assay indicated an expression pattern of THF1 similar to that of HCF106, with both genes mainly expressed in leaves (Fig. 3B; Supplemental Fig. S7A). The expression of THF1 was found to be down-regulated rapidly after 6 h of air drying in Col-0 (Supplemental Fig. S7B), which resembles the HCF106 gene in its gene expression response to drought (Fig. 3D). To dissect the function of THF1 in plant response to drought stress, an Arabidopsis thf1 null mutant, thf1-1 (Huang et al., 2006), was used in drought resistance assays. 21-d-old plants of Col-0, hcf106-1, hcf106-4, and thf1-1 grown under normal growth conditions were treated with drought stress for 10 d or 14 d and then rewatered (Fig. 6A). Approximately 81%, 83%, and 100% of the hcf106-1, hcf106-4, and thf1-1 mutant plants, respectively, survived following a subsequent 5 d recovery period after 10 d drought stress, compared with 8% of the wild-type Col-0 plants (Fig. 6B). After 14 d of drought treatment, approximately 95% of thf1-1 plants still survived after rewatering, only 21% for hcf106-1 and 36% for hcf106-4, whereas wild-type plants all died after this severe drought treatment (Fig. 6C). Furthermore, drought assays of thf1-1 complementation lines demonstrated that the THF1 gene complements the thf1-1 drought resistance phenotype (Supplemental Fig. S8). These results indicate that thf1-1 is the most drought-tolerant mutant among the tested genotypes.
Figure 6.
Knockout of THF1 leads to H2O2 accumulation, enhanced stomatal aperture closure, reduced water loss, and increased tolerance to drought stress. A, Drought resistance assay. Col-0, hcf106-1, hcf106-4, and thf1-1 plants grown under normal growth condition for 21 d were treated with drought stress for 10 d, or 14 d, and then rewatered for 5 d. Survival rate of Col-0, hcf106-1, hcf106-4, and thf1-1 after drought treatment for 10 d (B) and 14 d (C; n = 4 biological replicates, 16 plants of each replicate). Stomatal density (D) and stomatal aperture (E) of the middle leaves of Col-0, hcf106-1, hcf106-4, and thf1-1 after drought stress for 7 d. F, Water loss in Col-0, hcf106-1, hcf106-4, and thf1-1 leaves (n = 3, each containing five fully expanded leaves from 4-week-old plants following drought stress for 7 d). G, Quantitative analysis of H2O2 levels in guard cells of Col-0, hcf106-1, hcf106-4, and thf1-1 (n = 3 leaves, 15 stomata per leaf leaves of 4-week-old plants following drought stress for 7 d). Data represent means ± sd. H, H2O2 accumulation in guard cells of Col-0, hcf106-1, hcf106-4, and thf1-1 labeled with CM-H2DCFDA probe.
THF1 Resembles HCF106 in the Control of Stomatal Aperture and H2O2 Accumulation
The aperture and density of stomata in Col-0, hcf106-1, hcf106-4, and thf1-1 plants were measured and analyzed to determine whether THF1 and HCF106 function in the same way in drought response. There was no significant difference in stomatal density among thf1-1, hcf106-1, hcf106-4, and Col-0 (Fig. 6D). However, the stomatal aperture in response to drought treatment exhibited a clear difference among these genotypes. The values of stomatal aperture were approximately 15.9% in thf1-1, 19.5% in hcf106-1, 19.1% in hcf106-4, and 33.1% in Col-0 (Fig. 6E). Consistent with the stomatal aperture results, the rate of water loss in the detached leaves of thf1-1 was lower than that in hcf106-1 and hcf106-4, and the wild-type Col-0 displayed highest water loss rate (Fig. 6F). These results indicate that the enhanced drought resistance of thf1-1 mutant is also likely due to a decreased stomatal aperture and reduced water loss under drought conditions. Similar to hcf106 mutants, the endogenous ABA content in thf1-1 leaves did not show significant difference when compared with that in the wild type (Supplemental Fig. S5A), which also suggests that the decreased stomatal aperture in thf1-1 mutant is not due to excessive ABA. Resembling the hcf106 mutants, the thf1-1 mutant also accumulated a higher level of H2O2 in leaves than the wild type (Supplemental Fig. S9A). Furthermore, the assay using CM-H2DCFDA fluorescence dye showed that the guard cells of thf1-1 accumulated more H2O2 than in the Col-0 wild-type plants (Fig. 6, G and H). The role of ROS in promoting stomatal closure was further studied by applying the commonly used cell-permeable ROS scavenger N-acetyl-Cys (NAC; Wang et al., 2016). After applying NAC, ROS accumulation in the guard cells and stomatal aperture were measured in the leaves of drought-stressed hcf106-4, thf1-1, and Col-0. The results showed that, without NAC treatment, the guard cells of hcf106-4 and thf1-1 accumulated more H2O2 than that of wild type as expected, while comparable levels of H2O2 in all these three genotypes were observed after NAC treatment (Supplemental Fig. S10A). Consequently, the stomatal apertures of hcf106-4 and thf1-1 reached a similar size with that of the wild type after applying NAC (Supplemental Fig. S10B). These results suggest that higher accumulation of H2O2 in the guard cells of hcf106 and thf1-1 mutants may result in decreased stomatal aperture, reduced water loss, and thus enhanced drought resistance.
HCF106 and THF1 Interact Genetically
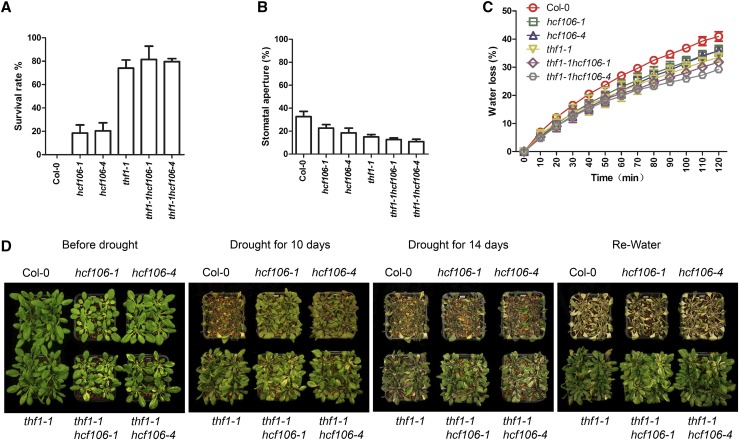
To further corroborate the observation that HCF106 and THF1 form a complex and function together in the control of stomatal aperture and drought resistance, we constructed double mutants thf1-1hcf106-1 and thf1-1hcf106-4 through genetic crosses (Supplemental Fig. S11) and then tested the drought resistance of the double and single mutants. All phenotypic analysis revealed that the double mutants behaved similarly as the thf1-1 single mutant (Fig. 7D). Under severe drought stress, the double mutants exhibited an approximately 80% survival rate, compared with approximately 74% in thf1-1 single mutant, which are significantly higher than 18%, 20%, and 0% in hcf106-1, hcf106-4, and Col-0, respectively (Fig. 7A). Consistent with the survival rates, the stomatal apertures in the leaves of 21-d-old single and double mutants after 7 d drought treatment were approximately 10.9% in thf1-1hcf106-1, 12.6% in thf1-1hcf106-4, 15.0% in thf1-1, 22.6% in hcf106-1, 18.4% in hcf106-4, and 32.7% in Col-0 (Fig. 7B), and the rate of water loss in the detached leaves of the double mutants was lower than that in the single mutants and the wild type (Fig. 7C). Collectively, these results indicate that the HCF106 and THF1 genes are likely to genetically interact and function in the same pathway to mediate drought resistance. However, both proteins may also act independently to modulate stomatal opening and water loss under drought conditions since the double mutants showed slightly stronger phenotypes than the thf1-1 single mutant (Fig. 7, B and C).
Figure 7.
The thf1hcf106 double mutants resemble the thf1-1 single mutant. A, Survival rate after drought treatment for 14 d (n = 3 biological replicates, 18 plants in each replicate). Data represent means ± sd. B, Stomatal aperture of the mature leaves of 3-week-old Col-0, hcf106-1, hcf106-4, thf1-1, thf1-1hcf106-1, and thf1-1hcf106-4 after drought stress for 7 d. C, Water loss in Col-0, hcf106-1, hcf106-4, thf1-1, thf1-1hcf106-1, and thf1-1hcf106-4 leaves (n = 5, each containing three fully expanded leaves from 4-week-old plants). Data represent means ± sd (n = 3). D, Drought resistance assay. Col-0, hcf106-1, hcf106-4, thf1-1 plants grown under normal growth condition for 21 d were treated with drought stress for 10 or 15 d and then rewatered for 5 d.
DISCUSSION
Chloroplast Function Is Crucial for Stomatal Regulation
The stoma is a multicellular complex on the leaf surface that is crucial for many important physiological processes such as photosynthesis and response to environmental changes. The stomatal complex consists of a pair of guard cells and subsidiary cells that are differentiated from epidermal cells. Interestingly, guard cells are the only epidermal cells possessing chloroplasts in many plant species, but guard cells have fewer and less-developed chloroplasts than mesophyll cells (Allaway and Setterfield, 1972). Guard cell chloroplasts may be involved in regulating stomatal movement. A strong argument against this notion came from the observation that the guard cells from plant species such as Paphiopedilum species lack chloroplasts but are still functional in controlling stomatal movement (Damelio and Zeiger, 1988). However, many studies on stomatal guard cells with chloroplasts suggested that chloroplasts do contribute to stomatal opening and closing. Although the underlying mechanisms are still unclear, guard cell chloroplasts could participate in stomatal movement via generating sugars as osmolytes, either by converting starch or by photosynthetic carbon reduction and/or via providing ATP for the activity of the plasma membrane H+-ATPase, which drives the influx of K+ (Lawson, 2009). A recent study showed that an Arabidopsis mutant having guard cells without chloroplasts has a significant reduction in stomatal opening when compared with the wild-type Arabidopsis (Wang et al., 2014), which supports the involvement of chloroplasts in stomatal function. Guard cell chloroplasts were also found to be essential for blue light-dependent stomatal opening in Arabidopsis (Suetsugu et al., 2014). In our study, we found that mutations in the nuclear-encoded chloroplast gene HCF106 promote stomatal closing under drought condition (Fig. 4C). The HCF106 gene is an essential gene for chloroplast development, and null mutants of this gene are lethal at the seedling stage (Supplemental Figs. S2A and S6A). Furthermore, the THF1 gene is an essential gene for chloroplast formation, and the thf1-1 mutation also promotes stomatal closing (Fig. 6E). HCF106 and THF1 physically interact in the chloroplast (Fig. 5). These results further support the important role of chloroplasts in guard cells in stomatal regulation.
Chloroplast Originated ROS and Stomatal Movement
Among several signaling molecules such as Ca2+, ABA, and other phytohormones, ROS have been well documented to play a signaling role in stomatal regulation (Song et al., 2014; Murata et al., 2015). Multiple sources of ROS have been proposed in guard cells in response to stress conditions. Apoplastic ROS can be generated through the activity of plasma membrane NADPH oxidases and cell wall-associated enzymes such as peroxidases, amine oxidases, and quinone reductases, while intracellular ROS are produced in several locations including chloroplasts, mitochondria, and peroxisomes (Marino et al., 2012; Kärkönen and Kuchitsu, 2015). The apoplastic ROS, particularly those produced via the plasma membrane NADPH oxidases and cell wall peroxidases, have been thought to be the major source of ROS in response to both biotic and abiotic stress. The Arabidopsis respiratory burst oxidase homologs (AtRbohs) are the plasma membrane NADPH oxidases that have been extensively studied. As the name indicated, Rboh enzymes are responsible for the fast production of ROS to form a ROS burst during the early stage of the responses. AtRbohs play key roles in stomatal closure in response to diverse factors, including ABA and other phytohormones, abiotic stress, and pathogenic elicitors by generating a burst of ROS and triggering downstream signaling events. In addition, AtRbohs are also involved in many other developmental processes, such as root hair formation and pollen tube growth (Marino et al., 2012; Baxter et al., 2014; Murata et al., 2015). Although intracellular organelles such as chloroplasts, mitochondria, and peroxisomes are well-known sites of ROS production, whether and how the ROS from these intracellular sources contribute to the regulation of stomatal movement has not been well understood.
Chloroplasts generate ROS under both normal and stress conditions. Various forms of ROS including singlet oxygen, oxygen anion, and H2O2 can be generated in chloroplasts, and stress conditions such as drought and heat enhance the production of chloroplastic ROS (Galvez-Valdivieso and Mullineaux, 2010; Miller et al., 2010). Although chloroplasts are major sites of ROS production (Asada, 2006), its role in ROS-mediated stomatal closure is unclear. In this study, we found that mutations in the nuclear encoded chloroplastic proteins HCF106 and THF1 resulted in an increase in ROS production in guard cells (Fig. 4, E and F; Fig. 6, G and H), and the increased ROS in guard cells coincided with the enhanced stomatal closure in these mutants in response to drought stress (Figs. 4C and 6E). The increased stomatal closure in the mutants under drought stress was abolished by exogenously applied ROS scavenger NAC (Supplemental Fig. S10). We deduce that disruption of HCF106 or THF1 may have resulted in malfunction of the photosystems in chloroplasts in guard cells, which then causes excessive ROS production especially under drought stress conditions, and the elevated ROS in the chloroplasts, together with apoplastic ROS, enhance stomatal closure under drought stress. The chloroplastic ROS in guard cells may provide the basal level of ROS that modulates the intensity and duration of stomatal closure in response to relatively long-term stress conditions such as drought.
How chloroplastic ROS regulate stomatal movement is still an unanswered question. A recent study indicated that ROS in guard cell chloroplasts were tripled after ABA and methyl jasmonate treatments, and the increased ROS were observed to coincide with starch grain accumulation (Leshem and Levine, 2013). Sugars from chloroplasts have long been proposed to serve as osmolytes to regulate water potential in guard cells thus control stomatal movement (Lawson, 2009). H2O2 accumulation was shown to inhibit α-amylase activity required for starch degradation to produce sugars (Sparla et al., 2006). These lines of evidence support the notion that increased chloroplastic ROS in hcf106 and thf1-1 mutants could inhibit starch degradation and thus reduce sugar accumulation, and reduced sugar content in guard cells results in higher water potential and water efflux from the guard cells, and as a result, the stomata close. It is also possible that higher chloroplastic ROS work together with apoplastic ROS to mediate stronger signaling response in guard cells to promote stomatal closure. Another possibility is that reduced photosynthetic capacity in guard cells of the mutants may result in reduced photosynthetic production of sugars thus causing stomatal closure. All these possibilities need to be experimentally examined.
Manipulating Nuclear Genes Encoding Chloroplast Proteins in Guard Cells Could Be a Way to Improve Drought Resistance in Crops
Both conventional breeding and modern genetic manipulation have been extensively attempted to create drought-resistant crops. Drought resistance is a complex trait that can be attributed to root morphological characters such as deep and large root system and/or shoot-related traits such as stomatal conductance. Major efforts on improving drought resistance have been made on utilizing key genes in drought response pathways through genetic engineering (Hu and Xiong, 2014). ABA as the vital phytohormone for drought stress response in plants has been the focal molecule for manipulating drought resistance in crops, and key genes in the ABA biosynthetic and signaling pathways have been tested in conferring drought resistance. Also, systems approaches including various omics analyses have recently been used for identifying drought resistance-related genes. The identified candidate genes are valuable for evaluation of natural variations between resistant and sensitive varieties and could be eventually used for breeding drought-resistant crops (Jogaiah et al., 2013; Krannich et al., 2015; Zhu et al., 2016).
Although plants have evolved diverse mechanisms to resist drought stress, control of stomatal movement is a common strategy for most plants to respond to drought conditions. Stomata close in order to preserve water under drought conditions, which is modulated by a battery of signaling events, including ABA signaling. Stomata are the sites of water transpiration and account for most of the water loss in plants. Thus, manipulation of stomata in leaves is one of the favorable strategies for improving drought resistance. One of the ways to modulate stomatal movement is to express genes specifically in guard cells in a spatial and temporal manner to promote stomatal closure in response to drought stress. This approach would be useful for improving drought resistance via specifically targeting stomata while minimizing potential yield penalty by avoiding side effects in other cells. Chimeric promoters that are drought inducible and guard cell specific have been developed (Rusconi et al., 2013; Na and Metzger, 2014), but application of these promoters in drought resistance has yet to be tested. Another way of manipulating stomata is to alter stomatal density, thus reducing water transpiration. In Arabidopsis, genetic manipulation of the epidermal patterning factors has generated plant lines with a range of stomatal density (Richardson and Torii, 2013). Reduced stomatal density was found to increase plant resistance to water deficit (Doheny-Adams et al., 2012; Hepworth et al., 2015).
In our study, we found that partial loss-of-function of HCF106 gene conferred drought resistance in Arabidopsis (Fig. 1, D and E). Consistent with THF1 being an interacting partner of HCF106, mutation in THF1 gene also caused drought-resistant phenotype (Fig. 6, A–C). These results suggest that modulating chloroplast function via changing the chloroplast proteins could be a way to enhance drought resistance in plants. However, many genes encoding chloroplastic proteins, including HCF106 and THF1, are essential genes or important for chloroplast functions, and null mutants of some of these genes are seedling lethal. Therefore, application of this strategy to crops requires targeted knockdown of the nuclear-encoded chloroplast genes in guard cells. Utilizing guard-cell-specific promoters and the recently developed CRISPR/Cas9 technology to specifically manipulate chloroplasts in guard cells is a promising approach to creating drought-resistant plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All the Arabidopsis (Arabidopsis thaliana) genetic materials used in this study were in the Columbia-0 background. The T-DNA insertion mutants, hcf106-1, hcf106-2, hcf106-3, hcf106-4, and hcf106-5 were obtained from the ABRC (stock numbers SALK_067017C, SALK_044421C, SALK_020680, SAIL_760_H06, and SAIL_831_E01). The thf1-1 was provided by Dr. Jinrong Huang (Huang et al., 2006). The thf1-1hcf106-1 and thf1-1hcf106-4 double mutants were generated by genetic crossing and subsequent PCR-based genotyping in the F2 population. For seedling growth in agar plate, seeds were surface-sterilized and stored in sterile water at 4°C for 48 h for stratification, followed by germination at 22°C in 0.5× Murashige and Skoog medium (pH 5.8) with 1% Suc and 0.6% (w/v) agar. Seven-day-old seedlings were then transplanted in soil and grow in a growth room at 22°C with 16 h light/8 h dark (long day) for observations of normal growth and development, seeds proliferation, and water loss experiments in detached leaves. For drought resistance test, 8-d-old seedlings grown in agar plates were transferred to soil and placed at 22°C in a growth room with 10 h light/14 h dark (short day) cycle and continue growing for 2 weeks; the plants were then subjected to drought treatment by withholding water for indicated days described in the figures.
To obtain the HCF106pro:GUS fusion construct, the 1645-bp promoter fragment of HCF106 gene was cloned into the pMDC162 binary vector (Curtis and Grossniklaus, 2003). For the HCF106pro:HCF106-GFP fusion, the genomic region containing the HCF106 gene with the 1645-bp promoter was cloned into the pMDC110 binary vector (Curtis and Grossniklaus, 2003). The 35S:HCF106-GFP fusion was constructed by inserting the HCF106 coding sequence (CDS) into the pGWB5 binary vector (Nakagawa et al., 2007). All constructs were confirmed by sequencing and then introduced into Col-0 by Agrobacterium tumefaciens GV3101 using the floral dip method (Clough and Bent, 1998). For the HCF106pro:HCF106-3×FLAG and THF1pro:THF1-3×FLAG, the genomic regions containing the HCF106 and THF1 genes with at least 1.5 kb native promoters were cloned into the pCAMBIA1305 binary vector (He et al., 2009). These constructs were introduced into hcf106-1, hcf106-4, or thf1-1 mutants for molecular complementation and co-IP experiments.
Measurement of Stomatal Aperture
For stomatal aperture measurement, 3-week-old plants of Col-0, hcf106-1, hcf106-4, hcf106-5, and thf1-1 grown in short day (10 h light/14 h dark) were withheld water for 1 week, and then the mature rosette leaves were sampled. To assess the effect of NAC on stomatal aperture, the mature rosette leaves were applied with 10 mm NAC separately for 0, 0.5, 1, 1.5, and 2 h and then sampled for analysis. The leaf surface imprint method was performed as described previously (Yu et al., 2008). Briefly, a glass slide was coated with glue, and the leaf abaxial side was pressed on the glue-coated slide for 20 s. The leaf was torn off, and the imprint on the glass slide was examined under a microscope. For statistical analysis of stomatal aperture, five leaves per line and five fields per leaves were analyzed.
Measurement of Water Loss in Detached Leaves
To measure water loss of detached leaves, mature leaves were harvested from 4-week-old Col-0, hcf106-1, hcf106-4, hcf106-5, and thf1-1 plants grown in a growth room with 10-h-light/14-h-dark period under normal growth conditions. The detached leaves were placed on an electronic balance and periodically weighed. Water loss was calculated as a percentage of the decreased fresh weight to the initial fresh weight of the detached leaves. The experiment was performed three times, each time with two replicate leaves per genotype.
Co-IP
The chloroplasts of mature leaves of 5-week-old Col-0 and transgenic HCF106pro:HCF106-3×FLAG plants were isolated (Huang et al., 2013) and incubated in the binding buffer (40 mm HEPES, pH 7.4, 2 mm EDTA, 10 mm sodium pyrophosphate tetrabasic, 10 mm β-glycerophosphate disodium salt hydrate, and 0.3% CHAPS hydrate) containing Proteinase inhibitor Cocktail (Roche) for 30 min at 4°C. After centrifugation, the supernatant of cell lysates was incubated with anti-FLAG antibodies coupled to magnetic Dynabeads Protein G (Life Technologies) for 3 h at 4°C. The magnetic beads were then collected by centrifugation and washed three times with the low-salt washing buffer (40 mm HEPES, pH 7.4, 2 mm EDTA, 10 mm sodium pyrophosphate tetrabasic, 10 mm β-glycerophosphate disodium salt hydrate, 0.3% CHAPS hydrate, and 150 mm NaCl). The immunoprecipitated proteins were digested with trypsin (Promega), and then analyzed by mass spectrometry using nanoAcquity ultraperformance LC (Waters) coupled with an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific).
BiFC Assays
The full-length CDS of HCF106 and THF1 genes were cloned into pSAT4-CYFP and pSAT4-NYFP BiFC system vectors, respectively, to create HCF106-CYFP and THF1-NYFP. Two micrograms of each of these two plasmids were mixed and cotransformed into Arabidopsis mesophyll protoplasts using polyethylene glycol-mediated transient transformation method (Yoo et al., 2007). After culturing in light at 22°C for 20 h, the fluorescence in the transformed cells were imaged using a Nikon A1 spectral confocal microscope imaging system.
LCI Assay
The full-length CDS of HCF106 and THF1 were constructed into pCAMBIA-NLuc and pCAMBIA-CLuc system vectors, respectively, to produce CLuc-HCF106 and THF1-NLuc. The constructs were cotransferred into Nicotiana benthamiana leaves by using A. tumefaciens-mediated transient transformation method as described previously (Chen et al., 2008). One millimolar luciferin was infiltrated into leaves, and the leaves were kept in dark for 10 min to quench the fluorescence. The LUC images were captured using a low-light cooled CCD imaging system (Lumazone; Roper Scientific).
Protein Expression and Pull-Down Assay
The HCF106 CDS was cloned into the vector pET28a, and the THF1 CDS was inserted into the vector pMAL-c5x. The constructs were verified by sequencing and transformed into BL21 competent cells. The pull-down assay was performed according to Cui et al. (2015) with some modifications. Briefly, Escherichia coli cells expressing His or His-HCF106 recombinant proteins were lysed with ultrasonic cell disruptor in 1× PBS followed by centrifugation. The supernatant was incubated with 100 μL nickel magnetic beads (Biotool) in 1 mL binding buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, and 50 mm imidazole) at 4°C for 2 h and washed three times with 1 mL washing buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, and 100 mm imidazole). After washing, the supernatants containing MBP or MBP-THF1 recombinant proteins were added to the beads and incubated for additional 2 h at 4°C. The beads were collected by centrifugation and washed three times with the binding buffer. The His pull-down proteins were then resuspended in elution buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, and 500 mm imidazole) and eluted for 30 min. The eluted proteins were separated by 12% SDS-PAGE and detected by immunoblotting with anti-His antibodies (Abmart) and anti-MBP antibodies (Abmart).
H2O2 Measurement in Guard Cells
CM-H2DCFDA was used as the molecule probe to detect H2O2 content in guard cells. The abaxial epidermal strips of the leaves from 4-week-old plants were floated in 0.1 m potassium phosphate buffer (pH 7.2) for 30 min. Two micromolar (final concentration) of CM-H2DCFDA (Sigma-Aldrich) was added to the solution, and the strips were incubated for 20 min at 22°C in dark. To study the effect of NAC on ROS level, epidermal strips were transferred to 0.1 m potassium phosphate buffer (pH 7.0) containing 1 mm NAC for 0, 10, 20, and 40 min. Two micromolar (final concentration) of CM-H2DCFDA was then added, and the strips were incubated for 30 min at room temperature in dark. The strips were washed twice with 0.1 mm KCl, 0.1 mm MgCl2 for 10 min to remove excess staining buffer, and the guard cells in the strips were observed under a fluorescent microscope (Olympus DP72). All images were acquired under identical conditions. The fluorescence emission of the guard cells was analyzed using the software equipped for the microscope. Each sample contained strips from ten independent leaves, and at least seven randomly selected guard cells from each strip were analyzed.
Histochemical Staining and Confocal Microscopy Analysis
Hydrogen peroxide and superoxide in the expended leaves of 4-week-old plants were detected by using DAB and NBT staining as previously described (Jin et al., 2014). Staining for GUS activity was performed as previously described (Wang et al., 2015) with some modifications. The samples were incubated in GUS staining solution (0.1 m potassium phosphate buffer, pH 7.0, 1 mmol/L ferrocyanide, 1 mmol/L ferricyanide, and 0.1% Triton X-100) containing 0.5 mg/mL X-gluc at 37°C in dark for 2 h. GUS images were taken by using the Olympus DP72 and SZX7 microscopes. For detection of GFP, 7-d-old transgenic seedlings harboring 35S:HCF106-GFP or HCF106pro:HCF106-GFP were used for detection of GFP and chlorophyll autofluorescence using a confocal microscope. The images were captured using Leica SP8 confocal microscope, and at least five independent transgenic lines were examined.
qRT-PCR Analysis
Total RNA was extracted from the shoots of 18-d-old seedlings using TRIzol reagent (Invitrogen) and reverse transcription was performed by using the iScript cDNA synthesis kit (Bio-Rad). After inactivation of the enzymes by heating, a 0.5-μL aliquot was used for real-time quantitative PCR. All quantitative real-time PCR analyses were performed using the AceQ qPCR SYBR Green Master Mix (Vazyme) according to the manufacturer’s protocol. UBQ5, ACT2, and ACT7 were used as internal controls in qRT-PCR. Each analysis consisted of three biological replicates. Each sample had three qPCR reactions. The primers used in this study were listed in Supplemental Table S1.
Analysis of Chloroplast Ultrastructure
Cotyledons of 7-d-old seedlings were collected for transmission electron microscopy. The cotyledons were fixed in 2.5% glutaraldehyde and postfixed overnight at 4°C in 1% OsO4. After dehydration using an ethanol series, the samples were gradually infiltrated with a series of epoxy resin in epoxy propane, and then embedded in Epon 812 resin. The embedded materials were thin sectioned, stained in uranium acetate followed by lead citrate, and photographed with a Phillips CM120 transmission electron microscope.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: HCF106 (AT5G52440), THF1 (AT2G20890), ACT2 (AT3G18780), ACT7 (AT5G09810), and UBQ5 (AT3G62250).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Drought resistance assay of SALK_067017C.
Supplemental Figure S2. Developmental phenotype of the hcf106 mutants.
Supplemental Figure S3. Subcellular localization of HCF106-GFP and root expression pattern of HCF106.
Supplemental Figure S4. Drought assay of the overexpression lines of HCF106.
Supplemental Figure S5. ABA contents and germination assay of hcf106 and thf1 mutants.
Supplemental Figure S6. Ultrastructure of chloroplasts in the hcf106 and thf1 mutants.
Supplemental Figure S7. Expression pattern of THF1.
Supplemental Figure S8. Drought assay of thf1-1 complementation lines.
Supplemental Figure S9. Determination of ROS and guard cell size in hcf106 and thf1 mutants.
Supplemental Figure S10. ROS affects the stomatal aperture in hcf106 and thf1 mutants.
Supplemental Figure S11. Identification of the double mutant thf1hcf106.
Supplemental Table S1. Proteins interacting with HCF106 as identified by mass spectrometry in chloroplast.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center at Ohio State University for seed stocks, Dr. Yimin She for assistance with mass spectrometry, and Dr. Minjie Cao for his constructive discussion and help.
Glossary
- ROS
reactive oxygen species
- NAC
N-acetyl-Cysteine
Footnotes
This work was supported by the Chinese Academy of Sciences.
References
- Acharya BR, Jeon BW, Zhang W, Assmann SM (2013) Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol 200: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Allaway WG, Setterfield G (1972) Ultrastructural observations on guard cells of vicia-faba and allium-porrum. Can J Bot 50: 1405–1413 [Google Scholar]
- Asada K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65: 1229–1240 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui LG, Shan JX, Shi M, Gao JP, Lin HX (2015) DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet 11: e1005617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amelio ED, Zeiger E (1988) Diversity in guard-cell plastids of the orchidaceae—a structural and functional study. Canadian Journal of Botany 66: 257–271 [Google Scholar]
- Doheny-Adams T, Hunt L, Franks PJ, Beerling DJ, Gray JE (2012) Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Philos Trans R Soc Lond B Biol Sci 367: 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Mullineaux PM (2010) The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol Plant 138: 430–439 [DOI] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK (2009) An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth C, Doheny-Adams T, Hunt L, Cameron DD, Gray JE (2015) Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol 208: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiong L (2014) Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol 65: 715–741 [DOI] [PubMed] [Google Scholar]
- Hua D, Wang C, He J, Liao H, Duan Y, Zhu Z, Guo Y, Chen Z, Gong Z (2012) A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24: 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J (2013) Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol Plant 6: 1673–1691 [DOI] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Liu B, Luo L, Feng D, Wang P, Liu J, Da Q, He Y, Qi K, Wang J, et al. (2014) HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell 26: 1213–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogaiah S, Govind SR, Tran LS (2013) Systems biology-based approaches toward understanding drought tolerance in food crops. Crit Rev Biotechnol 33: 23–39 [DOI] [PubMed] [Google Scholar]
- Kärkönen A, Kuchitsu K (2015) Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 112: 22–32 [DOI] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krannich CT, Maletzki L, Kurowsky C, Horn R (2015) Network candidate genes in breeding for drought tolerant crops. Int J Mol Sci 16: 16378–16400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T. (2009) Guard cell photosynthesis and stomatal function. New Phytol 181: 13–34 [DOI] [PubMed] [Google Scholar]
- Leshem Y, Levine A. 2013. Zooming into sub-organellar localization of reactive oxygen species in guard cell chloroplasts during abscisic acid and methyl jasmonate treatments. Plant Signal Behav 8: 10.4161/psb.25689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N (2012) A burst of plant NADPH oxidases. Trends Plant Sci 17: 9–15 [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP (2006) An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Motohashi R, Nagata N, Ito T, Takahashi S, Hobo T, Yoshida S, Shinozaki K (2001) An essential role of a TatC homologue of a Delta pH-dependent protein transporter in thylakoid membrane formation during chloroplast development in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 10499–10504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Mori IC, Munemasa S (2015) Diverse stomatal signaling and the signal integration mechanism. Annu Rev Plant Biol 66: 369–392 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na JK, Metzger JD (2014) Chimeric promoter mediates guard cell-specific gene expression in tobacco under water deficit. Biotechnol Lett 36: 1893–1899 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Richardson LG, Torii KU (2013) Take a deep breath: peptide signalling in stomatal patterning and differentiation. J Exp Bot 64: 5243–5251 [DOI] [PubMed] [Google Scholar]
- Robinson C, Bolhuis A (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta 1694: 135–147 [DOI] [PubMed] [Google Scholar]
- Rusconi F, Simeoni F, Francia P, Cominelli E, Conti L, Riboni M, Simoni L, Martin CR, Tonelli C, Galbiati M (2013) The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J Exp Bot 64: 3361–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles AM, Yonetani A, Baron A, Bush DR, Cline K, Martienssen R (1997) Sec-independent protein translocation by the maize Hcf106 protein. Science 278: 1467–1470 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, et al. (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Song Y, Miao Y, Song CP (2014) Behind the scenes: the roles of reactive oxygen species in guard cells. New Phytol 201: 1121–1140 [DOI] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P (2006) Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol 141: 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Takami T, Ebisu Y, Watanabe H, Iiboshi C, Doi M, Shimazaki K (2014) Guard cell chloroplasts are essential for blue light-dependent stomatal opening in Arabidopsis. PLoS One 9: e108374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R, Barkan A (1995) Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J 14: 3905–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WH, He EM, Chen J, Guo Y, Chen J, Liu X, Zheng HL (2016) The reduced state of the plastoquinone pool is required for chloroplast-mediated stomatal closure in response to calcium stimulation. Plant J 86: 132–144 [DOI] [PubMed] [Google Scholar]
- Wang SW, Li Y, Zhang XL, Yang HQ, Han XF, Liu ZH, Shang ZL, Asano T, Yoshioka Y, Zhang CG, et al. (2014) Lacking chloroplasts in guard cells of crumpled leaf attenuates stomatal opening: both guard cell chloroplasts and mesophyll contribute to guard cell ATP levels. Plant Cell Environ 37: 2201–2210 [DOI] [PubMed] [Google Scholar]
- Wang Z, Mao JL, Zhao YJ, Li CY, Xiang CB (2015) L-Cysteine inhibits root elongation through auxin/PLETHORA and SCR/SHR pathway in Arabidopsis thaliana. J Integr Plant Biol 57: 186–197 [DOI] [PubMed] [Google Scholar]
- Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL (2004) Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol 136: 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S, Held K, Schlücking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J (2008) A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Chen S, Harmon AC (2014) Protein phosphorylation in stomatal movement. Plant Signal Behav 9: e972845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wei Q, Wu W, Cheng Y, Hu G, Hu F, Sun Y, Zhu Y, Sakamoto W, Huang J (2009) Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidopsis. Plant J 58: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Zhu M, Monroe JG, Suhail Y, Villiers F, Mullen J, Pater D, Hauser F, Jeon BW, Bader JS, Kwak JM, et al. (2016) Molecular and systems approaches towards drought-tolerant canola crops. New Phytol 210: 1169–1189 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.