A systematic characterization of starch turnover in Arabidopsis during reproductive development unravels new starch deposits and sheds light on starch metabolism and transport in reproductive tissues.
Abstract
The accumulation of starch within photosynthetic tissues and within dedicated storage organs has been characterized extensively in many species, and a function in buffering carbon availability or in fueling later growth phases, respectively, has been proposed. However, developmentally regulated starch turnover within heterotrophic tissues other than dedicated storage organs is poorly characterized, and its function is not well understood. Here, we report on the characterization of starch turnover during flower, early embryo, and silique development in Arabidopsis (Arabidopsis thaliana) using a combined clearing-staining technique on whole-mount tissue. Besides the two previously documented waves of transient starch accumulation in the stamen envelope, occurring during meiosis and pollen mitosis I, we identified a novel, third wave of starch amylogenesis/amylolysis during the last stages of stamen development. To gain insights into the underlying molecular mechanisms, we analyzed publicly available microarray data, which revealed a developmentally coordinated expression of carbohydrate transport and metabolism genes during these waves of transient starch accumulation. Based on this analysis, we characterized starch dynamics in mutants affecting hexose phosphate metabolism and translocation, and identified the Glc-6-phosphate/phosphate antiporter GPT1 as the putative translocator of Glc-6-phosphate for starch biosynthesis in reproductive tissues. Based on these results, we propose a model of starch synthesis within the pollen grain and discuss the nutrient transport route feeding the embryo within the developing seed.
Starch is the major storage carbohydrate in plants. It exists as transitory starch or, more or less permanently, as storage starch (Streb and Zeeman, 2012). Transitory starch accumulates during the day within chloroplasts of photosynthetic tissues and is degraded during the night. In contrast, storage starch accumulates within dedicated organs, where it is stored for longer periods. Transitory starch is considered an important integrator of plant growth, buffering recurrent changes in carbon availability that result from the diurnal light/dark rhythm (Geigenberger, 2011; Streb and Zeeman, 2012). Storage starch covers long-term carbon needs, fueling germination or regrowth after certain dormancy periods (Smith, 2012). However, throughout plant development, other types of starch, which do not necessarily fall into these broad categories, accumulate within cells of sink tissues, such as most reproductive tissues. Various waves of amylogenesis/amylolysis have been reported during flower and seed development in different species. The best-characterized waves of transient starch accumulation are those that take place during stamen and pollen development. One to two cycles of starch amylogenesis/amylolysis occur either in the sporogenous cells or the anther wall layers (Feijó and Pais, 1988; Clément and Pacini, 2001). Much less is known about starch dynamics during carpel and embryo development. The only systematic characterization of starch turnover during fruit and seed development was done more than half a century ago in Dianthus chinensis (Buell, 1952). Recently, starch dynamics during seed development in Brassica napus and Arabidopsis (Arabidopsis thaliana), both of which accumulate oils in their mature seeds, have been characterized (Baud et al., 2002; Andriotis et al., 2010b). However, the developmental and physiological significance of this starch accumulation is not well understood.
Despite the importance of starch turnover for plant development and agriculture, the molecular mechanisms underlying starch metabolism are still not completely resolved. A fully chloroplast-based biosynthesis using photosynthetic metabolites is the widely accepted pathway in mesophyll cells (Streb and Zeeman, 2012). Three plastidic enzymes convert the Calvin-Benson cycle-derived Fru-6-P to ADP-Glc. The phosphoglucose isomerase (PGI) converts Fru-6-P to Glc-6-P; then, Glc-6-P is converted to Glc-1-P by phosphoglucomutase (PGM); Glc-1-P is then metabolized to ADP-Glc by ADP-Glc pyrophosphorylase (AGPase). However, there is ongoing discussion over the origin of the residual starch (3%–15%) in some null mutants of the plastidic PGI-PGM-AGPase pathway (Niewiadomski et al., 2005; Streb et al., 2009; Kunz et al., 2010; Bahaji et al., 2015), the significance of the observed uptake of exogenously applied Glc-1-P to isolated chloroplasts and its conversion to starch (Fettke et al., 2011), the disputed possibility of an alternative cytosolic route of ADP-Glc synthesis by Suc synthase (SUS) and its potential translocation to the chloroplast (Muñoz et al., 2005; Barratt et al., 2009; Baroja-Fernández et al., 2012; Smith et al., 2012), and the recently suggested PGI1-independent starch synthesis (Bahaji et al., 2015). Starch synthesis in nonphotosynthetic plastids relies on the translocation of carbon from the cytosol, mainly in the form of hexose phosphate (Flügge, 1999). Accumulating evidence in all dicotyledonous plant species studied so far (e.g. Arabidopsis, broad bean [Vicia faba], and potato [Solanum tuberosum]) points to Glc-6-P as the preferred translocated hexose phosphate and the Glc-6-P/phosphate antiporter GPT1 as the active translocator (Kammerer et al., 1998; Geigenberger et al., 2004; Niewiadomski et al., 2005; Rolletschek et al., 2007; Zhang et al., 2008). This is further supported by the fact that the loss of plastidial PGI1 activity impacts starch accumulation only in autotrophic Arabidopsis cells (Yu et al., 2000; Tsai et al., 2009), while mutations affecting plastidial PGM1 result in a low-starch phenotype in both autotrophic and heterotrophic tissues (Caspar et al., 1985). Starchless amyloplasts were reported in gpt1 mutant pollen grains, and a dual function of GPT1 in starch synthesis and the oxidative pentose phosphate pathway was suggested (Kammerer et al., 1998; Niewiadomski et al., 2005; Kunz et al., 2010). However, a seed-specific knockdown of GPT1 activity revealed that starch grains still accumulated in arrested embryos (Andriotis et al., 2010a). Thus, it is still unclear whether starch metabolism in heterotrophic cells of Arabidopsis is based on GPT1-mediated Glc-6-P translocation. Arabidopsis does not accumulate starch in its mature seeds; hence, seeds are not the best system in which to characterize long-term storage starch. However, given the extensive molecular knowledge in this species and the good understanding of transitory starch metabolism in its leaves, Arabidopsis is an excellent system in which to characterize developmentally regulated starch accumulation.
Here, we characterized starch dynamics throughout flower development as well as early embryo and silique development in Arabidopsis using a combined clearing-staining technique on whole-mount tissues (Herr, 1972). We analyzed publicly available microarray data of different stages and organs of flowers and seeds and related the transcriptional dynamics to the observed starch turnover in the flower and young embryo. This analysis indicated that GPT1 may be the translocator of hexose phosphate for starch biosynthesis in heterotrophic tissues. An analysis of starch dynamics in gpt1, pgi1, and pgm1 mutants, which are related to Glc-6-P transport and metabolism, provided evidence for the role of GPT1 in this process. Our work provides new insights into starch synthesis in the pollen grain and the nutrient transport route feeding the embryo during seed development. Understanding starch dynamics and metabolism during sexual reproduction is an important step toward a better understanding of the function of developmentally regulated starch metabolism.
RESULTS
A Previously Undescribed, Third Starch Synthesis Wave Occurs in Stamen Just before Anthesis
Using iodine staining, we could observe a single wave of starch accumulation/degradation in sporogenous and gametophytic cells and three waves within the sporophytic anther wall layers. The developmental delay of short stamens with respect to long stamens was critically helpful in tracking the highly dynamic starch turnover, especially in revealing the existence of a third wave in the staminal envelope.
During early flower development, starch accumulated mainly in the peduncle and the receptacle (Fig. 1A). As soon as the flower organ primordia developed, starch accumulated to high amounts in their basal parts (Fig. 1, B–D). Paralleling the development of the stamen filament and the connective tissue of the anther, amylogenesis extended to these two tissues (Fig. 1E). At the completion of anther wall layer formation, starch accumulation extended beyond the filament and the connective tissue into the newly differentiated endothecium (Fig. 1F). Starch accumulation reached its first maximum in the stamen at the tetrad stage (Fig. 1, G–I). Interestingly, concomitant with the release of the microspores from the tetrad, starch practically vanished from the stamens (Fig. 1J).
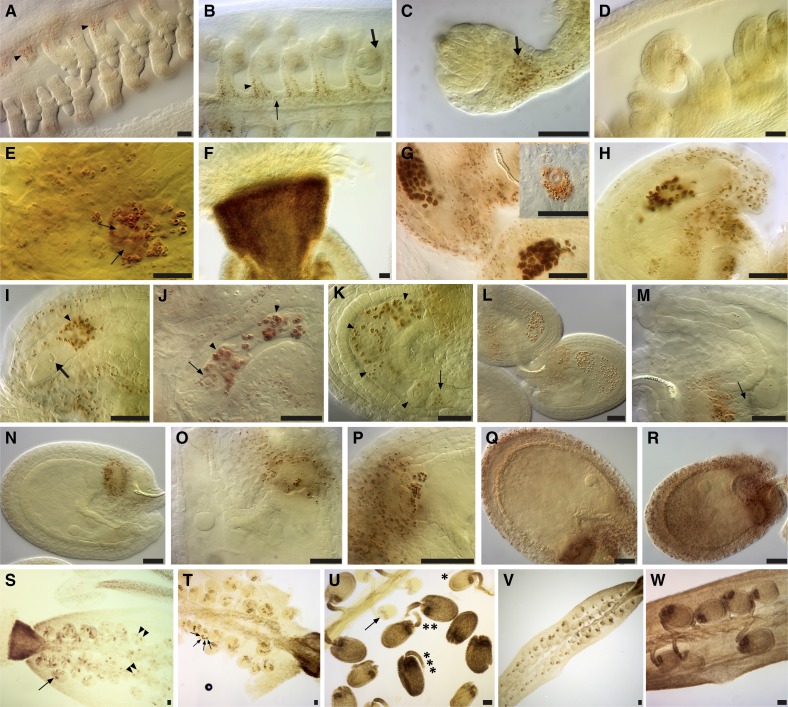
Figure 1.
The three starch synthesis waves in the stamen envelope. A to J, First wave. J to O, Second wave. P to Z, Third wave. A, Starch accumulates within the peduncle and the receptacle of young buds. B and C, Closeup images of buds at stages 3 and 7, respectively, where starch accumulates at their basal part and within the peduncle and the receptacle. D, Stage 8 bud where some starch deposits develop within the stamen (arrowhead) and the basal region of the carpel (arrow). E, A flower bud at completion of the development of the four anther wall layers. Starch accumulation extends to the filament (thick arrow) and the connective tissue (thin arrow) and is still seen within the lateral wall of the carpel (arrowheads). F, During male meiosis, starch grain deposits extend to the newly formed endothecium (arrowheads). G to I, Tetrad stage of male meiosis, the first peak of starch amylogenesis in the stamen. In the carpel, starch amylogenesis switched from the lateral wall to the placenta-septum region (arrowheads). J and K, Early microspore development. J, The short stamen (asterisk) just released microspores and is practically devoid of starch grains. The long stamen (double asterisk) have rounded microspores with a much more prominent exine. K, Starch starts to build up again in the filament (thin arrow), the connective tissue (thick arrow), and the staminal envelope (arrowheads). L and M, Bicellular stage (short stamens removed). This stage denotes the peak of the second starch wave in the staminal envelope. Pollen grains show a high starch accumulation. N and O, Late bicellular-early tricellular stage. While most of the starch grains in the staminal envelope are consumed, the pollen grains reach the peak of the unique starch wave. Starch is frequently seen surrounding the vegetative nucleus (inset in O). P to U, Successive developmental stages from the same inflorescence, illustrating the formation of large starch deposits within the sporophytic tissues of the stamen and its subsequent consumption just before anthesis. P, While a short stamen (arrowhead) is still at the end of the second wave, long stamens (arrows) have already started the third amylogenesis wave. Q, All long stamens show high starch deposition and are at the peak of the third starch accumulation wave. R and S, While a short stamen is still at the peak of its third wave, starch deposits already vanished from the anthers of long stamens. T, All stamens are at the same starch deposition stage, maintaining conspicuous starch deposits in the filaments. U, Anthesis. While starch vanishes progressively from the filaments, large deposits appear in the parenchyma of the style. V to Y, Starch accumulation/consumption in the anther during the third starch synthesis wave. V and W, Two optical sections illustrating strong starch accumulation in the endothecium and the connective tissue. X and Y, Two optical sections illustrating how starch disappears from the entire anther except for the stomata; pollen grains continue their amylolytic activity. Z, Mature pollen grains showing an intense Periodic acid-Schiff-positive staining; very few and small starch grains might persist (arrowheads within inset). Flower stages are according to Smyth et al. (1990). Bars = 20 μm.
As microspore development proceeded to pollen mitosis I, a new wave of starch accumulation occurred in the sporophytic tissues of the stamen (Fig. 1, J–M). During or just after pollen mitosis I, while starch deposits started to build up in the bicellular pollen grain, a concomitant increase in starch accumulation occurred in the staminal envelope, reaching a second amylogenesis peak that was higher than that of the first wave (Fig. 1, L and M). Soon thereafter, encompassing pollen mitosis II, starch accumulation in the pollen grain reached its maximum, while drastic amylolysis occurred in the anther wall layers (Fig. 1, N and O).
Shortly after the amylogenesis peak in the pollen grain, prominent starch deposition was observed in all sporophytic tissues of the stamen (Fig. 1, P, Q, V, and W). Compared with the previous two staminal envelope waves, this wave showed the strongest starch accumulation. Then, just before the final extension of the stamen to the level of the stigma, starch deposits vanished completely from the anther (except stomata), while conspicuous deposits accumulated in the filament (Fig. 1, T, X, and Y). Very few, small starch grains could be seen in maturing pollen grains (Fig. 1X). The remaining starch deposits in the filament were largely consumed by the time the stamen reached the stigma and the flower opened (Fig. 1U).
Starch Accumulates in Proliferating Tissues during Early Zygote and Embryo Development
In contrast to the staminal envelope, starch metabolism followed sequential amylogenesis and amylolysis in different tissues of the carpel/silique, ovule, and embryo.
Starch accumulated first in the lateral walls of young carpels (Fig. 1, D and E). Concomitant with the development of ovule primordia, starch deposits appeared in the placenta-septum region at the basal part of ovule primordia and were progressively lost from the lateral carpel wall (Fig. 1, G and I). During meiosis, more prominent starch accumulation occurred in the placenta-funiculus region and in the distal funiculus joining the chalaza (Fig. 2, A–C). These accumulations extended to the integuments surrounding the developing female gametophyte (Fig. 2D). During the last stages of flower development, while the progressive depletion of starch was observed in the placenta-septum region, conspicuous starch deposition occurred in both the style (Fig. 2E; compare Fig. 1, T and U) and the ovule. The largest deposits in the ovule were seen in the central cell, where starch accumulation started before the fusion of the two polar nuclei (Fig. 2F) and increased further after the formation of the secondary endosperm nucleus (Fig. 2G). However, the egg cell, the distal part of the funiculus, the chalazal proliferating tissue, and the micropylar part of the inner and outer integuments also had starch deposits.
Figure 2.
Starch turnover during ovule and early embryo and silique development. A to C, During meiosis. The megaspore mother cell (A) and functional megaspore stages (B and C) are illustrated. Starch accumulated in the placenta (thin arrow), the funiculus (arrowheads), and the chalazal region below the nucellus (thick arrows). D, Division and elongation of the embryo sac, showing an extension of starch deposits to the integuments. E, Starch starts to build up in the central cell when the two polar nuclei lie next to each other (arrows). F, Strong starch accumulation in the parenchymatic cells of the style at anthesis. G and H, Ovules at anthesis (G) and during fertilization (H), showing more conspicuous starch accumulation in the central cell (surrounding the secondary endosperm nucleus; inset in G), the distal portion of the funiculus, the micropylar portion of the integuments, and the chalazal proliferating tissue. I to L, Starch accumulation is maintained in the dividing zygote (arrows) and endosperm (arrowheads: uninuclear in I, binuclear in J, four- or eight-nuclear stage in K and L). Note the trace of a discharged pollen tube in I (thick arrow). M to P, Starch disappears from all tissues but the chalazal proliferating tissue. Note the crushed nucellar cells (arrow) between the chalazal cyst of the endosperm and the starch-accumulating proliferating tissue in M, the pronounced ring-like shape of starch accumulation in N, the depletion of starch at the micropylar part of the seed in O, and the cells forming an inverted cup-like shape that are filled with starch and in close contact with the chalazal cyst in P. Q and R, A new wave of starch amylogenesis occurs at the octant embryo stage (Q) and is more conspicuous at the dermatogen stage (R); the endosperm is starchless. S to U, Low-magnification views of different postfertilization stages that are easily recognizable by their starch accumulation pattern and/or intensity. Uninuclear (arrows) and binuclear (arrowheads) endosperm stages are shown in S; four-nuclear endosperm stages (arrows) are shown in T; and unfertilized starchless ovule (arrow), quadrant (asterisk), octant (double asterisk), and dermatogen embryo stages (triple asterisk) are shown in U. Note the persistence of starch in the style and the beginning of basipetal starch accumulation in the silique walls and the placenta-septum region in S. V, A low-magnification view of a silique illustrating the basipetal starch accumulation in the valves. W, A middle silique portion with octant and dermatogen embryos, where conspicuous starch accumulation occurs in the valves and the placenta-septum region. Bars = 20 μm.
During the progamic phase (i.e. the period from pollination to fertilization), the conspicuous starch deposits in the ovule were maintained (Fig. 2H). However, while starch gradually disappeared from the micropylar integuments after fertilization, it was maintained in both the uninuclear endosperm and the zygote (Fig. 2I). Starch persisted in the apical cell after the division of the zygote (single-cell stage of the embryo proper) and in the endosperm until approximately the eight-nuclear stage (Fig. 2, J–M). Interestingly, starch was consumed gradually in the endosperm along the micropylar-chalazal axis (Fig. 2L). Concomitant with the cellular and nuclear divisions of zygote and endosperm, respectively, starch gradually built up in the proliferating tissues of the ovule (Fig. 2, L and M). At the single-cell stage of the embryo proper, the only conspicuous starch deposits in the seed were seen as a ring-like structure in the sporophytic tissues of the ovule at the chalazal pole of the embryo sac (Fig. 2N). While the tightly packed and stretched cells in the center of this region had barely any starch, peripherally located cells accumulated a high quantity (Fig. 2, O and P). The micropylar-most cells of this structure had an inverted cup-like shape and were in direct contact with the chalazal cyst of the endosperm. At the octant embryo stage, the whole seed coat accumulated starch in all layers, with even stronger staining detected at the dermatogen stage (Fig. 2, Q and R). Interestingly, the ovary wall, the placenta-septum region, and the funiculus showed a gradual basipetal starch accumulation paralleling that of seed development (Fig. 2, S, V, and W).
In Silico Analysis of Sugar- and Starch-Related Gene Expression Profiles during Flower Development
To check whether this highly dynamic starch turnover pattern is related to the transcriptional activities of carbohydrate transport and metabolism genes, we analyzed publicly available microarray data from flower and silique tissues at different developmental stages. Based on the microscopic characterization of flower and early seed development, and to cover a wide variety of reproductive tissues, we chose to focus our analyses on two developmental processes: (1) pollen development from the microspore stage to maturation (haploid male gametophyte); and (2) the chalazal seed coat (diploid maternal tissue) and chalazal endosperm (fertilization product) regions of the developing seed.
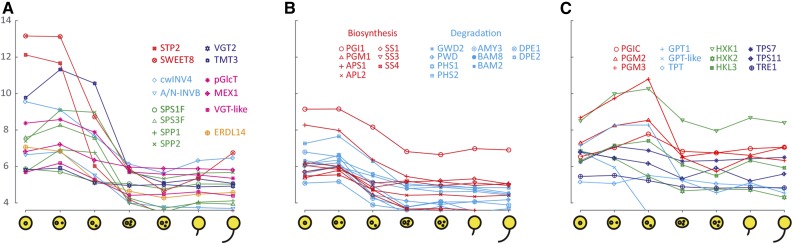
In developing pollen, we could distinguish three well-defined gene expression clusters (Fig. 3; Supplemental Fig. S1). The first cluster (Fig. 3) was characterized by high transcription during early microspore development, peaking in uninucleate or bicellular pollen, followed by a sharp or gradual decrease during later stages of development. This corresponds to the stage when we observed the unique starch amylogenesis wave in the developing pollen grain. Most of the genes for starch metabolism as well as the GPT1 gene encoding a translocator belong to this cluster (for the full names of the genes, see Table I). Interestingly, the gene coding for the AGPase regulatory subunit APL2, typical of mesophyll cells, was the only APL isoform present in this cluster. All three expressed starch synthase genes (SS1, SS3, and SS4) also belong to this cluster. Except for the genes encoding the glucan water dikinase GWD1 and the β-amylases BAM1, BAM4, and BAM9, most of the genes within the degradation pathway were also in the first cluster.
Figure 3.
The gene expression cluster during early microspore development. Coexpressed (log2 scale of expression values) sugar (A), starch (B), and sugar-phosphate (C) transport and metabolism genes are represented during pollen development (the first four x axis ticks indicate microspore, bicellular, tricellular, and mature pollen grains) and during in vitro pollen tube growth (the last three x axis ticks indicate hydrated pollen grain and 30 and 240 min after germination).
Table I. Carbohydrate metabolism and transport genes included in this study.
For a full list of all analyzed genes and the corresponding references, see Supplemental Table S1.
| Metabolic/Transport Group | Gene Name | Enzyme | Locus |
|---|---|---|---|
| Suc synthesis | SPS1F | Suc phosphate synthase1F | At5g20280 |
| SPS3F | Suc phosphate synthase3F | At1g04920 | |
| SPP1 | Suc phosphatase1 | At1g51420 | |
| SPP2 | Suc phosphatase2 | At2g35840 | |
| Cell wall acid invertase | AtcwINV3 | Cell wall invertase3 | At1g55120 |
| AtcwINV4 | Cell wall invertase4 | At2g36190 | |
| Cytoplasmic neutral invertase | A/N-INVI | Alkaline/neutral invertase I (CINV2) | At4g09510 |
| A/N-INVB | Alkaline/neutral invertase B | At4g34860 | |
| A/N-INVF | Alkaline/neutral invertase F | At1g72000 | |
| Sugar transport protein | AtSTP2 | Monosaccharide-H+ symporter | At1g07340 |
| AtSTP8 | Monosaccharide-H+ symporter | At5g26250 | |
| AtSTP14 | Monosaccharide-H+ symporter | At1g77210 | |
| Vacuolar Glc transporter-like | AtVGT2 | Tonoplast sugar transport protein | At5g17010 |
| AtVGTL | Tonoplast sugar transport protein | At5g59250 | |
| Tonoplast monosaccharide transporter | AtTMT3 | Tonoplast monosaccharide transporter | At3g51490 |
| Plastidic sugar transporter | pGlcT | Putative monosaccharide transporter | At5g16150 |
| MEX1 | Maltose excess1 | At5g17520 | |
| SWEET sugar transporter | SWEET4 | Sugars will eventually be exported transporter4 | At3g28007 |
| SWEET8 (RPG1) | Sugars will eventually be exported transporter8 | At5g40260 | |
| SWEET10 | Sugars will eventually be exported transporter10 | At5g50790 | |
| Early responsive to dehydration6-like | AtERDL8 | Putative sugar transport protein (ERD group) | At3g05150 |
| AtERDL14 | Putative sugar transport protein (ERD group) | At4g04760 | |
| Starch biosynthesis | PGI1 (PGI) | Phosphoglucoisomerase | At4g24620 |
| PGM1 (PGM) | Phosphoglucomutase | At5g51820 | |
| APS1 (ADG1) | AGPase small subunit1 | At5g48300 | |
| APL2 | AGPase large subunit2 | At1g27680 | |
| GBS1 | Granule-bound starch synthase1 | At1g32900 | |
| SS1 | Soluble starch synthase1 | At5g24300 | |
| SS2 | Soluble starch synthase2 | At3g01180 | |
| SS3 | Soluble starch synthase3 | At1g11720 | |
| SS4 | Soluble starch synthase4 | At4g18240 | |
| BE2 | Starch-branching enzyme2 | At5g03650 | |
| BE3 | Starch-branching enzyme3 | At2g36390 | |
| ISA1 | Isoamylase1 | At2g39930 | |
| ISA2 | Isoamylase2 | At1g03310 | |
| Starch breakdown | GWD1 (SEX1) | Glucan water dikinase1 | At1g10760 |
| GWD2 | Glucan water dikinase2 | At4g24450 | |
| PWD (GWD3) | Phosphoglucan water dikinase | At5g26570 | |
| SEX4 | Starch excess4 | At3g52180 | |
| BAM1 (BMY7/TR-BYM) | β-Amylase1 | At3g23920 | |
| BAM2 (BMY9) | β-Amylase2 | At4g00490 | |
| BAM5 (BMY1/RAM1) | β-Amylase5 | At4g15210 | |
| BAM8 | β-Amylase8 | At5g45300 | |
| BAM9 (BMY3) | β-Amylase9 | At5g18670 | |
| AMY1 | α-Amylase1 | At4g25000 | |
| ISA3 | Isoamylase3 | At4g09020 | |
| LDA (PU1) | Limit dextrinase | At5g04360 | |
| PHS1 | Glucan phosphorylase | At3g29320 | |
| PHS2 | Glucan phosphorylase | At3g46970 | |
| DPE1 | Disproportionating enzyme | At5g64860 | |
| DPE2 | Maltose transglucosidase | At2g40840 | |
| Cytoplasmic hexose phosphate metabolism | PGIC | Phosphoglucoisomerase | At5g42740 |
| PGM2 | Phosphoglucomutase2 | At170730 | |
| PGM3 | Phosphoglucomutase3 | At1g23190 | |
| HXK1 | Hexokinase1 | At4g29130 | |
| HXK2 | Hexokinase2 | At2g19860 | |
| HKL3 | Hexokinase-like3 | At4g37840 | |
| AtTPS7 | T6P synthase | At1g06410 | |
| AtTPS11 | T6P synthase | At2g18700 | |
| AtTRE1 | Trehalase1 | At4g24040 | |
| Plastidic translocator and translocator-like | AtGPT1 | 6-Phosphate/phosphate translocator1 | At5g54800 |
| AtGPT-like | Glc-6-P/phosphate translocator-like protein | At5g17630 | |
| TPT (TPT1) | Phosphate/triose phosphate translocator precursor | At5g46110 |
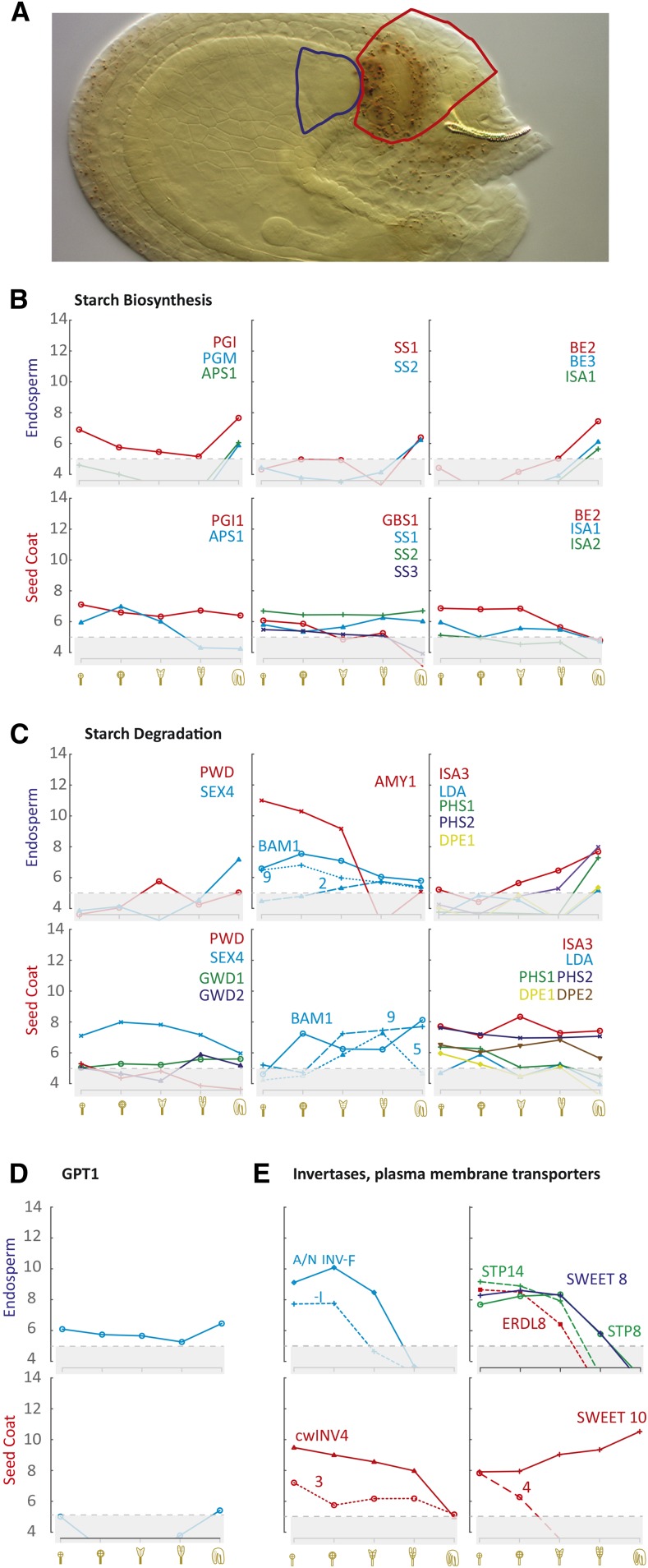
Microscopic analyses revealed high starch accumulation in the chalazal seed coat throughout early seed development (Fig. 2, M–R). Paralleling this pattern, we identified significant expression of genes for starch metabolism (Fig. 4, B and C) and GPT1 (Fig. 4D). Strikingly, neither PGM1 nor any APL isoforms of the biosynthetic pathway were expressed significantly above threshold. In contrast to the situation in the pollen grain, starch degradation pathway genes showed a high expression level, including the starch glucan phosphatase gene SEX4, the genes encoding the catalytic plastidial and cytosolic isoforms BAM1 and BAM5, respectively, and the uncharacterized BAM9 gene. The absence of any significant expression of many starch metabolism genes in the chalazal endosperm is congruent with our finding that no starch accumulated in this region. Comparing the expression level of sugar metabolism and transport genes in different tissues of the seed (Fig. 4E; Supplemental Figs. S3 and S4), the chalazal region (seed coat and endosperm; Fig. 4E) showed the highest expression of invertase and sugar transporter genes during early embryo development.
Figure 4.
Carbohydrate transport to and metabolism within the chalazal region of the seed. A, A seed at the single-cell stage of the embryo proper is shown to illustrate the two chalazal regions (chalazal seed coat in red and chalazal endosperm in blue) used in transcriptome analyses. B to E, Expression values (log2 scale) for starch synthesis (B), starch degradation (C), GPT1 (D), and highly expressed invertase and monosaccharide transporters (E) during early embryo development. For a full expression analysis of the 197 genes in these and other tissues, see Supplemental Figures S2 and S3; for the full names of the genes, see Table I.
Down-Regulation of GPT1 Expression Induces Tissue-Specific Deregulation of Starch Biosynthesis
Given the intriguing expression pattern of GPT1, we investigated whether GPT1 is indeed the translocator of Glc-6-P for starch biosynthesis in reproductive tissues. To do so, we analyzed starch dynamics in three mutants potentially affecting the plastidial Glc-6-P content: the homozygous pgi1-1, pgm1-1, and gpt1-3 mutants (highlighted with red boxes in Fig. 7). In pgm1-1 homozygotes, starch was undetectable in both photosynthetic and reproductive tissues, including inflorescence stalks and flower peduncles (Fig. 5, E, F, J1–J3, N, and T1–T3). In contrast, most inflorescences of pgi1-1 homozygotes showed starch dynamics comparable to the wild type in all reproductive tissues except for the sepals, where an appreciable reduction was observed (Fig. 5, C, D, I1–I3, M, and S1–S3). In some inflorescences, even higher starch accumulation was observed than in the wild type (compare Fig. 5, B and D). Compared with pgm1-1, a slightly higher residual starch accumulation was observed in mesophyll cells of pgi1-1 cauline leaves, and much higher starch accumulation was seen lining the vascular bundles (compare Fig. 5, C and E). Mutations affecting GPT1 were reported to be embryo lethal; however, there are alleles available as homozygous insertion lines (gpt1-3, gpt1-5, and gpt1-6) that did not reduce GPT1 transcription and, thus, had not been physiologically characterized (Niewiadomski et al., 2005). Our analysis of starch turnover within gpt1-3 homozygotes revealed a reduction in starch accumulation in the carpel and during early silique development. Compared with the wild type, while lower starch accumulation was observed during early stages of flower development (Fig. 5H), starch dynamics during stamen development appeared unaffected (Fig. 5, K1, K2, and O). At anthesis and during the early stages of silique and seed development, a strong reduction in starch accumulation was observed in the pistil (Fig. 5O) and the ovule (Fig. 5Q). The conspicuous starch accumulation usually seen in the central cell of wild-type ovules (Fig. 5, P and R1) was either strongly reduced (Fig. 5R2) or absent in the gpt1-3 mutant (Fig. 5R3). After fertilization, starch accumulation also was affected in young seeds (Fig. 5R4), the style, and the silique valve (Fig. 5R5). Starch amylogenesis in the seed after the octant embryo stage appeared unaffected (Fig. 5R5).
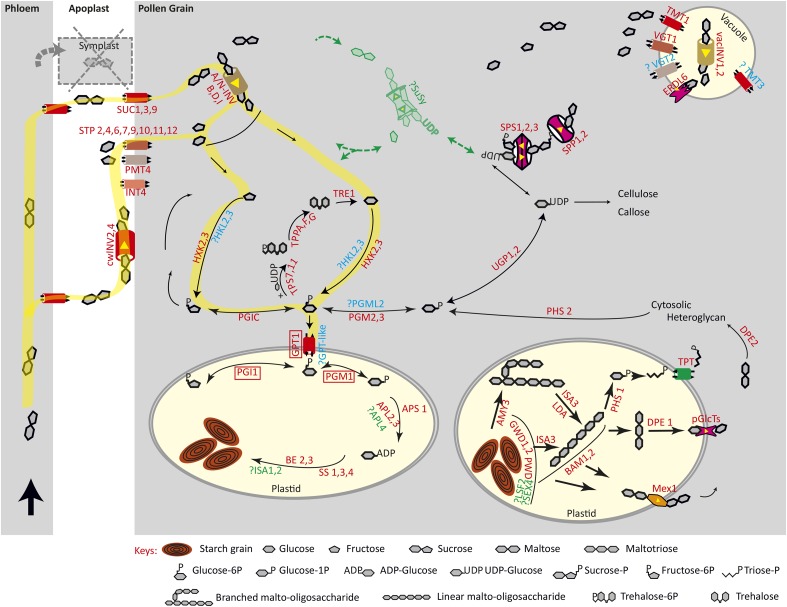
Figure 7.
Model of starch synthesis in pollen grains. A schematic representation is shown for carbohydrate metabolism and transport in the pollen grain. All expressed isoforms during pollen development are indicated in red. Sugar uptake by the pollen occurs only via the apoplast, because developing microspores are symplastically isolated. Suc is either transported via the apoplast (SUC transporters) or metabolized first by cell wall invertases, and the resulting monosaccharides are transported by STP, PMT, and INT transporters. In the cytosol, expressed genes of the different metabolic routes for Suc, monosaccharide, hexose phosphate, and starch metabolism are depicted. Not or lowly expressed but important genes are indicated in green; expressed genes with unknown functions or with unknown substrate specificities are in blue; and the three genes for which we analyzed insertional mutants are highlighted by red boxes. Based on these expression patterns, the most likely pathway of starch biosynthesis within the developing pollen grain is highlighted in yellow. For the full names of enzymes and genes, see Table I.
Figure 5.
Starch turnover in starch biosynthesis mutant lines. A, Strong starch accumulation in a wild-type (WT) cauline leaf at the end of the light phase. B, Starch accumulates in the peduncle (arrow) and the receptacle (arrowhead) of young wild-type buds. C, Highly reduced starch accumulation in mesophyll cells of a pgi1-1 cauline leaf at the end of the light phase. Higher starch accumulation, however, is maintained around vascular tissues (arrow). D, Starch accumulates in the peduncle (arrow) and the receptacle (arrowhead) of young pgi1-1 mutant buds. E and F, No starch accumulation is observed in any tissue of cauline leaves (E) or young buds (F) of the pgm1-1 mutant at the end of the light phase. G and H, While no strong effect is observed on starch accumulation in cauline leaves (G), an appreciable reduction is seen in young gpt1-3 buds (H) at the end of the light phase. I1 to I3, Different stages of pgi1-1 stamen development illustrating that the second and third waves of starch accumulation were not affected. J1 to J3, pgm1-1 stamen at different stages illustrating the absence of starch accumulation during anther development. K1 and K2, Unaffected second (K1) and third (K2) starch synthesis waves in gpt1-3 stamen. L to O, Starch distribution in wild-type (L), pgi1-1 (M), pgm1-1 (N), and gpt1-3 (O) flowers just before anthesis. Note the reduced starch accumulation in pgi1-1 sepals, the absence of starch accumulation in the whole pgm1-1 flower, and the reduction of starch accumulation in the gpt1-3 pistil. P, Typical starch accumulation in wild-type ovules at anthesis. Q, Highly reduced starch accumulation in gpt1-3 ovules. Note the absence of conspicuous starch accumulation in the central cell. R1, Closeup image of starch accumulation in wild-type ovules. R2 and R3, Closeup images of reduced (R2) and highly reduced (R3) starch accumulation in gpt1-3 ovules. R4 and R5, Reduced starch accumulation in gpt1-3 during early embryo development (arrow in R4) and in the silique valves (R5). Note the unaffected starch accumulation in more advanced seeds. S1 to S3, Unaffected starch accumulation during different stages of ovule and embryo development in pgi1-1. T1 to T3, Absence of starch accumulation at different stages of ovule and embryo development in pgm1-1. Bars = 20 μm.
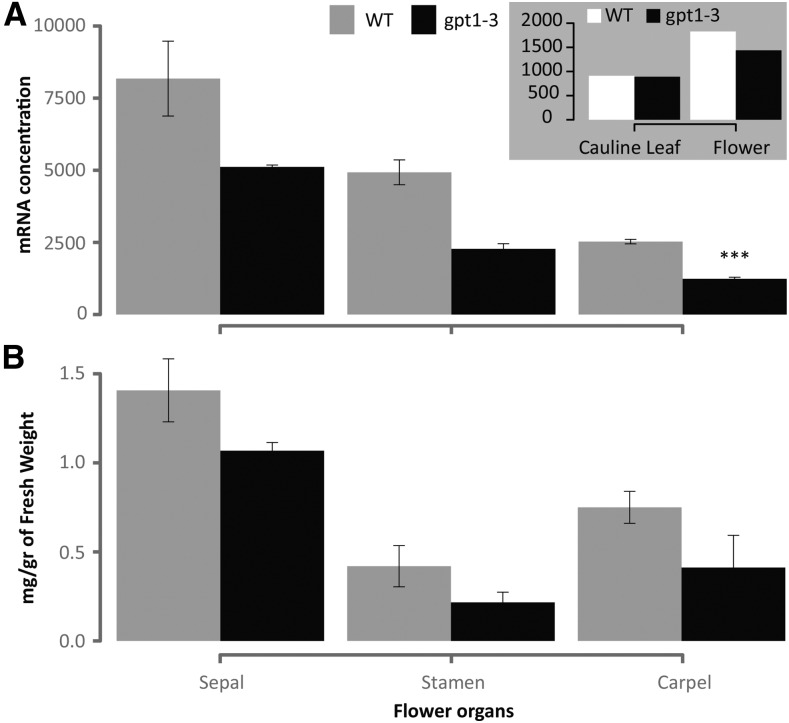
To check whether GPT1 transcript level was affected in an organ-specific manner in gpt1-3 homozygotes, we quantified the concentration of GPT1 mRNA using the highly sensitive droplet digital PCR (ddPCR) method using RNA isolated from sepals, stamens, and carpels. Compared with the wild type, mutant carpels showed a highly significant reduction in GPT1 transcript levels at anthesis (Fig. 6A), correlating with the impact of the mutation on starch accumulation in the carpel. The enzymatic quantification of starch content also revealed a slight but nonsignificant reduction in all organs (P > 0.05; Fig. 6B).
Figure 6.
ddPCR analysis of GPT1 transcript levels (A) and starch content (B) in sepals, stamen, and carpels of wild-type (WT) and gpt1-3 flowers at opening. A, ddPCR values are means of measurements of three biological replicates normalized to 10,000 molecules of IPP2. B, Starch content values are means of measurements of four biological replicates. Error bars indicate se. Asterisks denote a highly significant value (***, P < 0.01). The inset in A is a ddPCR analysis of GPT1 transcript levels in single biological samples of cauline leaves and whole flowers.
DISCUSSION
Starch Turnover during Flower and Early Silique Development Is Highly Dynamic
In many plant species characterized so far, a single wave of conspicuous starch accumulation is usually observed during microgametogenesis, while one to two amylogenesis/amylolysis waves are commonly observed in the staminal envelope (for review, see Clément and Pacini, 2001). These starch waves were suggested to sustain growth and differentiation of the anther wall layers, to translocate nutrients to the highly active tapetum and to the loculus, to provide carbon skeletons for the formation of lipid droplets within the pollen grain, and to store excess food during the anther maturation phase (Reznickova and Willemse, 1980; Miki-Hiroshige and Nakamura, 1983; Pacini and Franchi, 1983; Clément et al., 1994; Clément and Pacini, 2001). In this work, apart from confirming the occurrence of these waves in Arabidopsis, we unraveled the existence of a third wave of starch amylogenesis/amylolysis in the staminal envelope just before anthesis. Its occurrence close to anthesis suggests that products of starch degradation may sustain final maturation, elongation of the filaments to reach the level of the stigma, dehiscence, and/or other functions yet to be discovered. Whether the presence of this third wave is specific to Arabidopsis, species producing tricellular pollen grains, or, more generally, species with filament elongation prior to anthesis deserves further investigation.
During carpel development before anthesis, rather than in a strict wave in a given tissue, starch accumulated sequentially in different tissues and organs, such as the lateral carpel walls in young carpels, the placenta-septum and funiculus-chalaza regions during early ovule development, and, especially, the parenchymatic tissue of the style and the central cell at flower opening. These starch deposits may buffer the growth and development not only of reproductive cells but also of the sporophytic tissues that are growing and maturing at the same time (Buell, 1952). After pollination, while starch vanished progressively from the integuments, the embryo, and the endosperm, conspicuous starch deposits built up in the proliferating tissues of ovule, which, at the one-cell stage of the embryo proper, had the only conspicuous starch deposits in the seed, forming a ring-like structure surrounding the chalazal cyst. This chalazal proliferating tissue, which has an inverted cup-like shape, lies between the terminus of the vascular strand and the chalazal end of the embryo sac and has a high metabolic activity in the closely related species Capsella bursa-pastoris (Schulz and Jensen, 1971). Moreover, the elongating embryo sac, while crushing nucellar cells during its growth toward this proliferating tissue, develops ingrowths to absorb nutrients from these tissues. Soon after, the chalazal cyst of the endosperm develops a haustorial base that penetrates into the proliferating tissue (Nguyen et al., 2000). Taken together, although the integuments might ensure some nutrient flux to the growing zygote/embryo (Schulz and Jensen, 1969; Mansfield and Briarty, 1991), the following observations suggest a different nutrient transport route for endosperm and embryo: (1) the extensive starch deposits surrounding the chalazal cyst, (2) the high transfer activity characterizing the chalazal end of the embryo sac, (3) the absence of a membrane system during the syncytial phase of endosperm development, and (4) the gradual micropylar-chalazal starch consumption in the endosperm all indicate that assimilates are transported along the following pathway: vascular tissue > proliferating tissue > endosperm > embryo. Although starch accumulation per se does not represent a carbohydrate translocation process, correlations in starch turnover between adjacent tissues or specific patterns of starch accumulation suggest potential starch degradation, sugar translocation, and nutrient transport processes (Buell, 1952).
The analysis of starch dynamics in three homozygous mutants affecting Glc-6-P metabolism or transport identified GPT1 as the likely translocator related to starch biosynthesis in heterotrophic floral tissues. While all reproductive tissues were starchless in pgm1-1, no major effects on starch dynamics were reported for pgi1-1. Only sepals showed an appreciable reduction in starch accumulation in pgi1-1, suggesting that sepals have a starch metabolism pathway similar to that of leaf mesophyll cells. Although mutations in GPT1 were documented to be embryo lethal, we found that a homozygous viable allele showed reduced starch accumulation during early flower development and an effect on starch dynamics in female tissues during anthesis and early embryo/silique development. In particular, starch accumulation in the central cell was absent in gpt1-3 mutants. Indeed, GPT1 transcript levels were reduced significantly at anthesis in mutant carpels compared with stamens and sepals. Although these results show that GPT1 plays an important role in the starch metabolism of reproductive tissues, the unaffected starch dynamics in stamen indicates either a tissue-specific effect of the T-DNA insertion on the regulation of GTP1 or the existence of an alternative pathway for hexose phosphate transport in this organ.
In Silico Analyses of Gene Expression at Specific Reproductive Stages with High Starch Turnover Unravel Potential Carbohydrate Metabolic Pathways
Developing microspores and growing pollen tubes are symplastically isolated, and sugar transport to these heterotrophic organisms occurs mainly via the apoplast from the locular fluid of the stamen and the extracellular matrix of the pistilar transmitting tissue, respectively. The analysis of gene expression related to sugar, hexose phosphate, and starch metabolism during pollen development revealed highly coordinated gene expression, with three clusters of transcriptional activities. The first cluster, which includes most of the genes for starch synthesis and degradation, characterized the early stages of microspore development and coincided with the starch wave during microgametogenesis. This transcriptional coregulation of the three processes suggests that sugar and hexose phosphate transport and metabolism genes expressed during this stage might be related to starch metabolism. They also suggest that starch synthesis/degradation transcriptional activities are not discrete and temporally separated metabolic pathways, in agreement with similar findings during seed development (Andriotis et al., 2010b). Based on these transcriptional patterns, and the GPT1-mediated hexose phosphate transport to amyloplasts for starch biosynthesis, a model of sugar-starch metabolism is proposed for the starch wave occurring during microspore development (Fig. 7, yellow path). Suc could be metabolized in the apoplast by cwINV4, and monosaccharides, either derived from invertase activity or the locular fluid, are likely transported by the highly expressed STP2 and SWEET8 transporters, the latter a member of the SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTER family. While STP2 was suggested to have a role in Glc uptake (Truernit et al., 1999), the plasma membrane sugar transporter SWEET8, which showed the highest expression level, has been shown to mediate at least Glc transport (Chen et al., 2010). The rpg1 mutant disrupting SWEET8 revealed its importance for cell integrity and exine pattern formation (Guan et al., 2008), but whether it fulfills an uptake activity is unknown. Within the cytosol, the expression of two isoforms derived from each of the SPS and SPP genes suggests high Suc biosynthetic activities paralleling starch biosynthesis. SUS gene expression levels remained unaltered during all stages of pollen development and in vitro pollen tube growth, pointing toward SUS-independent starch biosynthesis, a finding that contrasts with the SUS-based starch biosynthesis suggested for other tissues (Déjardin et al., 1997; Angeles-Núñez and Tiessen, 2010). Low SUS and high invertase expression levels imply a higher abundance of Glc and Fru compared with UDP-Glc. Based on this observation, hexokinases and cytoplasmic PGI would play an important role in the pathway producing Fru-6-P and Glu-6-P (Fig. 7); Glc-6-P is then translocated to plastids by GPT1.
Comparing starch synthesis and degradation between different reproductive tissues and organs revealed a variation in the presence/absence or in the specific expressed isoform for the step-limiting AGPase (whether it is based on autotrophic or heterotrophic APL subunits), starch synthase, glucan phosphorylating/dephosphorylating, and exoamylase genes. While the prevalence of the specific autotrophic or heterotrophic APL regulatory subunit might reflect distinct regulatory starch biosynthesis needs in different tissues (Crevillén et al., 2003, 2005), the variation in the expressed isoforms for the starch synthase, and of some members within the degradation pathway, suggests that the different starch deposits might be structurally distinct.
Starch turnover during early seed development suggests an important role for the vascular tissue > chalazal proliferating tissue > chalazal endosperm > embryo nutrient transport route to nourish endosperm and embryo, at least during early seed development. A comparison of transcriptional activities within different territories of the seed reinforces this view. While globally similar transcriptional activities in the three regions of the endosperm can be appreciated, the highest expression levels of cell wall and cytoplasmic invertases occur in the chalazal seed coat and the chalazal endosperm, respectively (Fig. 4E; Supplemental Figs. S3 and S4). But the high expression level of so far uncharacterized genes of the SWEET and ERDL families, either at the whole-seed level or within the chalazal region, adds a challenging layer of complexity to traditionally simplified models (Baud et al., 2002; Fallahi et al., 2008; Wang and Ruan, 2012). Knowledge about the substrate specificities, subcellular localization, and function of the members of these families would certainly improve these models.
Our systematic microscopic and transcriptomic analyses of starch and sugar metabolism during flower and early silique development sets the foundation for further research on metabolic pathways using more specific microscopic and molecular approaches, in particular organ-, tissue-, or cell-specific transcriptomic and gene-silencing techniques (Andriotis et al., 2010a; Schmid et al., 2015). However, the simple, effective, and quantitative microscopic technique used here will help to distinguish between models by analyzing mutants, either for genes already known to be essential for sugar and starch metabolism or, to circumvent genetic redundancy, for multiple mutants representing coexpressed genes in specific pathways.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Sampling
Arabidopsis (Arabidopsis thaliana) variety Columbia-0 plants and homozygous lines for the mutants pgm1-1, pgi1-1, and gpt1-3 were used to characterize starch turnover during flower and early silique development, to quantify starch content, and for transcript level analyses. Both wild-type and mutant Arabidopsis seedlings were grown on Murashige and Skoog plates for 8 d, transferred to ED73 soil (Einheitserde), and grown under 16 h of light and 8 h of darkness at 21°C and 18°C, respectively. The homozygosity of insertions was confirmed using the oligonucleotide primers listed in Supplemental Table S1. To increase the homogeneity between samples, 40 plants with the first flower opening on the same day were selected, the primary inflorescence of 10 randomly selected plants was fixed toward the end of the light phase (after approximately 10 h of light) on that day, and 10 additional primary inflorescences each were fixed on the subsequent 3 d at the same time.
Histological Detection of Starch
To be able to make a systematic analysis of starch turnover during all flower stages and the early phase of silique development, we used a recipe developed by Herr (1972), which combines his 41/2 clearing solution (Herr, 1971) with lugol (lactic acid:chloral hydrate:phenol:clove oil:xylene:I2:KI, 2:2:2:2:1:0.1:0.5, w/w). Inflorescences from at least 10 plants were fixed either in Carnoy fixative (ethanol:acetic acid, 3:1, v/v) or in FPA50 (formalin:propionic acid:ethanol [55%], 5:5:90, v/v) for 24 h and then moved to 70% ethanol and stored at 4°C. The experiment was repeated three times. While Carnoy-fixed samples were ideal for tracking starch turnover, allowing much stronger starch staining, FPA50-fixed samples were used whenever higher structural details were needed at the expense of a lower starch staining sensitivity. Individual flower buds were dissected on the slide using the stereomicroscope within a minimal amount of 70% ethanol, taking care to avoid drying out of the sample and removing sepals and petals. Excess alcohol was absorbed with a blotting paper, and 20 to 30 µL of clearing-staining solution was added before placing the cover slip and gently squashing it. Periodic acid-Schiff staining of insoluble polysaccharides was done as described previously (Weigel and Glazebrook, 2002). For a more precise tracking of microspore developmental stages in relation to starch dynamics, one to two stamens of selected flowers were stained separately with 4′,6-diamidino-2-phenylindole (following Park et al., 1998; data not shown). Microscopic observations were made using differential interference contrast on either a Leica DMR or a Leica DM6000 microscope (Leica Microsystems), and images were captured with a digital camera (Magnafire S99802 [Optronics] or Leica DFC450 [Leica Microsystems]). Images were minimally and homogenously processed in Adobe Photoshop Lightroom 6 (Adobe Systems).
Microarray Data Analysis
To gain more insights into dynamic starch turnover, we analyzed the transcriptional dynamics of genes known to be related to carbohydrate transport and metabolism using publicly available microarray data (Honys and Twell, 2004; Pina et al., 2005; Schmid et al., 2005; Yu et al., 2005; Borges et al., 2008; Wang et al., 2008; Kram et al., 2009; Qin et al., 2009; Wuest et al., 2010; Boavida et al., 2011; Schmidt et al., 2011; Belmonte et al., 2013). We curated five sets of microarrays comprising a variety of reproductive tissues and cell types at different stages: (1) specific cell types of germline lineages; (2) individual organs of the flower before fertilization; (3) flowers, pistils, siliques, and seeds; (4) various developmental stages of pollen; and (5) ovules before fertilization, as well as several developmental stages of (6) embryo and endosperm, (7) seed coat, and (8) whole seeds (Supplemental Table S1). For each set, arrays were RMA normalized (Irizarry et al., 2003) using an updated annotation of the ATH1 microarray (brainarray.mbni.med.umich.edu; TAIRG, version 14). We defined genes with a log2-transformed expression value above 5 in a given tissue to be expressed. For a given plot, we removed genes that were (1) not expressed in any of the tissues shown or (2) expressed stably at a low level (defined as an average expression level below 5 and a coefficient of variation below 0.15). We first analyzed candidate genes known to be active in sugar metabolism and transport within autotrophic source tissues and all their family members as well as the SUC gene family for Suc transport. Tracking the metabolic pathway, we then analyzed candidate gene families for the conversion of Glc, Fru, and UDP-Glc to the different cytosolic hexose phosphate forms and for the translocation of Glc-6-P into plastids. Finally, we examined genes implicated in starch synthesis and degradation and in the transport and metabolism of degradation products within the cytoplasm (Supplemental Table S1).
ddPCR
All sepals, stamens, and carpels were harvested separately from two flowers per plant (stage 12 or 13 with flowers where petals were longer than sepals and just-opened flowers; Smyth et al., 1990) from a total of 36 plants (six plants for each of the three biological replicates of Columbia-0 and gpt1-3). Flowers were all chosen from positions corresponding to the first 10 flowers of the first inflorescence (avoiding the first two flowers), and each individual organ was dissected swiftly using a stereomicroscope and immediately placed in a liquid N2-cooled Eppendorf tube and stored at −80°C. Total RNA was extracted using the NucleoSpin RNA plant kit (Macherey-Nagel) according to the manufacturer’s instructions. RNA concentration and purity were determined using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). Total RNA (350 ng) was used for cDNA synthesis using the SuperScript II/oligo(dT) Reverse Transcriptase kit (Invitrogen) according to the manufacturer’s instructions. The ddPCR assay using the QX200 Droplet Digital PCR System (Bio-Rad) was performed according to the manufacturer’s instructions. Different reference genes (ACTIN2, UBC9, UBC21, PP2A, and IPP2) were assayed using both regular semiquantitative RT-PCR and ddPCR, and the IPP2 gene was chosen as the ideal reference gene (primers and amplicon lengths are given in Supplemental Table S1). Similar results were obtained using PP2A as a reference gene (data not shown). PCRs (25 µL) were prepared with 1× EvaGreen ddPCR Supermix (Bio-Rad), primers at a final concentration of 100 nm, and 5 µL of cDNA. Droplets were generated according to the manufacturer’s recommendation. PCR products were amplified under the following conditions: 95°C for 5 min, 40 cycles of 96°C for 30s and 60°C for 60s, followed by 4°C for 5 min, 90°C for 5 min, and held at 4°C until processing. PCR-amplified droplets were read individually with the QX200 droplet reader, and the results were analyzed with QuantaSoft software, version 1.4 (Bio-Rad).
Starch Quantification
To quantify the starch content in the three flower organs assayed by ddPCR, a similar separate flower organ sampling procedure was used; we sampled four biological replicates for each line with 10 flowers per replicate. Sepals, stamens, and carpels were harvested into liquid N2 and extracted in 200 µL of 0.7 m perchloric acid as described (Hostettler et al., 2011). Starch in the insoluble fraction was resuspended in 195 µL of 0.22 m sodium acetate, pH 4.8, and determined by measuring the amount of Glc released by treatment with α-amylase (Roche) and amyloglucosidase (Roche) as described (Hostettler et al., 2011).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries and in the in The Arabidopsis Information Resource under accession numbers At5g54800 (GPT1), At4g24620 (PGI/PGI1), and At5g51820 (PGM1/STF1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression values (log2 scale) for carbohydrate transport and metabolism genes during pollen development.
Supplemental Figure S2. Expression values (log2 scale) for hexose phosphate and starch metabolism genes during the development of different flower and silique organs/tissues/cells.
Supplemental Figure S3. Full list of sugar and sugar phosphate transport and metabolism genes expressed in the chalazal region of the seed.
Supplemental Figure S4. Expression values (log2 scale) for sugar transport and metabolism genes during the development of different flower and silique organs/tissues/cells.
Supplemental Table S1. List of the carbohydrate metabolism and transport genes included in this study.
Supplementary Material
Acknowledgments
We thank Arianna Nigro for providing homozygous gpt1-3 seeds and Arturo Bolaños, Christoph Eichenberger, Daniela Guthörl, and Peter Kopf for general laboratory support.
Glossary
- ddPCR
droplet digital PCR
Footnotes
This work was supported by the University of Zurich, an IEF Marie Curie Grant (grant no. Transepigen-254797 to A.H.), an ERC Advanced Grant (grant no. MEDEA-250358 to U.G.), a Research and Technology Development project from SystemsX.ch (grant MecanX to U.G.), and the Swiss National Science Foundation (grant no. SNSF 31003A_147074 to D.S.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Andriotis VME, Pike MJ, Bunnewell S, Hills MJ, Smith AM (2010a) The plastidial glucose-6-phosphate/phosphate antiporter GPT1 is essential for morphogenesis in Arabidopsis embryos. Plant J 64: 128–139 [DOI] [PubMed] [Google Scholar]
- Andriotis VME, Pike MJ, Kular B, Rawsthorne S, Smith AM (2010b) Starch turnover in developing oilseed embryos. New Phytol 187: 791–804 [DOI] [PubMed] [Google Scholar]
- Angeles-Núñez JG, Tiessen A (2010) Arabidopsis sucrose synthase 2 and 3 modulate metabolic homeostasis and direct carbon towards starch synthesis in developing seeds. Planta 232: 701–718 [DOI] [PubMed] [Google Scholar]
- Bahaji A, Sánchez-López AM, De Diego N, Muñoz FJ, Baroja-Fernández E, Li J, Ricarte-Bermejo A, Baslam M, Aranjuelo I, Almagro G, et al. (2015) Plastidic phosphoglucose isomerase is an important determinant of starch accumulation in mesophyll cells, growth, photosynthetic capacity, and biosynthesis of plastidic cytokinins in Arabidopsis. PLoS ONE 10: e0119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Fernández E, Muñoz FJ, Bahaji A, Almagro G, Pozueta-Romero J (2012) Letter. Reply to Smith et al.: No evidence to challenge the current paradigm on starch and cellulose biosynthesis involving sucrose synthase activity. Proc Natl Acad Sci USA 109: E777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DHP, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM (2009) Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci USA 106: 13124–13129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin J, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, Borges F, Becker JD, Feijó JA (2011) Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol 155: 2066–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell K. (1952) Developmental morphology in Dianthus. II. Starch accumulation in ovule and seed. Am J Bot 39: 458–467 [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément C, Chavant L, Burrus M, Audran JC (1994) Anther starch variations in Lilium during pollen development. Sex Plant Reprod 7: 347–356 [Google Scholar]
- Clément C, Pacini E (2001) Anther plastids in angiosperms. Bot Rev 67: 54–73 [Google Scholar]
- Crevillén P, Ballicora MA, Mérida A, Preiss J, Romero JM (2003) The different large subunit isoforms of Arabidopsis thaliana ADP-glucose pyrophosphorylase confer distinct kinetic and regulatory properties to the heterotetrameric enzyme. J Biol Chem 278: 28508–28515 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Ventriglia T, Pinto F, Orea A, Mérida A, Romero JM (2005) Differential pattern of expression and sugar regulation of Arabidopsis thaliana ADP-glucose pyrophosphorylase-encoding genes. J Biol Chem 280: 8143–8149 [DOI] [PubMed] [Google Scholar]
- Déjardin A, Rochat C, Wuillème S, Boutin JP (1997) Contribution of sucrose synthase, ADP-glucose pyrophosphorylase and starch synthase to starch synthesis in developing pea seeds. Plant Cell Environ 20: 1421–1430 [Google Scholar]
- Fallahi H, Scofield GN, Badger MR, Chow WS, Furbank RT, Ruan YL (2008) Localization of sucrose synthase in developing seed and siliques of Arabidopsis thaliana reveals diverse roles for SUS during development. J Exp Bot 59: 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijó JA, Pais MSS (1988) Ultrastructural modifications of plastids and starch metabolism during the microsporogenesis of Ophrys lutea (Orchidaceae). Ann Bot (Lond) 61: 215–219 [Google Scholar]
- Fettke J, Malinova I, Albrecht T, Hejazi M, Steup M (2011) Glucose-1-phosphate transport into protoplasts and chloroplasts from leaves of Arabidopsis. Plant Physiol 155: 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge UI. (1999) Phosphate translocators in plastids. Annu Rev Plant Physiol Plant Mol Biol 50: 27–45 [DOI] [PubMed] [Google Scholar]
- Geigenberger P. (2011) Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol 155: 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M, Fernie AR (2004) Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ 27: 655–673 [Google Scholar]
- Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr JM., Jr (1971) A new clearing-squash technique for the study of ovule development in angiosperms. Am J Bot 58: 785–790 [Google Scholar]
- Herr JM., Jr (1972) Applications of a new clearing technique for the investigation of vascular plant morphology. J Elisha Mitchell Sci Soc 88: 137–143 [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostettler C, Kölling K, Santelia D, Streb S, Kötting O, Zeeman SC (2011) Analysis of starch metabolism in chloroplasts. Methods Mol Biol 775: 387–410 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B, Fischer K, Hilpert B, Schubert S, Gutensohn M, Weber A, Flügge UI (1998) Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 10: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram BW, Xu WW, Carter CJ (2009) Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues. BMC Plant Biol 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz HH, Häusler RE, Fettke J, Herbst K, Niewiadomski P, Gierth M, Bell K, Steup M, Flügge UI, Schneider A (2010) The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol (Stuttg) (Suppl 1) 12: 115–128 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot 69: 461–476 [Google Scholar]
- Miki-Hiroshige H, Nakamura S (1983) Growth and differentiation of amyloplasts during male gamete development in Lilium longiflorum. In Mulcahy DL, Ottaviano E, eds, Pollen: Biology and Implications for Plant Breeding. Elsevier, Amsterdam, pp 141–147 [Google Scholar]
- Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Viale AM, Etxeberria E, Alonso-Casajús N, Pozueta-Romero J (2005) Sucrose synthase controls both intracellular ADP glucose levels and transitory starch biosynthesis in source leaves. Plant Cell Physiol 46: 1366–1376 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Brown RC, Lemmon BE (2000) The specialized chalazal endosperm in Arabidopsis thaliana and Lepidium virginicum (Brassicaceae). Protoplasma 212: 99–110 [Google Scholar]
- Niewiadomski P, Knappe S, Geimer S, Fischer K, Schulz B, Unte US, Rosso MG, Ache P, Flügge UI, Schneider A (2005) The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini E, Franchi GG (1983) Pollen grain development in Smilax aspersa L. and possible function of the loculus. In Mulcahy DL, Ottaviano E, eds, Pollen: Biology and Implications for Plant Breeding. Elsevier, Amsterdam, pp 183–190 [Google Scholar]
- Park SK, Howden R, Twell D (1998) The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznickova SA, Willemse MTM (1980) Formation of pollen in the anther of Lilium. II. The function of the surrounding tissues in the formation of pollen and pollen wall. Acta Bot Neerl 29: 141–156 [Google Scholar]
- Rolletschek H, Nguyen TH, Häusler RE, Rutten T, Göbel C, Feussner I, Radchuk R, Tewes A, Claus B, Klukas C, et al. (2007) Antisense inhibition of the plastidial glucose-6-phosphate/phosphate translocator in Vicia seeds shifts cellular differentiation and promotes protein storage. Plant J 51: 468–484 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmid MW, Schmidt A, Grossniklaus U (2015) The female gametophyte: an emerging model for cell type-specific systems biology in plant development. Front Plant Sci 6: 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wüest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U (2011) Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol 9: e1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz P, Jensen WA (1971) Capsella embryogenesis: the chalazal proliferating tissue. J Cell Sci 8: 201–227 [DOI] [PubMed] [Google Scholar]
- Schulz SP, Jensen WA (1969) Capsella embryogenesis: the suspensor and the basal cell. Protoplasma 67: 139–163 [Google Scholar]
- Smith AM. (2012) Starch in the Arabidopsis plant. Starke 64: 421–434 [Google Scholar]
- Smith AM, Kruger NJ, Lunn JE (2012) Letter. Source of sugar nucleotides for starch and cellulose synthesis. Proc Natl Acad Sci USA 109: E77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Egli B, Eicke S, Zeeman SC (2009) The debate on the pathway of starch synthesis: a closer look at low-starch mutants lacking plastidial phosphoglucomutase supports the chloroplast-localized pathway. Plant Physiol 151: 1769–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Zeeman SC (2012) Starch metabolism in Arabidopsis. The Arabidopsis Book 10: e0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Stadler R, Baier K, Sauer N (1999) A male gametophyte-specific monosaccharide transporter in Arabidopsis. Plant J 17: 191–201 [DOI] [PubMed] [Google Scholar]
- Tsai HL, Lue WL, Lu KJ, Hsieh MH, Wang SM, Chen J (2009) Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol 151: 1582–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ruan YL (2012) New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol 160: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yu HJ, Hogan P, Sundaresan V (2005) Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol 139: 1853–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TS, Lue WL, Wang SM, Chen J (2000) Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol 123: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Häusler RE, Greiten C, Hajirezaei MR, Haferkamp I, Neuhaus HE, Flügge UI, Ludewig F (2008) Overriding the co-limiting import of carbon and energy into tuber amyloplasts increases the starch content and yield of transgenic potato plants. Plant Biotechnol J 6: 453–464 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.