WOX11/12 directly activate WOX5/7 to induce root primordium development during de novo regeneration.
Abstract
De novo organogenesis, which gives rise to adventitious roots and shoots, is a type of plant regeneration for survival after wounding. In Arabidopsis (Arabidopsis thaliana), two main cell fate transition steps are required to establish the root primordium during de novo root organogenesis from leaf explants. The first step from regeneration-competent cells to root founder cells involves activation of WUSCHEL-RELATED HOMEOBOX11 (WOX11) and WOX12 (WOX11/12) expression by auxin. However, the molecular mechanism controlling the second step of fate transition from root founder cells to root primordium is poorly understood. In this study, we show that the expression levels of WOX11/12 decrease while those of WOX5 and 7 (WOX5/7) increase during the transition from root founder cells to the root primordium. WOX11/12 function genetically upstream of WOX5/7, and the WOX11/12 proteins directly bind to the promoters of WOX5/7 to activate their transcription. Mutations in WOX5/7 result in defective primordium formation. Overall, our data indicate that the expression switch from WOX11/12 to WOX5/7 is critical for initiation of the root primordium during de novo root organogenesis.
Plants have powerful regenerative abilities that allow them to reproduce vegetatively by forming a whole plant from somatic cells (Sugimoto et al., 2011; Xu and Huang, 2014; Ikeuchi et al., 2016). De novo organogenesis, a process in which adventitious roots and shoots regenerate from wounded or detached organs, is a type of vegetative reproduction that is widely exploited in modern agricultural applications (Sussex, 2008; Duclercq et al., 2011; Xu and Huang, 2014). De novo root organogenesis commonly occurs in nature, because wounded or detached plant organs can rapidly regenerate adventitious roots to ensure their survival (Bellini et al., 2014; Xu and Huang, 2014; Steffens and Rasmussen, 2016).
Previously, we established a simple method of de novo root organogenesis by culturing leaf explants of Arabidopsis (Arabidopsis thaliana) on B5 medium without exogenous hormones (Chen et al., 2014; Liu et al., 2014). In this system, endogenous hormones are sufficient to induce adventitious root formation from leaf explants. Using this method, we revealed that there are at least two steps of cell fate transition required for the formation of the newly regenerated root primordia. In the first step, regeneration-competent cells (i.e. procambium and vascular parenchyma cells) become root founder cells, which are marked by WUSCHEL-RELATED HOMEOBOX 11 and 12 (hereafter abbreviated as WOX11/12). In the second step, root primordium cells, which are marked by WOX5, initiate from root founder cells, and this step involves cell division.
The molecular mechanism of the first-step cell fate transition (i.e. from competent cells to root founder cells) involves the activation of WOX11/12 expression by auxin (Liu et al., 2014), a major hormone that triggers adventitious rooting (Greenwood et al., 2001; De Klerk, 2002; Ahkami et al., 2009; Correa et al., 2012; Liu et al., 2014). Endogenous free auxin is quickly produced in mesophyll cells upon wounding, and then polar transported into competent cells near the wound (Liu et al., 2014). In competent cells, auxin directly up-regulates transcription of WOX11/12 for priming root founder cells (Liu et al., 2014). Therefore, WOX11/12 seem to be pivotal genes that turn on the molecular pathway for adventitious rooting. Blocking of WOX11/12 resulted in rooting defects, and overexpression of WOX11/12 greatly enhanced the rooting process (Liu et al., 2014).
However, the molecular mechanism of the second-step cell fate transition (i.e. from root founder cells to root primordium) is barely understood. A key question is what molecular mechanism underlies the ability of WOX11/12 to convey the rooting process from root founder cells to root primordium cells. WOX5 is a molecular marker in the root primordium; however, whether it functions in controlling root primordium formation and how its transcription is activated in the root primordium are unclear. In this study, we show that WOX11/12 directly activates the transcription of WOX5 and its most closely related gene, WOX7 (hereafter, WOX5/7; Supplemental Fig. S1A) and that this is essential for the initiation of a normal root primordium. WOX11/12 and WOX5/7 belong to two different clades of the WOX family. WOX11/12 are intermediate-clade WOX genes, while WOX5/7 are in the WUS clade (Supplemental Fig. S1A; Haecker et al., 2004; Sarkar et al., 2007; Mukherjee et al., 2009; Nardmann et al., 2009; van der Graaff et al., 2009; Zhao et al., 2009; Nardmann and Werr, 2013; Lian et al., 2014; Pi et al., 2015; Ge et al., 2016; Zeng et al., 2016). Therefore, we propose that the switch between the two clades of WOX genes represents the fate transition from root founder cells to root primordium cells in de novo root organogenesis.
RESULTS
WOX11/12-to-WOX5/7 Expression Switch during Root Primordium Initiation
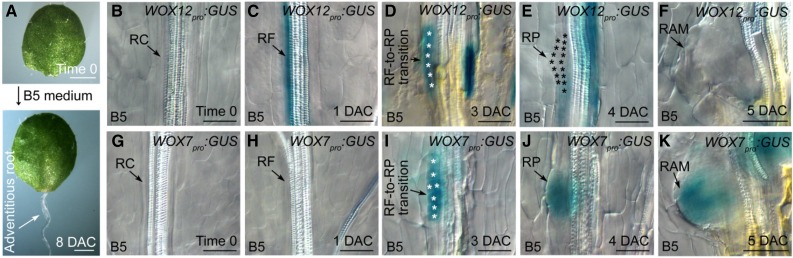
We cultured the Arabidopsis leaf explants on B5 medium without added hormones, and adventitious roots regenerated from the wounded site on the leaf explants (Fig. 1A; Chen et al., 2014; Liu et al., 2014). To analyze the mechanism by which the root primordium initiates from root founder cells, we first analyzed the expression patterns of WOX genes during adventitious rooting.
Figure 1.
Expression patterns of WOX12 and WOX7 during adventitious rooting. A, System of de novo root organogenesis used in this study (Chen et al., 2014). B to F, GUS staining of WOX12pro:GUS in leaf explants at time 0 (B), 1 DAC (C), 3 DAC (D), 4 DAC (E), and 5 DAC (F). G to K, GUS staining of WOX7pro:GUS in leaf explants at time 0 (G), 1 DAC (H), 3 DAC (I), 4 DAC (J), and 5 DAC (K). White asterisks in D and I indicate transient step in which root founder cells are undergoing cell division to form root primordium. Black asterisks in E indicate dome-shaped root primordia. RC, regeneration-competent cell; RF, root founder cell; RP, root primordium. Bars = 1 mm (A) and 50 μm (B–K).
Using WOX11pro:GUS (Liu et al., 2014) and WOX12pro:GUS reporter lines, in which the promoters of WOX11 and 12 were fused to the gene encoding GUS, we found that the expression of WOX11/12 was activated at the very beginning of adventitious rooting from the leaf explant, when competent cells became root founder cells at 1 d after culture (DAC; Fig. 1, B and C; Supplemental Fig. S2, A and B; Liu et al., 2014). Their expression could be observed during the initial cell divisions at 3 DAC (Fig. 1D; Supplemental Fig. S2C), but decreased when the dome-shaped root primordium formed at 4 DAC (Fig. 1E; Supplemental Fig. S2D; Liu et al., 2014). No WOX11/12 expression was detected in the newly formed root apical meristem (RAM) at 5 DAC (Fig. 1F; Supplemental Fig. S2E).
We then analyzed the expression patterns of WOX5/7 in adventitious rooting using WOX5pro:GUS (Liu et al., 2014) and WOX7pro:GUS reporter lines. No WOX5/7 expression was detected during root founder cell formation at 1 DAC (Fig. 1, G and H; Supplemental Fig. S2, F and G). However, WOX5/7 expression increased during the initial cell divisions at 3 DAC (Fig. 1I; Supplemental Fig. S2H). This expression pattern of WOX5/7 overlapped with that of WOX11/12. The expression of WOX5/7 continued in the dome-shaped root primordium at 4 DAC (Fig. 1J; Supplemental Fig. S2I), when WOX11/12 expression decreased.
During the formation of the RAM, WOX5 expression was gradually confined to the center region (Supplemental Fig. S2J). Expression of WOX7 was also gradually restricted to the inner region of the newly formed RAM (Fig. 1K). After formation of the RAM, the root tip emerged from the leaf explant. As it emerged, the expression of WOX5 and WOX7 continued to be gradually restricted to the stem cell niche (Supplemental Fig. S3), which comprises the quiescent center (QC) and initial cells (Scheres, 2007).
Together, these results show that WOX11/12 function during the very first event in root founder cell establishment and that WOX5/7 start to function when root founder cells divide to form root primordium cells.
WOX5/7 Are Involved in Root Primordium Formation
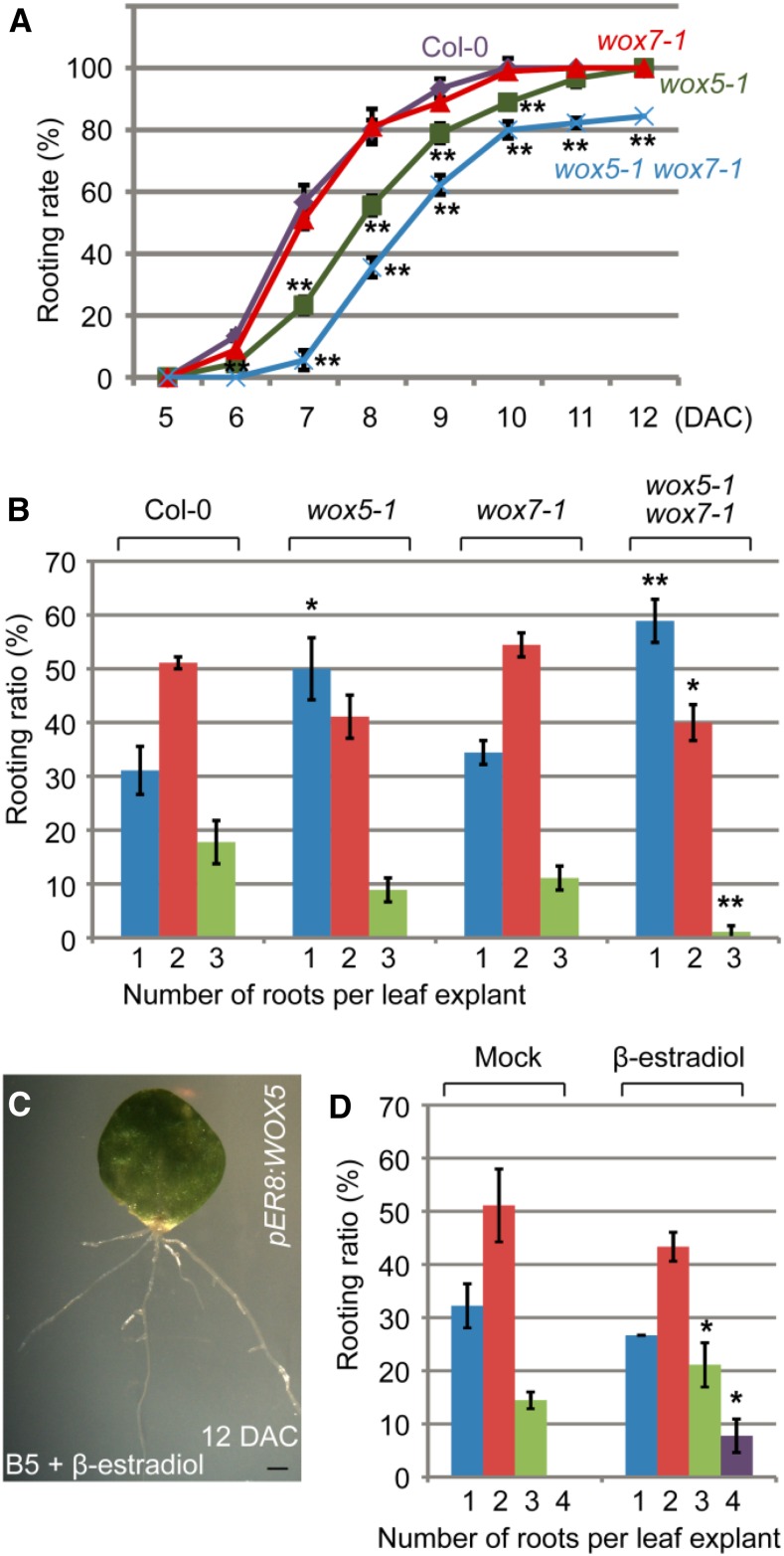
To explore the developmental role of WOX5/7 in de novo root organogenesis, we first analyzed the phenotypes of adventitious roots formed from leaf explants of the wox5-1 and wox7-1 single mutants and the double mutant. The rooting rate was reduced in the wox5-1 single mutant and further reduced in the wox5-1 wox7-1 double mutant (Fig. 2A), suggesting that mutations in WOX5/7 result in slow development of adventitious roots. In addition, the number of adventitious roots that regenerated from leaf explants was reduced in the wox5-1 single mutant, and this number was further decreased in the wox5-1 wox7-1 double mutant (Fig. 2B). In pER8:WOX5 transgenic lines, β-estradiol-induced overexpression of WOX5 increased the number of roots produced per leaf explant (Fig. 2, C and D). These results indicate that WOX5 and WOX7 are involved in de novo root organogenesis.
Figure 2.
WOX5/7 are involved in adventitious rooting. A, Rooting rate of leaf explants (percentage of leaf explants with regenerated adventitious roots) from wild-type Col-0, wox5-1 single mutant, wox7-1 single mutant, and wox5-1 wox7-1 double mutant on B5 medium. B, Quantitative analyses of ratio of adventitious root number per 15-DAC leaf explant from Col-0, wox5-1, wox7-1, and wox5-1 wox7-1 cultured on B5 medium. C, Leaf explant from pER8:WOX5 at 12 DAC on B5 medium containing 10 μm β-estradiol. D, Ratio of adventitious root number per 15-DAC leaf explant from pER8:WOX5 cultured on B5 medium without (mock) or with 10 μm β-estradiol. Bars in A, B, and D show sd from three biological repeats (n = 30 per repeat). *P < 0.05 and **P < 0.01 in two-sample t tests, compared with Col-0 (A and B) or mock (D). Bar = 1 mm (C).
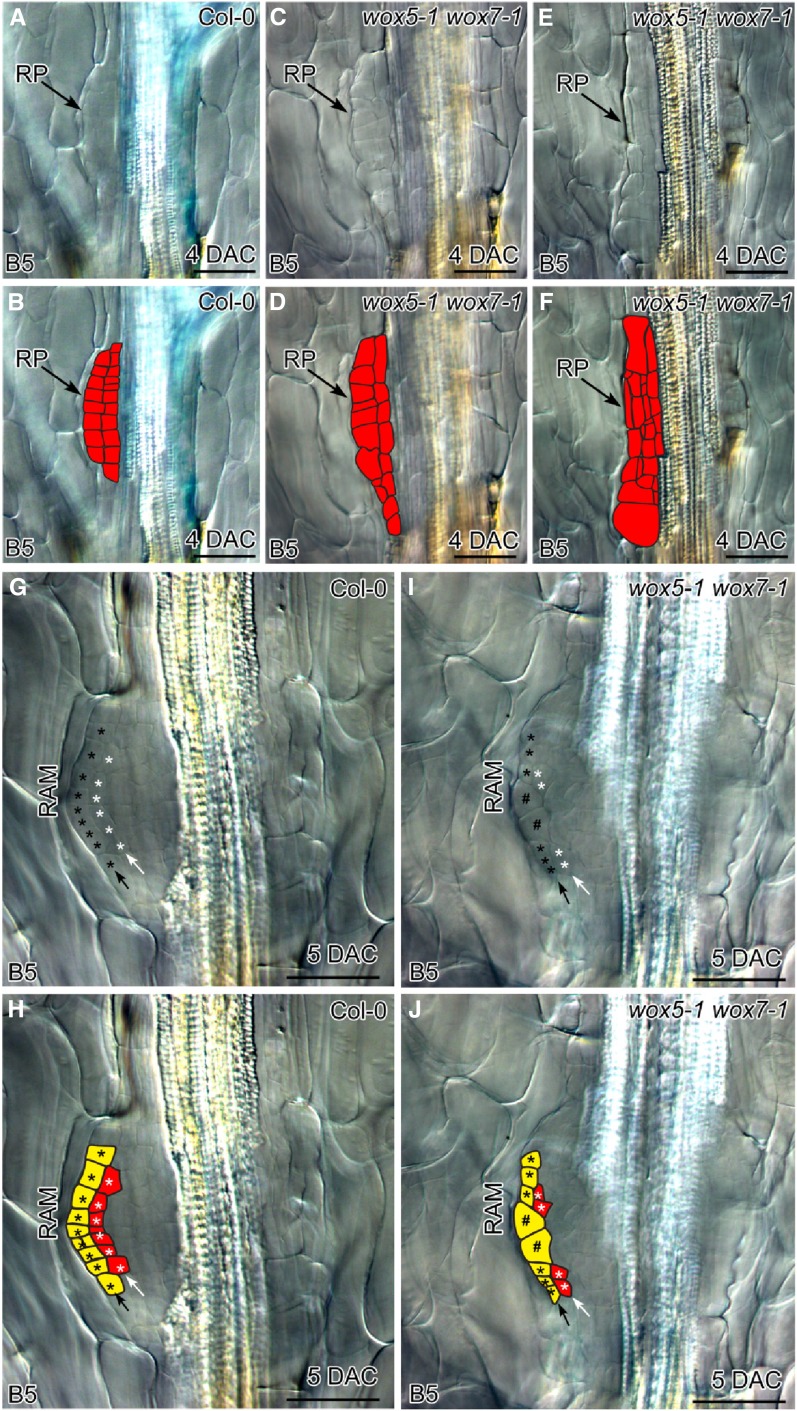
To further analyze the role of WOX5/7 at the cellular level, we carefully compared the root primordia in the wox5-1 wox7-1 double mutant with those in the wild type. In the dome-shaped root primordium with two cell layers from wild-type leaf explants, cells were regularly arranged with coordinated cell division between the two layers (Fig. 3, A and B). However, all root primordia in the wox5-1 wox7-1 double mutant showed a disorganized and irregularly arranged cell pattern, although cell division had occurred. Some mildly defective root primordia of wox5-1 wox7-1 showed uncoordinated cell division between the two layers (Fig. 3, C and D). In some severely defective root primordia of wox5-1 wox7-1, both anticlinal and periclinal cell divisions were disorganized and the shape of the root primordium was lost (Fig. 3, E and F).
Figure 3.
WOX5/7 are involved in cell division and tissue organization during adventitious root primordium formation. A and B, Adventitious root primordium in 4-DAC wild-type Col-0 leaf explant showing two layers of cells constituting the typical dome-shaped primordium. Note that cells in the primordium were approximately the same size, suggesting that cell division is coordinated between the two layers. C and D, Adventitious root primordium in 4-DAC wox5-1 wox7-1 leaf explant, showing mildly defective root primordium organization. Note that cell sizes differed within the primordium, indicating that cell division between the two layers was not properly coordinated. E and F, Adventitious root primordium in 4-DAC wox5-1 wox7-1 leaf explant, showing strongly defective root primordium organization. Note severely disordered cell division and complete loss of dome shape. G and H, Newly formed RAM in 5-DAC wild-type Col-0 leaf explant. Black arrow shows outermost layer that will develop into the root cap. White arrow shows the second layer, which will harbor the QC of the stem cell niche at its center. I and J, Newly formed RAM in 5-DAC wox5-1 wox7-1 leaf explant. Note enlarged cells in the center of the outermost layer (indicated by #) and absence of stem cell niche in the second layer. B, D, F, H, and J are schematics of each DIC image in A, C, E, G, and I, respectively. We analyzed 54 leaf explants from wox5-1 wox7-1 at 4 DAC; 29 showed mildly defective root primordia as shown in C and D, and 25 showed strongly defective root primordia as shown in E and F. We analyzed 30 leaf explants from wox5-1 wox7-1 at 5 DAC; 26 showed defective RAM organization as shown in I and J and four showed arrested development at the primordium stage with no RAM formation. RP, root primordium. Bars = 50 μm (A–J).
Although most of the root primordia in wox5-1 wox7-1 were able to form RAMs and finally root tips, the newly formed RAMs in wox5-1 wox7-1 showed defective stem cell niche formation (Fig. 3, G–J). In the newly formed RAMs in wild-type leaf explants, cells in the outermost layer (Fig. 3, G and H, black asterisks) were destined to form the root cap, and those in the center of the second layer (Fig. 3, G and H, white asterisks) were destined to form the stem cell niche, including QC during the next stage of development (Malamy and Benfey, 1997; Goh et al., 2016). In the newly formed RAMs in wox5-1 wox7-1 leaf explants, we observed defects with irregular cell division in both the outermost layer and the region where the stem cell niche was going to form (Fig. 3, I and J). The defective RAM patterning resulted in the loss of the QC in the stem cell niche in adventitious root tips regenerated from wox5-1 wox7-1 leaf explants (Supplemental Fig. S4).
The results of these phenotype analyses indicate that WOX5/7 are required for proper organization of adventitious root primordium and subsequent stem cell niche establishment in the newly formed RAM.
WOX11/12 Promote WOX5/7 Expression for Root Primordium Initiation
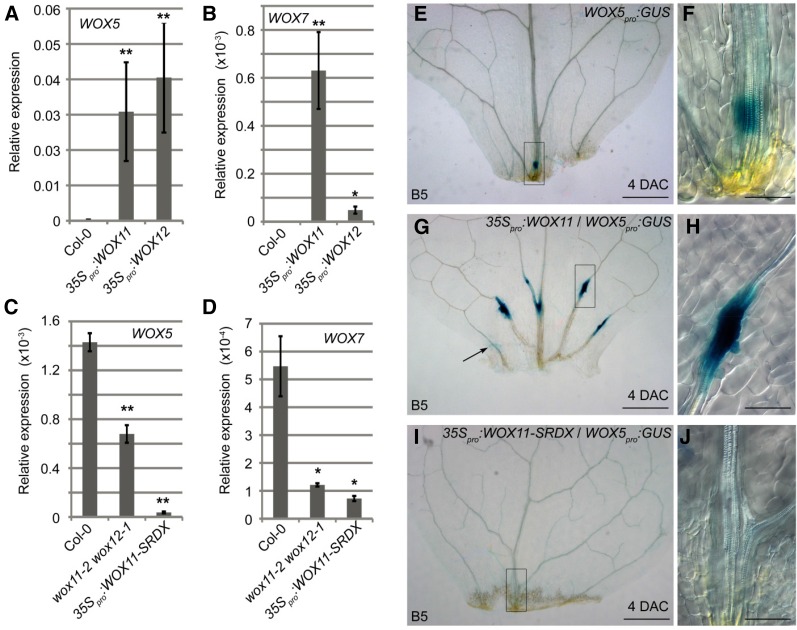
To test the genetic relationship between WOX11/12 and WOX5/7, we first performed quantitative reverse transcription-PCR (qRT-PCR) analyses to determine the transcript levels of WOX5/7 in 35Spro:WOX11 and 35Spro:WOX12 transgenic lines, which overexpressed WOX11 and WOX12, respectively. Transcripts of WOX5 and WOX7 could not be detected in wild-type leaves, but both WOX5 and WOX7 were detected in leaves of the WOX11- and WOX12-overexpression lines (Fig. 4, A and B), suggesting that WOX11/12 can activate the transcription of WOX5/7.
Figure 4.
WOX11/12 up-regulate WOX5/7 expression. A and B, qRT-PCR analyses showing transcripts of WOX5 (A) and WOX7 (B) ectopically expressed in leaves of 35Spro:WOX11 and 35Spro:WOX12. C and D, qRT-PCR analyses of transcripts of WOX5 (C) and WOX7 (D) in 4-DAC leaf explants from the wild-type Col-0, wox11-2 wox12-1, and 35Spro:WOX11-SRDX on B5 medium. Note that WOX5 expression in 35Spro:WOX11-SRDX was previously reported (Liu et al., 2014). E to J, GUS staining of leaf explants from WOX5pro:GUS in wild type (E and F), 35Spro:WOX11 (G and H), and 35Spro:WOX11-SRDX (I and J) backgrounds at 4 DAC on B5 medium. F, H, and J are close-ups of the boxed regions in E, G, and I, respectively. Arrow in G indicate GUS signal in mesophyll cells. Bars in A to D show se from three biological repetitions. Each biological repetition was performed with three technical repetitions. *P < 0.05 and **P < 0.01 (two-sample t test, compared with Col-0). Bars = 500 μm (E, G, and I) and 100 μm (F, H, and J).
Next, we analyzed the regulation of WOX5/7 by WOX11/12 during adventitious rooting from leaf explants. Expression of WOX5/7 could be detected in 4-DAC wild-type leaf explants by qRT-PCR. The transcript levels of WOX5/7 were reduced in 4-DAC leaf explants from the wox11-2 wox12-1 double mutant (Fig. 4, C and D), which shows a mild defect in adventitious rooting (Liu et al., 2014). WOX5/7 expression levels were severely reduced in 4-DAC leaf explants from the 35Spro:WOX11-SRDX lines (Fig. 4, C and D; Liu et al., 2014), in which a repression domain SRDX (Hiratsu et al., 2003) is fused to the WOX11 protein to block the WOX11 pathway and adventitious rooting (Liu et al., 2014). Using the WOX5pro:GUS reporter line, we observed that the WOX5-marked root primordium usually formed at the midvein of the leaf explants near the wound site (Fig. 4, E and F). In contrast, in the 35Spro:WOX11 leaf explants, WOX5-marked root primordia formed in many regions, including lateral veins (Fig. 4, G and H). Occasionally, WOX5 expression could be observed in mesophyll cells in the 35Spro:WOX11 leaf explants (Fig. 4G). In the 35Spro:WOX11-SRDX background, no WOX5-marked root primordia formed from the leaf explants (Fig. 4, I and J). Similar results were obtained using the WOX7pro:GUS reporter line (Supplemental Fig. S5). These data suggest that WOX11/12 play a role in activating WOX5/7 expression during de novo root organogenesis.
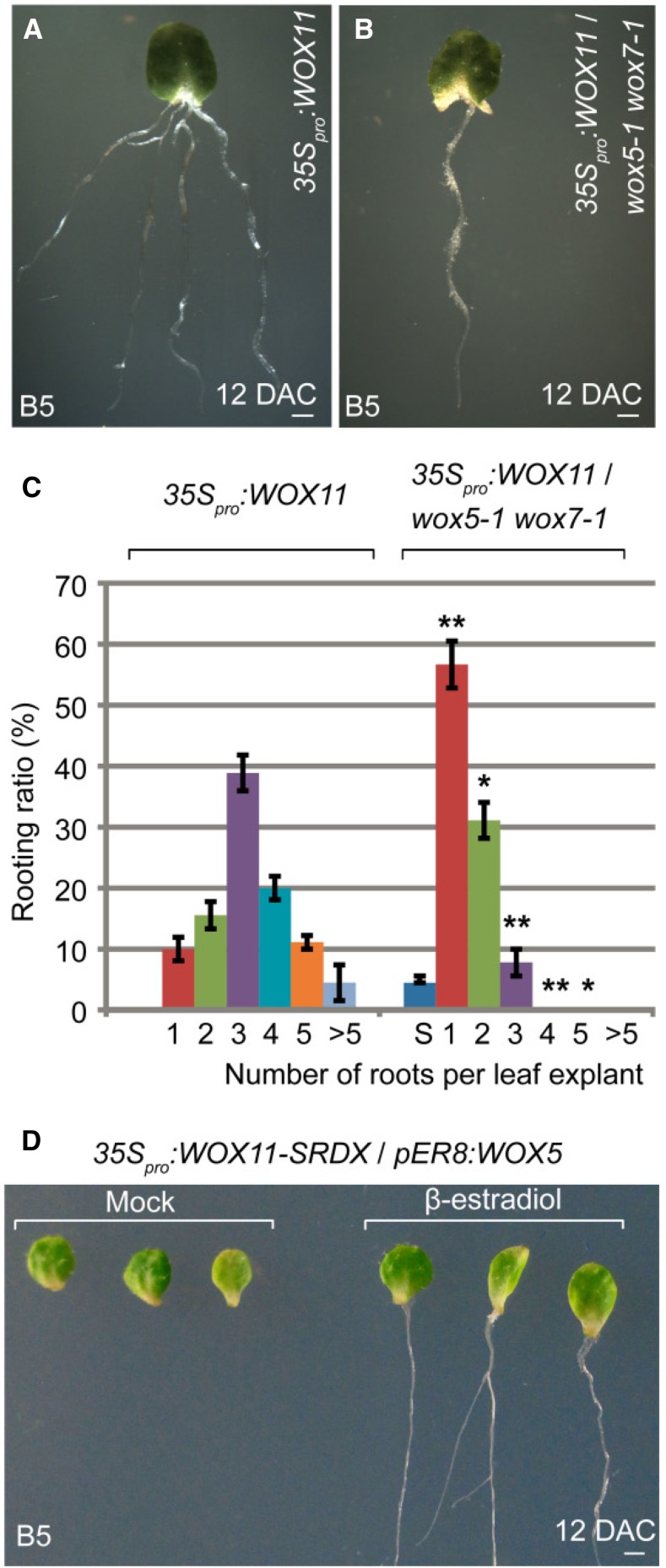
Leaf explants of 35Spro:WOX11 and 35Spro:WOX12 had enhanced rooting abilities and produced many more adventitious roots from the wound site than did leaf explants of wild type (Fig. 5A; Supplemental Fig. S6A; for comparison, see Fig. 1A; Liu et al., 2014). We introduced 35Spro:WOX11 and 35Spro:WOX12 into the wox5-1 wox7-1 double mutant background. The rooting abilities of 35Spro:WOX11 and 35Spro:WOX12 were reduced in the wox5-1 wox7-1 background compared with those in the wild-type background (Fig. 5, A–C; Supplemental Fig. S6). These data suggest that the rooting-promotion abilities of WOX11/12 at least partly depend on the function of WOX5/7.
Figure 5.
Genetic analysis of WOX11/12 and WOX5/7. A and B, Leaf explants from 35Spro:WOX11 (A) and 35Spro:WOX11/wox5-1 wox7-1 (B) at 12 DAC on B5 medium. C, Ratio of adventitious root number per 15-DAC leaf explants from 35Spro:WOX11 and 35Spro:WOX11/wox5-1 wox7-1 on B5 medium. S, a small portion of leaf explants turned yellow and became senescence. Bars show sd from three biological repeats (n = 30 per repeat). *P < 0.05 and **P < 0.01 (two-sample t test, compared with 35Spro:WOX11). D, Leaf explants from 35Spro:WOX11-SRDX/pER8:WOX5 at 12 DAC cultured on B5 medium without (left) or with (right) 10 μm β-estradiol. None of the 29 leaf explants cultured on B5 medium without β-estradiol formed adventitious roots. Seven of 29 leaf explants cultured on B5 medium with 10 μm β-estradiol regenerated adventitious roots. Bars = 1 mm (A, B, and D).
Rooting was severely blocked in 35Spro:WOX11-SRDX leaf explants (Fig. 5D; Liu et al., 2014). When we introduced a β-estradiol-induced WOX5-overexpression vector, pER8:WOX5, into the 35Spro:WOX11-SRDX background, the overexpression of WOX5 partly rescued the rooting defect caused by WOX11-SRDX (Fig. 5D). This result suggested that WOX5 is a functional downstream gene controlled by the WOX11 pathway.
Based on these genetic data, together with the expression analyses of WOX11/12 and WOX5/7, we propose that WOX11/12 promote WOX5/7 expression for the fate transition of root founder cells to root primordium cells.
WOX11/12 Directly Activate WOX5/7 Expression
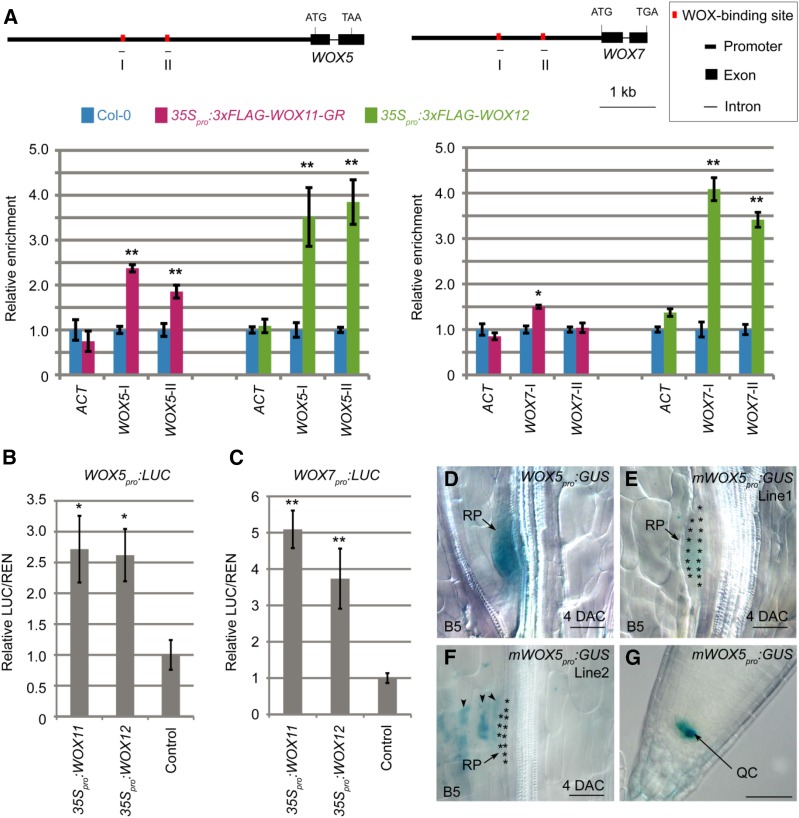
We conducted a chromatin immunoprecipitation (ChIP) experiment to test whether WOX11/12 proteins bind to the WOX5/7 loci. Using the anti-FLAG antibody, we detected that the 3×FLAG-WOX11-GR and 3×FLAG-WOX12 fusion proteins directly bound to the predicted WOX-binding cis elements (TTAATGG; Lohmann et al., 2001; Leibfried et al., 2005; Zhao et al., 2009) on the promoters of WOX5 and WOX7 (Fig. 6A).
Figure 6.
WOX11/12 directly regulate WOX5/7 expression. A, ChIP analysis showing enrichment of 3×FLAG-WOX11-GR and 3×FLAG-WOX12 in promoters of WOX5 and WOX7. Schematics of WOX5 and WOX7 gene structures are shown above ChIP data. Horizontal lines below genes show positions of PCR fragments in ChIP analysis. Bars show sd from three PCR repetitions. Results were confirmed with two independent biological repetitions. Values in Col-0 were arbitrarily fixed at 1.0. ACTIN (ACT) locus served as the negative control. *P < 0.05 and **P < 0.01 (two-sample t test, compared with Col-0 control). B and C, Relative ratio of firefly luciferase (LUC) to Renilla luciferase (REN) activity in tobacco leaves cotransformed with 35Spro:WOX11 or 35Spro:WOX12 and WOX5pro:LUC (B), and cotransformed with 35Spro:WOX11 or 35Spro:WOX12 and WOX7pro:LUC (C). Sole transformation with WOX5pro:LUC (B) or WOX7pro:LUC (C) served as the control. Bars show se from three biological repeats. Each biological repetition was performed with three experimental repetitions. *P < 0.05 and **P < 0.01 (two-sample t test, compared with control). D to F, GUS staining of 4-DAC leaf explants from WOX5pro:GUS (D) and two independent mWOX5pro:GUS lines (E and F) on B5 medium. Asterisks in E and F indicate dome-shaped root primordia. WOX5pro:GUS (D) served as the control. GUS signal could be occasionally observed in mesophyll in mWOX5pro:GUS (arrowheads in F). G, GUS staining of the primary root tip from mWOX5pro:GUS (line 1). Bars = 50 μm (D–G).
Next, we tested the direct activation of WOX5/7 by WOX11/12 using a transient expression system in tobacco (Nicotiana tabacum) leaves (Hellens et al., 2005). We coexpressed WOX11 or WOX12 together with WOX5pro:LUC or WOX7pro:LUC, in which the luciferase reporter gene is fused downstream of the WOX5 or WOX7 promoter. The results showed that WOX11/12 activated the luciferase response in tobacco leaves (Fig. 6, B and C), confirming that WOX11/12 directly activate WOX5/7 expression in planta.
To obtain genetic evidence that the WOX11/12-binding elements on the WOX5 locus are required for activation of WOX5, we constructed mWOX5pro:GUS reporter lines in which the WOX11/12-binding elements in the WOX5 promoter were mutated. GUS signals were barely detected in adventitious root primordia from leaf explants of two independent mWOX5pro:GUS lines (Fig. 6, D–F). In contrast, GUS signals could be clearly observed in the tip of the primary root (Fig. 6G). These results indicate that the WOX11/12-binding elements are specifically required for activation of WOX5 during the adventitious root primordium initiation, and they are not involved in maintenance of WOX5 expression in the stem cell niche within the primary root tip.
Auxin Is Required for WOX5/7 Activation
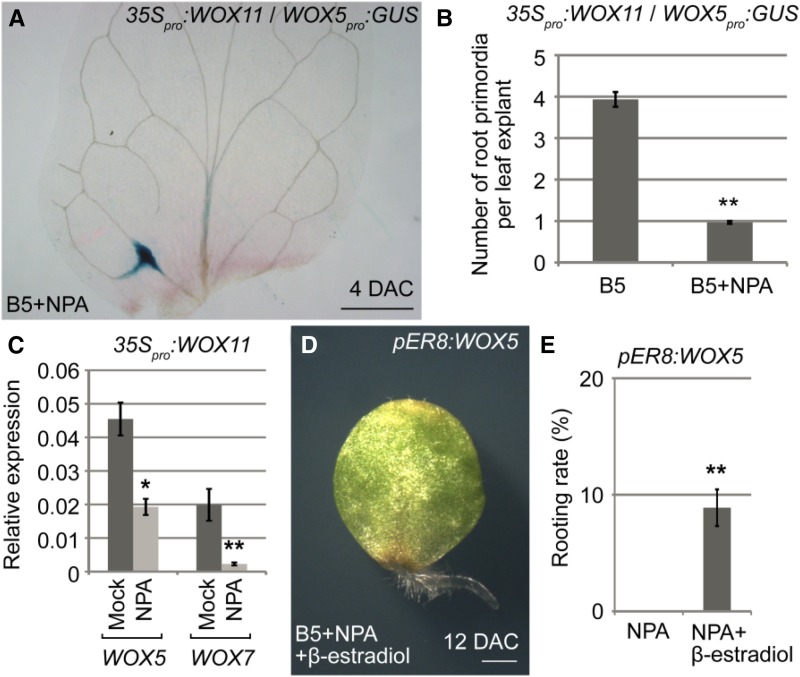
Auxin is the major hormone controlling cell fate transition during adventitious rooting (Greenwood et al., 2001; De Klerk, 2002; Ahkami et al., 2009; Correa et al., 2012; Liu et al., 2014; Xu and Huang, 2014). Therefore, we tested whether auxin is required for activation of WOX5/7 by WOX11/12. We cultured leaf explants on B5 medium containing naphthylphthalamic acid (NPA; a polar auxin transport inhibitor), which has been shown to block rooting by inhibition of auxin transport from mesophyll cells to competent cells near the wound (Liu et al., 2014). Using 35Spro:WOX11/WOX5pro:GUS and 35Spro:WOX11/WOX7pro:GUS, we found that the formation of WOX5- or WOX7-marked root primordia was severely blocked, even when these GUS reporter constructs were expressed in the WOX11-overexpression genetic background (Fig. 7, A and B; Supplemental Fig. S7; for comparison, see Fig. 4G and Supplemental Fig. S5B). Consistent with the GUS observations, qRT-PCR analyses of 35Spro:WOX11 leaf explants showed that the transcript levels of WOX5 and WOX7 were lower in leaf explants treated with NPA (Fig. 7C). In addition, overexpression of WOX5 partially rescued the rooting defects of leaf explants cultured on B5 medium containing NPA (Fig. 7, D and E). Together, these data suggest that auxin acts upstream of WOX5/7, and auxin polar transport to the wounded region is required for activation of WOX5/7 by WOX11/12.
Figure 7.
WOX5/7 activation requires auxin. A, GUS staining of 35Spro:WOX11/WOX5pro:GUS in leaf explant at 4 DAC on B5 medium containing 1 μm NPA. For comparison, see Figure 4G. B, Quantitative analyses of WOX5-marked root primordia in 35Spro:WOX11/WOX5pro:GUS leaf explants at 4 DAC on B5 medium without or with 1 μm NPA. Bars show se from three biological repeats. Each repeat comprised 10 leaf explants. **P < 0.01 (two-sample t test). C, qRT-PCR analyses of transcript levels of WOX5 and WOX7 in 35Spro:WOX11 leaf explants at 4 DAC on B5 medium without (mock) or with 1 μm NPA. Bars show se from three biological repetitions. Each biological repetition was performed with three technical repetitions. *P < 0.05 and **P < 0.01 (two-sample t test, compared with mock). D, Leaf explants from pER8:WOX5 at 12 DAC on B5 medium containing 1 μm NPA and 10 μm β-estradiol. Note that leaf explants on medium containing NPA could not regenerate adventitious roots (Liu et al., 2014), but overexpression of WOX5 partially rescued this rooting defect. E, Statistical analysis of rooting rates shown in D. Bars show sd from three biological repeats (n = 30 per repeat). **P < 0.01 (two-sample t test). Bar = 500 μm (A) and 1 mm (D).
DISCUSSION
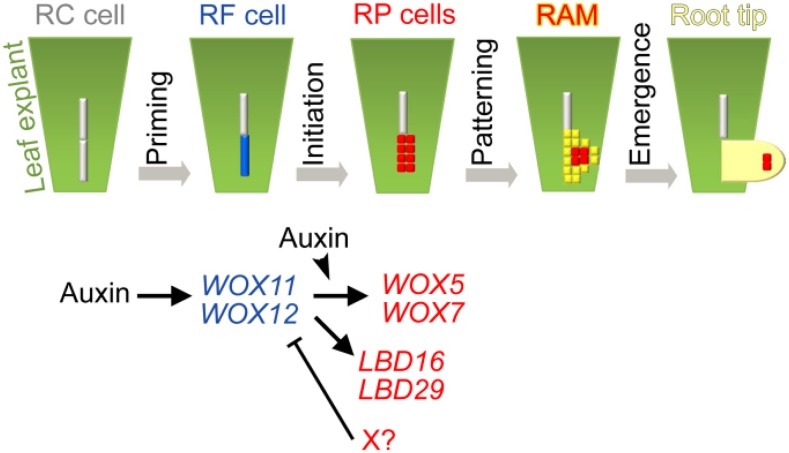
Plants have powerful abilities in regeneration of roots (Xu et al., 2006; Sena et al., 2009; Liu et al., 2014; Efroni et al., 2016). De novo root organogenesis from leaf explants requires consecutive cell fate transition steps to finally form an adventitious root. Here, we borrowed the cellular framework concept of lateral root formation (Péret et al., 2009; Lavenus et al., 2013; Goh et al., 2016) to summarize the consecutive steps in adventitious rooting (Fig. 8, model). The first step of cell fate transition is priming, which results in the formation of adventitious root founder cells. WOX11/12 serve as molecular markers for founder cells. However, wound signals that trigger the priming step are not clear (León et al., 2001; Maffei et al., 2007; Iwase et al., 2011; Xu and Huang, 2014; Chen et al., 2016a; Chen et al., 2016b). The second step of cell fate transition is initiation, which results in the formation of the dome-shaped root primordium via cell division. The expression levels of WOX11/12 decrease and those of WOX5/7 increase as the root founder cells transition into the root primordium. Currently, it is unclear how WOX11/12 expression is repressed in the root primordium cells (Fig. 8, X factor with a question mark). WOX11 is normally expressed in the wox5-1 wox7-1 background, suggesting that WOX5/7 might not serve as the X factor (Supplemental Fig. S8). The third step is patterning, which results in the formation of the preliminary RAM. In this step, the expression of WOX5/7 is gradually restricted toward the stem cell niche, suggesting that the stem cell niche is forming at this stage. The fourth step is emergence, when the newly formed adventitious root tip emerges from the leaf explant. In this step, a mature RAM with a well-organized stem cell niche forms and undergoes rapid cell division, cell differentiation, and elongation.
Figure 8.
Model of de novo root organogenesis from leaf explant. De novo regeneration of adventitious root comprises priming, initiation, patterning, and emergence steps. Founder-cell-specific WOX11/12 function as a valve to activate downstream gene expression, such as the WOX5/7 and LBD pathways, which function in root primordium formation. Activation of WOX11/12 and WOX5/7 requires auxin. An unknown factor (X with question mark) might shut down WOX11/12 expression in the root primordium. RC cell, regeneration-competent cell; RF cell, root founder cell; RP, root primordium.
Based on the above cellular and molecular framework of de novo root organogenesis, the founder-cell markers WOX11/12 act at the very beginning of adventitious rooting and therefore could serve as a valve for all of the following events in adventitious rooting. In 35Spro:WOX11-SRDX, no root primordium was observed in 4-DAC leaf explants (Supplemental Fig. S9), suggesting that there was a complete block of root primordium initiation. In this study, we revealed that WOX5/7 are direct downstream targets of WOX11/12 and that this molecular regulation delivers the fate transition from root founder cells to root primordium cells.
The results of our previous study suggested that WOX11/12 can also activate the expression of LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16) and LBD29 (Liu et al., 2014). Therefore, LBDs could be another downstream pathway regulated by WOX11/12 (Fig. 8). LBD genes are required for rooting and are likely involved in regulating cell division and cell wall metabolism (Lee et al., 2009; Berckmans et al., 2011; Feng et al., 2012a, 2012b; Goh et al., 2012; Lee et al., 2013). Although cell organization within the root primordium was defective in the wox5-1 wox7-1 double mutant, cell division still occurred. One possibility is that the LBD pathway still functions in the wox5-1 wox7-1 double mutant background and controls cell division to form abnormally organized root primordia.
Although both LBDs and WOX5/7 function downstream of WOX11/12, they have different expression patterns. We observed that LBD16 was expressed in dividing root founder cells and the two-layer root primordium cells, like WOX5/7, but it was not expressed in the newly formed RAM (Supplemental Fig. S10). Therefore, unlike the WOX5/7 pathway, the LBD pathway does not continue to function after root primordium initiation.
In the tip of a normal root, WOX5 and WOX7 are expressed in the stem cell niche, which comprises the QC and initial cells (Scheres, 2007); WOX5 is specifically expressed in the QC located in the center of the stem cell niche (Sarkar et al., 2007) and WOX7 is specifically expressed in the endodermis-cortical initial cell, which is adjacent to the QC (Cui et al., 2011). The stem cell niche is the source of all tissues within the root (Scheres, 2007; Aichinger et al., 2012). In this study, we show that WOX5/7 are ubiquitously expressed throughout the whole adventitious root primordium. Therefore, we hypothesize that the adventitious root primordium consists of precursor cells of QC and initials and is able to differentiate into a RAM. In addition, WOX5/7 may also have a role in primary root and lateral root development, suggesting that the function of WOX5/7 is not only restricted in adventitious root formation (Supplemental Fig. S11; Sarkar et al., 2007; Scheres, 2007; Chu et al., 2013; Tian et al., 2014; Ji et al., 2015; Pi et al., 2015; Zhou et al., 2015; Kong et al., 2016).
Overall, our study shows that the molecular mechanism of the second step of cell fate transition from root founder cells to root primordium initiation involves the direct activation of WOX5/WOX7 by WOX11/12 during de novo root organogenesis.
MATERIALS AND METHODS
Plant Materials and Culture Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 was used as the wild type in this study. To produce WOX12pro:GUS and WOX7pro:GUS transgenic plants, 5.3-kb WOX12 and 3.4-kb WOX7 promoters were PCR amplified and each inserted into the pBI101 vector. 35Spro:3×FLAG-WOX12 was constructed by insertion of the cDNA encoding the 3×FLAG-WOX12 protein into the pMON530 vector. For mWOX5pro:GUS, a 4.6-kb mutated WOX5 promoter with mutations at the WOX-binding sites was inserted into the pBI101 vector. To construct pER8:WOX5, a cDNA encoding the full-length WOX5 protein was PCR amplified and inserted into the pER8 vector (Zuo et al., 2000). Transgenic plants were obtained by Agrobacterium tumefaciens-mediated transformation of each of these constructs into Col-0. The wox5-1 (SALK_038262; Sarkar et al., 2007) and wox7-1 (SALK_065801; Kong et al., 2016) mutants and the wox11-2 wox12-1 double mutant (Liu et al., 2014) were described previously. The WOX11pro:GUS, WOX5pro:GUS, 35Spro:WOX11, 35Spro:WOX12, and 35Spro:WOX11-SRDX transgenic plants were described previously (He et al., 2012; Liu et al., 2014). The primers used for plasmid construction are listed in Supplemental Table S1.
Culture conditions were as described previously (Chen et al., 2014). Briefly, Arabidopsis seedlings were first cultured on 0.5× Murashige and Skoog medium at 22°C under a 16-h-light/8-h-dark photoperiod. Leaf explants from 12-d-old seedlings were cultured on B5 medium with Suc in the dark for de novo regeneration of adventitious roots.
RT-PCR, qRT-PCR, and ChIP Analyses
The extraction of RNAs and reverse transcription was conducted as described previously (He et al., 2012). The gene-specific primers used for PCR and real-time PCR are listed in Supplemental Table S1. The qRT-PCR results are shown as relative transcript levels, which were normalized against that produced using ACT-specific primers.
The ChIP experiment was carried out as described previously (He et al., 2012; Li et al., 2012). Briefly, 15-d-old seedlings of 35Spro:3×FLAG-WOX11-GR and the control Col-0 were treated with 10 μm dexamethasone for 4 h and leaves were harvested for chromatin extraction. Chromatin was also extracted from leaves of 15-d-old seedlings of 35Spro:3×FLAG-WOX12 and the control Col-0. The anti-FLAG antibody (Sigma-Aldrich, F1804) was used for immunoprecipitation. The ChIP results were normalized against the input control. The primers used for real-time PCR are listed in Supplemental Table S1.
Dual Luciferase Assay
To construct WOX5pro:LUC and WOX7pro:LUC, the promoter of WOX5 or WOX7 was inserted into the pGreenII-0800 vector (Hellens et al., 2005). The dual luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega).
Histology
GUS staining was performed as previously described (Chen et al., 2014; Zeng et al., 2016). Differential interference contrast observations were performed using a Nikon ECLIPSE 80i microscope (Nikon) as described previously (Chen et al., 2014).
Accession Numbers
Sequence data are listed in the Arabidopsis Genome Initiative under the following accession numbers: WOX11 (AT3G03660), WOX12 (AT5G17810), WOX5 (AT3G11260), and WOX7 (AT5G05770).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of wox5-1 wox7-1 double mutant.
Supplemental Figure S2. Expression patterns of WOX11 and WOX5 during adventitious rooting.
Supplemental Figure S3. WOX5/7 expression in adventitious root tip.
Supplemental Figure S4. WOX5/7 are involved in stem cell niche formation in the adventitious root rip.
Supplemental Figure S5. WOX11 up-regulates WOX7 expression.
Supplemental Figure S6. Function of WOX12 is dependent on WOX5/7.
Supplemental Figure S7. WOX7 expression is dependent on auxin.
Supplemental Figure S8. WOX11 expression in wox5-1 wox7-1.
Supplemental Figure S9. Primordium defect in 35Spro:WOX11-SRDX.
Supplemental Figure S10. LBD16 expression in de novo root organogenesis.
Supplemental Figure S11. WOX5/7 in primary root and lateral root development.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank H. Huang and Y. Du for discussion on this study, C. He for construction of pER8:WOX5, L. Sheng for construction of LBD16pro:LBD16-GUS and 35Spro:3×FLAG-WOX11-GR and preparation of the plant materials for ChIP analysis, and G. Zhang for assistance in ChIP experiment. We thank the ABRC for Arabidopsis seeds used in this work.
Glossary
- ChIP
chromatin immunoprecipitation
- DAC
days after culture
- NPA
naphthylphthalamic acid
- QC
quiescent center
- qRT-PCR
quantitative reverse transcription-PCR
- RAM
root apical meristem
References
- Ahkami AH, Lischewski S, Haensch KT, Porfirova S, Hofmann J, Rolletschek H, Melzer M, Franken P, Hause B, Druege U, et al. (2009) Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytol 181: 613–625 [DOI] [PubMed] [Google Scholar]
- Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666 [DOI] [PubMed] [Google Scholar]
- Berckmans B, Vassileva V, Schmid SP, Maes S, Parizot B, Naramoto S, Magyar Z, Alvim Kamei CL, Koncz C, Bögre L, et al. (2011) Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tong J, Xiao L, Ruan Y, Liu J, Zeng M, Huang H, Wang JW, Xu L (2016a) YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J Exp Bot 67: 4273–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cheng J, Chen L, Zhang G, Huang H, Zhang Y, Xu L (2016b) Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol 170: 2136–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qu Y, Sheng L, Liu J, Huang H, Xu L (2014) A simple method suitable to study de novo root organogenesis. Front Plant Sci 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Liang W, Li J, Hong F, Wu Y, Wang L, Wang J, Wu P, Liu C, Zhang Q, et al. (2013) A CLE-WOX signalling module regulates root meristem maintenance and vascular tissue development in rice. J Exp Bot 64: 5359–5369 [DOI] [PubMed] [Google Scholar]
- Correa Lda R, Troleis J, Mastroberti AA, Mariath JE, Fett-Neto AG (2012) Distinct modes of adventitious rooting in Arabidopsis thaliana. Plant Biol (Stuttg) 14: 100–109 [DOI] [PubMed] [Google Scholar]
- Cui H, Hao Y, Kovtun M, Stolc V, Deng XW, Sakakibara H, Kojima M (2011) Genome-wide direct target analysis reveals a role for SHORT-ROOT in root vascular patterning through cytokinin homeostasis. Plant Physiol 157: 1221–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Klerk G-J. (2002) Rooting of microcuttings: theory and practice. In Vitro Cell Dev Biol Plant 38: 415–422 [Google Scholar]
- Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16: 597–606 [DOI] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Sun X, Wang G, Liu H, Zhu J (2012a) LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann Bot (Lond) 110: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhu J, Du X, Cui X (2012b) Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 236: 1227–1237 [DOI] [PubMed] [Google Scholar]
- Ge Y, Liu J, Zeng M, He J, Qin P, Huang H, Xu L (2016) Identification of WOX family genes in Selaginella kraussiana for studies on stem cells and regeneration in lycophytes. Front Plant Sci 7: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893 [DOI] [PubMed] [Google Scholar]
- Goh T, Toyokura K, Wells DM, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett MJ, et al. (2016) Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 143: 3363–3371 [DOI] [PubMed] [Google Scholar]
- Greenwood MS, Cui X, Xu F (2001) Response to auxin changes during maturation-related loss of adventitious rooting competence in loblolly pine (Pinus taeda) stem cuttings. Physiol Plant 111: 373–380 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L (2012) Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Ji H, Wang S, Li K, Szakonyi D, Koncz C, Li X (2015) PRL1 modulates root stem cell niche activity and meristem size through WOX5 and PLTs in Arabidopsis. Plant J 81: 399–412 [DOI] [PubMed] [Google Scholar]
- Kong D, Hao Y, Cui H (2016) The WUSCHEL related homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana. Mol Plant 9: 261–270 [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim MJ, Kim NY, Lee SH, Kim J (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J 73: 212–224 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol 151: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Li G, Zhang J, Li J, Yang Z, Huang H, Xu L (2012) Imitation Switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant J 72: 261–270 [DOI] [PubMed] [Google Scholar]
- Lian G, Ding Z, Wang Q, Zhang D, Xu J (2014) Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. ScientificWorldJournal 2014: 534140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Maffei ME, Mithöfer A, Boland W (2007) Before gene expression: early events in plant-insect interaction. Trends Plant Sci 12: 310–316 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Bürglin TR (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26: 2775–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Reisewitz P, Werr W (2009) Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol Biol Evol 26: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W (2013) Symplesiomorphies in the WUSCHEL clade suggest that the last common ancestor of seed plants contained at least four independent stem cell niches. New Phytol 199: 1081–1092 [DOI] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Scheres B. (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8: 345–354 [DOI] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD (2009) Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B, Rasmussen A (2016) The physiology of adventitious roots. Plant Physiol 170: 603–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM (2011) Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol 21: 212–218 [DOI] [PubMed] [Google Scholar]
- Sussex IM. (2008) The scientific roots of modern plant biotechnology. Plant Cell 20: 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Wabnik K, Niu T, Li H, Yu Q, Pollmann S, Vanneste S, Govaerts W, Rolcík J, Geisler M, et al. (2014) WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol Plant 7: 277–289 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B (2006) A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Xu L, Huang H (2014) Genetic and epigenetic controls of plant regeneration. Curr Top Dev Biol 108: 1–33 [DOI] [PubMed] [Google Scholar]
- Zeng M, Hu B, Li J, Zhang G, Ruan Y, Huang H, Wang H, Xu L (2016) Stem cell lineage in body layer specialization and vascular patterning of rice root and leaf. Sci Bull 61: 847–858 [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX (2009) The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.