tRNA import into plant mitochondria requires the outer mitochondrial membrane PRAT domain-containing proteins Tric1 and Tric2 that are orthologous to the protein import translocases of the inner membrane.
Abstract
A variety of eukaryotes, in particular plants, do not contain the required number of tRNAs to support the translation of mitochondria-encoded genes and thus need to import tRNAs from the cytosol. This study identified two Arabidopsis (Arabidopsis thaliana) proteins, Tric1 and Tric2 (for tRNA import component), which on simultaneous inactivation by T-DNA insertion lines displayed a severely delayed and chlorotic growth phenotype and significantly reduced tRNA import capacity into isolated mitochondria. The predicted tRNA-binding domain of Tric1 and Tric2, a sterile-α-motif at the C-terminal end of the protein, was required to restore tRNA uptake ability in mitochondria of complemented plants. The purified predicted tRNA-binding domain binds the T-arm of the tRNA for alanine with conserved lysine residues required for binding. T-DNA inactivation of both Tric proteins further resulted in an increase in the in vitro rate of in organello protein synthesis, which was mediated by a reorganization of the nuclear transcriptome, in particular of genes encoding a variety of proteins required for mitochondrial gene expression at both the transcriptional and translational levels. The characterization of Tric1/2 provides mechanistic insight into the process of tRNA import into mitochondria and supports the theory that the tRNA import pathway resulted from the repurposing of a preexisting protein import apparatus.
Mitochondria are essential organelles found in almost all eukaryotic cells and are involved in multiple critical functions. The event that gave rise to mitochondria took place over 1 billion years ago, and the symbiogenesis scenario is favored, where an archaebacterium and a eubacterium formed a symbiosis, leading to the evolution of mitochondria (Martin and Müller, 1998). The loss or transfer of thousands of genes from the mitochondrial progenitor to the nucleus of the host cell resulted in mitochondria that have a small though essential protein-coding capacity (Adams and Palmer, 2003). The vast majority of the mitochondrial proteome is nucleus encoded, translated in the cytosol, and subsequently imported into the mitochondria. The mitochondrial protein import pathways have been studied intensively in plants, animals, and fungi to the extent that detailed structures of the import machinery, intramitochondrial sorting routes, and regulatory processes are well characterized (Dudek et al., 2013; Murcha et al., 2014a; Neupert, 2015).
Additional factors such as mRNA, tRNA, and rRNA are also are encoded in the mitochondrial genome (Gray, 2012), with many eukaryotes lacking complete sets of tRNA genes required for efficient mitochondrial translation (Schneider, 2011; Huot et al., 2014). Consequently, the import of cytosolic tRNA is required; interestingly, in species that do encode a complete set of tRNA, the import of cytosolic tRNA can still occur and is proposed to be an additional point of regulation of mitochondrial biogenesis (Tarassov et al., 1995; Kamenski et al., 2007). Current knowledge regarding the mechanisms of how tRNA is imported into mitochondria is limited. The uptake of tRNA was first shown in vivo in plant mitochondria in 1992 (Small et al., 1992). Subsequently, an in vitro tRNA uptake assay was established, which showed the uptake of cytosolic tRNA into isolated mitochondria (Sieber et al., 2011). Further studies have suggested that there is cooperation among components of the protein import translocase of the outer membrane (TOM) and Porin/voltage-dependent anion channel (VDAC), although specific proteins dedicated for the import of tRNA have yet to be identified (Salinas et al., 2006).
The mechanisms of tRNA import are highly variable between species (Salinas-Giegé et al., 2015). In yeast (Saccharomyces cerevisiae), the cytosolic tRNA for Lys is bound specifically by glycolytic enolase and transferred to the precursor of the mitochondrial lysyl-tRNA synthetase at the mitochondrial surface, after which it is cotransported through the protein import apparatus (Tarassov et al., 1995; Sepuri et al., 2012). In plants, the VDAC and components of the protein import apparatus such as Tom20 and Tom40 have been shown to be involved in tRNA import (Salinas et al., 2006). Furthermore, the addition of nucleic acid-binding proteins improves tRNA import rates in vitro, suggesting the involvement of as yet unidentified cytosolic factors. In Trypanosoma brucei, where almost all tRNAs are required to be imported, a role for the cytosolic elongation factor eEF1α has been shown, in addition to components of the protein import apparatus (Bouzaidi-Tiali et al., 2007; Tschopp et al., 2011). A common theme is that various components of the protein import apparatus or other proteins may have been coopted and serve two roles. Also, it is possible that these proteins evolved rapidly in a short time to facilitate tRNA import, and this may have occurred several times independently (Schneider, 2011; Salinas-Giegé et al., 2015).
The PRAT (for preprotein and amino acid transporters) family of proteins represents the largest protein import family, consisting of both mitochondrial and plastid components and characterized by the presence of four transmembrane regions and a conserved PRAT domain (Rassow et al., 1999). Of the 16 members in the Arabidopsis (Arabidopsis thaliana) PRAT family, 10 are located in the mitochondria, with eight genes encoding for inner membrane protein transporters, specifically the Translocase of the Inner Membrane17 (Tim17), Tim23, and Tim22 (Murcha et al., 2007). In chloroplasts, three PRAT proteins, HP20, HP30-1, and HP30-2, have been proposed to be involved in the import of proteins that do not contain a cleavable transit peptide (Rossig et al., 2013), while the outer envelope PRAT proteins OEP16.1 and OEP16.2 have been shown to be involved in amino acid transport (Philippar et al., 2007; Pudelski et al., 2009, 2010, 2012). In plants, in comparison with yeast and mammalian systems, the PRAT family appears to have undergone neofunctionalization in that, while all members are predicted to contain four transmembrane regions and a conserved/degenerate PRAT domain, additional domains also have been acquired (Murcha et al., 2007). Specifically, plant Tim17 proteins contain a C-terminal extension that protrudes into the outer membrane, increasing the efficiency of protein import for some precursor proteins (Murcha et al., 2005a; Pudelski et al., 2010).
Here, we show that two Arabidopsis PRAT members encoded by At3g49560 and At5g24650 (as outlined above, known as HP30-1 and HP30-2; Rossig et al., 2013), which we named Tric1 and Tric2 (for tRNA import component), respectively, are located in the mitochondrial outer membrane and have a role in mitochondrial tRNA import. Both proteins contain a sterile-α-motif (SAM) domain, known to bind RNA with high affinity in a variety of proteins from various organisms, and we show that this domain is required to mediate efficient tRNA uptake. Abolition of these proteins results in a significantly altered growth phenotype as well as modification in mitochondrial ultrastructure and substantial changes to the nucleus- and mitochondria-encoded transcriptomes.
RESULTS
Tric1 and Tric2 Are Predicted RNA-Binding Proteins
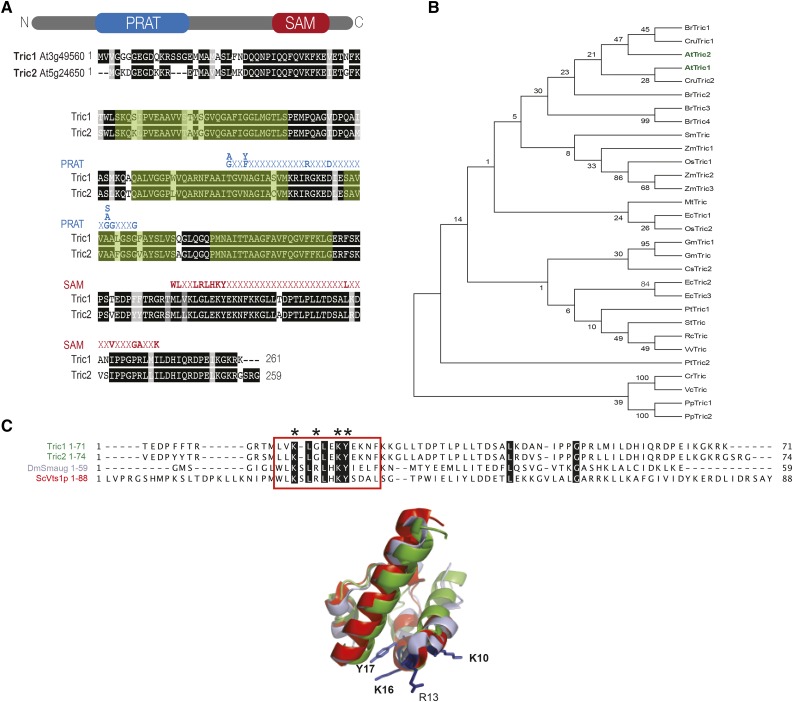
Sequence analysis of At3g49560 (Tric1) and At5g24650 (Tric2) indicates that, in addition to the predicted four transmembrane regions and the PRAT domain, a SAM domain also is present (Fig. 1A; Pudelski et al., 2010; Murcha et al., 2015). Both proteins display over 91% sequence identity, and phylogenetic analysis indicates that the occurrence of both domains is unique to plants (also present in the unicellular green alga Chlamydomonas reinhardtii) and is present in a wide variety of lineages (Fig. 1B; Supplemental Fig. S1). The SAM domain, which varies in size from 70 to 100 amino acids, was defined from a diverse range of proteins, mostly functionally related to signal transduction and transcriptional regulation (Kim and Bowie, 2003). Despite being initially identified as a protein-protein interaction module, the SAM domain present in the RNA-binding proteins Smaug from Drosophila melanogaster and Vts1 from S. cerevisiae has been shown to specifically bind RNA with high affinity (Aviv et al., 2003). Homology modeling suggests that the SAM domain from Tric1 and Tric2 can form the characteristic five-α-helix bundle structure with a striking conservation of basic (Lys) and hydrophobic (Leu and Tyr) residues involved in RNA binding by Smaug and Vts1 (Fig. 1C).
Figure 1.
Identification of novel plant preprotein and amino acid transporter proteins with a putative RNA-binding domain. A, Pairwise sequence alignment of Arabidopsis Tric1 (At3g49560) and Tric2 (At5g24650). Both proteins are predicted to contain a PRAT domain and a SAM domain, indicated in blue and red, respectively. The four predicted transmembrane domains are highlighted in green as determined by TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). B, Phylogenetic analysis of all Tric1/2 orthologs from 17 plant species and algae (red, green, and brown), with plant species chosen as representatives of each evolutionary clade from Physcomitrella patens (Pp) to Eucalyptus grandis (Eg). At, Arabidopsis; Br, Brassica rapa; Cre, Chlamydomonas reinhardtii; Cru, Capsella rubella; Cs, Cucumis sativus; Gm, Glycine max; Mt, Medicago truncatula; Os, Oryza sativa; Pt, Populus trichocarpa; Rc, Ricinus communis; Sm, Selaginella moellendorfii; St, Solanum tuberosum; Vc, Volvox carteri; Vv, Vitis vinifera; Zm, Zea mays. C, Comparison of the SAM domains of Tric1 and Tric2 from Arabidopsis with Vts1p (Uniprot Q08831) from Saccharomyces cerevisiae (Sc) and Smaug (Uniprot Q23972) from Drosophila melanogaster (Dm), which have been demonstrated to bind RNA. Smaug variants that display reduced RNA binding are indicated, according to Aviv et al. (2003). Amino acid residues (Y, L, and K) involved in RNA binding are conserved in both Tric1 and Tric2 proteins (indicated by asterisks). Superimposition of the three-dimensional model of Tric1 (green) with the structures of Smaug (light blue) and Vts1p (red) is shown at bottom, and the conserved residues are indicated (numbering corresponds to the SAM domain of Smaug). The structural prediction for Tric1 was generated using I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The image was generated using PyMOL (http://www.pymol.org/).
Tric1 and Tric2 Interact with Components of the TOM and TIM Protein Import Apparatus and Have Exposed Domains on the Mitochondrial Outer Membrane
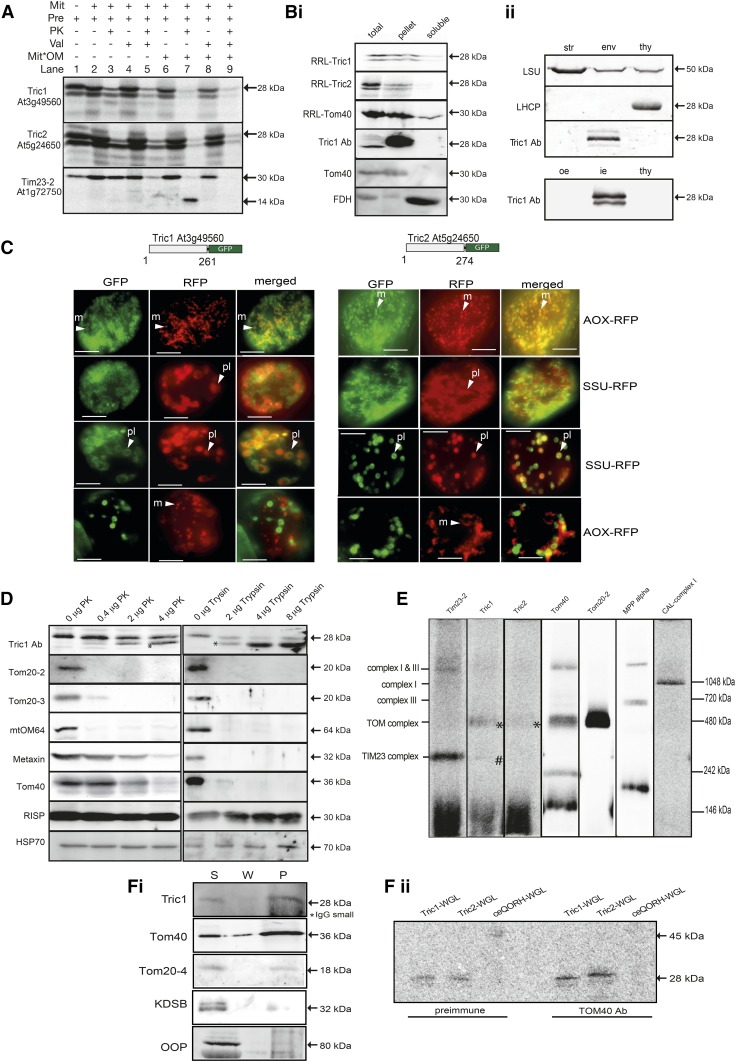
A previous study has suggested that Tric1 and Tric2 are dually located in mitochondria and chloroplasts (Murcha et al., 2007), whereas another study reported an exclusive chloroplastic localization for Tric1 and Tric2 (Rossig et al., 2013). To comprehensively address the localization of Tric1 and Tric2, a number of independent approaches were undertaken to investigate the targeting and accumulation of these proteins (Millar et al., 2009). In vitro protein uptake assays with radiolabeled precursor proteins revealed that both proteins bind to isolated mitochondria (Fig. 2A). In vitro translation of Tric1 and Tric2 results in proteins with an apparent molecular mass of 28 kD (and a secondary product of 26 kD resulting from translation at Met position 18/19; see Fig. 1A). In the in vitro import assay, the proteins bind to mitochondria and the subsequent addition of PK generates a protease-insensitive band (Fig. 2A, lanes 2 and 3). This import was not affected by the addition of valinomycin, which dissipates the inner membrane potential, suggesting that import occurs into the outer membrane but not into the inner membrane (Fig. 2A, lanes 4 and 5). Rupture of the outer mitochondrial membrane, following the import reaction, results in Tric1/2 being sensitive to protease treatment, irrespective of the presence of valinomycin (Fig. 2A, lanes 6–9). These results suggest either an outer membrane or intermembrane space location for these proteins. As a control, the mitochondrial inner membrane protein Tim23-2 was used (Murcha et al., 2003). Rupture of the outer membrane following import and prior to protease digestion resulted in a protease-insensitive band of 14 kD, representing the portion of Tim23-2 that is inserted into the inner membrane (Fig. 2A). This product was not observed when valinomycin was added to the import assay, confirming that the isolated mitochondria were intact and import competent. Taken together, these results suggest that Tric1 and Tric2 are not located within the inner membrane but, rather, are located in the outer membrane or intermembrane space. Carbonate extractions were carried out following the import of radiolabeled reticulocyte lysate (RRL) proteins, confirming that both RRL-Tric1 and RRL-Tric2 are incorporated as integral membrane proteins (Fig. 2Bi). Similarly imported into the membrane fraction is the integral outer membrane protein Tom40 (Fig. 2Bi). Immunodetection of carbonate-extracted mitochondria confirms endogenous Tric1/2 protein within the membrane pellet along with Tom40, while soluble FDH (Colas des Francs-Small et al., 1993) is located within the soluble fraction as expected (Fig. 2Bi). Investigation into the localization of Tric proteins in chloroplasts reveals that Tric proteins are located within the envelope protein subfraction, specifically within the inner envelope fraction (Fig. 2Bii).
Figure 2.
Tric1 and Tric2 are dual targeted proteins. A, In vitro uptake of Tric1 and Tric2 into mitochondria. In vitro translated and radiolabeled Tric1 and Tric2 proteins were incubated with isolated mitochondria under conditions that support the uptake of proteins. Lane 1, Precursor protein alone showing a product with an apparent molecular mass of 28 kD; a lower band with a molecular mass of 26 kD represents translation from a Met residue at position 18/19 (see Fig. 1A). Lane 2, Incubation of precursor protein with mitochondria under conditions that support import. Lane 3, as for lane 2 with proteinase K (PK) added to 0.4 µg mL−1 to digest all protein still exposed on the outer membrane. Note the increase in signal intensity of the 26-kD band compared with lane 2. Lane 4, as for lane 2 with the addition of valinomycin (Val) prior to the commencement of the import uptake assay. Lane 5, as for lane 4 with PK added following the import uptake assay. Lanes 6 and 7, as for lanes 2 and 3 except that, prior to the addition of PK, the outer mitochondrial membrane was ruptured (Mit*OM). Lanes 8 and 9, as for lanes 6 and 7 with the addition of valinomycin prior to the commencement of the import uptake assay. Tim23-2 was used to test mitochondrial import ability and successful rupture of the outer membrane, as evidenced by the generation of a 14-kD inner membrane-located, PK-protected band. Bi, Carbonate extractions following the import of RRL-Tric1, RRL-Tric2, and Tom40 into isolated mitochondria and immunodetection of carbonate-extracted mitochondria followed by immunodetection against endogenous Tric1, Tric2, Tom40, and formate dehydrogenase (FDH). Bii, Immunoblot analysis of Tric protein in chloroplast subfractions. Equal protein amounts of Arabidopsis chloroplast stroma (str), envelopes (env), and thylakoids (thy) were resolved by SDS-PAGE, blotted to nitrocellulose membranes, and probed with antibodies, as indicated. The bottom gel shows pea chloroplast outer envelope (oe), inner envelope (ie), and thylakoids (thy). Marker proteins are stromal LSU (large subunit of ribulose-1,5-bisphosphate carboxylase) and thylakoid LHCP (light-harvesting chlorophyll protein). C, In vivo targeting analysis of Tric1 and Tric2 using fluorescent protein tagging. The full-length coding sequences of Tric1 and Tric2 were fused in frame with GFP and cotransformed into Arabidopsis suspension cell cultures using biolistic transformation. For both Tric proteins, targeting to mitochondria and chloroplasts was observed, although observation of dual targeting within the same cell was rare. Close examination of the GFP fluorescence revealed that it forms halo- or doughnut-type patterns in both mitochondria and chloroplasts. Superimposition of the RFP pattern for mitochondrial targeting (alternative oxidase [AOX]) or chloroplast (SSU) revealed that it was on the periphery of the structures. Bars = 20 mm. m, Mitochondria; pl, plastid. D, Protease accessibility of Tric1 and Tric2 on the mitochondrial outer membrane by protease digestion (PK and trypsin) of Columbia-0 (Col-0) mitochondria followed by immunodetection. Purified mitochondria were subjected to protease treatment as indicated and resolved by SDS-PAGE, blotted to nitrocellulose membranes, and probed with antibodies as indicated. The Tric1 antibody detected at 28 kD produced a breakdown product (asterisk) at the lowest levels of protease treatment, increasing in intensity with increasing concentrations. As controls, the mitochondrial outer membrane proteins Tom20-2, Tom20-3, mtOM64, metaxin, Tom40, the inner membrane protein RISP, and matrix protein (HSP70) also were detected. E, In vitro import of radiolabeled Tim23-2, Tric1, Tric2, Tom40, Tom20-2, MPP-α, and complex I subunit CAL into isolated mitochondria followed by BN-PAGE analysis. Fi, Coimmunoprecipitation analysis of purified mitochondria with Tom40 antibody. Digitonin-ruptured mitochondria were incubated with Tom40 antibody and Protein A-Sepharose. Supernatant (S), wash (W), and pellet (P) fractions were resolved by SDS-PAGE, blotted to nitrocellulose membranes, and probed with antibodies as indicated. The asterisk indicates cross-reactivity toward the small subunit of IgG resolving at 25 kD. Fii, Coimmunoprecipitation analysis as in Fi, except that wheat germ lysate (WGL)-translated radiolabeled Tric1, Tric2, and ceQORH was added to the immunoprecipitation reaction containing either preimmune serum or Tom40 antibody. Pellet fractions were resolved by SDS-PAGE, dried, and exposed to a phosphor-imaging screen.
To further investigate the dual localization of Tric1 and Tric2, GFP tagging was carried out alongside known mitochondria- and chloroplast-targeted red fluorescent protein (RFP) controls. Both Tric1 and Tric2 have the ability to target a C-terminal fusion of GFP to both organelles, as determined by overlapping with the fluorescent marker protein (Fig. 2C). Close examination revealed that, for mitochondria, the GFP pattern formed a halo or doughnut shape, with the GFP encircling the RFP control (Fig. 2C). In the case of chloroplasts, the GFP pattern appeared as a crescent shape partially encircling the RFP chloroplast control (Fig. 2C). These localization patterns are consistent with what has been observed for other known mitochondrial outer membrane (Duncan et al., 2011) and chloroplast envelope (Breuers et al., 2012) proteins. Moreover, as observed in previous studies on dual targeted proteins (Beardslee et al., 2002; Xu et al., 2013), in individual cells targeting was frequently only detectable in one of the organelles.
To verify the mitochondrial outer membrane localization, protease sensitivity experiments using PK and trypsin were carried out. The addition of protease to purified mitochondria followed by immunodetection with antibodies raised against Tric1 led to the detection of Tric1/2 at a molecular mass of 28 kD (Fig. 2D), with Tric1 antibody confirmed to equally cross-react against Tric1 and Tric2 (Supplemental Fig. S2). Upon the addition of PK or trypsin, a lower molecular mass band was readily detected (Fig. 2D, asterisk). Immunodetection against known cytosol facing the mitochondrial outer membrane protein import receptors Tom20 and OM64 revealed almost complete digestion of these proteins at 0.4 μg of PK and 2 μg of trypsin. For Tric1, increasing the PK amount increased the abundance of the proteolytic breakdown product, revealing an overall protease sensitivity that was similar to the integral outer membrane proteins metaxin and Tom40 (Lister et al., 2007; Fig. 2D). In the case of trypsin, a 26-kD protein fragment was generated with the lowest levels of trypsin, which was stable, while almost complete digestion of Tom40 and metaxin was observed. Immunodetection against the inner membrane-located Rieske iron-sulfur protein (RISP) and the matrix-located heat shock protein70 (HSP70) confirmed that the inner membrane had remained intact throughout the digestion process with both PK and trypsin (Fig. 2D). Thus, in vitro and in vivo targeting studies, as well as western-blot analyses, reveal that Tric1 and Tric2 can target and accumulate in mitochondria and chloroplasts. Additionally, the proteomic identification of Tric2 in outer mitochondrial membrane fractions (Duncan et al., 2011) further supports a mitochondrial (dual) localization.
To identify if Tric1 and Tric2 associate within a larger protein complex in mitochondria, radiolabeled Tric1 and Tric2 were imported into isolated mitochondria and analyzed by blue native-PAGE (BN-PAGE) alongside Tim23-2 (Wang et al., 2012), Tom40, Tom20-2, and respiratory complex I (carbonic anhydrase-like [CAL1]) and complex III (mitochondrial processing peptidase [MPP]-α) subunits (Murcha et al., 2014b; Fig. 2E). Incorporation of Tric1 and Tric2 was evident in a protein complex of approximately 400 kD, similar in size to that of TOM complex subunits as identified by the import of radiolabeled Tom40 and Tom20-2 (Fig. 2E, asterisks). A smaller weak band also was evident for Tric1 (Fig. 2E, number sign), which is of a similar size to the TIM17:23 complex as identified by the import and assembly of radiolabeled Tim23-2. Radiolabeled signal also was detected in the dye front, as observed for Tim23-2, suggesting that a portion of Tric1/2 (and Tim23-2) has not assembled into a larger complex (Fig. 2E). This is likely due to the in vitro nature of this assembly assay.
To further test the interaction of Tric1/2 with mitochondrial import complexes in vivo, immunoprecipitation assays were carried out. Digitonin-treated mitochondria were incubated with Tom40 antibody and Protein A-Sepharose. The supernatant and wash and pellet samples were subsequently immunodetected with antibodies against Tric1 and the TOM subunits Tom40 and Tom20-4, showing that Tric1 protein could be pulled down by Tom40 antibodies along with the TOM complex subunits Tom40 and Tom20-4 (Fig. 2Fi). The outer membrane protein 3-deoxy-manno-octulosonate cytidylyltransferase (KDSB; Duncan et al., 2011) and the matrix-located organellar oligopeptidase (OOP; Kmiec et al., 2013) were not detected in the pellet fraction (Fig. 2Fi). An additional immunoprecipitation assay also was carried out this time with the addition of radiolabeled Tric1, Tric2, and the chloroplast envelope protein quinone oxidoreductase (ceQORH) translated in a wheat germ lysate. It was observed that Tom40 antibody had the ability to interact with Tric1 and Tric2 at a greater intensity than preimmune serum. A weak signal for ceQORH also was detected using the Tom40 antibody, although as the intensity of this signal is similar to that observed when using preimmune serum, this band most likely represents nonspecific binding to the Sepharose A beads.
To further test the putative interaction of Tric1/2 with mitochondrial protein import components, a yeast two-hybrid assay was undertaken. Full-length Tric1 and Tric2, along with the SAM domain of Tric1 (Tric1Δ1-150), were tested against a variety of known TOM complex components (Tom40, Tom20-2, Tom20-3, Tom20-4, Tom5, Tom6, and Tom7), TIM complex components (Tim17, Tim23, Tim50, Tim21, and Tim44), and outer membrane proteins (OM64, SAM, metaxin, and VDAC1; Supplemental Fig. S3). Positive interactions (as determined by growth on a quadruple dropout medium, with and without the presence of X-α-Gal) of the full-length Tric1 and Tric2 were observed with the cytosolic domain of Tom20-3, and Tric1 was seen to interact with Tom40 (Supplemental Fig. S3). Additionally, the small TOM proteins Tom5, Tom6, and Tom7, known to associate with the TOM complex and maintain its stability, were observed to interact with the SAM domain. The full-length Tric1 protein also exhibited a positive interaction with the TIM subunits Tim17-3 and Tim50. Known interacting protein pairs Tom40-Tom20-3 and SV40-p53 both exhibited growth on selection medium as expected. All diploid strains were viable on double dropout medium, confirming that yeast mating was successful (Supplemental Fig. S3). Therefore, Tric1 has the ability to interact with several subunits of the TOM complex, such as Tom40, the cytosolic domain of Tom20-3, and several of the small TOM proteins, with this interaction occurring via the SAM domain.
Tric1 and Tric2 Are Required for tRNA Uptake, But Not Protein Uptake, into Mitochondria
To determine the function of Tric1 and Tric2, two independent T-DNA insertion lines were obtained for each gene (Supplemental Fig. S4A). Double knockout lines (tric1tric2) were obtained by crossing (Supplemental Fig. S4B) and verified by RT-PCR and immunodetection, which confirmed that no transcript and protein could be detected in total protein and mitochondria/chloroplasts isolated from these lines (Supplemental Fig. S4C). Phenotypic analysis on both Murashige and Skoog (MS) medium and soil revealed that the single knockout lines exhibited only slightly delayed growth phenotypes, whereas the double knockout line tric1tric2 exhibited severe growth delays and a chlorotic phenotype (Supplemental Fig. S4D).
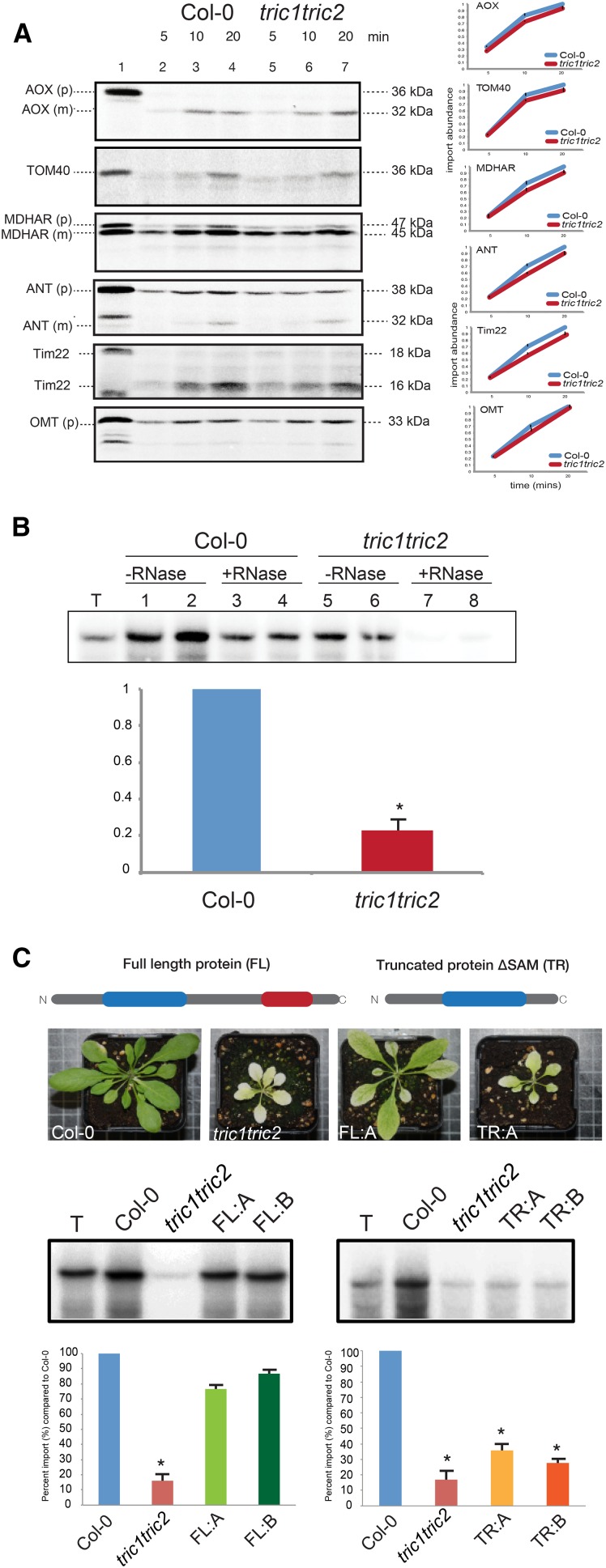
Due to the observed interactions of Tric1/2 with the TOM complex in mitochondria and a previously ascribed role in the import of proteins without transit peptides in chloroplast (Rossig et al., 2013), tric1tric2 plants were tested for protein import ability using a variety of mitochondrial precursor proteins (Fig. 3A). These included the alternative oxidase protein (AOX) targeted to the mitochondrial matrix via the general import pathway, the outer membrane protein Tom40, the dual targeted protein monodihydroascorbate reductase (MDHAR), and the inner membrane carrier protein adenine nucleotide translocator (ANT), which contains a cleavable presequence. Additionally, two mitochondrial carrier proteins that do not contain a cleavable presequence were tested, Tim22 and the OMT. Quantification reveals that all proteins were imported at rates comparable to Col-0 (Fig. 3B), suggesting that all known protein import pathways are not compromised in tric1tric2 plants.
Figure 3.
Tric proteins are involved in tRNA but not in protein import into isolated mitochondria. A, In vitro protein uptake assays into mitochondria isolated from wild-type (Col-0) and tric1tric2 plants. Lane 1, Precursor protein alone. Lanes 2 to 4, Incubation of precursor protein with isolated mitochondria for the time period indicated followed by treatment with PK (0.4 μg mL−1). Lanes 5 to 7, As for lanes 2 to 4 except mitochondria were isolated from tric1tric2 plants and used in the protein uptake assays. For precursor proteins without a cleavable presequence, Tim22 and oxaloacetate malate translocator (OMT) were used. Mitoplasts were prepared following import and prior to PK digestion to visualize the incorporation of proteins into the inner mitochondrial membrane. The apparent molecular mass of the precursor (p) and mature (m) proteins are indicated at right in kD. The graphs show the quantification of import from independent assays (n = 3, P < 0.05 by Student’s t test). B, 32P-labeled plant cytosolic tRNAAla was incubated with mitochondria isolated from Col-0 and tric1tric2 plants under conditions that support tRNA uptake into mitochondria. Lane T, The radiolabeled probe represents 1% of that added to the uptake assay. Lanes 1 and 2, Labeled probed that was precipitated after the uptake assay in mitochondria isolated from Arabidopsis wild-type (Col-0) plants. Lanes 3 and 4, As for lanes 1 and 2 except that mitochondria were treated with RNase after the uptake assay to remove all labeled probe outside mitochondria. Lanes 5 to 8, As for lanes 1 to 4 except that mitochondria were isolated from tric1tric2 mutants and used in the uptake assays. Quantification of the probe intensity from RNase-treated (import) tric1tric2 mitochondria relative to the wild type is graphed below (as a percentage relative to Col-0, where Col-0 is 100%). C, Complementation of the tric1tric2 mutant with full-length Tric1 and truncated Tric1ΔSAM. Top, Diagrammatic representation of the Tric proteins used in the complementation. Representative images of the full-length (FL) Tric1 and truncated (TR) Tric1ΔSAM used in the complemented lines are shown. Bottom, In vitro tRNA uptake assays into isolated mitochondria from various lines: Col-0 = wild type plants; tric1tric2 = double knockout T-DNA mutant; FL:A = complementation line 1 with full-length Tric1; FL:B = complementation line 2 with full-length Tric1; TR:A = complementation line 1 with truncated Tric1; and TR:B = complementation line 2 with Tric1. Gel images represent the tRNA probe alone and the RNase-treated samples from the various lines. Shown below is the quantification of probe intensity relative to Col-0 (shown as a percentage). Quantification of three independent assays (n = 3, P < 0.05) is shown. Asterisks indicate significant differences (Student’s t test).
As no defects were detected with protein uptake, and prompted by the presence of a putative RNA-binding domain in Tric1 and Tric2, in vitro tRNA import assays were carried out using mitochondria isolated from Col-0 and the double knockout line tric1tric2 (Fig. 3B). This assay involves the import of radiolabeled tRNA for Ala (tRNAAla), a well-characterized nucleus-encoded tRNA shown to be imported in vivo into plant mitochondria (Small et al., 1992). tRNA import assays are carried out with the addition of a carrier protein, Su9-DHFR (the presequence of subunit 9 of mitochondrial ATP synthase from Neurospora crassa linked to dihydrofolate reductase), that has been shown to allow for the efficient uptake of tRNAAla in isolated mitochondria (Sieber et al., 2011). It was observed that the uptake of radiolabeled tRNAAla into mitochondria isolated from tric1tric2 was reduced significantly by approximately 80% compared with mitochondria from wild-type (Col-0) plants (Fig. 3B, lanes 7 and 8). Notably, protein uptake into mitochondria of the carrier protein Su9-DHFR remained unchanged in mitochondria isolated from tric1tric2 compared with Col-0 (Supplemental Fig. S5A). tRNA import also was tested in mitochondria isolated from the single lines tric1 and tric2, exhibiting a decrease in import of approximately 30% (Supplemental Fig. S5B).
To determine if the SAM domain was a requirement for tRNA import, tric1tric2 was complemented with the full-length Tric1 or a truncated Tric1 (amino acids 1–150) without the SAM domain under the control of a strong constitutive promoter (35S CaMV). A comparable partial restoration of the growth phenotype could be observed with all independent full-length Tric1 complemented lines but not in the lines harboring the truncated proteins (approximately 10 T1 lines each), suggesting that the SAM domain is required for normal growth (Fig. 3C). Subsequent import of tRNA into mitochondria isolated from these complemented lines was largely restored in lines expressing the full-length Tric1 lines (green bars; Fig. 3C) but not in complementation lines expressing the truncated protein (Fig. 3C, orange/yellow bars), confirming that the SAM domain plays a role in tRNA uptake.
To test whether Tric1 and Tric2 are able to form homooligomers or heterooligomers, as seen for other members of the PRAT superfamily (Pudelski et al., 2010; Murcha et al., 2014a), a yeast (split-ubiquitin) interaction assay was carried out. Here, it was observed that the bait protein Tric1 was able to interact with itself and with Tric2 (Supplemental Fig. S5C). However, without the SAM domain, the interaction did not occur (Supplemental Fig. S5C), indicating that, besides mediating tRNA import, the SAM domain mediates for the homooligomerization and heterooligomerization of the Tric proteins.
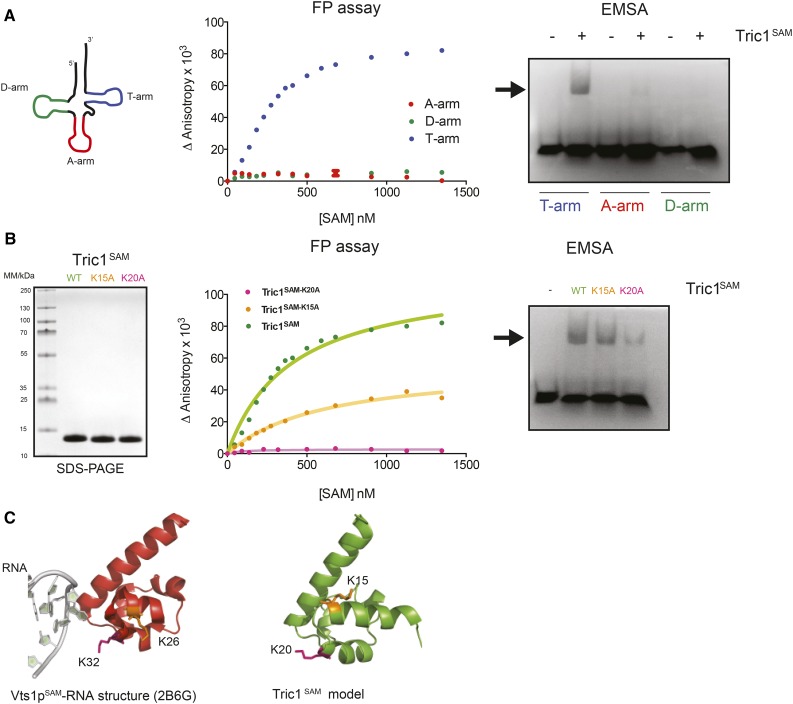
Given that our previous experiments suggested that the SAM domain plays a role in tRNA import, the ability of the SAM domain to bind directly to tRNA was tested. Ribooligonucleotides representing the A-, D-, and T-arms of tRNAAla were synthesized and tested for binding with the expressed and purified Tric1 SAM domain (Tric1SAM; Fig. 4, A and B). Specifically, both a fluorescence polarization assay and an EMSA were employed, and the results indicated that only the T-arm of the tRNA has the ability to bind to Tric1SAM (Fig. 4A). Variants of Tric1SAM also were generated, in which the positive Lys residues were substituted by Ala (marked with asterisks in Fig. 1), and these also were tested for binding (Fig. 4B). Both variants were seen to have reduced binding affinity to the T-arm ribooligonucleotide, with the variant K20A showing almost no binding (Fig. 4B). This analysis, combined with sequence alignments and homology modeling of Tric1SAM (Fig. 4C), suggests that the RNA-binding surface is conserved between Vts1SAM and Tric1SAM, whereby the substitution of the conserved Lys residue (Lys-32 in Vts1SAM and Lys-20 in Tric1SAM) abolished binding to the RNA-binding surface in both cases (Aviv et al., 2003). Thus, the two proteins were named Tric1 and Tric2.
Figure 4.
The SAM domain of Tric1 binds RNA. A, Structure of the A-, D-, and T-arms of the tRNA for Ala (left). Binding analysis of Tric1SAM and RNA oligonucleotides corresponding to the three arms of tRNAAla (15-base A-arm, 16-base D-arm, and 17-base T-arm) was carried out. Fluorescence polarization (FP) assay (middle) and output from electrophoretic mobility shift assay (EMSA; right) also are shown. The arrow indicates the position of the shifted band. B, Binding analysis of Tric1SAM (wild type [WT], K15A, and K20A) to the T-arm RNA oligonucleotide. The SDS-PAGE gel, Coomassie Blue stained, shows the purity of Tric1SAM variants (left). The FP assay (middle) and output from EMSA (right) also are shown. The arrow indicates the position of the shifted band. C, Three-dimensional digital representations of the crystal structure of Vts1p in complex with RNA (left) and a model for Tric1SAM structure (right) highlighting the key Lys residues involved in RNA binding. These images were generated using PyMOL (http://www.pymol.org/).
tric1tric2 Plants Have Altered Mitochondrial Protein Content and Morphology
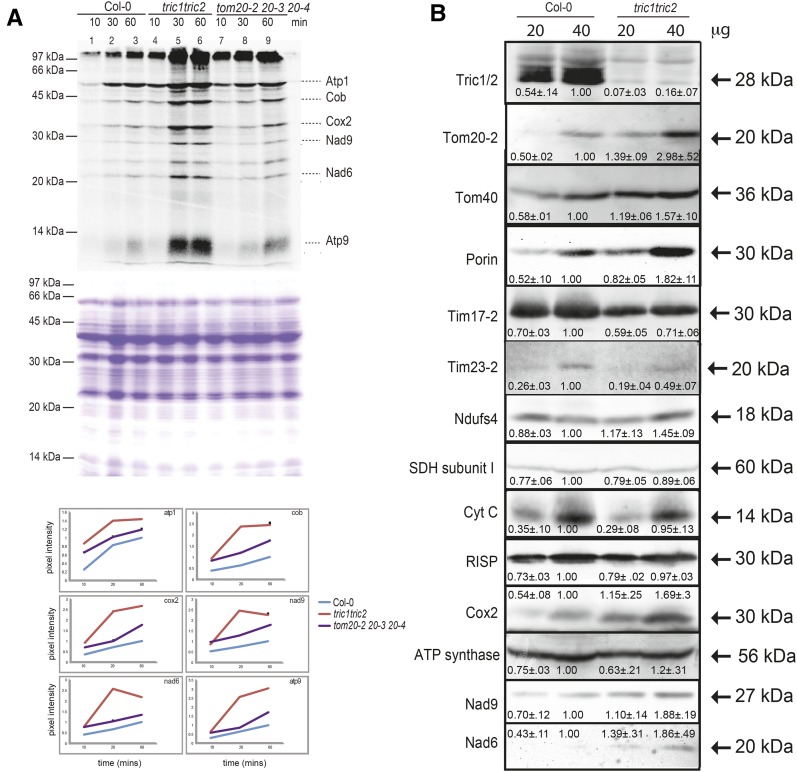
Given this role in tRNA import into mitochondria, tric1tric2 plants were investigated with regard to mitochondrial biogenesis. Paradoxically, in organello protein synthesis analyses revealed that there was a higher rate of mitochondrial protein synthesis in tric1tric2 mitochondria compared with the mitochondria isolated from Col-0 and tom20-2 tom20-3 tom20-4 plants, which were shown previously to import proteins at only 20% efficiency (Lister et al., 2007; Fig. 5A). Analyses of the mitochondrial protein abundance of various protein import and respiratory chain components revealed that the loss of Tric proteins resulted in an increase in the abundance of several mitochondrial proteins, including Tom20-2 and Porin/VDAC by nearly 2-fold and, to a lesser extent, Tom40 (Fig. 5B). In contrast, the inner membrane protein import components Tim17-2 and Tim23-2 decreased in abundance by nearly 50% (Fig. 5B). While several other nucleus-encoded subunits of the mitochondrial respiratory chain, NdufS4 complex I (Meyer et al., 2009), succinate dehydrogenase subunit II (Peters et al., 2012), cytochrome c, and RISP complex III (Duncan et al., 2011), did not change in abundance, the mitochondria-encoded proteins Cox2, Nad6, and Nad9 all increased in abundance from tric1tric2 mitochondria compared with the wild type (Fig. 5B). This increase in the abundance of mitochondria-encoded proteins is consistent with the increase in the rates of in organello protein synthesis outlined above. This suggests that, to compensate for a decrease in tRNA import ability, components associated with the expression of mitochondria-encoded genes are up-regulated in abundance. Furthermore, components identified previously to be involved with tRNA import, such as Tom20 and Porin/VDAC (Salinas et al., 2006), also were observed to increase in abundance.
Figure 5.
Deleting Tric1 and Tric2 affects mitochondrial translation and protein abundance. A, In organello translation in mitochondria isolated from wild-type (Col-0), tric1tric2, and tom20-2tom20-3tom20-4 plants. The positions of several mitochondria-encoded proteins are indicated with the apparent molecular mass (kD) and quantification (top). Coomassie Blue staining is shown in the middle to confirm equal loading. At bottom is a quantification of band intensity as identified previously by apparent molecular mass values (Giegé et al., 2005): atp1, α-subunit of ATP synthase; cob, cytochrome b; cox2, subunit 2 of cytochrome oxidase; nad9, subunit 9 of complex I (NADH ubiquinone oxidoreductase); nad6, subunit 6 of complex I; atp9, subunit 9 of ATP synthase. B, Immunodetection of mitochondrial proteins associated with protein import and respiration.
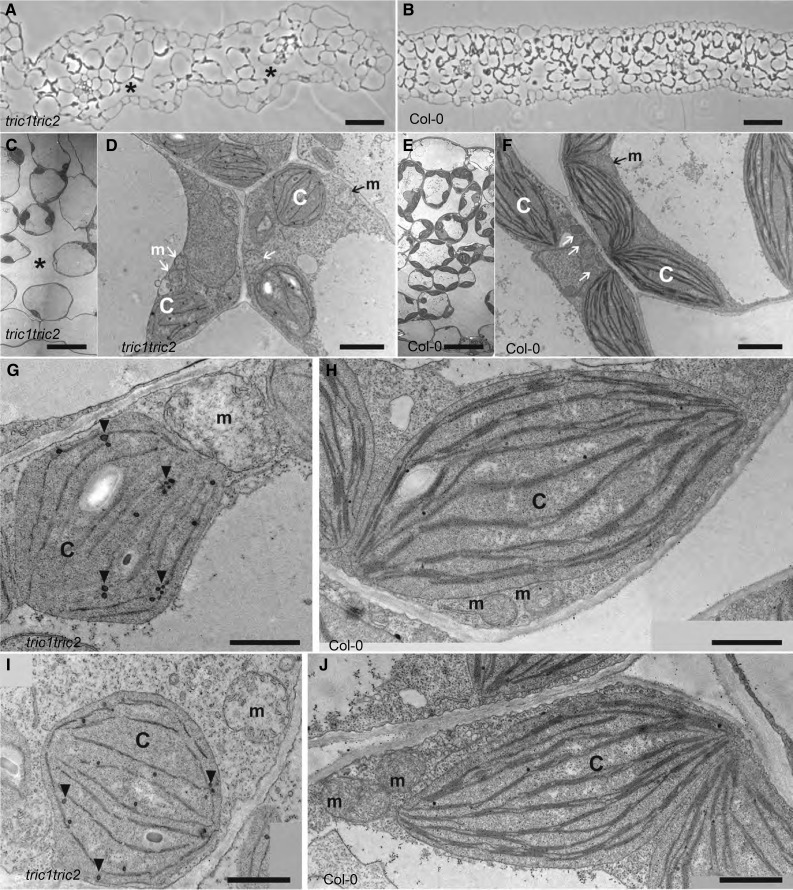
As well as the molecular changes at the protein level, examination with transmission electron microscopy identified changes to both the cellular and organelle morphology of tric1tric2 plants compared with wild-type plants (Fig. 6). When comparing the leaf structure, we found large intracellular spaces and disordered palisade and spongy parenchyma cells (Fig. 6, A and B). Furthermore, in line with the dual localization of Tric proteins, altered chloroplastic and mitochondrial morphologies also were observed (Fig. 6, C–J; Supplemental Table S1). Mitochondria in the tric1tric2 plants appeared larger compared with Col-0 (Fig. 6, G and I), with 46.7% mitochondria in tric1tric2 of at least double the common size and with a spheroidal shape (442 mitochondria analyzed from 161 cells; Supplemental Table S1). No similar abnormalities were observed in Col-0 (512 mitochondria analyzed from 279 cells). tric1tric2 mitochondria also were characterized by a decrease in visible cristae invaginations (Fig. 6, G–J). The majority of mitochondria from tric1tric2 had cristae observable only at the periphery (45 of 64) and 11% of mitochondria had no observable cristae at all, while in Col-0, all mitochondria showed well-developed cristae crossing the entire organelle (Supplemental Table S1). The profound morphological changes, such as fewer cristae junctions and an enlarged swollen mitochondrion, had been observed previously in a variety of mutants in various species, commonly in mutants of transcription and translation components that display altered mitochondrial translation rates (Arbustini et al., 1998; Zick et al., 2009; Guitart et al., 2013). Vice versa, mutants to components involved in maintaining mitochondrial morphology, such as OpaI (an inner membrane protein involved in mitochondrial cristae formation), which are characterized by disorganized mitochondria with rounded cristae, interestingly exhibit increased mitochondrial translation levels (Cogliati et al., 2013). The tric1tric2 mutant line does exhibit significant changes in the expression of some genes involved in maintaining mitochondrial morphology (Supplemental Tables S2 and S3), such as PMD2 (At1g06530), a peroxisomal and mitochondrial division factor that plays a role in mitochondrial morphology (Aung and Hu, 2011), and MIRO1 (At5g27540), involved in morphology and dynamics (Yamaoka and Leaver, 2008).
Figure 6.
The loss of function of Tric proteins alters cellular morphology. A and B, Semithin cross sections of rosette leaves from 17-d-old tric1tric2 plants (A) and Col-0 wild-type plants (B). Asterisks indicate large intracellular spaces in tric1tric2. Bars = 50 µm. C to J, Transmission electron micrographs of mature rosette leaves from 44-d-old tric1tric2 plants (C, D, G, and I) and 22-d-old Col-0 wild-type plants (E, F, H, and J). C to F, Cross sections of leaves. The asterisk indicates a large intracellular space in tric1tric2. c, Chloroplast; m, mitochondria, indicated by arrows. Bars = 50 µm (C and E) and 2 µm (D and F). G to J, Closeup views of chloroplasts and mitochondria. Note that in tric1tric2, plastid thylakoid membranes are reduced while plastoglobuli (arrowheads) accumulate. The majority of the mitochondria from tric1tric2 are larger and spheroidal with fewer cristae and disrupted invaginations. Bars = 1 µm.
Transcriptome Changes in tric1tric2 Plants Reveal Direct and Indirect Perturbation of Mitochondrial Function
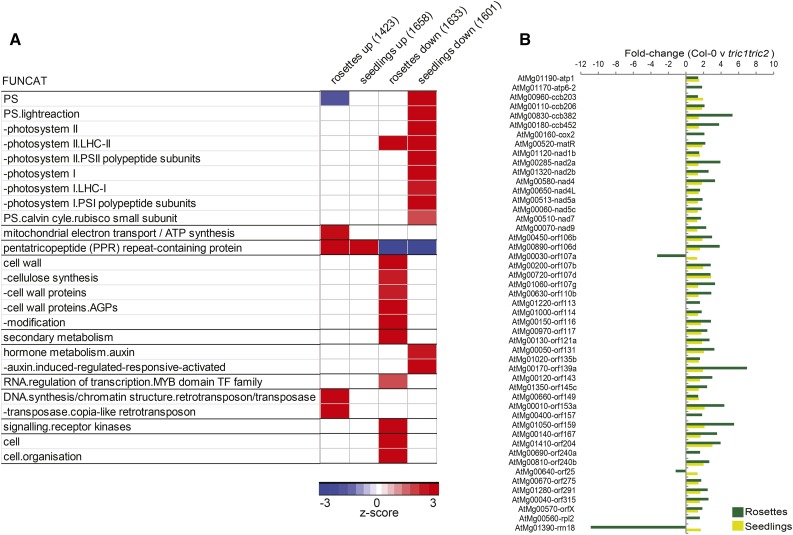
To further investigate the increase in the abundance of mitochondria-encoded proteins and morphology, whole-transcriptome comparisons also were carried out using RNA sequencing (RNA-seq) from Col-0 and tric1tric2 plants. Analyses of the changes in the transcriptome revealed that 3,072 and 3,267 genes were significantly differentially expressed (P < 0.05, q < 0.05, false discovery rate [FDR] < 5%) in the rosettes and seedlings, respectively, of tric1tric2 plants compared with Col-0. These included both mitochondria- and plastid-encoded genes (Supplemental Table S2). An analysis of the functional classifications of these differentially expressed genes (PageMan analysis using Fisher’s test, P < 0.05; Usadel et al., 2006) revealed that the down-regulated genes of tric1tric2 were significantly enriched in photosynthesis-related functions (Fig. 7A). Closer analyses revealed that these genes encode proteins from both PSI and PSII in chloroplasts as well as proteins associated with the Calvin cycle (Supplemental Fig. S6). Very similar changes in transcript abundance were observed previously when mitochondrial function was altered, either by chemical means with rotenone, antimycin A, or monofluoroacetate (Garmier et al., 2008; Umbach et al., 2012) or in T-DNA mutants that affect complex I (Meyer et al., 2009) or a combination of complex I and IV (Kühn et al., 2011, 2015).
Figure 7.
Transcriptome analyses of tric1tric2 plants. A, All differentially expressed genes in the rosettes and seedlings (Col-0 versus tric1tric2) were analyzed for overrepresented functional categories (FUNCAT) using the PageMan tool (over-representation analysis: Fisher’s test with Benjamini-Hochberg FDR correction). The number of genes in each set is indicated in parentheses. Only significantly overrepresented functional categories (P < 0.05) in one or more gene sets are shown (z score of 1.96 represents an FDR-corrected P value of 0.05). B, Bar graph showing the 49 significantly differentially expressed (FDR < 0.05) mitochondria-encoded genes in rosettes and/or seedlings of tric1tric2 mutants compared with Col-0.
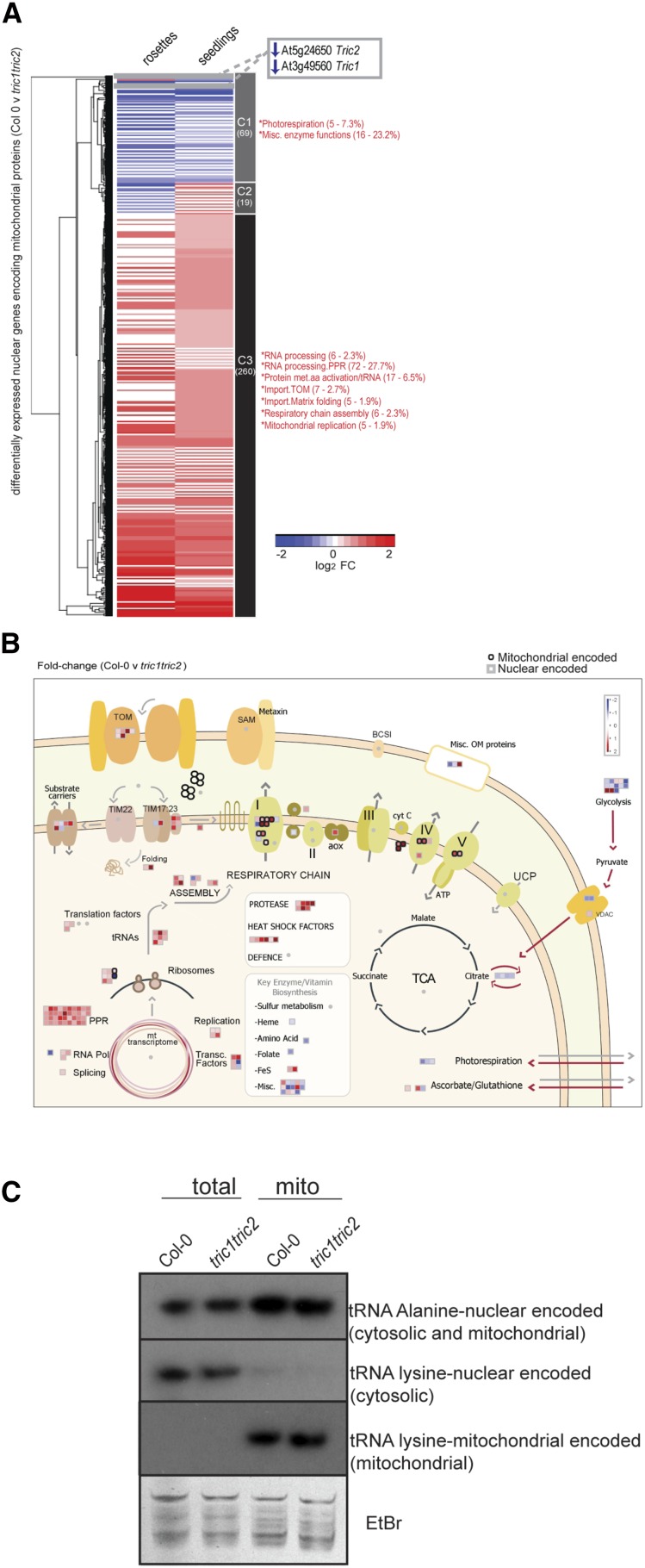
An up-regulation of transcript abundance in tric1tric2 plants was observed for genes encoding components of the mitochondrial electron transport chain/ATP synthase and dominated by mitochondria-encoded components of the respiratory chain. Of all functional categories, the most significant enrichment observed was for the genes encoding pentratricopeptide repeat (PPR) proteins, which are known to have roles in RNA processing and were highly overrepresented in the up-regulated transcripts of tric1tric2 plants (Fig. 7A). The association of PPR proteins with mitochondrial genome editing, splicing, and modification is documented extensively (Barkan and Small, 2014). Notably, of the 92 differentially expressed transcripts encoding PPR proteins in tric1tric2 seedlings, 51 are predicted to be mitochondrial and 19 are predicted to be plastidial. Similarly, of the 54 differentially expressed transcripts encoding PPR proteins in the rosettes, 34 are predicted to be mitochondrial while 15 are predicted to be plastidial, using the consensus algorithm for protein localization SUBAcon (Hooper et al., 2014). Of the mitochondria-encoded genes, 49 genes were observed to have significantly higher (P < 0.05) transcript abundance in tric1tric2 plants compared with Col-0 (Fig. 7B; Supplemental Fig. S7). This included Cox2 and Nad9 (Fig. 7B), which is consistent with the respective changes observed with the in organello synthesis and protein levels (Fig. 5). Investigation into the steady-state levels of tRNAAla by northern-blot analysis revealed little change in the steady-state abundance between Col-0 and tric1tric2 using whole tissue and isolated mitochondria (Fig. 8C). Likewise, there was no difference in the abundance of the mitochondria-encoded tRNALys or the nucleus-encoded cytosol-located tRNALys (cytosolic). This is consistent with both the in organellar translation assays (Fig. 5A) and the extensive transcriptome changes (Figs. 7 and 8), where the up-regulation in abundance of a variety of factors associated with mitochondrial gene expression compensates for the slower import rate of mitochondrial tRNAAla in tric1tric2 plants.
Figure 8.
In-depth analyses of the mitochondrial transcriptome (nucleus and mitochondria encoded) in tric1tric2 plants. A, All significantly differentially expressed (FDR < 0.05) nuclear genes encoding mitochondria-localized proteins in tric1tric2 compared with Col-0 in rosettes and seedlings grown under normal conditions. Three clusters were identified, and the number of genes in each cluster is shown below the cluster number (C1–C3). Genes in each cluster were analyzed for significantly overrepresented functional categories by z score analysis, and significantly enriched categories (P < 0.05) are indicated by asterisks, with the number of genes and the percentage in each cluster also shown. FC, Fold change. B, MapMan visualization of the changes in transcript abundance encoding mitochondrial proteins. The nucleus-encoded genes are represented by squares, while the mitochondria-encoded genes are represented by circles. The fold changes and functional classification of the proteins encoded by the genes are shown in Supplemental Table S2. TCA, Tricarboxylic acid cycle. C, Northern-blot analysis of total plant and isolated mitochondrial RNA from Col-0 and tric1tric2 using probes specific for nucleus-encoded tRNAAla, nucleus-encoded and cytosolic tRNALys, and mitochondria-encoded tRNALys. Ethidium bromide (EtBr) staining is shown to confirm equal RNA extraction between samples.
In-depth analysis of nuclear genes encoding mitochondrial proteins revealed 348 significantly (P < 0.05, q < 0.05, FDR < 5%) differentially expressed genes in the rosettes and/or seedlings of tric1tric2 plants compared with the wild type. Notably, nearly 75% of these showed significantly increased transcript abundance in tric1tric2 plants compared with the wild type (cluster 3; Fig. 8A). Close examination of the differentially expressed genes encoding mitochondrial proteins (Fig. 8B) revealed that five proteins associated with DNA replication were up-regulated, three encoding DNA gyrases, which suggests that unwinding of the DNA for transcription may be increased in tric1tric2 plants. Furthermore, almost 100 genes annotated as encoding proteins involved in mitochondrial transcription were increased significantly in transcript abundance in tric1tric2 plants; however, At2g34620, which is annotated as being involved in the termination of transcription, was down-regulated (Supplemental Table S3). Genes encoding a DNA polymerase (At5g15700) and Whirly2 (At1g71260) also were increased in abundance in tric1tric2 plants, with the latter shown previously to increase mitochondrial DNA copy number when expressed (Cai et al., 2015). Ninety-two genes encoding PPR proteins were significantly differentially expressed in tric1tric2 seedlings, with almost all of these being up-regulated (Fig. 8B; Supplemental Table S2). This is particularly notable given the well-documented association of these proteins with the editing of mitochondrial transcripts (Barkan and Small, 2014). At the translation level, 17 genes encoding proteins associated with amino activation of tRNA, four genes encoding proteins involved in elongation, and 18 genes encoding ribosomal proteins all were up-regulated in abundance in tric1tric2 plants compared with Col-0 (Fig. 8B). Taken together, these findings indicate that the transcripts of machinery involved in mitochondrial transcription, processing, and translation are up-regulated in tric1tric2 plants.
Finally, in terms of mitochondrial dysfunction, an increase in the transcript abundance of NAC13 transcription factor and OM66 has been shown to be associated with mitochondrial stress or retrograde signaling (Van Aken et al., 2009; De Clercq et al., 2013; Zhang et al., 2014), with NAC13 defined as one of the common markers of mitochondrial dysfunction (Schwarzländer et al., 2012). The AOX1c gene was up-regulated in tric1tric2 plants, but not the AOX1a gene, which is normally highly responsive to mitochondrial dysfunction (Clifton et al., 2006). Notably, this was similar to a mutant for metaxin, an outer mitochondrial membrane component of the SAM, in which AOX1c also was observed to increase (Lister et al., 2007).
Thus, altering mitochondrial biogenesis triggers transcriptome responses that are in part typical of what has been observed previously in terms of the impact on chloroplast function. In addition, there are additional changes in the transcriptome compared with that observed when mitochondrial electron transport is restricted (i.e. the expression of mitochondria-encoded genes and proteins associated with their synthesis that changed in transcript abundance; Fig. 8B). These responses at the protein (Fig. 5) and transcript (Fig. 7) levels are unusual and have not been observed previously with mutations in other genes affecting mitochondrial function.
DISCUSSION
Here, we have identified and characterized a component required for the import of tRNA into plant mitochondria. Tric1 and Tric2 are plant-specific members of the PRAT family of proteins, characterized by the presence of both PRAT and SAM domains. Tric1 and Tric2 are both dual targeted to mitochondria and chloroplasts (Murcha et al., 2007). While mitochondrial PRAT proteins are normally located in the inner membrane in mitochondria, multiple lines of evidence suggest a mitochondrial outer membrane localization for Tric1/2. First, Tric2 was identified in the mitochondrial outer membrane proteome (Duncan et al., 2011), and the results from both in vitro import and GFP localization patterns of Tric1/2 indicate an outer membrane localization (Fig. 2, A–C). Protease sensitivity experiments using PK and trypsin show that a portion of Tric1/2 is sensitive to externally added protease (Fig. 2D). Lastly, BN-PAGE analysis, immunoprecipitation assays, and yeast two-hybrid interaction assays suggest that Tric1/2 has the ability to interact with several mitochondrial outer membrane components (Fig. 2, E and F; Supplemental Fig. S3). However, the possible localization of Tric1/2 in the inner mitochondrial membrane or the ability to interact with subunits of the inner membrane cannot be ruled out, due to the fact that complete protease digestion was not observed and interactions with subunits of the TIM17:23 complex also were identified (Fig. 2D; Supplemental Fig. S3).
A large proportion of the PRAT proteins are involved in the process of protein import into mitochondria, including Tim22, Tim17, and Tim23. However, that is not the case for the PRAT homologs Tric1 and Tric2. Here, we show that deletion of Tric1 and Tric2 has no effect on the mitochondrial protein import ability by examining all known protein import pathways into plant mitochondria, including those for carrier proteins with and without cleavable targeting signals (Fig. 3A). Examination of the kinetics of protein import for the outer membrane protein (Tom40), inner membrane proteins typically imported via the carrier import pathway (ANT, Tim22, and OMT), the matrix-located protein AOX (on the matrix side of the inner membrane), and a dual targeted mitochondrial and chloroplastic protein (MDHAR) reveals no defects in the import ability and rates of these proteins (Fig. 3). It was evident that the Tric protein may interact with the components of the protein import machinery, notably Tom20 and Tom40 in the outer membrane. Previously, it was shown that antibodies to TOM components can inhibit the import of tRNA into mitochondria (Salinas et al., 2006). This suggests that, just like for protein import, where the receptor component binds the substrate, the transfer of the macromolecule (i.e. protein or tRNA) from the outside to the inside of mitochondria is achieved by a larger protein complex. A preexisting TOM complex may have facilitated the import of tRNA, with the receptor component providing specificity to this process by binding tRNA molecules.
In chloroplasts, PRAT protein family members are found in both membranes as well, OEP16 in the outer envelope (Pudelski et al., 2012) and Tric1/2 in the inner envelope membrane (Rossig et al., 2013; this study), further supporting a general PRAT protein function in both membrane systems of mitochondria and chloroplasts. A previous study has shown that, in chloroplasts, Tric1/2 is involved in the import of chloroplastic proteins that do not contain a cleavable transit peptide (Rossig et al., 2013). We have shown that the in vitro uptake of proteins does not differ between chloroplasts isolated from wild-type (Col-0) and tric1tric2 plants (Supplemental Fig. S8). In particular, the import of ceQORH, the protein defined previously to require Tric1/2 for uptake into chloroplasts (Rossig et al., 2013), displayed no difference in in vitro uptake. Our results suggest that Tric1 and Tric2 are not involved in the process of protein import in mitochondria or in chloroplast but, instead, are involved in the uptake of tRNA into mitochondria. Attempts to demonstrate the in vitro uptake of tRNA into isolated chloroplasts were not successful, although this may be a technical artifact of the assay used. Furthermore, in Arabidopsis, the chloroplast genome encodes for a full complement of tRNA molecules, suggesting that Tric1/2 is not involved in tRNA import in chloroplasts.
Inactivation of both isoforms of the Tric protein results in a severely chlorotic and developmentally delayed phenotype. While such a phenotype might suggest a chloroplast dysfunction, that may well be a secondary effect caused by mitochondrial perturbation. In fact, as outlined in “Results,” a number of specific studies reported that perturbation of mitochondrial function results in a transcriptomic response of genes encoding proteins involved in photosynthesis. A meta-analyses examining the common responses to perturbation of mitochondrial function under 11 different conditions (chemical and genetic) showed that alterations in the transcript abundances of genes encoding proteins involved in the light reactions of photosynthesis was a common category that was overrepresented (Schwarzländer et al., 2012). Additionally, the observation that nonchromosomal stripe mutants of maize (Zea mays), which are mutations in mitochondria-encoded subunits of cytochrome oxidase (Karpova et al., 2002) that were specifically shown to have a PSI biochemical deficiency (Jiao et al., 2005), provides an additional link that mitochondrial dysfunction directly impacts chloroplastic photosynthetic capacity. Thus, while the function of Tric1 and Tric2 in chloroplasts is not clear (see above), the chlorotic phenotype can be explained by altered mitochondrial function, specifically in the import of tRNAs. At the molecular level, an up-regulation of mitochondrial biogenesis is observed, with increased in organello translation rates accompanied by an increase in the abundance of several mitochondria-encoded proteins. While slowing the rate of import of tRNA may be expected to slow the rate of in organello protein synthesis, there are several observations that may account for this apparent discrepancy. It has been reported that both Porin/VDAC also may be involved in the import of tRNA into plant mitochondria (Salinas et al., 2006), and we observed an increase in abundance of these proteins in the tric1tric2 double mutant, possibly providing an alternative pathway for the import of tRNA. Removal of Tric1 and Tric2 results in a transcriptomic up-regulation of the machinery involved in the expression of mitochondrial genes that, in effect, may overcompensate for the defect in tRNA import. The analysis of the transcriptome encoding mitochondrial proteins reveals that the machinery associated with the expression of genes located in mitochondria is up-regulated at every level, from accessibility of DNA, transcription and processing, to translation. This is a unique response not observed previously in mutants with defect(s) in protein import into mitochondria or in a variety of mutants that encode mitochondrial proteins involved in metabolism or the respiratory chain (Lister et al., 2007; Murcha et al., 2014a). In fact, the only time a similar effect has been observed, for both the nuclear and organelle transcriptome, is when Tim23-2 (another mitochondrial PRAT protein) was overexpressed at the protein level (Wang et al., 2012). Here, an up-regulation of the components involved in mitochondrial biogenesis was observed in terms of DNA replication, transcription, and translation, and importantly, the mitochondria-encoded transcriptome was altered and in organello proteins synthesis was increased, similar to many of the effects observed in the tric1tric2 plants of this study. The up-regulation of mitochondrial biogenesis is a trend that also can be seen at the earliest stages of germination (Law et al., 2012). In-depth profiling of germination in Arabidopsis revealed that one of the earliest peaks in transcript abundance encodes for proteins associated with mitochondrial gene expression, with a predominance of genes encoding PPR proteins also observed (Narsai et al., 2011). Thus, it is proposed that altering mitochondrial biogenesis by altering the machinery that is required to import components (either protein or RNA) results in a similar response (i.e. that components associated with the expression of mitochondria-encoded genes at a variety of levels are up-regulated in order to compensate for the loss of function of Tric1 and Tric2).
The fact that mitochondrial ultrastructure also was altered was particularly notable, appearing swollen with decreased cristae in tric1tric2 plants. Similar morphological aberrations have been observed in yeast, mammals, and D. melanogaster upon mutation of tRNA, tRNA synthesis, and other components of the mitochondrial translation system (Arbustini et al., 1998; Zick et al., 2009; Guitart et al., 2013). Thus, it is likely that the morphological changes observed in the tric1tric2 line are consequently due to the dynamic up-regulation of mitochondrial transcription and translation rates resulting from impaired tRNA uptake ability in tric1tric2 plants.
MATERIALS AND METHODS
Phylogenetic and Structure Prediction Analyses
Tric1 (At3g49560) and Tric2 (At5g24650), also known as HP30-1/HP30-2 (Rossig et al., 2013), PRAT2.1/PRAT2.2 (Pudelski et al., 2010), and PRAT3/PRAT4 (Murcha et al., 2015), were selected for investigation. The presence of a PRAT domain (PF08294) and a SAM domain (PF00536) was identified using the Conserved Domains Database (Marchler-Bauer et al., 2011). Plant orthologs to Tric1 and Tric2 were identified by sequence homology using BLASTN (Altschul and Koonin, 1998) within each database representative of each evolutionary clade using Phytozome 9.1 (Goodstein et al., 2012) from Physcomitrella patens, Selaginella moellendorffii, Zea mays, Oryza sativa, Solanum tuberosum, Vitis vinifera, Eucalyptus grandis, Brassica rapa, Capsella rubella, Medicago truncatula, Glycine max, Ricinus communis, Populus trichocarpa, Cucumis sativus, Volvox carteri, and Chlamydomonas reinhardtii. Potential orthologs in nonplant genomic databases of Cyanidioschyzon merolae, Dictyostelium discoideum, Homo sapiens, Caenorhabditis elegans, Saccharomyces cerevisiae, Ectocarpus siliculosus, and Neurospora crassa also were searched, with no orthologs identified. All identified orthologs were further manually curated to exclude proteins with a similarity threshold above 1e−40 so as to confirm specific Tric orthologs and exclude orthologs to the other PRAT family members (Murcha et al., 2015). Protein alignments were performed using Clustal Omega (www.ebi.ac.uk; Sievers et al., 2011) and drawn using BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). Transmembrane domains were identified using TMHMM (Krogh et al., 2001). Phylogenetic analysis was carried out and drawn using MEGA 5.2.2 (Tamura et al., 2011) using the maximum likelihood tree method and the Jones-Thornton-Taylor model after 1,000 replications. Structural predictions were generated using I-TASSER ONLINE (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). PyMOL (http://www.pymol.org/) was used for visualization and comparison of RNA-binding domains.
cDNA Clones
Full-length cDNA was amplified using gene-specific primers flanked by Gateway recombination cassettes (Supplemental Table S4) and cloned into pDONR201. LR reactions were carried out with C-terminal GFP fusion vectors for GFP localization (Carrie et al., 2009), pDEST14 for in vitro transcription and translation, and pB2GW7 (35S CaMV promoter) for Agrobacterium tumefaciens-mediated complementation (Karimi et al., 2002). Yeast two-hybrid vectors were created by the amplification of cDNA using gene-specific primers flanked with appropriate restriction enzyme sites for subsequent directional cloning using standard techniques.
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertional knockout lines for Tric1 (SALK_031707, SALK_112126, and At3g49560) and Tric2 (SALK_136525, SALK_149871, and At5g24650) were obtained from ABRC and screened for homozygosity using primers as described in Supplemental Table S4 and Supplemental Figure S4A. Double knockout lines were generated and confirmed for loss of function by RT-PCR and immunodetection with antibodies raised against Tric1 and Tric2 (Supplemental Fig. S4, B and C). All germplasms where phenotyped on MS medium containing 0% and 3% (w/v) Suc and soil (peat/perlite/vermiculite) according to the parameters described previously (Boyes et al., 2001).
Seeds were sterilized with chlorine gas and sown on MS medium containing 3% (w/v) Suc followed by 48 h of stratification at 4°C. Plants were grown for 2 weeks at 22°C with a light intensity of 80 μmol quanta m−2 s−1 in a 16-h photoperiod.
Organelle Isolation
Mitochondria and chloroplasts were isolated from 14-d-old plate-grown plants according to protocols as outlined previously (Aronsson and Jarvis, 2011; Murcha and Whelan, 2015).
Protein Uptake Assays
Mitochondrial protein import assays were carried out according to Duncan et al. (2015). To investigate the intramitochondrial location of precursor proteins such as Tim23-2 (Murcha et al., 2003), OMT (GenBank accession no. X99853.1; Murcha et al., 2004), and Tim22 (Murcha et al., 2003), outer membrane ruptured mitochondria were prepared following the import reaction and prior to the addition of PK as described previously (Murcha et al., 2005b). For large-scale in vitro import experiments for analysis by BN-PAGE, 250 μL of mitochondria was incubated in 360 μL of the import master mix with 50 μL of 35S-labeled precursor protein. Chloroplast protein uptake assays were carried out as follows: 30 µg of protein was incubated in import buffer (3 mm ATP, 10 mm Met, 10 mm Cys, 20 mm potassium gluconate, 10 mm NaHCO3, 3 mm MgSO4, 330 mm sorbitol, 50 mm HEPES-KOH, pH 8, 0.2% BSA, and 50 mm ascorbic acid) with radiolabeled precursor protein at 25°C for 5 min. Chloroplasts were pelleted and washed in wash medium (50 mm HEPES-KOH, pH 8, 0.3 m sorbitol, and 3 mm MgSO2) followed by incubation with thermolysin (10 μg) and CaCl2 (5 mm) for 15 min on ice. EDTA was added, and chloroplasts were pelleted at 2,000g for 5 min. Samples were resolved by SDS-PAGE, dried, and exposed to a phosphor-imaging screen.
In Organello (Mitochondrial) Protein Synthesis Assays
Mitochondria were isolated as described above, and protein synthesis reactions were carried out as described previously with bands identified as described previously (Giegé et al., 2005).
Carbonate Extraction
Mitochondrial pellets were resuspended in 0.1 m Na2CO3 with 1 mm PMSF and incubated on ice for 30 min followed by centrifugation for 200,000g for 1 h at 20°C. The pellet was taken as the membrane fraction, while the supernatant was taken as the soluble fraction.
Chloroplast Fractionation
Chloroplast envelopes and total protein extracts from Arabidopsis plants were prepared as specified (Duy et al., 2007). Pea (Pisum sativum) chloroplasts were isolated from 10-d-old pea leaf and fractionated into outer and inner envelope membranes, stroma, and thylakoids as described previously (Waegemann et al., 1992).
Immunoprecipitation
Two hundred micrograms of freshly isolated mitochondria was pelleted at 14,000g for 5 min at 4°C. The mitochondrial pellet was resuspended in digitonin buffer (30 mm HEPES-KOH, pH 7.4, 150 mm potassium acetate, 10% [v/v] glycerol, and 1 mg of digitonin) and kept on ice for 30 min. Ten microliters of antibody serum, 50 μL of Protein A-Sepharose, 1% (w/v) BSA, and Complete protease inhibitor (Roche) were added to the mitochondria and incubated for 6 h at 4°C on a rotating wheel at 20 rpm. Beads were washed, and samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunodetected with the antibodies indicated. For immunoprecipitation assays using wheat germ lysate-translated protein, Tric1, Tric2, and ceQORH were translated in a wheat germ lysate transcription/translation kit (Promega); 10 μL of radiolabeled protein was added to the reaction, and samples were resolved by SDS-PAGE, dried, and exposed to a phosphor-imaging screen.
Protease Titration
One hundred micrograms of mitochondria was treated with PK (0–4 μg) or trypsin (0–8 μg) in 1 mL of mitochondrial wash buffer (0.3 m Suc, 10 mm TES, and 0.1% [w/v] BSA, pH 7.5 NaOH) for 30 min on ice. PMSF (1 mm) was added to mitochondria containing PK, and the mitochondria were pelleted by centrifugation at 10,000g for 10 min. The mitochondria were washed three times with 1 mL of wash buffer via resuspension and centrifugation. Mitochondria were resolved by SDS-PAGE and transferred to PVDF membranes for subsequent immunodetection.
Immunoblot Analysis
Mitochondrial and chloroplastic proteins were separated by SDS-PAGE and transferred to Hybond-C extra nitrocellulose or PVDF membranes, and immunodetection was performed as described previously (Wang et al., 2012). Previously published/commercially available antibodies used in immunodetection were as follows: Tric1 (Murcha et al., 2007), Tom40 (Carrie et al., 2010), FDH (Colas des Francs-Small et al., 1993), Tom20-2, Tom20-3, mtOM64, and metaxin (Lister et al., 2007), RISP (Carrie et al., 2010), Porin/VDAC (Elthon et al., 1989), Tim17-2 (Murcha et al., 2003), Tim23-2 (Wang et al., 2012), Ndufs4 (Meyer et al., 2009), Nad9 and Nad6 (Meyer et al., 2011), LSU and LHCP (Philippar et al., 2007; Li et al., 2015), KDSB (Duncan et al., 2011), and OOP (Kmiec et al., 2013). SDH-1, cytochrome c, COXII, α-ATP synthase, and HSP70 were obtained from Agrisera.
GFP Localization Assays
Biolistic cotransformation of GFP and mtCherry fusion vector (Nelson et al., 2007) or GFP and chloroplastic RFP (SSU-RFP; Carrie et al., 2009) was carried out on 5-d-old Arabidopsis (Col-0) cell suspensions as described previously (Carrie et al., 2009). Five micrograms of GFP and organelle marker control were coprecipitated onto gold particles and bombarded using the PDS-1000/He biolistic transformation system (Bio-Rad). GFP and mtCherry/SSU-RFP expression was visualized and captured at 100× magnification using the Olympus BX61 microscope at 460/480 nm (GFP) and 570/625 nm (mtCherry/SSU-RFP).
Split-Ubiquitin Assays
Split-ubiquitin assays in yeast (Saccharomyces cerevisiae) NMY51 cells were performed by using the DUALmembrane kit 3 (Dualsystems Biotech) according to the manufacturer’s instructions. For bait constructs, we used Tric1 and Tric2ΔSAM (amino acids 1–191) in the plasmid vector pBT3-SUC; for prey constructs, we used Tric1 and Tric2 in pPR3-N. Control preys were pAL-Alg5 (positive) and pDL2-Alg5 (negative).
Yeast Two-Hybrid Assays
Protein interaction assays were performed using the Matchmaker GAL4 Two Hybrid System (Clontech) according to the manufacturer’s instructions. Bait constructs were cloned into the plasmid vector pGBKT7 and transformed into Y187. Prey constructs were cloned into pGADT7 and transformed into AH109. The mating of bait and prey for all constructs was carried out on 96-well plates overnight and plated on double dropout (synthetic defined [SD]-Leu-Trp) and quadruple dropout (SD-Leu-Trp-Ade-His) media. Positive interactors were identified after 4 d of growth at 30°C. Only positive interacting diploid colonies were then serially diluted and plated out onto double dropout (SD-Leu-Trp) and quadruple dropout (SD-Leu-Trp-Ade-His) media with or without the addition of X-α-Gal. Immunodetection on yeast protein extracts from single transformants was carried as described previously (Kushnirov, 2000) using anti-HA and anti-c-myc antibodies.
tRNA Uptake Assays
In vitro import of radiolabeled tRNAAla was performed as described previously (Kubiszewski-Jakubiak et al., 2015). Briefly, 100 μg of freshly isolated mitochondria was incubated with 5 μL of radiolabeled tRNAAla in protein import buffer (0.3 m Suc, 100 mm KCl, 20 mm MOPS, pH 7.4, 10 mm KH2PO4, 0.2% [w/v] BSA, 1 mm MgCl2, 1 mm Met, 0.2 mm ADP, 0.75 mm ATP, 5 mm succinate, 5 mm DTT, 5 mm NADH, and 5 mm GTP). Five micrograms of thawed recombinant Su9-DHFR protein was added to the import reaction and incubated at 25°C for 25 min. Import was stopped by incubating on ice. RNase mix was added to the reaction and incubated further for 10 min. Inactivation of RNase was carried out by several washing steps with cold stop solution (5 mm EDTA, 5 mm EGTA, 0.3 m Suc, 10 mm TES, and 0.2% [w/v] BSA, pH 7.5), and the mitochondria were pelleted by centrifugation. Total RNA was extracted by resuspending the pellet in 10 mm Tris-HCl (pH 7.5), 10 mm MgCl2, and 1% (w/v) SDS and extracted with a 1:1 volume of water-saturated phenol. The aqueous phase was ethanol precipitated, and samples were resolved via electrophoresis through 7 m urea-15% polyacrylamide gels. Gels were dried and exposed to a phosphor-imaging plate for several hours and visualized using a BAS100 phosphor imager.
Purification of Tric1SAM
The gene fragment corresponding to the SAM domain of At3g49560/Tric1 (amino acids 191–261) was cloned in the pET15b vector (Novagen), yielding an N-terminal His tag fusion protein. Mutations corresponding to the Tric1SAM variants K15A and K20A were introduced by site-directed mutagenesis using the Quikchange II-XL kit (Agilent Technologies).
The expression vectors were transformed into Escherichia coli Rosetta 2 (DE3) (Millipore) and grown as 1-L batches in Luria-Bertani medium supplemented with 100 μg mL−1 carbenicillin and 30 μg mL−1 chloramphenicol. Bacteria were grown at 30°C until the optical density reached 0.8 to 0.9 followed by induction of protein expression by 400 μm isopropyl-β-d-1-thiogalactopyranoside. The temperature was adjusted to 20°C, and bacteria were grown for a further 6 h. Bacterial cells were recovered by centrifugation at 5,000g and frozen as pellets.
His-tagged TricSAM was purified by standard immobilized metal-affinity chromatography (GE Healthcare), with the protein recovered by elution with 200 mm imidazole. The elution samples were then concentrated and further purified by size-exclusion chromatography on a Sephadex S200 column (GE Healthcare) using as buffer 10 mm HEPES-KOH, pH 7.4, 50 mm NaCl, and 1 mm DTT.
Fluorescein-Labeled RNA Oligonucleotides
Binding analysis was performed using 5′-fluorescein-labeled RNA oligonucleotides (Supplemental Table S3) corresponding to the three tRNAAla arms, supplied deprotected and desalted (Thermo Fisher Scientific).
Fluorescence Polarization Assay
tRNA oligonucleotides were heated to 95°C for 5 min, immediately cooled on ice for 5 min, and subsequently diluted to 50 nm in binding buffer (10 mm HEPES-KOH, pH 7.4, 75 mm NaCl, and 10% glycerol). Anisotropy measurements (excitation wavelength, 494 nm; emission wavelength, 524 nm) were recorded at 22°C using a FluoroLog spectrofluorometer (Horiba Scientific) following successive additions of purified Tric1SAM (wild type or variants; concentration ranging from 0 to 1,350 nm). The binding reactions were allowed to stabilize for 200 s before measurements were recorded. All binding reactions were measured in triplicate. Dissociation constants were estimated by nonlinear regression assuming a single-site binding model using GraphPad Prism software.
EMSA
Fluorescein-labeled tRNAAla oligonucleotides (final concentration, 120 nm) were incubated with 80 μm purified TricSAM (wild type or variants) in a binding buffer (15 mm Tris-HCl, pH 7, 150 mm NaCl, 10% glycerol, and 2 units μL−1 RNase inhibitor RiboLock [Thermo Scientific]) for 15 min at 22°C. The reaction was resolved on a nondenaturing 10% polyacrylamide 0.5× Tris/borate/EDTA gel, run in 0.5× Tris/borate/EDTA buffer, and imaged on a LAS-1000 dark box (Fujifilm).
RNA-Seq
All steps in the RNA-seq analyses were performed on three independent biological replicates. Total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich) with an on-column DNase treatment according to the manufacturer’s instructions from 2-week-old seedlings grown on MS medium and 4-week-old rosette tissue collected from soil-grown plants under normal growth conditions as described above. The quantity and quality of the RNA were assessed using a NanoDrop 1000 spectrophotometer and agarose gel electrophoresis. RNA-seq libraries were generated using the TruSeq Stranded Total RNA with Ribo-Zero Plant kit according to the manufacturer’s instructions (Illumina). Sequencing was performed on an Illumina HiSeq 1500 instrument.
RNA-Seq Analysis
RNA-seq data were analyzed using the TopHat/Cufflinks pipeline (version 2) by estimating transcript abundances in fragments per kilobase of exon per million fragments mapped values (Trapnell et al., 2012). The TAIR10 genome assembly was used for read mapping, and significant differential expression for all genes in the Arabidopsis genome was determined with FDR < 0.05. For all genes, fold changes are presented as log2 fold change values.
Transmission Electron Microscopy
Transmission electron microscopy was carried out as described earlier (Li et al., 2015).
Northern-Blot Analysis
tRNA purification and northern-blot analysis were carried out as described previously (Duchêne and Maréchal-Drouard, 2001). tRNA was extracted from whole tissue (total) and purified mitochondria from 2-week-old seedlings. tRNA was separated on 15% polyacrylamide gels and transferred to Hybond nylon membranes. Hybridization was carried out using the oligonucleotides specific for the nucleus-encoded tRNAAla, located in the cytosol and mitochondria, the nucleus-encoded tRNALys, located in the cytosol, and the mitochondria-encoded tRNALys, located in the mitochondria, as listed in Supplemental Table S4.
Antibody Testing
For testing the antibody specificity, 6× His-tagged recombinant proteins of Tric1 and Tric2 were produced by Gateway cloning into pDEST17 and expressed in BL21 cells. Tric1SAM was expressed and purified as described above. The equal abundance of recombinant proteins was confirmed by detection with 6× His antiserum (Qiagen).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: Tric1 (At3g49560), Tric2 (At5g24650), Tim23-2 (At1g72750), Tim17-2 (At2g37410), Tim22 (At3g10110/At1g18320), RISP (At5g13430), MPP-α (At3g16480), and tRNAAla (At3g62245). Sequencing data are available from the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE76287.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Multiple sequence alignment (Clustal Omega) of all Tric orthologs from 17 plant species and algae.
Supplemental Figure S2. Testing specificity of Tric1 antibody.
Supplemental Figure S3. Tric1 and Tric2 interactions with subunits of the protein import apparatus.
Supplemental Figure S4. Characterization of single and double T-DNA insertion knockouts for Tric1 and Tric2.
Supplemental Figure S5. tRNA import assays into mitochondria isolated from the single lines tric1 and tric2.
Supplemental Figure S6. MapMan general metabolism overview of genes modulated in the tric1tric2 line compared with Col-0 from 2-week-old seedlings and 4-week-old rosette leaves.
Supplemental Figure S7. Fold change responses of 146 mitochondria-encoded genes.
Supplemental Figure S8. In vitro protein uptake assays into chloroplasts isolated from wild-type Col-0 and tric1tric2 plants.
Supplemental Table S1. Analysis of mitochondrial morphology in tric1tric2 mutant and Col-0 wild-type plants.
Supplemental Table S2. Differential expression of all nucleus-encoded genes in Col-0 and tric1tric2 from rosettes and seedlings.
Supplemental Table S3. Differentially expressed genes encoding mitochondrial proteins.
Supplemental Table S4. Primers used in this study.
Supplementary Material
Glossary
- SAM
sterile-α-motif
- BN-PAGE
blue native-PAGE
- MS
Murashige and Skoog
- Col-0
Columbia-0
- EMSA
electrophoretic mobility shift assay
- FDR
false discovery rate
- PPR
pentratricopeptide repeat
- SD
synthetic dextrose
Footnotes
This work was supported by the Australian Research Council (Future Fellowship Grant no. FT130100112 and Discovery Grant no. DP150100293 to M.W.M. and Discovery Grant no. DP130102384 to J.W.); by the French National Program Investissement d’Avenir administered by the Agence Nationale de la Recherche (grant no. ANR-10-IDEX-0002-02), the University of Strasbourg, and the Centre National de la Recherche Scientifique; by the Deutsche Forschungsgemeinschaft (Heisenberg fellowship no. PH73/6-1 to K.P.); and by the Swedish Foundation for International Cooperation in Research and Higher Education (to E.G., J.W., and M.W.M.).
Articles can be viewed without a subscription.
References
- Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Koonin EV (1998) Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem Sci 23: 444–447 [DOI] [PubMed] [Google Scholar]
- Arbustini E, Diegoli M, Fasani R, Grasso M, Morbini P, Banchieri N, Bellini O, Dal Bello B, Pilotto A, Magrini G, et al. (1998) Mitochondrial DNA mutations and mitochondrial abnormalities in dilated cardiomyopathy. Am J Pathol 153: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Jarvis RP (2011) Rapid isolation of Arabidopsis chloroplasts and their use for in vitro protein import assays. Methods Mol Biol 774: 281–305 [DOI] [PubMed] [Google Scholar]
- Aung K, Hu J (2011) The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23: 4446–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA (2003) The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 10: 614–621 [DOI] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Beardslee TA, Roy-Chowdhury S, Jaiswal P, Buhot L, Lerbs-Mache S, Stern DB, Allison LA (2002) A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts. Plant J 31: 199–209 [DOI] [PubMed] [Google Scholar]
- Bouzaidi-Tiali N, Aeby E, Charrière F, Pusnik M, Schneider A (2007) Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. EMBO J 26: 4302–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuers FK, Bräutigam A, Geimer S, Welzel UY, Stefano G, Renna L, Brandizzi F, Weber AP (2012) Dynamic remodeling of the plastid envelope membranes: a tool for chloroplast envelope in vivo localizations. Front Plant Sci 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Guo L, Shen ZR, Wang DY, Zhang Q, Sodmergen (2015) Elevation of pollen mitochondrial DNA copy number by WHIRLY2: altered respiration and pollen tube growth in Arabidopsis. Plant Physiol 169: 660–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Giraud E, Duncan O, Xu L, Wang Y, Huang S, Clifton R, Murcha M, Filipovska A, Rackham O, et al. (2010) Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J Biol Chem 285: 36138–36148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155: 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Rémy R (1993) Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol 102: 1171–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchêne AM, Maréchal-Drouard L (2001) The chloroplast-derived trnW and trnM-e genes are not expressed in Arabidopsis mitochondria. Biochem Biophys Res Commun 285: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Dudek J, Rehling P, van der Laan M (2013) Mitochondrial protein import: common principles and physiological networks. Biochim Biophys Acta 1833: 274–285 [DOI] [PubMed] [Google Scholar]
- Duncan O, Carrie C, Wang Y, Murcha MW (2015) In vitro and in vivo protein uptake studies in plant mitochondria. Methods Mol Biol 1305: 61–81 [DOI] [PubMed] [Google Scholar]
- Duncan O, Taylor NL, Carrie C, Eubel H, Kubiszewski-Jakubiak S, Zhang B, Narsai R, Millar AH, Whelan J (2011) Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol 157: 1093–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy D, Wanner G, Meda AR, von Wirén N, Soll J, Philippar K (2007) PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19: 986–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmier M, Carroll AJ, Delannoy E, Vallet C, Day DA, Small ID, Millar AH (2008) Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P, Sweetlove LJ, Cognat V, Leaver CJ (2005) Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell 17: 1497–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. (2012) Mitochondrial evolution. Cold Spring Harb Perspect Biol 4: a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitart T, Picchioni D, Piñeyro D, Ribas de Pouplana L (2013) Human mitochondrial disease-like symptoms caused by a reduced tRNA aminoacylation activity in flies. Nucleic Acids Res 41: 6595–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper CM, Tanz SK, Castleden IR, Vacher MA, Small ID, Millar AH (2014) SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30: 3356–3364 [DOI] [PubMed] [Google Scholar]
- Huot JL, Enkler L, Megel C, Karim L, Laporte D, Becker HD, Duchêne AM, Sissler M, Maréchal-Drouard L (2014) Idiosyncrasies in decoding mitochondrial genomes. Biochimie 100: 95–106 [DOI] [PubMed] [Google Scholar]
- Jiao S, Thornsberry JM, Elthon TE, Newton KJ (2005) Biochemical and molecular characterization of photosystem I deficiency in the NCS6 mitochondrial mutant of maize. Plant Mol Biol 57: 303–313 [DOI] [PubMed] [Google Scholar]
- Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I (2007) Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol Cell 26: 625–637 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ (2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Bowie JU (2003) SAM domains: uniform structure, diversity of function. Trends Biochem Sci 28: 625–628 [DOI] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Berntsson RP, Murcha MW, Branca RM, Radomiljac JD, Regberg J, Svensson LM, Bakali A, Langel U, et al. (2013) Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc Natl Acad Sci USA 110: E3761–E3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kubiszewski-Jakubiak S, Megel C, Ubrig E, Salinas T, Duchene A-M, Marechal-Drouard L (2015) In vitro RNA uptake studies in plant mitochondria. Methods Mol Biol 1305: 45–60 [DOI] [PubMed] [Google Scholar]
- Kühn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J (2011) The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J 67: 1067–1080 [DOI] [PubMed] [Google Scholar]
- Kühn K, Yin G, Duncan O, Law SR, Kubiszewski-Jakubiak S, Kaur P, Meyer E, Wang Y, Small CC, Giraud E, et al. (2015) Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol 167: 228–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV. (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol 158: 1610–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Gügel IL, Giavalisco P, Zeisler V, Schreiber L, Soll J, Philippar K (2015) FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol 13: e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Carrie C, Duncan O, Ho LH, Howell KA, Murcha MW, Whelan J (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19: 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Müller M (1998) The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Meyer EH, Solheim C, Tanz SK, Bonnard G, Millar AH (2011) Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J Biol Chem 286: 26081–26092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Carrie C, Pogson B, Whelan J (2009) Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Elhafez D, Lister R, Tonti-Filippini J, Baumgartner M, Philippar K, Carrie C, Mokranjac D, Soll J, Whelan J (2007) Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol 143: 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Elhafez D, Millar AH, Whelan J (2004) The N-terminal extension of plant mitochondrial carrier proteins is removed by two-step processing: the first cleavage is by the mitochondrial processing peptidase. J Mol Biol 344: 443–454 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Elhafez D, Millar AH, Whelan J (2005a) The C-terminal region of TIM17 links the outer and inner mitochondrial membranes in Arabidopsis and is essential for protein import. J Biol Chem 280: 16476–16483 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Kmiec B, Kubiszewski-Jakubiak S, Teixeira PF, Glaser E, Whelan J (2014a) Protein import into plant mitochondria: signals, machinery, processing, and regulation. J Exp Bot 65: 6301–6335 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Kubiszewski-Jakubiak S, Wang Y, Whelan J (2014b) Evidence for interactions between the mitochondrial import apparatus and respiratory chain complexes via Tim21-like proteins in Arabidopsis. Front Plant Sci 5: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Lister R, Ho AY, Whelan J (2003) Identification, expression, and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol 131: 1737–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcha MW, Millar AH, Whelan J (2005b) The N-terminal cleavable extension of plant carrier proteins is responsible for efficient insertion into the inner mitochondrial membrane. J Mol Biol 351: 16–25 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Narsai R, Devenish J, Kubiszewski-Jakubiak S, Whelan J (2015) MPIC: a mitochondrial protein import components database for plant and non-plant species. Plant Cell Physiol 56: e10. [DOI] [PubMed] [Google Scholar]
- Murcha MW, Whelan J (2015) Isolation of intact mitochondria from the model plant species Arabidopsis thaliana and Oryza sativa. Methods Mol Biol 1305: 1–12 [DOI] [PubMed] [Google Scholar]
- Narsai R, Law SR, Carrie C, Xu L, Whelan J (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157: 1342–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Neupert W. (2015) A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol 427: 1135–1158 [DOI] [PubMed] [Google Scholar]
- Peters K, Niessen M, Peterhänsel C, Späth B, Hölzle A, Binder S, Marchfelder A, Braun HP (2012) Complex I-complex II ratio strongly differs in various organs of Arabidopsis thaliana. Plant Mol Biol 79: 273–284 [DOI] [PubMed] [Google Scholar]
- Philippar K, Geis T, Ilkavets I, Oster U, Schwenkert S, Meurer J, Soll J (2007) Chloroplast biogenesis: the use of mutants to study the etioplast-chloroplast transition. Proc Natl Acad Sci USA 104: 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudelski B, Kraus S, Soll J, Philippar K (2010) The plant PRAT proteins: preprotein and amino acid transport in mitochondria and chloroplasts. Plant Biol (Stuttg) (Suppl 1) 12: 42–55 [DOI] [PubMed] [Google Scholar]
- Pudelski B, Schock A, Hoth S, Radchuk R, Weber H, Hofmann J, Sonnewald U, Soll J, Philippar K (2012) The plastid outer envelope protein OEP16 affects metabolic fluxes during ABA-controlled seed development and germination. J Exp Bot 63: 1919–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudelski B, Soll J, Philippar K (2009) A search for factors influencing etioplast-chloroplast transition. Proc Natl Acad Sci USA 106: 12201–12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol 286: 105–120 [DOI] [PubMed] [Google Scholar]
- Rossig C, Reinbothe C, Gray J, Valdes O, von Wettstein D, Reinbothe S (2013) Three proteins mediate import of transit sequence-less precursors into the inner envelope of chloroplasts in Arabidopsis thaliana. Proc Natl Acad Sci USA 110: 19962–19967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas T, Duchêne AM, Delage L, Nilsson S, Glaser E, Zaepfel M, Maréchal-Drouard L (2006) The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc Natl Acad Sci USA 103: 18362–18367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T, Giegé R, Giegé P (2015) tRNA biology in mitochondria. Int J Mol Sci 16: 4518–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A. (2011) Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem 80: 1033–1053 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, König AC, Sweetlove LJ, Finkemeier I (2012) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Sepuri NB, Gorla M, King MP (2012) Mitochondrial lysyl-tRNA synthetase independent import of tRNA lysine into yeast mitochondria. PLoS ONE 7: e35321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber F, Placido A, El Farouk-Ameqrane S, Duchêne AM, Maréchal-Drouard L (2011) A protein shuttle system to target RNA into mitochondria. Nucleic Acids Res 39: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Maréchal-Drouard L, Masson J, Pelletier G, Cosset A, Weil JH, Dietrich A (1992) In vivo import of a normal or mutagenized heterologous transfer RNA into the mitochondria of transgenic plants: towards novel ways of influencing mitochondrial gene expression? EMBO J 11: 1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Entelis N, Martin RP (1995) Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J 14: 3461–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp F, Charrière F, Schneider A (2011) In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep 12: 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Zarkovic J, Yu J, Ruckle ME, McIntosh L, Hock JJ, Bingham S, White SJ, George RM, Subbaiah CC, et al. (2012) Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS ONE 7: e44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J (2009) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Waegemann K, Eichacker S, Soll J (1992) Outer envelope membranes from chloroplasts are isolated as right-side-out vesicles. Planta 187: 89–94 [DOI] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Carrie C, Law SR, Murcha MW, Whelan J (2013) Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol 161: 644–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Leaver CJ (2008) EMB2473/MIRO1, an Arabidopsis Miro GTPase, is required for embryogenesis and influences mitochondrial morphology in pollen. Plant Cell 20: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Van Aken O, Thatcher L, De Clercq I, Duncan O, Law SR, Murcha MW, van der Merwe M, Seifi HS, Carrie C, et al. (2014) The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J 80: 709–727 [DOI] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS (2009) Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta 1793: 5–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.