SWI2/SNF2 chromatin remodeling ATPase BRAHMA controls vegetative phase change by regulation of miR156 expression at the level of nucleosome in Arabidopsis.
Abstract
Plants progress from a juvenile vegetative phase of development to an adult vegetative phase of development before they enter the reproductive phase. miR156 has been shown to be the master regulator of the juvenile-to-adult transition in plants. However, the mechanism of how miR156 is transcriptionally regulated still remains elusive. In a forward genetic screen, we identified that a mutation in the SWI2/SNF2 chromatin remodeling ATPase BRAHMA (BRM) exhibited an accelerated vegetative phase change phenotype by reducing the expression of miR156, which in turn caused a corresponding increase in the levels of SQUAMOSA PROMOTER BINDING PROTEIN LIKE genes. BRM regulates miR156 expression by directly binding to the MIR156A promoter. Mutations in BRM not only increased occupancy of the −2 and +1 nucleosomes proximal to the transcription start site at the MIR156A locus but also the levels of trimethylated histone H3 at Lys 27. The precocious phenotype of brm mutant was partially suppressed by a second mutation in SWINGER (SWN), but not by a mutation in CURLEY LEAF, both of which are key components of the Polycomb Group Repressive Complex 2 in plants. Our results indicate that BRM and SWN act antagonistically at the nucleosome level to fine-tune the temporal expression of miR156 to regulate vegetative phase change in Arabidopsis.
Genetic analyses in both animals and plants demonstrated that different developmental phases must be temporally coordinated in order for them to develop normally (Poethig, 2003; Moss, 2007). A representative example of this is the coordination of shoot development between juvenile, adult, and reproductive phases of development in plants. Changes in the relative timing of juvenile-to-adult shoot development can lead to considerable effects on traits like leaf morphology and the onset of flowering (Huijser and Schmid, 2011; Poethig, 2013). Therefore, the study of the mechanism of vegetative phase change is crucial for our understanding of plant ontogeny.
Shoot development in plants can be divided into a juvenile vegetative phase, an adult vegetative phase, and a reproductive phase (Poethig, 1990; Kerstetter and Poethig, 1998), and each developmental phase is marked by changes in a series of distinct phase-specific traits. The transition from vegetative to reproductive phase of development is relatively abrupt and is easy to recognize because of the absence or presence of flowers or other reproductive structures; the transition from juvenile to adult phases of development involves rather subtle changes and is therefore hard to recognize. In Arabidopsis (Arabidopsis thaliana), vegetative phase change is characterized by changes in the production of trichomes on the abaxial side of the leaf blade, an increase in the leaf length/width ratio, an increase in the degree of serration of the leaf margin, and a decrease in cell size (Telfer et al., 1997; Tsukaya et al., 2000; Usami et al., 2009). Genetic and molecular analyses of vegetative development in Arabidopsis demonstrated that miR156 negatively regulates the expression of a class of plant-specific SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes and functions as a master regulator of vegetative phase change by coordinating several functionally distinct pathways during vegetative development (Wu and Poethig, 2006; Gandikota et al., 2007; Wang et al., 2009; Wu et al., 2009). miR156 expression declines gradually, while the expression of some of its targets, such as SPL3, SPL9, increases during shoot development (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009). SPL proteins act as upstream activators of miR172, which in turn represses AP2-like transcription factors that regulate floral organ identity and flowering (Aukerman and Sakai, 2003; Chen, 2004); the sequential interaction between miR156 and miR172 constitutes a major pathway to regulate vegetative development in plants (Wu et al., 2009; Poethig, 2013). miR156 expression responds to a variety of exogenous cues such as ambient temperature (Lee et al., 2010; Xin et al., 2010; Yu et al., 2012), phosphate starvation (Hsieh et al., 2009), sugar (Yang et al., 2013; Yu et al., 2013), and CO2 treatment (May et al., 2013). However, the mechanism of how these extrinsic cues affect miR156 expression remains elusive. In Arabidopsis, FUSCA3 (FUS3), a B3 domain transcription factor, as well as two MADS box genes, AGL15 and AGL18, were shown to regulate miR156 expression by direct binding to the promoter regions of MIR156A and MIR156C loci (Wang and Perry, 2013; Serivichyaswat et al., 2015), but the biological function of direct binding remains unexplored. Recent work also indicated that the Arabidopsis BMI1 (AtBMI1) protein, a PRC1 component (Picó et al., 2015), and the CHD3 chromatin remodeler, PICKLE (PKL), promote the addition of Lys 27 on histone H3 (H3K27me3) to MIR156A/MIR156C (Xu et al., 2016), implying that epigenetic regulation also plays an important role in the regulation of miR156 expression.
Shoot development in plants also requires coordinated temporal and spatial gene expression at the genome level. Transcriptional regulation of gene expression at the chromatin level by covalent modification of histones and/or DNA by histone or DNA modifying enzymes (Li et al., 2007), or by noncovalent alteration of nucleosome position, occupancy, confirmation, and composition by chromatin remodeling ATPases (Hargreaves and Crabtree, 2011) is pivotal. In plants, one of the most important covalent modifications of histones is the trimethylation of H3K27me3 by two histone methyltransferase, CURLY LEAF (CLF) and SWINGER (SWN), two important components of the Polycomb group (PcG) proteins acting redundantly during the vegetative and reproductive stages of development to keep a repressive state for genes where they should be inactive in plants (Chanvivattana et al., 2004). Loss-of-function mutations in CLF resemble phenotypes caused by ectopic overexpression of AGAMOUS (Goodrich et al., 1997), and loss-of-function mutations in SWN were originally considered to cause no obvious phenotypes until recent work showed that swn and clf single mutants exhibited weak vegetative phase change phenotypes (Xu et al., 2016), while a double mutant between clf and swn forms somatic embryos (Chanvivattana et al., 2004; Schubert et al., 2005), indicating that PcG proteins play important regulatory roles in plant development. Noncovalent modification of histones is mainly mediated by the SWI/SNF-type chromatin remodeling protein complex, which uses the energy derived from ATP hydrolysis to change the histone octamer-DNA interaction, such as nucleosome shifting, conformational change, and histone loss or nucleosome disassembly to change the accessibility of DNA to transcription factors or other transcription regulators (Saha et al., 2006; Clapier and Cairns, 2009). Among the DNA-dependent ATPases, the SWI2/SNF2 subgroup is the most widely studied in plants (Kwon and Wagner, 2007). The Arabidopsis genome encodes three different types of SWI2/SNF2 subgroup proteins, including BRAHMA (BRM), SPLAYED, and MINUSCULE, and these proteins have been shown to participate in different aspects of plant growth and development as well as abscisic acid response (Flaus et al., 2006; Jerzmanowski, 2007; Kwon and Wagner, 2007; Han et al., 2012; Sang et al., 2012). A genome-wide analysis of H3K27me3 in brm mutant seedlings using chromatin immunoprecipitation (ChIP)-sequencing indicated that BRM directly binds to its targets to antagonize the function of PcG proteins at those loci (Li et al., 2015). The recruitment of BRM to its target genes was facilitated by a plant-specific H3K27 demethylase, RELATIVE OF EARLY FLOWERING 6 (Li et al., 2016). brm mutants exhibited pleiotropic phenotypes, such as reduced plant size (Farrona et al., 2004; Hurtado et al., 2006), curled-down leaf shape (Hurtado et al., 2006; Tang et al., 2008), defects in floral organ identity (Hurtado et al., 2006; Wu et al., 2012), hypersensitivity to abscisic acid treatment (Han et al., 2012), and early flowering (Farrona et al., 2004, 2011; Tang et al., 2008). However, whether BRM plays a role in vegetative phase change remains unknown.
In a forward genetic screen to identify upstream regulators of miR156, we identified that a loss-of-function mutation in the BRM ATPase enhanced the phenotype of plants with pSPL9::eGFP-SPL9 (translational fusion of eGFP to the SPL9 protein under the control of the SPL9 native promoter). The single brm mutant exhibited an accelerated juvenile-to-adult phase change phenotype. Molecular analyses revealed that BRM directly binds to the promoter region of MIR156A to regulate its expression; the brm mutant caused high levels of nucleosome occupancy and H3K27me3 at the proximal regions of the transcription start site (TSS) of the MIR156A locus compared with wild type. Contrary to the phenotype of the brm mutant, a loss-of-function mutation in SWN exhibited a delayed juvenile-to-adult phase change phenotype by reducing the degree of H3K27me3 at the MIR156A locus, and it partially rescued the phenotype of the brm mutant. BRM functions upstream of MIR156A to promote miR156 expression during vegetative phase change by restricting nucleosome occupancy at the MIR156A locus as well as by restricting the activity of SWN to reduce the level of H3K27me3 at the MIR156A locus.
RESULTS
Loss-of-Function Mutations in BRM Accelerate the Juvenile-to-Adult Transition in Arabidopsis
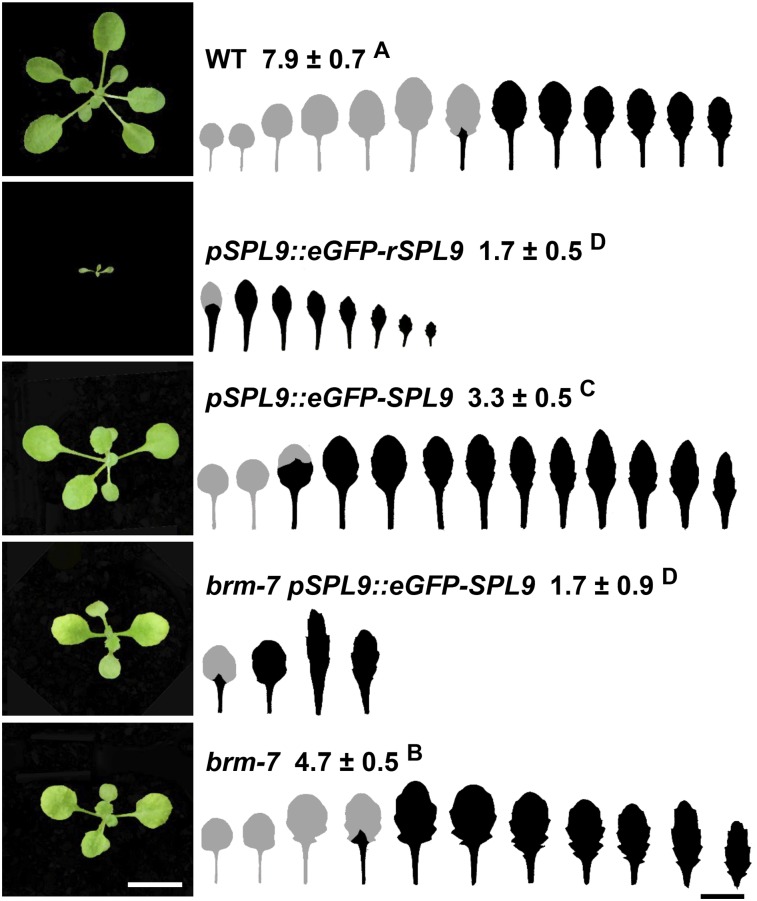
miR156 represses SPL gene expression posttranscriptionally to coordinate functionally different pathways to regulate vegetative phase change (Wu et al., 2009). To explore how miR156 itself is regulated, we did an ethyl methanesulfonate mutagenesis for a reporter line transformed with a pSPL9::eGFP-SPL9 construct, and a forward genetic screen was performed in the M2 population. Plants transformed with the pSPL9::eGFP-SPL9 construct sensitive to miR156 surprisingly had a precocious vegetative phase change phenotype with early abaxial trichomes on leaf 3 and leaves were more elongated compared with those of wild type, while plants with a pSPL9::eGFP-rSPL9 construct insensitive to miR156 gave rise to earlier abaxial trichomes on leaf 1 and the leaves were more elongated and smaller than those with pSPL9::eGFP-SPL9 (Fig. 1), indicating that SPL9 is under the regulation of miR156. We can expect to isolate factors regulating miR156 expression by screening for enhancers and suppressor of this pSPL9::eGFP-SPL9 reporter line. A forward genetic screen yielded a mutation that can dramatically enhance the phenotype of this reporter line. pSPL9::eGFP-SPL9 reporter line has early abaxial trichomes on leaf 3, whereas the enhancer has early abaxial trichomes on leaf 1 in the mutant background (Fig. 1). Isolation of the single mutant showed that the mutant resembled brm mutants (Farrona et al., 2004; Hurtado et al., 2006; Bezhani et al., 2007; Tang et al., 2008), which encode a SWI2/SNF2 ATPase. Sequencing of the BRM coding region showed that there was a substitution of a G by an A at the 5,663-bp position in the open reading frame of BRM, resulting in the replacement of a Gly by a Glu in the conserved ATPase domain in the BRM coding region. To confirm the identity of this mutation, we did a genetic complementation test by crossing this mutant to the brm-5 mutant (Tang et al., 2008). F1 plants basically showed exactly the brm-5 phenotype (Supplemental Fig. S1), indicating that this mutant is allelic to brm-5, and the phenotype we saw was attributable to the mutation in the BRM coding region. Based on the availability of different brm alleles, we renamed this mutant as brm-7. In addition to phenotypes described previously, the brm-7 single mutant typically exhibited an accelerated vegetative phase change phenotype, leaves were more serrated, and it had early abaxial trichomes on leaf 4 (Fig. 1), implying that BRM functions to repress vegetative development in Arabidopsis.
Figure 1.
Identification of the brm-7 mutant with a precocious vegetative phase change phenotype. The 18-d-old wild-type, pSPL9::eGFP-rSPL9, pSPL9::eGFP-rSPL9, brm-7 pSPL9::eGFP-SPL9, and brm-7 plants were grown in short days. The first leaf with abaxial trichomes was scored. Numbers indicate the first leaf with abaxial trichomes (n = 27, ±SD). Juvenile leaves are shown in gray and adult leaves in black in the heteroblastic analyses. Different letters indicate significant difference between genotypes using one way ANOVA at P < 0.01. Scale bar = 1 cm.
BRM Regulates Vegetative Phase Change by Modulating the Expression of miR156 and SPL Genes
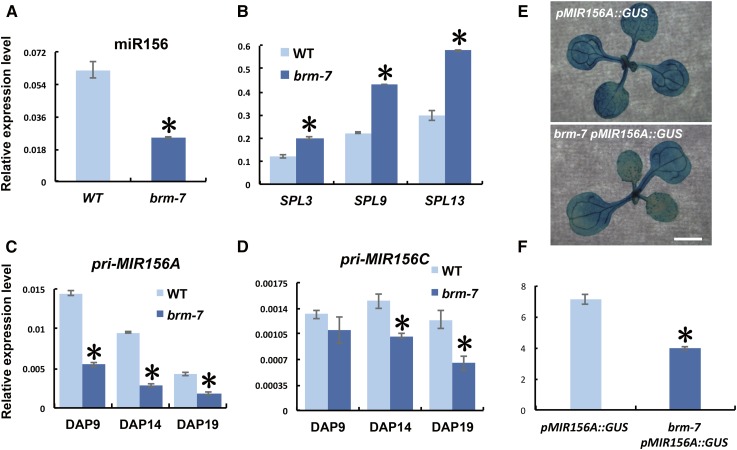
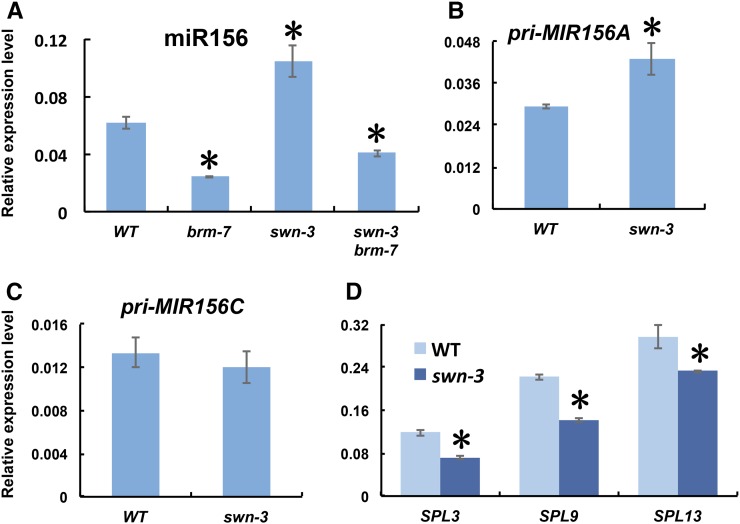
To gain insight into the molecular mechanism of the precocious vegetative phase change phenotypes conferred by brm-7, we measured the expression of miR156, SPL genes, and different primary miR156 transcripts in 14-d-old seedlings of wild type and the brm-7 mutant. miR156 was reduced significantly in brm-7, whereas the expression of its targets, SPL3, SPL9, and SPL13, was elevated significantly in brm-7 (Fig. 2, A and B). To see if the reduction in mature miR156 was caused by a corresponding reduction in the transcription of miR156 loci, we determined primary miR156 transcript levels, especially the levels of primary MIR156A (pri-MIR156A) and primary MIR156C (pri-MIR156C), both of which are highly expressed major miR156 loci contributing to the level of mature miR156 (Yang et al., 2013; Yu et al., 2013). Real-time quantitative reverse transcription-PCR (qRT-PCR) revealed that pri-MIR156A was down-regulated significantly in the brm-7 mutant at all developmental stages tested, while pri-MIR156C was down-regulated only at late stages (Fig. 2, C and D). To further validate that BRM affects the transcription of miR156, we took advantage of a pri-MIR156A GUS reporter line, pMIR156A::GUS, which could partially reflect the transcription of the MIR156A locus in that it could respond to exogenous cues normally (Yang et al., 2013). We crossed this reporter line to brm-7 and generated homozygous brm-7 pMIR156A::GUS plants. GUS staining and qRT-PCR showed a significant reduction in the expression level of GUS in brm-7 compared with that in the wild-type background (Fig. 2, E and F). To see if pMIR156A::GUS also exhibited a similar temporal expression pattern to miR156, we did GUS staining for 10- and 13-d-old pMIR156A::GUS line in both wild type and brm-7 background. Neither line exhibited a temporal decrease in GUS expression, whereas the GUS expression level was consistently reduced significantly in brm-7 (Supplemental Fig. S2), suggesting that BRM activates miR156 expression by promoting the transcription of the MIR156A locus.
Figure 2.
BRM activates miR156 expression transcriptionally during vegetative phase change. A, The level of mature miR156 in 14-d-old wild-type and brm-7 seedlings in short days. B, Expression of SPL3, SPL9, and SPL13 in 14-d-old wild-type and brm-7 seedlings in short days. C, The temporal expression pattern of pri-MIR156A in wild type and brm-7 in short days. D, The temporal expression pattern of pri-MIR156C in wild type and brm-7 in short days. E, GUS staining of 11-d-old pMIR156A::GUS and pMIR156A::GUS brm-7 plants in long days. Scale bar = 2 mm. F, Relative expression of GUS transcript in 11-d-old pMIR156A::GUS and pMIR156A::GUS brm-7 plants as shown in E. DAP, Days after planting. Asterisk denotes statistical significance from wild type at P < 0.01 using Student’s t test.
BRM Interacts with the miR156-SPLs Pathway Genetically
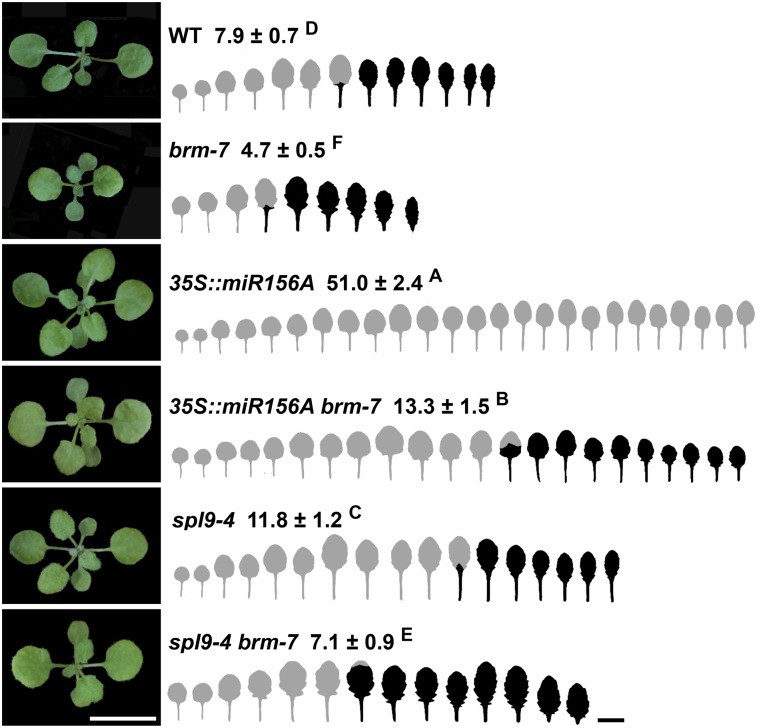
To study the genetic interaction between BRM and the miR156-SPLs pathway, we crossed 35S::miR156A and spl9-4 to brm-7. Under short-day conditions, the brm-7 mutant produced early abaxial trichomes on leaf 4 and 35S::miR156A produced abaxial trichomes on leaf 51, while 35S::miR156A brm-7 produced abaxial trichomes on leaf 13 (Fig. 3). Overexpression of miR156 in the brm-7 background could significantly rescue the precocious vegetative phase change phenotype of the brm-7 mutant, and the leaf shape of 35S::miR156A brm-7 was more like those of 35S::miR156A. This result, together with the gene expression data that miR156 was down-regulated in the brm-7 mutant (Fig. 2A), strongly indicated that miR156 functions downstream of BRM. To further validate the genetic interaction between BRM and SPL genes, we generated a double mutant of spl9-4 brm-7. spl9-4 produced abaxial trichomes on leaf 11, while spl9-4 brm-7 had abaxial trichomes on leaf 7; spl9-4 can significantly delay the precocious vegetative phase change phenotype of brm-7 (Fig. 3). This fact, plus that SPL9 expression was up-regulated in brm-7, suggested that SPL9 functions downstream of BRM as well.
Figure 3.
BRM interacts genetically with the miR156-SPLs pathway. Plants were grown in short days. The first leaf with abaxial trichomes was scored. Numbers indicate the first leaf with abaxial trichomes (n = 27, ±SD). Juvenile leaves are shown in gray and adult leaves in black in the heteroblastic analyses. Different letters indicate significant difference between genotypes using one-way ANOVA at P < 0.01. Scale bar = 1 cm.
BRM Regulates miR156 Expression by Directly Binding to Its Promoter
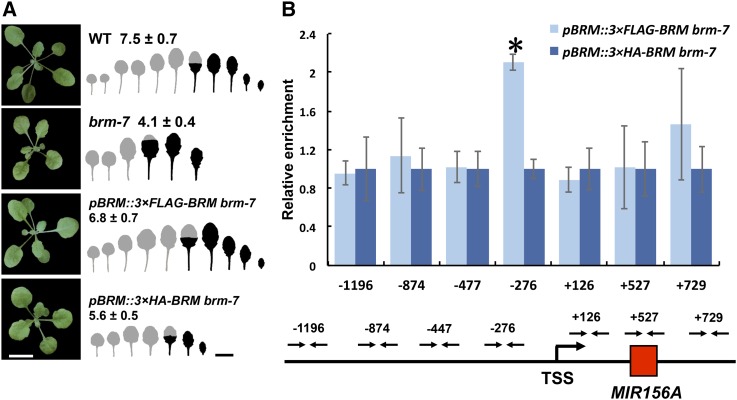
BRM has been shown to regulate gene expression by directly binding to the promoter or coding regions of its targets (Han et al., 2012; Wu et al., 2012; Li et al., 2015). To test if BRM also regulates miR156 expression by directly binding to miR156, we generated 3×FLAG and 3×HA-tagged versions of BRM (pBRM::3×FLAG-BRM and pBRM::3×HA-BRM) constructs under the control of the BRM native promoter and transformed into the brm-7 mutant background. Either construct could fully or partially rescue the brm-7 mutant phenotype (Fig. 4A), implying that both epitope-tagged proteins were biologically functional in transgenic plants. Thirty homozygous lines with single T-DNA insertions of pBRM::3×FLAG-BRM or pBRM::3×HA-BRM were isolated in the progeny by selecting plants on 0.5× Murashige and Skoog medium containing 30 mg/L hygromycin. Semiquantitative RT-PCR analysis of total RNA from pBRM::3×FLAG-BRM and pBRM::3×HA-BRM showed that both 3×FLAG-BRM and 3×HA-BRM were expressed in the rescued transgenic plants (Supplemental Fig. S3A). Western blotting using a polyclonal anti-FLAG antibody detected a specific band at about the 245-kD position, similar to the size of full-length 3×FLAG-BRM fusion protein (Supplemental Fig. S3B), implying that the 3×FLAG-BRM protein was normally expressed in the transgenic plants. ChIP was carried out using a polyclonal anti-FLAG antibody for homozygous pBRM::3×FLAG-BRM plants, while homozygous pBRM::3×HA-BRM plants were served as a negative control. We detected binding of BRM to the −276-bp region upstream of the TSS in the MIR156A promoter (Fig. 4B), strongly suggesting that BRM also regulates miR156 expression by directly binding to the promoter region of MIR156A. To see if the down-regulation of miR156 in brm-7 was also attributable to the effect of brm-7 on the expression of FUS3, AGL15/18, and BMI1, we measured the expression of these genes in brm-7 and wild type. qRT-PCR results indicated that these genes were not obviously affected by brm-7 (Supplemental Fig. S4), excluding the possibility of the indirect effect of miR156 expression by brm-7.
Figure 4.
BRM binds to the MIR156A promoter region directly. A, The phenotype of wild type, brm-7, and brm-7 transformed with pBRM::3×FLAG-BRM and pBRM::3×HA-BRM in short days. Numbers indicate the first leaf with abaxial trichomes (n = 32, ±sd). Juvenile leaves are shown in gray and adult leaves in black in the heteroblastic analyses. Scale bar = 1 cm. B, Direct binding of BRM to the MIR156A promoter region shown by ChIP analysis. Red rectangle denotes the MIR156A stem loop structure region. TSS, TSS of MIR156A. Arrows denotes the promoter regions detected by ChIP. Asterisk denotes statistical significance at P < 0.01. ChIP results were averaged from three biological replicates with three technical replicates for each sample.
BRM Contributes to Proximal Nucleosome Occupancy but Not Positioning at the MIR156A Promoter Region
SWI/SNF remodeling complexes guide nucleosome movement via ATP-dependent alterations in histone-DNA interactions (Peterson and Workman, 2000). Nucleosome occupancy typically decreases upstream of transcriptionally induced genes and increases in regulatory regions of repressed genes. To see if mutations in BRM cause changes in nucleosome positioning or occupancy at the proximal regions at the MIR156A locus, we examined nucleosome positioning and occupancy at the MIR156A locus using the MNase mapping method (Chodavarapu et al., 2010; Rafati et al., 2011). We identified two nucleosomes (−2 and −1 nucleosomes) upstream and one nucleosome (+1 nucleosome) downstream of the MIR156A TSS both in wild type and brm-7 (Fig. 5; Supplemental Fig. S5). A significant difference in occupancy at the −2 and +1 nucleosomes was detected between wild type and brm-7 (Fig. 5; Supplemental Fig. S5), whereas no BRM-dependent shifting of nucleosome position was detected among these three nucleosomes. This result implies that BRM functions to lower occupancy at the −2 and +1 nucleosomes to activate MIR156A transcription, which is consistent with the qRT-PCR result that pri-MIR156A transcript was reduced in brm-7 (Fig. 2C).
Figure 5.
BRM is required to maintain low occupancy of the −2 and +1 nucleosome at the MIR156A locus. qPCR was used to detect the nucleosome positioning and occupancy at the MIR156A locus using MNase-digested DNA from 10-d-old wild-type and brm-7 plants in short days. The fraction of digested mononucleosome DNA amplified for each amplicon was normalized to that of the −73 position of the negative control locus of gypsy-like retrotransposon. Values represent mean ± sd from three technical replicates in a representative experiment. The number on the x axis denotes distance (bp) from the TSS (0 bp) of MIR156A. A diagram of the positioned nucleosomes is shown below the x axis. Blue ovals, nucleosomes; black arrow, TSS; black lines, genomic DNA; red rectangle, MIR156A stem loop structure region. Two figure insets show the detailed increase in occupancy at the −2 and +1 nucleosomes in brm-7 compared with wild type. Asterisk denotes statistical significance from wild type at P < 0.01.
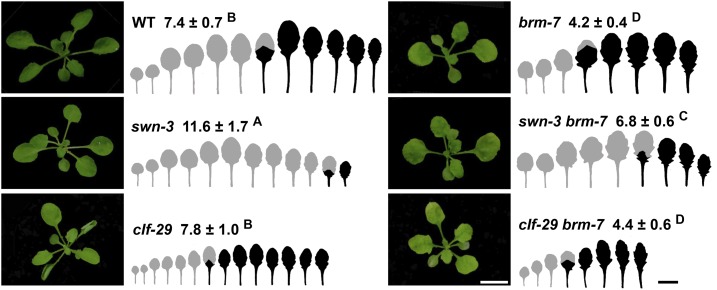
SWN, but Not CLF, Antagonizes BRM to Promote Vegetative Phase Change in Arabidopsis
BRM was initially identified as a Trithorax (TrxG) protein to antagonize the function of PcG proteins in fly development (Tamkun et al., 1992). Loss-of-function mutations in BRM accelerated vegetative phase change, and we expect that loss-of-function mutations in PcG proteins might exhibit some opposite vegetative phase change phenotypes. To see if PcG proteins play a role in vegetative phase change, we did phenotypic characterization of two null alleles of SWN and CLF, swn-3 and clf-29, both of which encode two important components of PcG complex with H3K27 methyltransferase activity during vegetative and reproductive development (Chanvivattana et al., 2004). In contrast to wild type with abaxial trichomes on leaf 7, swn-3 mutant exhibited significantly delayed vegetative phase change phenotypes with rounder leaves and later abaxial trichomes on leaf 11; clf-29, however, had similar trichome production and leaf shape to wild type (Fig. 6). The swn-3 brm-7 double mutant had abaxial trichomes on leaf 6 compared with brm-7 that had early abaxial trichomes on leaf 4; swn-3, but not clf-29, partially rescued the phenotype of brm-7 (Fig. 6). These genetic results imply that SWN, but not CLF, functions more importantly to promote vegetative phase change in Arabidopsis. Even though clf-29 did not exhibit a vegetative phase change phenotype and clf-29 brm-7 resembled brm-7 phenotype, we cannot rule out the possibility that CLF acts redundantly with SWN in regulating vegetative phase change. However, it is difficult to test this possibility in that clf and swn double mutant de-differentiated into callus in the early seedling stage (Chanvivattana et al., 2004; Aichinger et al., 2009; Xu et al., 2016), making it impossible to characterize the vegetative phase change phenotype of clf and swn double mutant.
Figure 6.
SWN, but not CLF, antagonizes BRM to promote vegetative phase change. Plants were grown in short days. The first leaf with abaxial trichomes was scored. Numbers indicate the first leaf with abaxial trichomes (n = 18, ±sd). Juvenile leaves are shown in gray and adult leaves in black in the heteroblastic analyses. Different letters indicate significant difference between genotypes using one-way ANOVA at P < 0.01. Scale bar = 1 cm.
SWN Integrates the miR156-SPLs Pathway to Regulate Vegetative Phase Change
To learn if SWN functions to promote vegetative phase change by integration into the miR156-SPLs pathway, we determined the level of miR156 in wild type, brm-7, swn-3, and swn-3 brm-7. qRT-PCR reproducibly showed that miR156 was significantly elevated in swn-3, and swn-3 brm-7 had an intermediate level of miR156 between brm-7 and swn-3 (Fig. 7A), consistent with the result that swn-3 partially rescued the brm-7 mutant phenotype (Fig. 6). To see if swn-3 affects the level of miR156 by affecting the level of primary miR156 transcripts, we determined the levels of pri-MIR156A and pri-MIR156C transcripts in swn-3 and wild type. We reproducibly saw a significant increase in the level of pri-MIR156A, whereas the level of pri-MIR156C transcript remained unchanged in swn-3 compared with that in wild type (Fig. 7, B and C). As expected, the levels of SPL3, SPL9, and SPL13 transcripts in swn-3 were significantly reduced (Fig. 7D). All these data implied that SWN functions by integration into the miR156-SPLs pathway to regulate vegetative phase change in Arabidopsis.
Figure 7.
swn-3 delays vegetative phase change by modulating the expression of genes in the miR156-SPLs pathway. A, The level of mature miR156 in 14-d-old wild-type, brm-7, swn-3, and swn-3 brm-7 seedlings in short days. B, The level of pri-MIR156A in 14-d-old swn-3 seedlings in short days. C, The level of pri-MIR156C in 14-d-old swn-3 seedlings in short days. D, The level of SPL3, SPL9, and SPL13 in 14-d-old swn-3 seedlings in short days. Asterisk denotes statistical significance from wild type at P < 0.01 using Student’s t test.
BRM and SWN Function to Regulate the Level of H3K27me3 at the MIR156A Locus
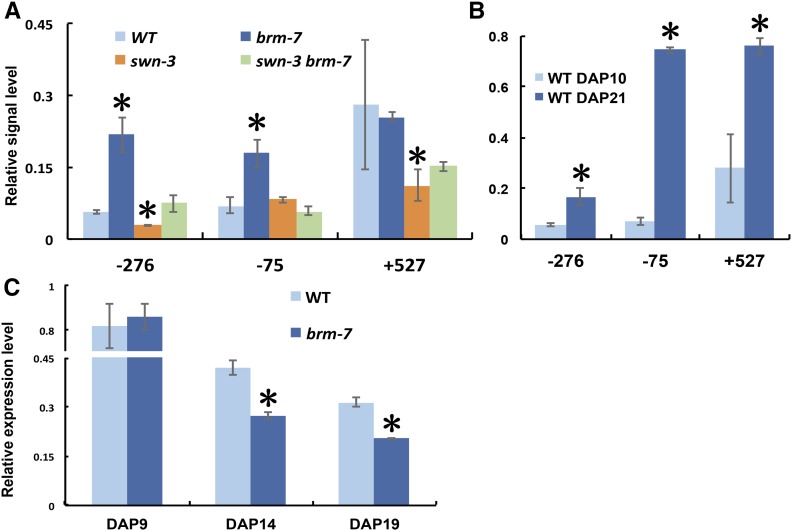
The fact that SWN and CLF function as H3K27me3 methyltransferases and that BRM antagonizes PcG function during fly and plant development (Tamkun et al., 1992; Wu et al., 2012; Li et al., 2015) prompted us to ask if BRM and SWN function to regulate the level of H3K27me3 at the MIR156A locus. We performed ChIP using an anti-H3K27me3 antibody in wild-type, brm-7, swn-3, and swn-3 brm-7 mutant plants. Levels of H3K27me3 were elevated in brm-7 at −276 bp and −75 bp upstream of MIR156A TSS (Fig. 8A), while swn-3 reduced levels of H3K27me3 at −276 bp upstream and +527 bp downstream of MIR156A TSS and ultimately resulted in intermediate levels of H3K27me3 in the swn-3 brm-7 mutant (Fig. 8A). This result demonstrated that H3K27me3 plays an important role in regulating miR156 expression. miR156 expression declines when plants age (Wu and Poethig, 2006), and whether this temporal expression pattern of miR156 is attributable to changes in the level of H3K27me3 during development remains unknown. To further explore the possibility that H3K27me3 may contribute to the temporal expression pattern of miR156, we harvested 10-d-old juvenile and 21-d-old adult wild-type seedlings to examine the levels of H3K27me3 at the MIR156A locus. ChIP revealed that the H3K27me3 level was significantly higher at all three regions examined in 21-d-old adult wild-type seedlings than in 10-d-old juvenile wild-type seedlings (Fig. 8B), implying that H3K27me3 may be responsible for shaping the temporal expression of miR156 when plants age. However, we saw similar temporal expression patterns of miR156 between wild type and the brm-7 mutant (Fig. 8C), indicating that BRM and H3K27me3 were not required for the temporal expression pattern of miR156 but for maintaining a relatively stable abundance of miR156 during vegetative phase change.
Figure 8.
The level of H3K27me3 at MIR156A and temporal expression of miR156 in wild type and brm-7. A, The level of H3K27me3 in the promoter and coding region of MIR156A in 14-d-old wild-type, brm-7, swn-3, and swn-3 brm-7 seedlings in short days. Values represent mean ± sd from two technical replicates in a representative experiment. B, The level of H3K27me3 in 10-d-old wild-type juvenile plants and 21-d-old wild adult plants in short days. Values represent mean ± sd from two technical replicates in a representative experiment. C, Temporal expression pattern of miR156 in wild type and brm-7 in short days. Wild-type and brm-7 plants were harvested for qRT-PCR analysis of mature miR156 levels at different time points. Asterisk denotes statistical significance from wild type at P < 0.01 using Student’s t test.
Interplay between H3K27me3 and Nucleosome Occupancy Regulates Vegetative Phase Change in Arabidopsis
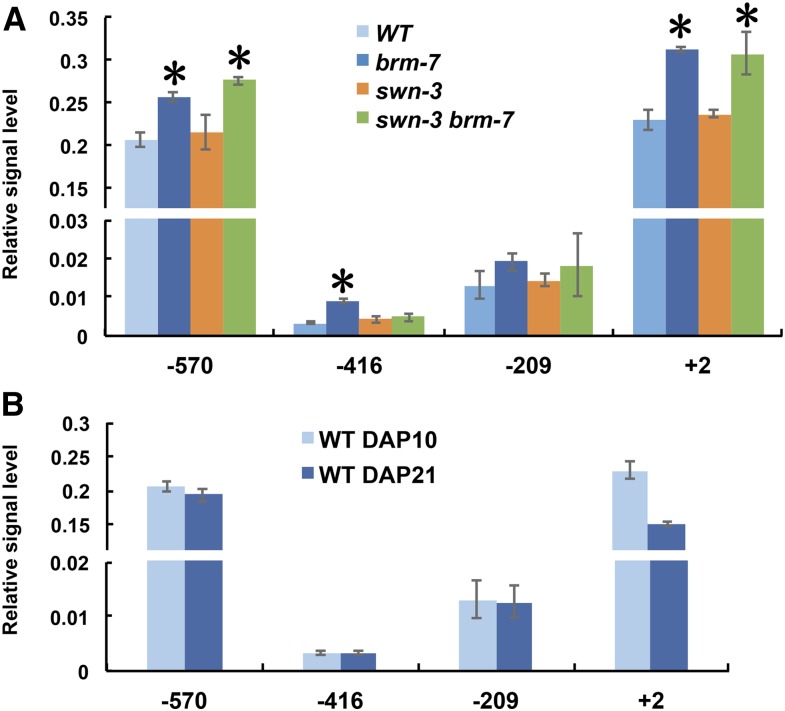
The phenotype of swn-3 brm-7 double mutant led us to ask the underlying cause for the intermediate phenotype (Fig. 6). We examined nucleosome occupancy in the promoter region of MIR156A and found that the swn-3 brm-7 double mutant had a similar level of nucleosome occupancy to the brm-7 single mutant for most regions examined except for the −416 region (Fig. 9A); furthermore, introduction of the swn-3 mutation to brm-7 gave rise to almost wild-type abaxial trichome production (Fig. 6). These results suggest that SWN-mediated H3K27me3 is partially responsible for the brm-7 vegetative phase change phenotype. As miR156 expression declines gradually during development, we asked if this temporal expression pattern is also related to changes in nucleosome occupancy during vegetative phase change. We examined nucleosome occupancy in 10-d-old juvenile and 21-d-old adult wild-type seedlings. No significant difference in nucleosome occupancy was detected between these two samples (Fig. 9B), together with the result that juvenile plants had lower levels of H3K27me3 than adult plants (Fig. 8B), implying that the gradual increase in the level of H3K27me3, but not changes in nucleosome occupancy, contributed to maintaining a relative stable abundance of miR156 during vegetative phase change in Arabidopsis. However, when we compared the phenotype of the swn-3 brm-7 double mutant to that of swn-3, the brm-7 mutation partially rescued the swn-3 mutant phenotype (Fig. 6) and the swn-3 brm-7 double mutant had higher levels of nucleosome occupancy at almost all regions of the MIR156A promoter than swn-3 (Fig. 9A), consistent with the reduced level of miR156 in the double mutant (Fig. 7A), implying that changes in nucleosome occupancy in the MIR156A promoter region in brm-7 also contributed to the swn-3 brm-7 mutant phenotype. We also noticed that the level of H3K27me3 at −75 upstream of TSS of MIR156A between wild type and swn-3 was similar, but not between wild type and brm-7 (Fig. 8A); likewise the level of H3K27me3 at +527 was similar between wild type and brm-7, but not between wild type and swn-3 (Fig. 8A), implying that BRM and SWN might function in an uncoupled way at regions away from the BRM-binding site in the MIR156A promoter. Therefore, interplay and coordination between H3K27me3 and nucleosome occupancy in the MIR156A promoter region regulate the transcription of miR156 to control vegetative phase change in Arabidopsis.
Figure 9.
Nucleosome occupancy at the MIR156A locus in different mutants. A, Nucleosomes occupancy at the MIR156A locus in 10-d-old wild-type, brm-7, swn-3, and swn-3 brm-7 plants in short days. B, Nucleosome occupancy at the MIR156A locus in juvenile and adult wild-type plants in short days. qPCR was used to detect the nucleosome positioning and occupancy at the MIR156A locus using MNase-digested DNA from wild-type, brm-7, swn-3, and swn-3 brm-7 plants (A) or from 10-d-old juvenile wild-type plants and 21-d-old adult wild-type plants (B). The fraction of digested mononucleosome DNA amplified for each amplicon was normalized to that of the −73 position of the negative control locus of gypsy-like retrotransposon. Values represent mean ± sd from three technical replicates in a representative experiment. Asterisk denotes the statistical significance from wild type at P < 0.01 using Student’s t test.
DISCUSSION
Both BRM and SWN Play an Important Regulatory Role in Vegetative Phase Change in Arabidopsis
BRM functions as a TrxG protein in plant development, and loss-of-function mutations in BRM caused pleiotropic phenotypes in Arabidopsis (Farrona et al., 2004, 2011; Hurtado et al., 2006; Tang et al., 2008; Wu et al., 2012; Li et al., 2015). SWN constitutes an important component of PRC2 whose role in vegetative phase change, however, has not been well described until recently (Xu et al., 2016). In this study, we discovered a role that BRM and SWN play to regulate vegetative phase change by modulating the expression of miR156 in Arabidopsis. Loss-of-function mutations in BRM accelerated vegetative phase change, while loss-of-function mutations in SWN delayed vegetative phase change in Arabidopsis. Therefore, BRM functions to delay vegetative phase change, whereas SWN functions to promote vegetative phase change in Arabidopsis. Genetics and molecular analyses demonstrated that both BRM and SWN function in the regulation of vegetative phase change by integration into the miR156-SPLs pathway. In this study, we demonstrated that BRM directly binds to the promoter of the MIR156A region to regulate its expression. A previous report showed that CLF and SWN function redundantly as histone methyltransferases in plant development (Chanvivattana et al., 2004). In our study, swn-3 delayed vegetative phase change, while clf-29 did not show any obvious vegetative phase change phenotypes, implying that SWN and CLF have diverse roles in vegetative phase change. The question of whether these two proteins act redundantly in regulating vegetative phase change still awaits further investigation.
Regulation of miR156 Expression by BRM and SWN
miR156 has been shown to be the master regulator of vegetative phase change in plants (Wu and Poethig, 2006; Poethig, 2009; Wu et al., 2009; Poethig, 2013), and it also controls a plethora of other different processes vital for plant growth and development (Jiao et al., 2010; Gou et al., 2011; Zhang et al., 2011; Fu et al., 2012; Aung et al., 2015; Bhogale et al., 2014; Ferreira e Silva et al., 2014; Stief et al., 2014; Wang et al., 2015). However, the question of how miR156 expression is regulated remains elusive. Recent work showed that FUS3, AGL15, and AGL18 regulate miR156 expression by directly binding to the promoter regions of miR156A and miR156C loci (Wang and Perry, 2013; Serivichyaswat et al., 2015), but the function of the direct binding remains unexplored. miR156 was also shown to be regulated by AtBMI1, a PRC1 component (Picó et al., 2015), and PKL, a CHD3 chromatin remodeler (Xu et al., 2016), to repress miR156 expression through modification of H3K27me3 marks at the miR156A/C loci. In this study, we demonstrated that MIR156A is a direct target of BRM, which promotes miR156 expression by directly binding to the promoter of the MIR156A locus. This direct binding confers functionality by promoting the expression of miR156 to delay vegetative phase change in Arabidopsis. BRM was shown to either activate or repress target gene expression by increasing or decreasing the accessibility of the target DNA to transcription factors or transcription regulators (Tang et al., 2008; Hargreaves and Crabtree, 2011; Han et al., 2012). In this study, BRM acted as an activator of miR156 expression. The brm-7 mutant had more elevated levels of occupancy than wild type at the −2 and +1 nucleosomes proximal to the TSS of MIR156A to repress its transcription. A previous study showed that SWI/SNF was largely depleted at the −1 nucleosome but enriched at the −2 nucleosome (Yen et al., 2012). This is consistent with our result of changes in occupancy at the −2 nucleosome, but not the −1 nucleosome upstream of the MIR156A promoter in this study (Fig. 5). The +1 nucleosome is a gateway to transcription, and therefore it is a preferable target of chromatin remodelers; the closer the +1 nucleosome to the TSS, the more repressive transcription would be (Yen et al., 2012). However, we did not detect any shifting of the +1 nucleosome proximal to the TSS of MIR156A; instead we saw an elevated level of +1 nucleosome occupancy in this region. Therefore, we favor a model that BRM normally functions to lower occupancy of the +1 nucleosome at the MIR156A locus to activate the expression of miR156. When BRM is absent, SWN functions as a histone methyltransferase to repress miR156 expression by establishing a repressive mark of H3K27me3 at the MIR156A promoter (Fig. 8A). Meanwhile, higher nucleosome occupancy also contributes to the repression of miR156 in this background. The level of miR156 declines, while H3K27me3 level at the MIR156A locus increases during vegetative phase change. This inverse pattern implies that H3K27me3 might be responsible for the temporal change in the level of miR156 during vegetative phase change, but this hypothesis was negated by the fact that the temporal expression pattern of miR156 was not affected in the brm-7 background with elevated levels of H3K27me3 at the MIR156A locus (Fig. 8, A and C). Therefore, the level of H3K27me3 at the MIR156A locus was important for maintaining the miR156 level during vegetative phase change but not essential for the temporal expression of miR156, and disruption of this relative stable abundance of miR156 contributes to defects in vegetative phase change manifested by brm-7 and swn-3 mutants.
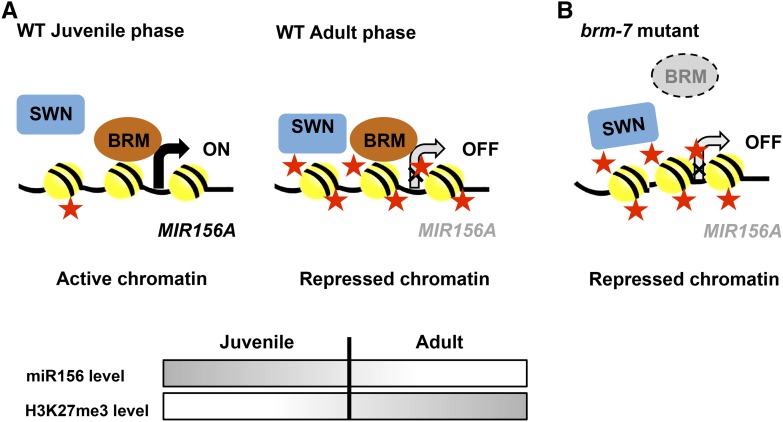
A Model for the Antagonistic Interaction between BRM and SWN during Vegetative Phase Change
The evidence that occupancy at the −2 and +1 nucleosome and levels of H3K27me3 were elevated in the promoter region of MIR156A in brm-7 suggested that both noncovalent and covalent histone modifications are the underlying mechanism involved in the regulation of miR156 expression in this background. Loss-of-function mutations in BRM were recently shown to cause increased deposition of H3K27me3 at several hundred genes, and this increase was partially suppressed by mutations in the histone methyltransferase CLF and SWN (Li et al., 2015). ChIP results showed that SWN occupancy was increased at the majority of BRM target sites when BRM is absent, resulting in elevated levels of H3K27me3 in brm mutants (Li et al., 2015). In our study, the brm-7 mutation caused an elevated level of H3K27me3, while the swn-3 mutation resulted in a reduced level of H3K27me3; brm-7 swn-3, however, had an intermediate level of H3K27me3 at the −276 region (Fig. 8A), implying that BRM and SWN function antagonistically in this region to regulate the level of H3K27me3 in the promoter region of MIR156A to fine tune the expression of miR156. However, the levels of H3K27me3 at the −75 and +527 regions were not complementary between brm-7 and swn-3 (Fig. 8A), implying that BRM and SWN do not always work antagonistically at some specific regions. However, the questions as to whether SWN directly binds to the promoter region of MIR156A and the increased H3K27me3 level in the promoter region of MIR156A in brm-7 is due to the increased SWN occupancy in this region remain to be investigated. Based on all results, we proposed a model for the antagonistic interaction between BRM and SWN in the regulation of vegetative phase change in Arabidopsis (Fig. 10). In wild-type plants, BRM and SWN function normally but antagonistically to balance the level of miR156; the level of H3K27me3 increases as plants age to maintain a relatively stable level of miR156 during vegetative phase change (Fig. 10A). When BRM is absent, SWN functions to increase H3K27me3 levels at the miR156 regulatory regions to down-regulate miR156 (Fig. 10B); in the meantime, the brm mutation also represses miR156 expression by increasing occupancy at the −2 and +1 nucleosomes close to the TSS at the MIR156A locus. This combinatorial mode of action reduces the miR156 level to accelerate vegetative phase change; when SWN is absent, levels of H3K27me3 are decreased, but BRM functions normally and reduced levels of H3K27me3 promote miR156 expression to delay vegetative phase change. When both SWN and BRM are absent, lower levels of H3K27me3 caused by swn promote miR156 expression, while higher levels of occupancy at the −2 and +1 nucleosomes in the brm-7 background repress miR156 expression. Therefore, the level of miR156 was maintained at an intermediate level between swn and brm to give rise to an intermediate phenotype. This is also consistent with our gene expression analysis (Fig. 7A) and phenotypic characterization of the swn-3 brm-7 mutant (Fig. 6).
Figure 10.
A model for the function of BRM and SWN in the regulation of MIR156A expression during vegetative phase change in Arabidopsis. A, Regulation of MIR156A by BRM and SWN during vegetative phase change in wild-type plants. At the juvenile phase, BRM binds to the MIR156A promoter to antagonize SWN function to reduce the level of H3K27me3 at the MIR156A locus as well as to reduce nucleosome occupancy to activate MIR156A expression. When plants enter the adult phase, BRM still functions normally, but SWN overtakes the function of BRM to increase the level of H3K27me3 at the MIR156A locus to repress MIR156A expression. Red star: H3K27me3 modification. Number of red stars denotes the degree of H3K27me3. Temporal changes in the level of miR156 and H3K27me3 were illustrated by the shaded bars during the juvenile-to-adult development. B, Regulation of MIR156A by BRM and SWN during vegetative phase change in the brm mutant plants. When BRM is mutated, SWN takes over BRM function to increase the level of H3K27me3, and the brm mutation increases nucleosome occupancy at the MIR156A locus to repress MIR156A expression.
Chromatin remodelers themselves do not have DNA binding ability. They are recruited to their target sites by interacting with specific transcription factors (cofactors; Bezhani et al., 2007; Wu et al., 2012). ChIP experiment showed that BRM physically binds to the −267-bp region upstream of the TSS in MIR156A promoter (Fig. 4B), implying that this region is critical for the direct binding of cofactors to recruit BRM to regulate the expression of miR156A. Identification of these cofactors will greatly facilitate our understanding of the regulation of miR156 in Arabidopsis in the future.
MATERIALS AND METHODS
Genetic Stocks, Genotyping, and Growth Conditions
All of the genetic stocks used in this study were in a Col genetic background. spl9-4 (CS807258), swn-3 (SALK_050195), and clf-29 (SALK_021003) were obtained from the Arabidopsis Biological Resource Center. pMIR156A::GUS and 35S::miR156A transgenic lines are the same seed stocks as described previously (Wu and Poethig, 2006; Yang et al., 2013). A specific DNA sequence encompassing the brm-7 mutation was PCR-amplified using a pair of dCAPS primers. The amplified DNA was then digested with HaeIII. DNA amplified from brm-7 was resistant to HaeIII cleavage, whereas that of wild type was susceptible to HaeIII cleavage. All T-DNA lines including spl9-4, swn-3, and clf-29 were genotyped using combinations of a T-DNA-specific LBb1.3/LB1 primer and gene-specific primers RP and LP. All genotyping results are summarized in Supplemental Figure S6. Seeds were grown in a mixture of soil and vermiculite (1:1) and left at 4°C for 2 d before transfer to the growth chamber. Plants were grown under short-day conditions (10 h light:14 h dark, 140 µmol/m2/s) at 22°C.
Gene Expression Analyses
For gene expression analyses, 9-, 14-, and 19-d-old aerial parts of whole seedlings were collected and stored at −80°C until use. Total RNA was extracted using TRIzol reagent (Invitrogen) and digested by RNase-Free DNase (Promega). Reverse transcription was done using PrimerScript II 1st Strand cDNA Synthesis Kit (TaKaRa). For the synthesis of the first cDNA strand of miRNA, the reverse transcription primers were designed as previously reported (Varkonyi-Gasic et al., 2007). Reverse transcription was done as follows: one cycle at 16°C for 30 min, 60 cycles at 30°C for 30 s, 42°C for 30 s, and 50°C for1 s, followed by incubation at 85°C for 5 min to inactivate the reverse transcriptase. Real-time PCR was performed using diluted cDNA on a Step One Plus (ABI) real time PCR machine. TUB2 and AtsnoR101 served as the control for mRNAs and miRNAs analyses, respectively. The sequences of primers used are listed in Supplemental Table S1.
GUS Staining
For GUS staining, 11-d-old plants as well as 10- and 13-d-old plants were collected and pretreated with 90% acetone on ice for 20 min. After washing three times with 100 mm phosphate buffer (pH 7.0), plants were submerged in X-Gluc solution and vacuumed twice and then incubated at 37°C for 3 h. Stained plants were decolorized in 70% ethanol and photographed.
Generation of Transgenic Lines
The whole intergenic region containing SPL9 as well as rSPL9 (insensitive to miR156 regulation) genomic sequence was PCR-amplified using primers as described previously (Wu et al., 2009). The PCR fragment was then fused in-frame to the eGFP sequence using overlapping PCR, and the fused pSPL9::eGFP-SPL9, pSPL9::eGFP-rSPL9 sequences were then cloned into pCAMBIA3300 for genetic transformation. To generate pBRM::3×FLAG-BRM and pBRM::3×HA-BRM constructs, the whole intergenic region of BRM was PCR-amplified; three copies of FLAG or HA epitope tags were fused in-frame to the N terminus of the BRM protein, and the fused sequences were then cloned into pCAMBIA1300. The primer sequences are listed in Supplemental Table S1. All constructs were transformed into the Agrobacterium tumefaciens GV3101 strain, and transformation was done using the floral dipping method. Transformants of pSPL9::eGFP-SPL9, pSPL9::eGFP-rSPL9 were selected in soil by spraying BASTA, and transformants of pBRM::3×FLAG-BRM and pBRM::3×HA-BRM were selected on 0.5× Murashige and Skoog medium containing 30 mg/L hygromycin. Lines containing single insertions were obtained on the basis of the segregation ratio of the resistant or susceptible plants to BASTA or hygromycin in the selfed progeny of these primary transformants, and homozygous stocks were established from these lines.
Nucleosome Occupancy Assay
A total of 1.0 g 10- and 21-d-old aerial parts of seedlings of wild type and brm-7 was harvested on ice and cross-linked with 1% formaldehyde for 15 min and then treated in 0.125 m Gly for 5 min. Samples were ground into fine power with liquid nitrogen, and the chromatins were isolated as previously described (Zhu et al., 2012). Chromatin was resuspended with 1× digestion buffer and digested with 0.05 unit/µL (final concentration) Micrococcal Nuclease (Takara) for 10 min at 37°C. The digestion was stopped with 20 mm EDTA, and reverse cross-linking was carried out in 0.3 m NaCl at 65°C overnight. Mononucleosome fragments were separated on 1.5% agarose gels and purified by a gel purification kit. The purified DNAs were used for qPCR analysis of nucleosome occupancy. The gypsy-like retrotransposon (At4g07700) −73 loci served as the control, and the data were calculated with 2−ΔCt (= 2−Ct(target)-Ct(control)). Primers used for nucleosome occupancy detection are listed in Supplemental Table S1.
ChIP Assays
ChIP was carried out with the same samples as nucleosome occupancy detection using the method as described previously (Zhu et al., 2012) with minor modifications. The chromatin DNA was sonicated into about 200- to 1,000-bp DNA fragments on ice, and the sonicated chromatin was then immunoprecipitated with 5 µL anti-FLAG polyclonal antibody (Sigma, F7425), or anti-H3K27me3 (Millipore 07-449) or anti-H3 antibodies (Abcam 1791), and then the precipitated DNAs were purified using a PCR Purification Kit (Sangon). To detect the BRM binding region, the exon region of retrotransposon TA3 was used as the internal control, and the data were calculated with 2−ΔCt (= 2−Ct(ChIP)-Ct(input)). For H3K27me3 modification detection, the SHOOT MERISTEMLESS (STM) gene was used as the internal control, and the data were calculated as the ratio of H3K27me3 target gene 2−ΔCt (= 2−Ct(H3K27me3 target gene-Ct(input H3 target gene)) to H3K27me3 STM 2−ΔCt (= 2−(Ct(H3K27me3 STM)-Ct(H3 STM)). Primer sequences used for ChIP-qPCR detection are listed in Supplemental Table S1.
Western Blot
The aerial parts of 2-week-old seedlings in short day conditions were collected and ground to isolate total protein. Equal amounts of protein samples (about 20 µg) were loaded onto an SDS-PAGE gel. After electrophoresis, protein in the gel was electrotransferred to a nitrocellulose membrane, stained with Ponceau, then incubated with an anti-FLAG antibody and subsequently with the secondary antibody. The signal was detected with an ECL kit (Beyo).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A genetic complementation test confirmed the precocious vegetative phase change phenotype was caused by the brm-7 mutation.
Supplemental Figure S2. GUS staining of pMIR156A::GUS and brm-7 pMIR156A::GUS reporter lines at different developmental stages in short-day conditions.
Supplemental Figure S3. Expression of FLAG-BRM and HA-BRM in brm-7 plants transformed with pBRM::3×FLAG-BRM and pBRM::3×HA-BRM constructs.
Supplemental Figure S4. Expression of BMI1, AGL15, AGL18, and FUS3 genes in 14-d-old wild-type and brm-7 plants in short days.
Supplemental Figure S5. The second biological replicate for nucleosome occupancy in wild type and brm-7 as shown in Figure 5.
Supplemental Figure S6. The genotyping results of different mutant plants.
Supplemental Table S1. Primers used in this study (from 5′ to 3′, F: forward, R: reverse).
Acknowledgments
We thank members of the Wu laboratory for useful comments on this manuscript.
Glossary
- ChIP
chromatin immunoprecipitation
- qRT-PCR
quantitative reverse transcription-PCR
- TSS
transcription start site
Footnotes
This work was supported by the National Natural Science Foundation of China (31271313 and 31470063), Zhejiang Natural Science Foundation (LR13C060001), and a grant for Innovative Research Team of Zhejiang Province of China (2013TD05) to G.W. This work was also partially supported by a start-up fund from Zhejiang Agriculture and Forestry University (2012FR025) and the 1000 Youth Talents Program in China (2034020065) to G.W. B. Zhou was supported by an Undergraduate Innovation Incubator Project from China (2015R412042). S.P. was supported by a grant from the National Institutes of Health (NIH GM051893).
These authors contributed equally to the article.
References
- Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Köhler C (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung B, Gruber MY, Amyot L, Omari K, Bertrand A, Hannoufa A (2015) MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol J 13: 779–790 [DOI] [PubMed] [Google Scholar]
- Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, Pfluger J, Su Y, Wagner D (2007) Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogale S, Mahajan AS, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee AK (2014) MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol 164: 1011–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, Bowman JL, Reyes JC (2004) The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975 [DOI] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, March-Díaz R, Schmitz RJ, Florencio FJ, Turck F, Amasino RM, Reyes JC (2011) Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS One 6: e17997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira e Silva GF, Silva EM, Azevedo MdaS, Guivin MA, Ramiro DA, Figueiredo CR, Carrer H, Peres LE, Nogueira FT (2014) microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J 78: 604–618 [DOI] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DG, Stewart CN Jr, Tang Y, et al. (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Sang Y, Rodrigues A, Wu MF, Rodriguez PL, Wagner D; BIOL425 F2010, Wu MF, Rodriguez PL, Wagner D (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24: 4892–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR (2011) ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21: 396–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L-C, Lin S-I, Shih AC-C, Chen J-W, Lin W-Y, Tseng C-Y, Li W-H, Chiou T-J (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Hurtado L, Farrona S, Reyes JC (2006) The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol 62: 291–304 [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A. (2007) SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta 1769: 330–345 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, et al. (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42: 541–544 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Poethig RS (1998) The specification of leaf identity during shoot development. Annu Rev Cell Dev Biol 14: 373–398 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Wagner D (2007) Unwinding chromatin for development and growth: a few genes at a time. Trends Genet 23: 403–412 [DOI] [PubMed] [Google Scholar]
- Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, Carrington JC, Ahn JH (2010) Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res 38: 3081–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li C, Chen C, Gao L, Yang S, Nguyen V, Shi X, Siminovitch K, Kohalmi SE, Huang S, Wu K, et al. (2015) The Arabidopsis SWI2/SNF2 chromatin remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP. PLoS Genet 11: e1004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q, Chien CW, Wang S, Jiang L, Ai LF, et al. (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May P, Liao W, Wu Y, Shuai B, McCombie WR, Zhang MQ, Liu QA (2013) The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat Commun 4: 2145. [DOI] [PubMed] [Google Scholar]
- Moss EG. (2007) Heterochronic genes and the nature of developmental time. Curr Biol 17: R425–R434 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev 10: 187–192 [DOI] [PubMed] [Google Scholar]
- Picó S, Ortiz-Marchena MI, Merini W, Calonje M (2015) Deciphering the role of Polycomb Repressive Complex 1 (PRC1) variants in regulating the acquisition of flowering competence in Arabidopsis. Plant Physiol 168: 1286–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. (1990) Phase change and the regulation of shoot morphogenesis in plants. Science 250: 923–930 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2003) Phase change and the regulation of developmental timing in plants. Science 301: 334–336 [DOI] [PubMed] [Google Scholar]
- Poethig RS. (2009) Small RNAs and developmental timing in plants. Curr Opin Genet Dev 19: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. (2013) Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol 105: 125–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T (2011) Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol 9: e1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Sang Y, Silva-Ortega CO, Wu S, Yamaguchi N, Wu MF, Pfluger J, Gillmor CS, Gallagher KL, Wagner D (2012) Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J 72: 1000–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Clarenz O, Goodrich J (2005) Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 8: 553–561 [DOI] [PubMed] [Google Scholar]
- Serivichyaswat P, Ryu HS, Kim W, Kim S, Chung KS, Kim JJ, Ahn JH (2015) Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol Cells 38: 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26: 1792–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA (1992) brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561–572 [DOI] [PubMed] [Google Scholar]
- Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, Reyes JC, Wang A, Keller WA, Harada JJ, et al. (2008) The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol 147: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Shoda K, Kim GT, Uchimiya H (2000) Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta 210: 536–542 [DOI] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Perry SE (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol 161: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Z, Amyot L, Tian L, Xu Z, Gruber MY, Hannoufa A (2015) Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol Genet Genomics 290: 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D (2012) SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Smith MR, Poethig RS (2016) Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28: 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu M, Koo Y, He J, Poethig RS (2013) Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF (2012) Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu J, Huang J, Wang G, Wang JW (2013) Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, Prins M, van Tunen A, He Y (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z (2011) Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett 585: 435–439 [DOI] [PubMed] [Google Scholar]
- Zhu J-Y, Sun Y, Wang Z-Y (2012) Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). In Wang Z-Y, Yang Z, eds, Plant Signalling Networks, Vol 876. Humana Press, New York, pp 173–188 [DOI] [PubMed] [Google Scholar]