Abstract
Histones are a group of proteins with a high number of post-translational modifications, including methylation, acetylation, phosphorylation, and monoubiquitination, which play critical roles in every chromatin-templated activity. The quantitative analysis of these modifications using mass spectrometry (MS) has seen significant improvements over the last decade. It is now possible to perform large-scale surveys of dozens of histone marks and hundreds of their combinations on global chromatin. Here, we review the development of three MS strategies for analyzing histone modifications that have come to be known as Bottom Up, Middle Down, and Top Down. We also discuss challenges and innovative solutions for characterizing and quantifying complicated isobaric species arising from multiple modifications on the same histone molecule.
Introduction
The fundamental repeating unit of eukaryotic chromatin, the nucleosome, is formed by wrapping 147 bp of DNA around a histone octamer, consisting of two copies of histones H2A, H2B, H3, and H4. Linker DNA between nucleosomes is further bound with histone H1 for hierarchical folding of chromatin. These histones are highly modified with posttranslational modifications (PTMs) that dynamically modulate local and global chromatin structure [1]. The large number of recurrent mutations identified in histone modifying enzymes has spurred great interest in cancer epigenetics [2]. A recent survey of literature has identified total of 519 distinct modifications from 237 sites in core and linker histones [3], creating a perception of vast combinatorial complexity. However, we focus here on methods to readout those PTMs that form the discrete marks and their combinations (often referred as ‘codes’ in literature) present at high levels, sharply curtailing the combinatorial explosion of histone forms theoretically possible.
Biochemical methods relying on modification-specific antibodies are the gold standards for analyzing histone modifications in biological systems. However, these approaches suffer from antibody cross-reactivity arising from the high similarities in both modification chemical structures themselves (e.g., mono-, di-, and trimethylation) and their surrounding amino acid sequences (e.g., H4 K5/8/12/16 acetylation, S1GRGKGGKGLGKGGAKR17) [4]. Antibody selectivity can be negatively affected by co-occurring histone modifications on the same molecule [5]. In addition to interrogating individual sites like the antibody-based approaches, liquid chromatography with MS (LC–MS) can directly measure multiple modifications on the same molecule (i.e., ‘codes’), which has proven central in chromatin biology [6••,7••,8••]. Here, we focus in discussing recent advances in analyzing histone marks and codes by quantitative MS and LC–MS. Other aspects of this topic are well covered elsewhere [9].
Development of mass spectrometric analysis of histone modifications
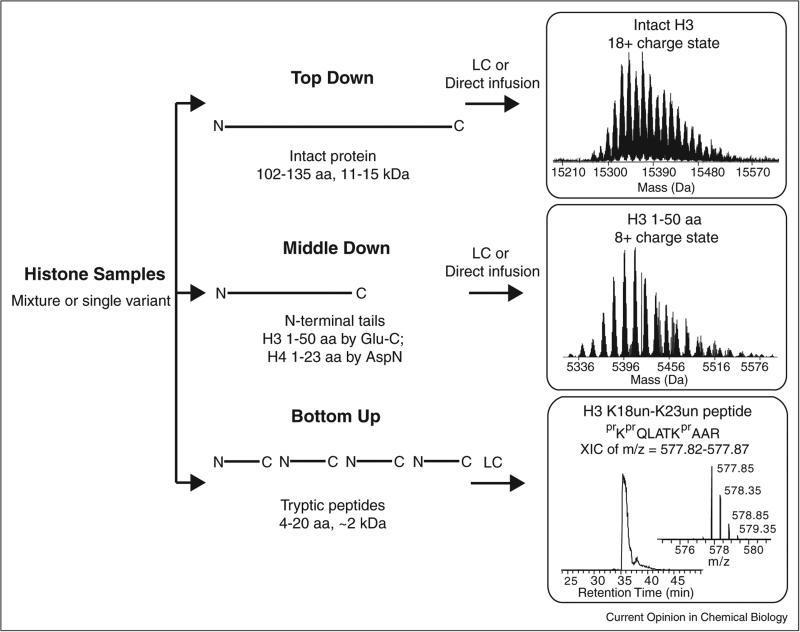
Over the past 15 years, three analytical modes for histone modifications have come to be known as Bottom Up, Middle Down, and Top Down (Figure 1). The first two terms refer to the use of proteases to create small- or medium-sized peptides, whereas Top Down designates use of no proteases prior to tandem MS (MS/MS). Early MS analyses of histones used MALDI-TOF [10,11] before the field shifted heavily toward LC–MS/MS. Because histones are highly enriched with arginine and lysine residues (especially on the N-termini where most PTMs are known to reside), trypsin digestion results in peptides <5 residues, most of which are too hydrophilic to be retained on the reverse phase LC columns in widespread use. The method of chemical derivatization followed by trypsin digestion developed in the Hunt laboratory [12] has now been widely adopted for Bottom Up analysis of histones [6••,13,14,15••]. In this method, free amine group on the N-termini and ε-amine group of unmodified and monomethylated lysine residues react with propionic anhydride to form propionyl amides [12]. This method produces Arg-C like peptides (cleaved only C-terminal to arginine residues, see Figure 2a) with high efficiency and reproducibility. The presence of small levels of acetic anhydride and low levels of endogenous arginine methylation are two analytical interferences that should be noted for this approach. The first causes the artifactual introduction of acetyl groups and the second can lead to fluctuation of the total amount of the peptides generated by cleavage C-terminal to modified arginine residues in this Bottom Up procedure. Such effects are generally operative at low levels (<3%) because the stoichiometry of arginine methylation is known to be relatively low in yeast, mammalian cells, and other model eukaryotes. For the most widely studied histone marks, the Bottom Up/propionylation procedure allows readout of ~20–50 of them in a single LC–MS run [15••,16•,17]. Most quantitative studies use a label-free roll-up of the data, where individual modified species are normalized against total signal from all Arg-C peptides sharing the same underlying sequence (Figure 2a) and standard Z-scores are used to create heat maps to survey for changes across many conditions [17].
Figure 1.
Three general strategies to analyze histone modifications using liquid chromatography and mass spectrometry (LC–MS): Top Down, Middle Down, and Bottom Up. Histones, usually enriched by acid extraction of isolated nuclei, or fractionated into sub-sets of variants by RP-HPLC can be used as starting material. Example MS data of histones from human cell lines are shown for all three approaches (at right). Top right: partial spectrum of the 18+ charge state of whole histone H3.1. The multiple species observed arise from methylation and acetylation (each peak differs by a nominal mass of 14 Da). Center right: partial spectrum of the 8+ charge state of 1-50 N-terminal tail from histone H3.1. Bottom right: an extracted ion chromatogram (XIC) of a LC–MS run is shown for 2+ ion of H3 K18un-K23un (peptide is referred by the highly-modified lysine residues within the sequence for clarity; un, unmodified) with its spectrum along the chromatogram. Chemical derivatization is widely adopted in the Bottom Up approach to block unmodified and monomethylated lysine residues to generate Arg-C like peptide. prX, propionylation in the N-terminal amino group of an amino acid; Kpr, propionylation in the ε-amino group of lysine.
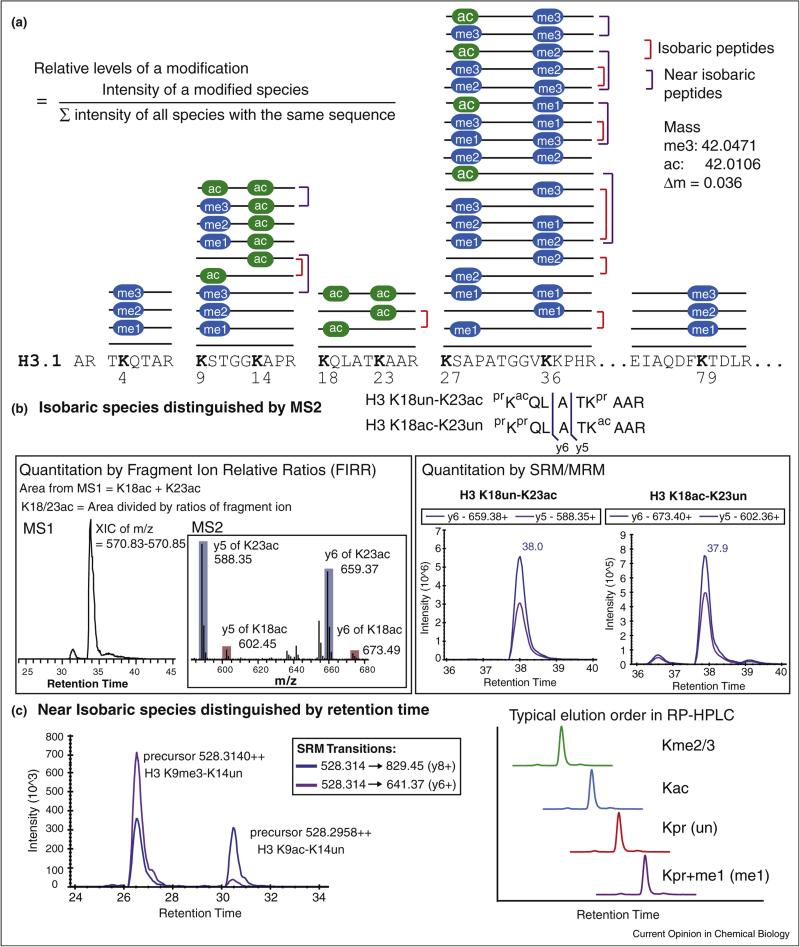
Figure 2.
Analytical challenges of analyzing histone modifications by Bottom Up and LC–MS. (a) 42 unique modified species are shown as sticks with modifications over specific residues in histone H3.1 (ac, acetylation; me, methylation). It represents a Bottom Up experiment outcome when unmodified and monomethylated lysines are blocked by chemical derivatization (e.g., propionic anhydride or deuterated acetic anhydride) before trypsin digestion. Using this workflow, eight highly-modified lysine residues in histone H3.1 (K4, K9, K18, K23, K27, K36, and K79) fall into five groups of peptides which share the same underlying sequence. The combination of modifications when two modified lysines are present in the same peptide generates isobaric species (red bracket). In addition, small mass difference between acetylation and trimethylation produce near isobaric species (purple bracket). (b) Fragment ions (such as y5 or y6) between K18 and K23 generated by tandem mass spectrometry (MS/MS or MS2) can be used to distinguish isobaric species of H3 K18un-K23ac from H3 K18ac-K23un. In the left panel, an extracted ion chromatogram of the most abundant monoisotopic peak from these doubly charged isobaric peptide species is used to determine the combined abundances, which can be further divided by fragment ion relative ratios obtained in multiplexed collision-induced dissociation (CID) tandem mass spectrum (shown in the inset) to quantify the abundance of individual isobaric species. In the right panel, SRM chromatograms of specific transitions (e.g., precursor → y5 and precursor → y6) can be used to quantify each isobaric species. Noting that data obtained by these two methods for a same sample (human multiple myeloma cell line, KMS11-TKO) is consistent to show that H3 K18un-K23ac is about 7-fold more abundant than the co-eluted H3 K18ac-K23un (retention time labeled along SRM peaks). (c) Trimethylation and acetylation are indistinguishable in low resolution instrument, such as triple quadrupole used for SRM. An example of SRM data from a human cell line (MCF-7) demonstrates that transitions designated to quantify H3 K9me3-K14un can detect two peaks. Using the retention time profile determined by synthetic peptides, these near isobaric specices (me3 vs. ac) can be distinguished.
Our laboratory has published extensively on the Top Down approach for all histones a decade ago, establishing a ‘basis set’ of histone proteoforms [18] that arise from multiple modifications and are present at approximately 5% levels and above [18,19,20,21•,22]. For histone H4, 42 proteoforms were described from 50% down to 0.05% relative abundance in bulk chromatin using weak cation exchange-hydrophilic interaction liquid chromatography (WCX-HILIC) to separate primarily by extent of acetylation before Top Down MS [23]. The labor intensive nature of this ‘off-line’ approach using direct infusion nanospray motivated the Garcia and Pasa-Tolic laboratories to automate ‘on-line’ LC solutions using ‘saltless’ pH gradient of WCX-HILIC [24•] or two dimensional LC [25•], respectively.
A compromise between Bottom Up and Top Down is the Middle Down approach. These strategies and terms are trending in proteomics generally, but were piloted first in ‘epiproteomics’ using histones where endoproteinase Glu-C or Asp-N clips the first 1-50 or 1-23 N-terminal residues off H3 or H4, respectively (Figure 1, middle). The rationale is that these peptides preserve the co-occurrence of most modifications in the same molecule because the majority of them occur on the N-terminal tails. One early Middle Down report focused on histone H3.2, and characterized >150 codes by direct infusion of WCX-HILIC fractions [26]. Shortly after, 72 unique forms of the H4 tail were profiled using on-line RP-HPLC separation of Asp-N digested histone H4 [27]. More recently, the feasibility of nanocapillary LC–MS/MS for faster Middle Down has been demonstrated for unfractionated histones digested by Glu-C or Asp-N using simple reverse phase HPLC [28] or innovative hybrid LC [29••]. A direct comparison has concluded that Bottom Up and Middle Down can achieve comparable accuracies of quantifying individual histone modifications [30].
Challenges and strategies of quantifying histone modifications using MS
Although a large number of histone proteoforms exist due to the combination of multiple modifications, it is not true that every random combination of modifications exists. Otherwise a cell would produce an astronomical number of proteoforms, far exceeding the ~30 million nucleosomes in a human cell. Nonetheless, many combinatorial forms of methylation and acetylation have been observed in peptides carrying two residues modified with high occupancy (e.g., K9-K14, K18-K23, and K27-K36 in H3 as shown Figure 2a) or four modified residues (e.g., K5, K8, K12, and K16 of H4 4-17 in Figure 3a and b). The complexity originating from large amount of isobaric or near isobaric species (Figure 2a) found in these combinatorially modified species presents a central challenge for MS-based epiproteomics.
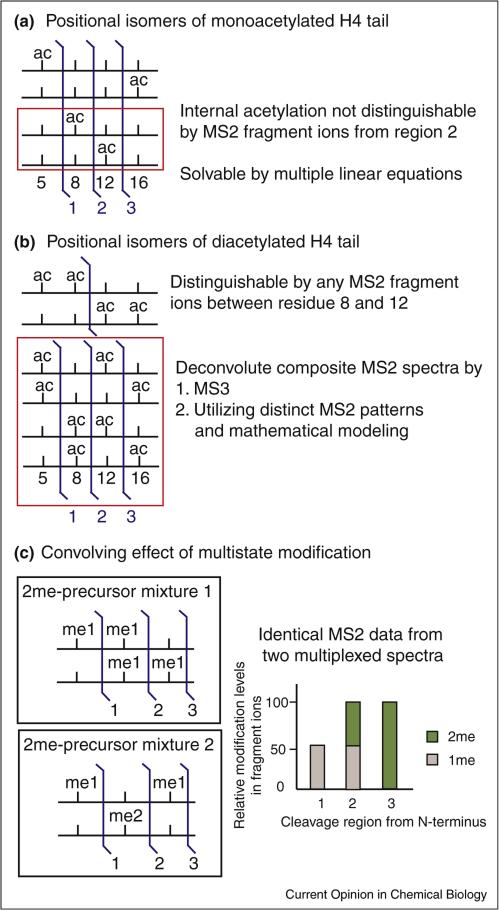
Figure 3.
The increasing analytical challenges encountered in a larger histone peptide or a whole histone protein (number of modified residues usually exceeds two) cannot be solved by a routine tandem mass spectrometry (MS2) strategy. (a) Four lysines (residue 5, 8, 12, and 16) in the N-terminal of histone H4 are known to be acetylated. In a monoacetylated peptide (residue 4–17) generated in Bottom Up/derivatization experiment, there are four possible positional isomers. K5ac and K16ac can be determined by MS2 fragment ions from regions 1 and 3, respectively (blue flag). However, fragment ions from region 2 can only determine the combined amount of K5ac + K8ac and K12ac + K16ac. Therefore, multiple linear equations are required to quantify these positional isomers. (b) Long-standing problem of positional isomers of diacetylated H4 4-17. The number of equations formulated from ratios of MS2 measurement is fewer than the parameters needed to be solved for ratios of these isomers. Recent efforts used MS3 (refragmentation of an isolated MS2 fragment ion) [38••] and unique fragment ion patterns associated with each isomer to deconvolute them [39••]. (c) When two methyl groups are detected in MS2 fragment ions with multiple lysines, it could be a dimethylation on a single lysine or two monomethylation on two residues. Such complication was called ‘convolving’ effect of multistate modification [7]. Two hypothetical scenarios of equal molar ratio mixture carrying two methyl groups are depicted here to illustrate this problem. The relative levels of measured methyl groups present in the fragment ions (N-termini to the cleavage site) are shown in the right panel. Because ‘meX’ (i.e., me1,2,3) is used in literature for mono-, di-, and trimethylation on a single residue, we use ‘Xme’ to indicate ‘X’ number of methyl groups is inferred from MS data. As shown here, identical fragment ion pattern is observed for both scenarios, which makes modification assignment ambiguous. However, both scenarios can pass current search algorithms relying on the presence of ‘site-determining’ MS2 fragment ions. Therefore, non-existed modification forms could be ‘created’ due to the false interpretation of MS2 evidences that identify both mixtures when only one of them actually existed.
The area under the curve, or simply peak area, in LC–MS has become a major quantitation strategy due to its high reproducibility, large linear dynamic range and multiplexed nature (i.e., hundreds of targets can be monitored in a single LC–MS/MS run). ‘Discovery’ and ‘targeted’ modes are two general data acquisition strategies for tandem MS. In ‘discovery’ mode, multiple precursor ions identified in the first MS survey scan (MS1) are selected for fragmentation in the subsequent MS events (MS2). This workflow, also called data dependent acquisition (DDA), is the current standard for peptide identification because its data collecting scheme enables powerful database searching, even in ‘error-tolerant’, discovery mode [31]. However, DDA has limited quantification capabilities due to stochastic precursor ion selection. More specifically, MS2 events are not collected frequently enough over the entire peptide elution window to generate the best quantitative metrics. As a result, DDA mainly provides MS1-based quantitation as shown for an example in Figure 2b. Therefore, Fragment Ion Relative Ratios (FIRRs) from discriminative fragment ions are required to further divide the total area determined in MS1 to quantify multiple isobaric species (see inset of left panel in Figure 2b) [21•]. However, due to the complexity of highly modified histone peptides, the implementation of such an approach is challenging, relegating it to expert laboratories to perform large-scale studies involving hundreds of modified peptides. In an elegant methodology developed by the Garcia laboratory, an entire set of modified histone peptides was generated for a precursor ion and a superposition problem was formulated using mixed integer linear optimization to determine the relative fractions of the isobaric species present in the multiplexed MS2 spectra [32••]. Recently, a software tool, EpiProfile, was further developed to facilitate this quantitation workflow [33••].
Selected or multiple reaction monitoring (SRM/MRM) is a ‘targeted’ mode of LC–MS running mostly on triple quadrupole mass spectrometers, in which transitions (i.e., a precursor transitioning into a specific fragment ion) are used to generate a chromatogram for a predetermined target. The target list has grown rapidly from a single mark (H3K56ac) in the first report [14] to 21 shortly after [34] and 93 in the most recent effort [16• ]. The development of software such as Skyline and Pinpoint has simplified the workflow for targeted proteomics using the venerable SRM/MRM approaches [35]. Examples of visualized SRM peaks in Skyline are shown in the right panel of Figure 2b. Skyline allows users to manually select peaks, which is useful for determining the correct peak in the presence of interfering signal, such as the trimethylation vs. acetylation example illustrated in Figure 2c. More recently, multiplexed parallel reaction monitoring (PRM) running on the Q-Exactive mass spectrometers achieves a SRM-like procedure to quantify histone modifications [15••,36••]. The simplification of data analysis afforded by SRM/PRM has allowed the large-scale screening of altered histone modifications in the burgeoning field of cancer epigenetics [15••,37].
When MS2 is not enough
When ≥3 modified residues are present in a peptide/protein, MS2 fragment ions are not adequate to rigorously localize and quantify all modifications. As shown in Figure 3a, internal acetylation (K8ac and K12ac) on a monoacetylated H4 4-17 peptide cannot always be cleanly distinguished by MS2 fragment ions. Instead, multiple linear equations determined by ratios of fragment ions from all three regions between modified residues are required to calculate their abundances [27]. Two strategies have been reported recently to solve the long-standing problem of four ‘positional isomers’ of a diacetylated H4 (Figure 3b, [27]). The first one used an MS3 approach [38••] and the second took advantage of the characteristic fragment ion pattern associated with each positional isomer [39••]. This isobaric problem grows more complicated when methylation is involved. For example, when two methyl groups are inferred in the fragment ion (Δm = 28 Da), a single dimethylation on a single lysine residue or two monomethylation on two separate residues are ambiguously indicated. This so called ‘convolving effect’ of methylation has been shown to complicate the analysis of codes harboring ≥3 marks of the same type, making MS3 or other approach necessary [7••]. A hypothetical example of the convolving effect is shown in Figure 3c to demonstrate the possibility of erroneous assignment of a code using current search algorithms for matching MS2 fragment ions.
Caveats of Bottom Up quantitation
As noted about Bottom Up above, sample processing can affect the readout of low abundance histone marks. Derivatization reagents other than propionic anhydride, such as deuterated anhydride [40] should be used when measuring endogenous lysine propionylation [41]. A hybrid method using phenyl isocyanate to derivatize the newly generated N-termini after trypsin digestion has been developed to avoid the selective losses of very hydrophilic peptides, such as H3K4me2/3 [42]. More significantly, chemical derivatization using acid anhydride can partially alkylate the hydroxyl group of serine and threonine [43] and methanol used in the procedure can lead to the methyl esterification of glutamic acid and aspartic acid residues [14]. However, a direct comparison of 12 commercially available anhydrides with different hydrophobicity suggests propionic anhydride is still the best choice for the overall performance [44] yet can introduce a few percent of acetylation from contaminating acetic anhydride. To reduce such deleterious side reactions, N-hydroxysuccinimide (NHS) propionate has been demonstrated for its specificity [43]. An evaluation of side reactions using propionic anhydride and NHS-propionate at various conditions has helped to identify proper procedures for accurate quantitation [45].
A comprehensive evaluation of the detection efficiencies performed recently demonstrated a wide spectrum of differences (>1700-fold) using MS1 quantitation by extracted ion chromatograms [46•]. Because these detection efficiencies are method and platform specific, they are not applicable to generally correct all quantitation methods. Luckily, the correction factors to reduce detection bias does not change the conclusions (i.e., up- or down-regulated) for most histone marks except some low abundant ones in a differential analysis [46•]. In addition, spiked-in internal standards, such as stable-isotopic-labeled synthetic peptides [47] and histones purified from SILAC labeled cells [15••], can be used to improve quantitation accuracy across the study. Top Down Epiproteomics using either LC–MS or direct infusion has fewer sample handling steps, introduces significantly fewer sources of bias and directly reads out (not inferring) proteoform-level dynamics and PTM crosstalk. Various free and commercial versions of a software, ProSight, have been developed to support a complete view of histone proteoforms by Top Down MS over the years [22,23,26].
Conclusions and outlook
With the steady advancements in three pillars of LC–MS analysis (instrumentation, chromatographic separation, and informatics), Middle/Top Down will play increasingly important roles for histone modification analysis because they can provide more comprehensive information that putatively links to function more directly (vide supra and Figure 4). The advantages go beyond the aforementioned readout of co-occurring, multiple modifications. Two very recent Middle/Top Down studies have identified unexpected results quite readily: the clipping of histone tails [48] and the hypermethylated trivalent marks in a histone methyltransferase overexpression system [7••]. However, more challenges arise from dissecting the increasing number of isobaric species in Middle/Top Down, which demands innovations in MS3 or other approach, such as internal fragmentation [49] to distinguish them. Finally, the use of native Top Down MS [50] on whole nucleosomes may eventually be able to readout intermolecular codes imprinted multiple histone tails non-covalently bound together.
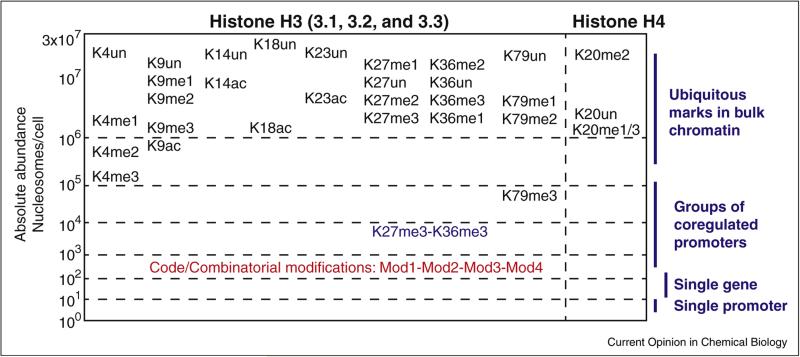
Figure 4.
Estimated levels of major H3/H4 modifications in the pool of bulk histones depicted on a log scale of absolute abundance. When properly calibrated, mass spectrometry can provide absolute abundance information, such as the numbers of nucleosomes per cell carrying a specific epigenetic mark. The ubiquitous presence of the major histone marks in their individual state (e.g., H3K27me3 present in >1 million of nucleosomes/cell) means their abundances are unlikely to change when transcriptions from a small subset of genes are altered (e.g., in response to the loss of UTX, a H3K27 demethylase). On the other hand, those combinatorial modifications (or ‘codes’) only present on 100–1000 nucleosomes, as an example shown in red, are more likely to correlate with the transcription activity from a small subset of genes. It is worth noting that comparing the frequency of combinatorial modification, consisting of two co-occurring marks, against the co-frequency of these two independent marks is a simple yet elegant method to identify positive and negative crosstalk between pairs of marks [8]. For example, the frequency of H3 K27me3-K36m3 is near 100-fold less than the co-frequency of H3K27me3 and H3K36me3, suggesting a strong negative crosstalk between these two marks.
Acknowledgements
This review was written with partial support from the National Resource for Translational and Developmental Proteomics under Grant P41 GM108569 from the National Institute of General Medical Sciences, National Institutes of Health.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peach SE, Rudomin EL, Udeshi ND, Carr SA, Jaffe JD. Quantitative assessment of chromatin immunoprecipitation grade antibodies directed against histone modifications reveals patterns of co-occurring marks on histone protein molecules. Mol Cell Proteomics. 2012;11:128–137. doi: 10.1074/mcp.M111.015941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2011;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Zheng Y, Sweet SM, Popovic R, Martinez-Garcia E, Tipton JD, Thomas PM, Licht JD, Kelleher NL. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci U S A. 2012;109:13549–13554. doi: 10.1073/pnas.1205707109. [The first use of the SRM approach to overcome multiple isobaric targets faced by Bottom Up MS analysis of histone modification. This nanocapillary LC-SRM-MS platform provided both reliable quantitation for 15 combinatorial modification forms arising from H3K27 and H3K36 methylation in the same peptide and implemented a dual stable isotope labeling scheme for the further development of MS-based measurement and modeling of histone methylation kinetics (M4K), which provides a kinetic readout of PTM crosstalk governing the reading, writing and maintenance of the histone code in living cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, Fornelli L, Compton PD, Sharma S, Canterbury J, Mullen C, Zabrouskov V, Fellers RT, Thomas PM, Licht JD, et al. Unabridged analysis of human histone H3 by differential top-down mass spectrometry reveals hypermethylated proteoforms from MMSET/NSD2 overexpression. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M115.053819. [This study assessed the feasibility of charactering and quantifying histone H3 proteoform using direct infusion so that MS analysis was not limited by LC time scale. It took the advantage of the small isolation window of 0.6 Th and improved ion fragmentation efficiency offered in improved MS-technology and successfully identified the potential disease associated proteo-forms (i.e., ‘trivalent’ hypermethylated histone H3 in a differential study).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Schwämmle V, Aspalter C-M, Sidoli S, Jensen ON. Large scale analysis of co-existing post-translational modifications in histone tails reveals global fine structure of cross-talk. Mol Cell Proteomics. 2014;13:1855–1865. doi: 10.1074/mcp.O113.036335. [Developed the database, CROSSTALKDB, to facilitate the reporting and scoring of co-occurring histone PTMs measured by quantitative MS experiments. In essence, two completely independent marks 1 and 2 would co-occur with probability f1*f2, where f1 and f2 are the probabilities of marks 1 and 2. Frequencies of PTM co-occurrence that are larger or smaller than this co-frequency ( f1*f2) indicate positive or negative crosstalk between two marks.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H, Lin S, Garcia BA, Zhao Y. Quantitative proteomic analysis of histone modifications. Chem Rev. 2015;115:2376–2418. doi: 10.1021/cr500491u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Williams KE, Huang L, Yau P, Siino JS, Bradbury EM, Jones PR, Minch MJ, Burlingame AL. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Mol Cell Proteomics. 2002;1:500–508. doi: 10.1074/mcp.m200031-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Bonaldi T, Imhof A, Regula JT. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4:1382–1396. doi: 10.1002/pmic.200300743. [DOI] [PubMed] [Google Scholar]
- 12.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protocols. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drogaris P, Wurtele H, Masumoto H, Verreault A, Thibault P. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure S function relationships. Anal Chem. 2008;80:6698–6707. doi: 10.1021/ac800739d. [DOI] [PubMed] [Google Scholar]
- 15••.Jaffe JD, Wang Y, Chan HM, Zhang J, Huether R, Kryukov GV, Bhang H-eC, Taylor JE, Hu M, Englund NP, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–1391. doi: 10.1038/ng.2777. [A large-scale screen of global levels of histone modification using mass spectrometry. Reported a targeted quantitative LC–MS assay using parallel reaction monitoring (PRM) mode running in the Q-Exative to quantify large number of histone modification. This Bottom Up approach allowed screening of 115 cell lines from the Cancer Cell Line Encyclopedia (CCLE) collection.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun. 2013;4 doi: 10.1038/ncomms3203. [Reported a quantitative nanocapillary LC-SRM-MS assay for total of 93 histone peptides using Bottom Up/propionylation approach; subsequently combined the assay with metabolic labeling with stable-isotope-labeled glucose to determine the turnover rate of histone acetylation in a site-specific manner.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leroy G, Dimaggio PA, Chan EY, Zee BM, Blanco MA, Bryant B, Flaniken IZ, Liu S, Kang Y, Trojer P, et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin. 2013;6:20. doi: 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. Gene-specific characterization of human histone H2B by electron capture dissociation. J Proteome Res. 2006;5:233–239. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 19.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J Proteome Res. 2006;5:248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 20.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J Proteome Res. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 21•.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [Demonstration that Fragment Ion Relative Ratios (FIRRs) from discriminative fragment ions can be used to quantify isobaric peptides and proteoforms in multiplexed electron capture dissociation (ECD) spectra; also quantified the levels of MS-1 bias (or lack of bias for whole histones).] [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, John S, Pesavento JJ, Schultz-Norton JR, Schiltz RL, Baek S, Nardulli AM, Hager GL, Kelleher NL, Mizzen CA. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol. 2010;189:407–415. doi: 10.1083/jcb.201001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Young NL, DiMaggio PA, Plazas-Mayorca MD, Baliban RC, Floudas CA, Garcia BA. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–2284. doi: 10.1074/mcp.M900238-MCP200. [The presence of high concentration salts, such as NaClO4, in the elution buffer of WCX-HILIC is not compatible with electrospray ionization (ESI). This study developed the ‘saltless’ WCX-HILIC buffer/pH gradient, which is compatible with ESI and can be directly coupled for LC–MS. Using this on-line, Middle Down approach, the characterization and quantification of over 200 codes in H3.2 (1-50 piece) and 70 in H4 (1-23 piece) from a human cell line. Compared with previous off-line approach, it greatly reduced sample amount requirement and analysis time.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Tian Z, Tolic N, Zhao R, Moore RJ, Hengel SM, Robinson EW, Stenoien DL, Wu S, Smith RD, Pasa-Tolic L. Enhanced top-down characterization of histone post-translational modifications. Genome Biol. 2012;13:R86. doi: 10.1186/gb-2012-13-10-r86. [Reported a two-dimensional LC workflow consisting of separating unfractionated intact histones with reverse phase and followed by ‘saltless’ WCX-HILIC, which provided better throughput and sensitivity for the comprehensive histone proteoform characterization. Total of 708 his-tone proteoforms (H4: 105; H2B: 110; H2A: 77; H3: 416) were unambiguously identified in a single 2D-LC-MS/MS run.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 27.Phanstiel D, Brumbaugh J, Berggren WT, Conard K, Feng X, Levenstein ME, McAlister GC, Thomson JA, Coon JJ. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalli A, Sweredoski MJ, Hess S. Data-dependent middle-down nano-liquid chromatography–electron capture dissociation-tandem mass spectrometry: an application for the analysis of unfractionated histones. Anal Chem. 2013;85:3501–3507. doi: 10.1021/ac303103b. [DOI] [PubMed] [Google Scholar]
- 29••.Sidoli S, Schwammle V, Ruminowicz C, Hansen TA, Wu X, Helin K, Jensen ON. Middle-down hybrid chromatography/tandem mass spectrometry workflow for characterization of combinatorial post-translational modifications in histones. Proteomics. 2014;14:2200–2211. doi: 10.1002/pmic.201400084. [Presented an integrated Middle Down proteomics platform for histone analysis. The practical LC workflow included loading Glu-C-digested, unfractionated histones onto a reverse phase trap column followed by the separation in analytical WCX-HILIC column. The collected electron transfer dissociation (ETD) tandem mass spectra were deconvolved using XTRACT and searched in MASCOT. Then authors developed HISTONE CODER software to evaluate the PTM assignments from MASCOT search results to eliminate false positive identification. Essentially, the presence of ‘site-determining ions’ in both directions of the candidate modification site are required to localize a modification. In addition, ISOSCALE software was developed to quantify co-fragmented isobaric species. Using this platform, the quantification of 713 unique codes in H3 1-50 piece using purified H3 as input and 256 codes from large peptide of histones H3, H4, and H2A when crude histone extract was used as input.] [DOI] [PubMed] [Google Scholar]
- 30.Sidoli S, Lin S, Karch KR, Garcia BA. Bottom-up and middle-down proteomics have comparable accuracies in defining histone post-translational modification relative abundance and stoichiometry. Anal Chem. 2015;87:3129–3133. doi: 10.1021/acs.analchem.5b00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan ZF, Lin S, Molden RC, Garcia BA. Evaluation of proteomic search engines for the analysis of histone modifications. J Proteome Res. 2014;13:4470–4478. doi: 10.1021/pr5008015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.DiMaggio PA, Jr, Young NL, Baliban RC, Garcia BA, Floudas CA. A mixed integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed electron transfer dissociation tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2527–2543. doi: 10.1074/mcp.M900144-MCP200. [Authors used histone H3.2 as model system to drive the development of a computational framework adopting mixed integer linear optimization (MILP) for the identification and quantification of highly modified proteins using LC–MS and ETD tandem mass spectrometry. The framework first enumerated the entire set of PTMs that satisfy a precursor mass for a given primary sequence. For example, 227 H3 proteoforms were enumerated that satisfied the precursor m/z of 610.5 (with assumption of phosphorylation at residue 3, 8, and 28; acetylation at 14, 18, and 23; methylation/acetylation at 4, 9, 27, and 36; theoretical combination increased when more candidate modifications considered). An MILP superposition problem was then solved to calculate the relative fractions of the proteoforms present in multiplexed tandem mass spectra.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Yuan ZF, Lin S, Molden RC, Cao XJ, Bhanu NV, Wang X, Sidoli S, Liu S, Garcia BA. EpiProfile quantifies histone peptides with modifications by extracting retention time and intensity in high-resolution mass spectra. Mol Cell Proteomics. 2015;14:1696–1707. doi: 10.1074/mcp.M114.046011. [Reported the quantification software, EPIPROFILE, to solve the challenges of isobaric histone peptide (e.g., H3 K9ac-K14un vs. K9un-K14ac). It extracted ion chromatography for known histone peptide using prior knowledge of retention time and resolved the isobaric species by solving series of linear equations derived from peak heights of unique MS2 ions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darwanto A, Curtis MP, Schrag M, Kirsch W, Liu P, Xu GL, Neidigh JW, Zhang KL. A modified “Cross-talk” between histone H2B Lys-120 ubiquitination and H3 Lys-79 methylation. J Biol Chem. 2010;285:21868–21876. doi: 10.1074/jbc.M110.126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Tang H, Fang H, Yin E, Brasier AR, Sowers LC, Zhang K. Multiplexed parallel reaction monitoring targeting histone modifications on the Q-Exactive mass spectrometer. Anal Chem. 2014;86:5526–5534. doi: 10.1021/ac500972x. [Reported the development of PRM mode on the Q-Exactive mass spectrometer for the targeted analysis of trypsin digested histones (no derivatization to avoid side reactions). Total of 55 modified peptides were detected from 24 targeted precursor ions in the inclusion list. Also demonstrated the utility of using the characteristic immonium ions in higher-energy c-trap dissociation (HCD) MS2 spectra to determine the modification.] [DOI] [PubMed] [Google Scholar]
- 37.Oyer JA, Huang X, Zheng Y, Shim J, Ezponda T, Carpenter Z, Allegretta M, Okot-Kotber CI, Patel JP, Melnick A, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28:198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Feller C, Forne I, Imhof A, Becker PB. Global and specific responses of the histone acetylome to systematic perturbation. Mol Cell. 2015;57:559–571. doi: 10.1016/j.molcel.2014.12.008. [Optimized a LC-MS workflow for quantifying histone modification using Bottom Up MS with deuterated acetic anhydride (D3AA) derivatization. One key achievement was the implementation of iterative loops of targeted MS1-MS2-MS3 scans to utilize the information from the refragmented (MS3) ions to solve the positional isomer problem of histone H4 diacetylated across 4 possible sites (K5, K8, K12, and K16). Synthetic peptides were used to correct for LC–MS bias and systematic examination of the relationship between histone modifying enzyme and its substrate through RNAi knockdown of 23 histone acetyltransferase (HAT) and 8 histone deacetylase (HDAC) in cultured Drosophila cells. Monitored global levels of 28 combinatorial modifications, in which the complete collection of all 14 combinatorial acetylations in H4 4-17 was included. Demonstrated that depleting single HAT activities leads to complex alterations of histone modification pattern. Notable was the ability to discriminate unique combinatorial modification (e.g., H4 K8acK16ac vs. K12acK16ac) critical to many interesting observations reported.] [DOI] [PubMed] [Google Scholar]
- 39••.Abshiru N, Caron-Lizotte O, Rajan RE, Jamai A, Pomies C, Verreault A, Thibault P. Discovery of protein acetylation patterns by deconvolution of peptide isomer mass spectra. Nat Commun. 2015;6 doi: 10.1038/ncomms9648. [This paper presented ISO-PEPTIDACE, a software that enables deconvolution of multiplexed MS2 spectra of isomeric peptides based on features associated with their characteristic fragment ion patterns obtained from the corresponding synthetic peptides. In essence, fragment ion patterns were reduced to a set of maximum network flow problems and then solved by well-established algorithms. Because it did not rely on unique fragment ions to distinguish isomers, it was able to distinguish very challenging positional isomers (e.g., diacetyalted H4 4-17) without the need of additional information from MS3, which can be difficult to obtain in robust fashion. Included a validation using defined mixtures of synthetic acetylated isomers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316:23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maile TM, Izrael-Tomasevic A, Cheung T, Guler GD, Tindell C, Masselot A, Liang J, Zhao F, Trojer P, Classon M, et al. Mass spectrometric quantification of histone post-translational modifications by a hybrid chemical labeling method. Mol Cell Proteomics. 2015;14:1148–1158. doi: 10.1074/mcp.O114.046573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao R, Wu H, Deng H, Yu Y, Hu M, Zhai H, Yang P, Zhou S, Yi W. Specific and efficient N-propionylation of histones with propionic acid N-hydroxysuccinimide ester for histone marks characterization by LC-MS. Anal Chem. 2013;85:2253–2259. doi: 10.1021/ac303171h. [DOI] [PubMed] [Google Scholar]
- 44.Sidoli S, Yuan ZF, Lin S, Karch K, Wang X, Bhanu N, Arnaudo AM, Britton LM, Cao XJ, Gonzales-Cope M, et al. Drawbacks in the use of unconventional hydrophobic anhydrides for histone derivatization in bottom-up proteomics PTM analysis. Proteomics. 2015;15:1459–1469. doi: 10.1002/pmic.201400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meert P, Govaert E, Scheerlinck E, Dhaenens M, Deforce D. Pitfalls in histone propionylation during bottom-up mass spectrometry analysis. Proteomics. 2015;15:2966–2971. doi: 10.1002/pmic.201400569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Lin S, Wein S, Gonzales-Cope M, Otte GL, Yuan ZF, Afjehi-Sadat L, Maile T, Berger SL, Rush J, Lill JR, et al. Stable-isotope-labeled histone peptide library for histone post-translational modification and variant quantification by mass spectrometry. Mol Cell Proteomics. 2014;13:2450–2466. doi: 10.1074/mcp.O113.036459. [Systematically examined the Bottom Up LC-MS bias using a library containing 93 stable-isotope-labeled synthetic histone peptides. Reported large range of detection efficiencies, with >1700-fold difference between the peptides with the lowest and highest efficiencies using MS1-based quantification.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Liao R, Yu Y, Zhai H, Wang Y, Sack R, Peters AHFM, Chen J, Wu H, Huang Z, et al. Absolute quantification of histone PTM marks by MRM-based LC-MS/MS. Anal Chem. 2014;86:9679–9686. doi: 10.1021/ac502333a. [DOI] [PubMed] [Google Scholar]
- 48.Tvardovskiy A, Wrzesinski K, Sidoli S, Fey SJ, Rogowska-Wrzesinska A, Jensen ON. Top-down and middle-down protein analysis reveals that intact and clipped human histones differ in post-translational modification patterns. Mol Cell Proteomics. 2015;14:3142–3153. doi: 10.1074/mcp.M115.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durbin KR, Skinner OS, Fellers RT, Kelleher NL. Analyzing internal fragmentation of electrosprayed ubiquitin ions during beam-type collisional dissociation. J Am Soc Mass Spectrom. 2015;26:782–787. doi: 10.1007/s13361-015-1078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner OS, Havugimana PC, Haverland NA, Fornelli L, Early BP, Greer JB, Fellers RT, Durbin KR, Do Vale LHF, Melani RD, et al. An informatic framework for decoding protein complexes by top-down mass spectrometry. Nat Meth. 2016 doi: 10.1038/nmeth.3731. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]