Abstract
Background:
MiR-214 is aberrantly regulated in several tumours, but its underlying mechanisms in colorectal cancer (CRC) metastasis remain largely unknown. This study aimed to demonstrate the function and potential mechanism of miR-214 in regulating invasion and metastasis of CRC.
Methods:
The transcription factor and targets of miR-214 were predicted by bioinformatics and validated using ChIP and dual-luciferase reporter assay. DNA methylation status was explored using bisulphite sequencing PCR. The in vitro and in vivo function of miR-214 in CRC was evaluated using MTT, plate colony formation, Matrigel invasion and animal models. Real-time PCR or western blotting was performed to detect FOXD3, miR-214 and MED19 expressions in CRC cells and clinical specimens.
Results:
MiR-214 was downregulated in CRC and was significantly correlated with lymphatic metastasis. Downregulation of miR-214 might due to promoter hypermethylation in CRC. FOXD3 was validated as a transcription factor of miR-214 by ChIP assay. Dual-luciferase assay identified MED19 as a target of miR-214 in CRC. In vitro and in vivo experiments showed that miR-214 mediated the inhibiting effect of FOXD3 on proliferation, invasion and metastasis by targeting MED19. Spearman's correlation analysis showed a positive correlation between FOXD3 and miR-214, and negative correlations between FOXD3 and MED19, miR-214 and MED19 in CRC cells and clinical specimens.
Conclusions:
FOXD3/miR-214/MED19 axis is important for the regulation of growth, invasion and metastasis of CRC. Targeting the miR-214-mediated axis might be helpful for the treatment of CRC.
Keywords: miR-214, FOXD3, MED19, metastasis, methylation, colorectal cancer
Colorectal cancer (CRC) is a common malignancy in the world, its incidence and mortality rise gradually (Torre et al, 2015). Considering its high mortality, elucidate the molecular mechanisms of CRC metastasis and provide theoretical and experimental basis for clinical treatment are of vital importance. MiRNAs are a non-protein coding class of short regulatory RNAs (22-nucleotides long) involved in the regulation of a variety of physiological and pathological progresses through post-transcriptionally modulating gene expression, such as tumour development and progression, cell proliferation, apoptosis and basal metabolism (Bueno et al, 2008; Nicoloso et al, 2009; Inui et al, 2010; Voorhoeve, 2010; Hao et al, 2014). Dynamin-3 gene (DNM3) is a member of Dynamin family that has a key role in endocytosis and possessing mechanochemical properties of tabulating and severing membranes (Zhang et al, 2016). MiR-214 is located in the chromosomal region 1q24.3, 14th intron of DNM3 and encoded within DNM3 opposite strand and has vital roles in the regulation of cancer onset, growth, and progression (Penna et al, 2015). MiR-214 is downregulated in several human tumours including breast, cervical, rhabdomyosarcoma, and hepatocellular carcinomas (Derfoul et al, 2011; Shih et al, 2012; Huang et al, 2014; Wen et al, 2014). The pleiotropic and tumour-specific of miR-214 contributes to various cancer formation and progression via its specific target genes. Moreover, miR-214 is a critical component involved in many fundamental signalling pathways such as PTEN/Akt, β-catenin, and tyrosine kinase receptor pathways (Wang et al, 2012a, b; Momose et al, 2013). Interestingly, recent reports have identified that miR-214 alterations in tumour cells lead to negative regulation of CRC liver metastasis (Chen et al, 2014). On the basis of this, miR-214 is considered to be a potential target for tumour diagnosis, treatment and prognosis.
In this study, we report the suppressive role of the FOXD3/miR-214/MED19 axis in CRC cells. We provide evidence that miR-214 induced by its upstream transcription factor FOXD3, can suppress tumour growth and metastasis in CRC by targeting MED19.
Materials and Methods
Cell lines, human tissue samples, and animals
Human CRC cell lines LOVO, SW620, SW480, HCT116, HT-29, LS174T and human embryonal kidney 293 cells were purchased from Shanghai Cell Bank of Type Culture Collection. The cell lines were freshly authenticated in last year. The cell lines were cultured in DMEM medium (GIBCO, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) in 5% CO2 at 37 °C. Images of CRC cells were taken by Olympus inverted microscope and were outputted by CellSens Dimension software (Olympus, Shinjuku, Japan). Paired fresh CRC tissues were collected from 30 patients who underwent CRC resection without prior radiotherapy and chemotherapy in Nanfang Hospital in 2010. These samples were snap-frozen in liquid nitrogen immediately after resection, and then stored at −80 °C until needed. Four- to 6-week-old male athymic BALB/c-nu/nu mice were purchased from the Central Laboratory of Animal Science of Southern Medical University (Guangzhou, China), and maintained in a specific pathogen free environment. All protocols for animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of Southern Medical University.
Construction of plasmids and transfection
Lentiviral constructs expressing miR-214 (Lenti-miR microRNA precursor clone collection; System Biosciences, Carlsbad, CA, USA) were packaged using the pPACKH1 lenti-vector Packaging Kit (System Biosciences). ShRNAs towards FOXD3 (System Biosciences) were cloned into pSuper-retro-puro. Lentiviral constructs were used to infect CRC cells to establish cells stably expressing miR-214 and MED19 or repressing FOXD3. In the rescue experiments, FOXD3-depleting cells were transfected with miR-214 vector. MiR-214 inhibitor and its negative control were antisense oligos obtained from Genechem Company (Shanghai, China), and was used to transfect indicated cells according to the manufacturer.
DNA methylation analysis
DNA methylation analysis of DNM3 was performed as previously descriped (He et al, 2015). In brief, genomic DNAs of paired CRC tissues were obtained using Promega wizard genomic DNA purification kit (Promega, Salt Lake City, UT, USA) and then bisulfite-modified using the EpiTect Bisulfite Kit (Qiagen, Valencia, CA, USA). The CpG island of DNM3 gene was predicted online UCSC Genome Bioinformatics (http://www.genome.ucsc.edu/). The primers used in bisulfite genomic-sequencing PCR (BSP) detection were designed as following (F: 5′-TTGTATATGTTTGATGTGGTTTTAG-3′ R: 5′-TTCCTCTAAAATAAATTCCATAATCC-3'). The PCR reaction was performed at 95 °C for 5 min, then 40 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 60 s, followed by an extra extension at 72 °C for 5 min. The BSP products were confirmed by electrophoresis on a 1% agarose gel. Finally, they were cloned into a pMD19-T (TaKaRa, Osaka, Japan), and sequenced (Taihegene Biotechnology Co Ltd, Beijing, China).
MTT, plate colony formation, cell invasion assays in vitro
The MTT, plate colony formation, cell invasion assays of transfected CRC cells were determined as previously described (Liang et al, 2013).
Animal models
To evaluate the in vivo tumorigenic effects, 4 × 106 cells were injected subcutaneously into the flank of nude mice (n=5 per group). Tumour size was measured with calipers to estimate volume every 6–7 days until day 28 after injection. The mice were sacrificed and tumors were collected 28 days later. For tail vein metastasis assay, 4 × 106 cells were injected into the tail vein of nude mice. After 2 months, mice were sacrificed and various organs from the thoracic, peritoneal and retroperitoneal cavities were removed, rinsed, fixed and subjected to pathological examination. The number of tumour colonies was determined by using a dissecting microscope. All animal experiments were conducted in strict accordance with the principles and procedures approved by the Committee on the Ethics of Animal Experiments of Southern Medical University.
Luciferase activity assay
For luciferase reporter assays, the 3′ untranslated region (3′ UTR) segment or promoter of MED19 gene was amplified by PCR and inserted into the vector. Co-transfections of MED19 3′ UTR plasmid with miR-214 lentivirus vector or MED19 promoter plasmid with FOXD3 vector into indicated cells were accomplished by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). For the binding of FOXD3 to miR-214 promoter or MED19 promoter, the coding region of FOXD3 and the 2 kb region directly upstream of miR-214 or the 1.3 kb region directly upstream of MED19 transcription binding site were amplified by PCR and then inserted into the vectors respectively. Luciferase activity was measured 48 hours after transfection by the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Each assay was repeated in three independent experiments.
Chromatin immunoprecipitation (ChIP) assay
According to the ChIP Assay Kit (Millipore, Darmstadt, Germany) protocol, SW620 and HT-29 cells were lysed using SDS lysis buffer and DNA was sheared by sonication to lengths between 200 bp and 1000 bp. Protein-DNA complexes were precipitated by anti-FODX3 (Abcam, Cambridge, MA, USA) and anti-IgG antibody respectively. Crosslinks in protein-DNA complexes were then reversed by NaCl. The immunoprecipitated DNA was amplified by PCR for specific sequences (R1) containing putative FOXD3 binding sites.
Immunohistochemical staining (IHC)
Four-micrometer-thick histology sections from xenograft tumours were cut, deparaffinised using xylene, and hydrated through graded alcohol to water. Antigen retrieval was performed by boiling at 100 °C for 10 min in 10 mmol/l citrate buffer (pH=6.0). In brief, these sections were incubated in polyclonal antibody against human Ki-67 (Abnova, Taiwan) overnight at 4 °C. Subsequently, the horseradish-peroxidase-conjugated anti-goat secondary antibody (DakoCytomation, Glostrup, Denmark) was applied and incubated for 1 h at room temperature. The visualisation signal was developed with 3, 3-diaminobenzidine tetra hydrochloride staining, and the slides were counterstained in hematoxylin.
Statistical analysis
All statistical analyses were performed using SPSS 13.0 statistical software and were calculated from 3 independent experiments. Statistical significance was determined using Student t test, Fisher's exact test, or one way analysis of variance (ANOVA) as appropriate. Spearman's correlation coefficient was used to measure the degree of the linear relationship of gene expression levels. Growth curves were generated using the log-rank test. P<0.05 was considered to be statistically significant.
Results
Identification of FOXD3/miR-214/MED19 axis in CRC
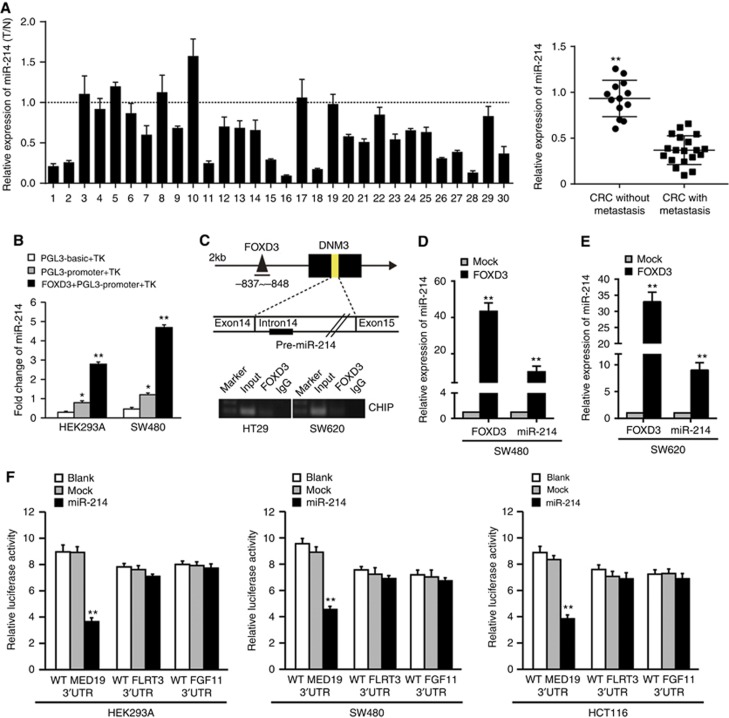
In our previous study, a miRNA microarray assay was used to screen potential metastasis associated miRNAs in CRC tissues with lymphatic metastasis and those without lymphatic metastasis (Li et al, 2015). From the list of differentially expressed miRNAs, we focused on miR-214 because it was one of the most downregulated miRNAs in CRC tissues with lymphatic metastasis and its underlying mechanisms in CRC metastasis remain unclear (Supplementary Figure S1A). Real-time PCR analysis showed that miR-214 was significantly downregulated in 30 paired fresh CRC tissues compared to normal mucosa (Figure 1A). Meanwhile, miR-214 expression was markedly lower in primary CRC tissues with lymphatic metastasis than those without lymphatic metastasis (Figure 1A). These results indicate that miR-214 is downregulated in CRC, which might be associated with CRC metastasis.
Figure 1.
Identification of a FOXD3/miR-214/MED19 axis in human colorectal cancer. (A) Real-time PCR analysis of miR-214 expression in 30 paired cases of the primary CRC tissues with or without metastasis and matched adjacent normal mucosa. The expression of miR-214 in normal mucosa were normalised to 1. (B) Luciferase activity of PGL3-miR-214-promoter construct after transfection of FOXD3 plasmid in HEK293A and SW480 cells. (C) Consite and TFsearch predicted the 2000 bp upstream region of miR-214 for putative transcriptional factors binding site. Chromatin immunoprecipitation assay was performed in HT-29 and SW620 cells transfected with FOXD3-expressing vector. (D and E) Real-time PCR analysis of FOXD3 expression in FOXD3 overexpressing SW480 and SW620 cells. (F) Luciferase activities of wild-type 3′-UTR-MED19-luc, 3′UTR-FLRT3-luc and 3′UTR-FGF11-luc constructs in HEK293A, SW480 and HCT116 cells after transfection of miR-214 plasmid. *P<0.05, **P<0.01. Data represent the mean±SD.
To investigate the potential molecular mechanism of miR-214 in CRC, we analysed transcription factors that are located within 2 kb region directly upstream of the transcription start site of miR-214 using Consite and TFsearch databases (Supplementary Figure S1B). One binding motif was found for FOXD3 within −837 bp to −848 bp regions in the promoter of miR-214. Luciferase reporter assay showed that FOXD3 effectively elevated the luciferase activity of miR-214 promoter in HEK293A and SW480 cells (Figure 1B). ChIP assay was performed to further investigate the combination of FOXD3 with miR-214 promoter. Results showed that FOXD3 could directly bind the region of −837 bp to −848 bp in the promoter of miR-214 in HT-29 and SW620 cells (Figure 1C). After that, we analysed the expression of miR-214 in FOXD3-expressing SW480 and SW620 cells. FOXD3 markedly increased the expression of miR-214 (Figures 1D–E). These data suggest that FOXD3 binds to specific promoter of miR-214 and activates transcription.
MicroRNAs suppress gene expression through interacting with the 3′ UTRs of target mRNAs (Ioshikhes et al, 2007). To identify the downstream targets of miR-214, we used five miRNA target-predicting databases including miRanda, TargetScan, Picta, RNAhybrid, miRBase. MED19, FLRT3, and FGF11 were selected as potential targets due to their overlap among all databases and metastasis-related functions (Hu et al, 2007; Chen et al, 2009; Wen et al, 2013). We cloned the 3' UTRs of MED19, FLRT3 and FGF11 into a luciferase plasmid psiCHEKTM-2 in HEK293A, SW480 and HCT116 cells. Among all cell lines, only the luciferase activity of 3' UTR of MED19 was significantly suppressed by miR-214 (Figure 1F). To determine whether MED19 is transcriptionally regulated by FOXD3, MED19 was ectopically overexpressed using plasmid of MED19 promoter. We found that dual-luciferase activity of MED19 promoter was not significantly affected by FOXD3 compared to mock group (Supplementary Figure S1C). These above data indicate that transcription factor FOXD3 induces miR-214 transcription, subsequently down-regulates MED19 expression, which constitutes a potential FOXD3/miR-214/MED19 axis to play a role in CRC cells.
MiR-214-mediated suppression of MED19 inhibits cell proliferation of CRC
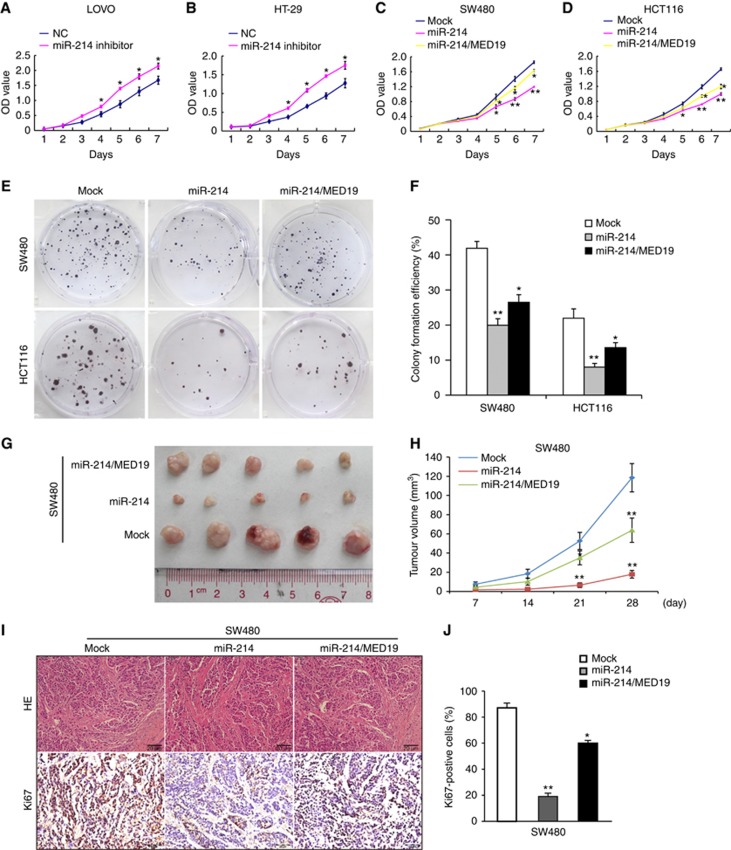
Gain-of-function and loss-of-function assays were performed to confirm whether miR-214 functionally regulating the proliferation of CRC cells by targeting MED19. According to endogenous expression of miR-214 in six CRC cell lines (Supplementary Figure S1D), we transfected miR-214-expressing lentivirus vector into SW480 and HCT116 cells (Supplementary Figure S1E) and knocked down miR-214 with inhibitor in LOVO and HT-29 cells (Supplementary Figure S1F). To perform the rescue experiments, we transfected a construct that encodes entire MED19 coding domains, but lacking the 3′ UTR region into miR-214-expressing cells. Reintroduction of MED19 rescued MED19 expression in miR-214-expressing cells, but did not change the expression of miR-214 (Supplementary Figure S1G–H). MTT assay showed that miR-214 inhibitor significantly increased the growth rate of LOVO and HT-29 cells compared with control group (Figure 2A–B). On the contrary, ectopic miR-214 expression inhibited the proliferation of SW480 and HCT116 cells (Figure 2C–D). Consistently, miR-214 obviously suppressed colony formation of SW480 and HCT116 cells (Figure 2E–F). MTT assay, Boyden chamber, and Matrigel invasion assay showed that ectopic MED19 expression could significantly promote SW480 and LOVO cell proliferation, migration and invasion in vitro (Supplementary Figure S2A–C). Constitutive MED19 expression rescued the proliferation and colony formation phenotypes in miR-214-expressing cells (Figure 2C–F).
Figure 2.
miR-214 suppresses tumour proliferation by target gene MED19 in CRC. (A and B) Effect of miR-214 inhibitor on proliferation of LOVO and HT-29 cells by MTT assay. (C and D) Effects of miR-214 and miR-214/MED19 on proliferation in SW480 and HCT116 cells by MTT assay. (E and F) Effects of miR-214 and miR-214/MED19 on cell proliferation in SW480 and HCT116 cells by colony formation assay. (G and H) Subcutaneous tumours of nude mice injected with SW480/Mock, SW480/miR-214 and SW480/miR-214/MED19 cells. The nodules volume of subcutaneous tumours was measured. (I and J) Hematoxylin–eosin (HE) staining and immunohistochemical (IHC) staining of Ki-67 expression in subcutaneous tumours of nude mice injected with SW480/Mock, SW480/miR-214 and SW480/miR-214/MED19 cells. Ki-67 positive expression rate was measured. Scare bars represent 20 μm. *P<0.05, **P<0.01. Data represent the mean±SD.
To further investigate the effect of miR-214 on cell proliferation in vivo, we subcutaneously injected SW480/miR-214, SW480/miR-214/MED19 and control cells into nude mice. Subcutaneous tumour volume in miR-214 group was significantly decreased compared with mock group, whereas co-expression of MED19 reversed the suppression induced by miR-214 (Figure 2G–H). IHC analysis showed that Ki-67 expression in miR-214-expressing tumours was lower than that in control group, while reintroduction of MED19 displayed increased Ki-67 index (Figure 2I–J, Supplementary Figure S3A–B). Our results thus reveal that miR-214 inhibits the proliferation of CRC cells by targeting MED19.
MiR-214-mediated suppression of MED19 inhibits invasion and metastasis of CRC
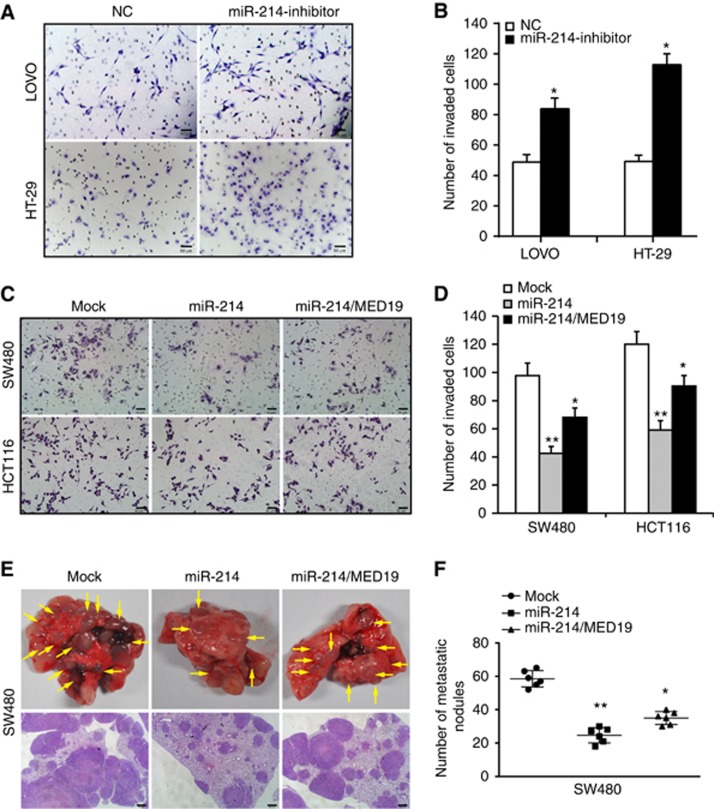
We also assessed the effect of miR-214 on invasion and metastasis of CRC cells. Matrigel invasion assay showed that miR-214 knockdown in LOVO and HT-29 cells obviously increased the number of invaded cells (Figure 3A and B), whereas ectopic miR-214 showed the opposite effect in SW480 and HCT116 cells (Figure 3C and D). Reintroduction of MED19 could partly reverse the suppression of miR-214 on invasion (Figure 3C and D). To test the effect of miR-214 on CRC metastasis, SW480/miR-214 cells, SW480/miR-214/MED19 cells and control cells were injected into tail vein of nude mice. The lung metastatic rate of SW480/mock, SW480/miR-214, and SW480/miR-214/MED19 group in nude mice was 100% (6/6), 33.33% (2/6), and 66.67% (4/6), respectively. Large lung metastatic nodules could be detected in SW480/miR-214/MED19 and SW480/mock groups, while only few small nodules were observed in SW480/miR-214 group (Figure 3E). Moreover, the number of metastatic nodules was obviously reduced in mice injected with SW480/miR-214 cells compared to mock cells, whereas MED19 and miR-214 co-expressing cells caused increased metastatic nodules compared to SW480/miR-214-expressing cells (Figure 3F). Therefore, these results demonstrate that miR-214 suppresses invasion and metastasis of CRC via downregulation of MED19.
Figure 3.
miR-214 suppresses invasion and metastasis of CRC by targeting MED19. (A and B) Effect of miR-214 inhibitor on the invasion of LOVO and HT-29 cells by Boyden chamber. Morphologic comparison of cells penetrating the artificial basement membrane was shown. Scale bars represent 50 μm. (C and D) Effects of miR-214 and miR-214/MED19 on cell invasion of SW480 and HCT116 cells by Boyden chamber. Morphologic comparison of cells penetrating the artificial basement membrane was shown. Scale bars represent 50 μm. (E) SW480/Mock, SW480/miR-214 and SW480/miR-214/MED19 cells were injected into the tail vein of nude mice. Yellow arrows in top panels point at lung metastatic nodules. Scale bars in bottom panels represent 500 μm. (F) The number of lung metastatic nodules per mouse was counted under the microscope. *P<0.05, **P<0.01. Data represent the mean±SD.
Promoter hypermethylation might contribute to the transcriptional silencing of miR-214 in CRC
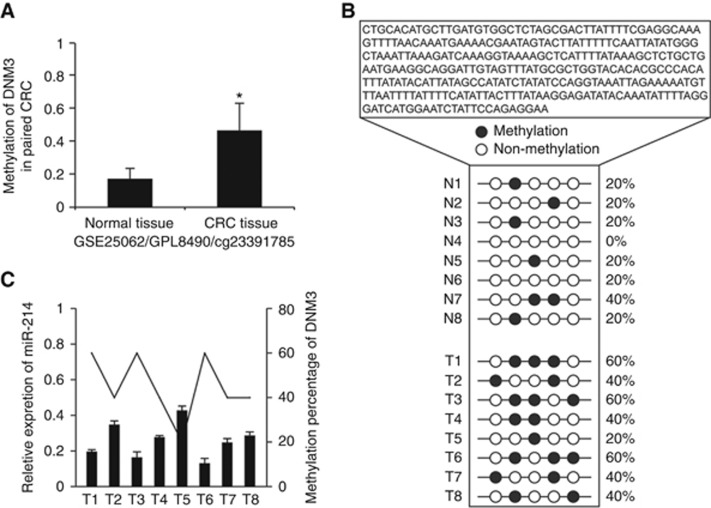
Recent studies indicate that miRNA expression can be deregulated in cancer by different epigenetic mechanisms, including aberrant methylation of the promoter regions or histone modifications (Scott et al, 2006; Suzuki et al, 2013). DNA methylation of promoter associated CpG islands has been reported for several microRNAs, such as miR-137 (Bier et al, 2013); miR-145 (Lee et al, 2013), miR-204 (Ying et al, 2013). Interestingly, miR-214 was an intronic miRNA located between exons 14 and 15 of DNM3 gene, so we investigated the methylation status of DNM3 promoter. Gene Expression Omnibus (GEO) database was used to analyse the CpG island methylation of DNM3 promoter and results indicated that the methylation status of DNM3 promoter in CRC was higher than in normal mucosa (Figure 4A). We next analysed the methylation status of DNM3 promoter in eight cases of paired CRC tissues by BSP. BSP results showed increased CpG island methylation of DNM3 promoter in CRC tissues (Figure 4B). Combining the results of real-time PCR and BSP, we found that miR-214 expression was decreased in CRC tissues with hypermethylation status of DNM3 promoter (Figure 4C). These results indicate, to a certain extent, that hypermethylation of DNM3 promoter might lead to transcriptional silence of miR-214 in CRC.
Figure 4.
Promoter hypermethylation contributes to the transcriptional silencing of miR-214 in CRC. (A) The analysis results of GEO showed that the degree of CpG Island methylation in CRC was higher than that in normal mucosa. (B) Bisulphite genomic-sequencing analysis of the CpG island of DNM3 promoter in paired CRC tissue. (C) The relationship between the expression of miR-214 and the degree of CpG Island methylation in eight cases of CRC tissues and corresponding normal mucosa. miR-214 expression was determined by real-time PCR. The methylation percentage was calculated using the BSP results from B. *P<0.05, **P<0.01. Data represent the mean±SD.
FOXD3 upregulates miR-214 transcription and suppresses CRC cell proliferation in vitro and in vivo
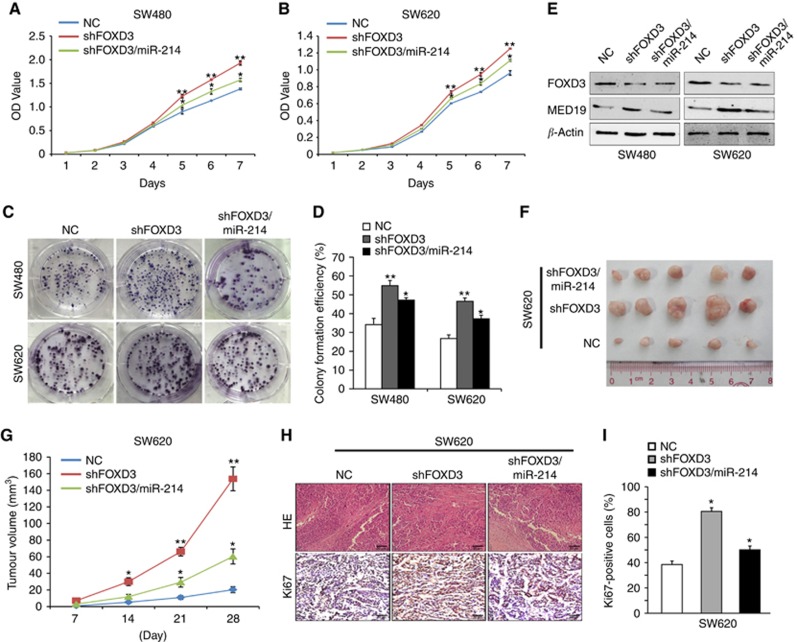
To further investigate whether miR-214 affects the function of FOXD3 in the progression of CRC, we silenced FOXD3 with three siRNAs in SW480 and SW620 cells according to endogenous expression of FOXD3 in six CRC cell lines. According to transfection efficiency, we chose siRNA3 to perform following experiment (Supplementary Figure S3C and D). Subsequently, we established stable FOXD3-depleting SW480 and SW620 cells and then stably transfected FOXD3-depleting cells with miR-214 and confirmed its over-expression (Supplementary Figure S3E). Compared with NC group, knockdown of FOXD3 promoted cell proliferation and colony formation in vitro, while reintroduction of miR-214 incompletely reversed the promotion (Figures 5A and D). Moreover, ectopic miR-214 expression reduced the expression of MED19. Knockdown of FOXD3 led to increased expression of MED19, whereas enhanced expression of miR-214 abolished FOXD3 responsiveness (Figure 5E).
Figure 5.
FOXD3 upregulates miR-214 expression and suppresses tumorigenesis. (A, B) Effects of shFOXD3 and shFOXD3/miR-214 on cell proliferation in SW480 and SW620 cells by MTT assay. (C, D) Effects of shFOXD3 and shFOXD3/miR-214 on cell proliferation in SW480 and SW620 cells by colony formation assay. (E) MED19 expression in cells treated with NC, shFOXD3 or shFOXD3/miR-214 by Western blot. Expression levels were normalised to β-actin. (F, G) Subcutaneous tumours of nude mice injected with SW620/NC, SW620/shFOXD3 and SW620/shFOXD3/miR-214 cells. The nodules volume of subcutaneous tumours was measured. (H, I) Hematoxylin–eosin(HE) staining and immunohistochemical (IHC) staining of Ki-67 expression in subcutaneous tumours of nude mice injected with SW620/NC, SW620/shFOXD3 and SW620/shFOXD3/miR-214 cells. Ki-67 positive expression rate was measured. Scale bars represent 20 μm. *P<0.05, **P<0.01. Data represent the mean±SD.
We further examined the effect of FOXD3 on tumour growth in vivo. SW620/shFOXD3/miR-214, SW620/shFOXD3 and control cells were subcutaneously injected to nude mice respectively. Mice injected with SW620/shFOXD3 cells had increased subcutaneous tumour volume compared with those injected with SW620/NC cells, whereas constitutive miR-214 rescued tumour growth induced by FOXD3 knockdown (Figures 5F–G). Immunohistochemistry staining exhibited higher positive rate of Ki-67 index in FOXD3-depleting tumour than control group, whereas miR-214 could partly abrogated the Ki-67 expression induced by FOXD3 knockdown (Figures 5H and I, Supplementary Figure S3F and G). These results indicate that FOXD3 upregulates miR-214 transcription, then suppresses the proliferation of CRC cells.
FOXD3 upregulates miR-214 transcription and suppresses CRC invasion and metastasis in vitro and in vivo
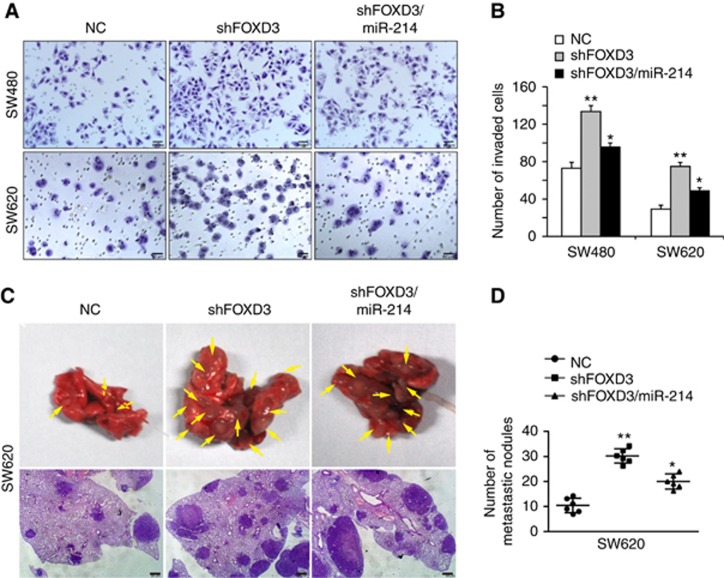
We next investigated the role of FOXD3 in CRC invasiveness and metastases. Results of Matrigel invasion assay showed that knockdown of FOXD3 enhanced invasiveness of CRC cells in vitro, while reintroduction of miR-214 attenuated the promoting effects of FOXD3 knockdown (Figures 6A–B). To confirm whether downregulation of FOXD3 is associated with CRC metastasis in vivo, SW620/NC, SW620/shFOXD3 and SW620/shFOXD3/miR-214 cells were injected into tail vein to seed lung metastases. In the group of mice injected with SW620/NC cells, only 16.667% (1 of 6) of mice had lung metastases. However, lung metastatic rate of mice injected with SW620/shFOXD3 and SW620/shFOXD3/miR-214 was 83.333% and 50%, respectively (Figure 6C). The number and volume of metastatic nodules in the lung were significantly increased in mice injected with FOXD3-depleting cells compared with those injected with control cells. Moreover, less lung metastatic nodules were observed in shFOXD3/miR-214 group than shFOXD3 group (Figure 6D). On the basis of these results, it would be reasonable to conclude that FOXD3 suppresses invasion and metastasis of CRC by upregulating miR-214.
Figure 6.
FOXD3 upregulates miR-214 expression and suppresses CRC invasion and metastasis. (A) Effects of shFOXD3 and shFOXD3/miR-214 on cell invasion of SW480 and SW620 cells by Boyden chamber. Morphologic comparison of cells penetrating the artificial basement membrane was shown. Scale bars represent 50 μm. (B) The number of invaded SW480 and SW620 cells was measured under the microscope. (C) SW620/NC, SW620/shFOXD3 and SW620/shFOXD3/miR-214 cells were injected into the tail vein of nude mice. Yellow arrows in top panels point at lung metastatic nodules. Scale bars in bottom panels represent 500 μm. (D) The number of lung metastatic nodules per mouse was counted under the microscope. *P<0.05, **P<0.01. Data represent the mean±SD.
Correlations of miR-214 with MED19, FOXD3 expressions in CRC cell lines and clinical specimens
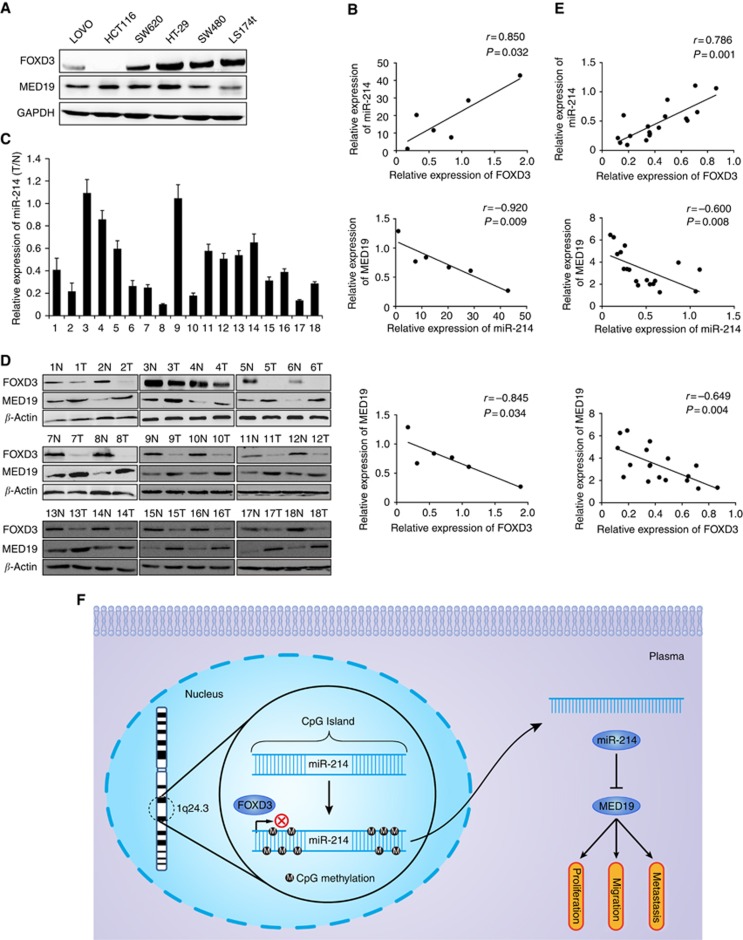
To demonstrate the relationship between miR-214, MED19 and FOXD3 expressions in CRC cell lines and tissues, we detected expressions of miR-214, MED19 and FOXD3 in 6 CRC cell lines and 18 paired cases of human CRC clinical specimens. Spearman's correlation analyses showed a positive correlation between FOXD3 and miR-214, and negative correlations between miR-214 and MED19, FOXD3 and MED19 expressions in six CRC cell lines (Figures 7A and B). In addition, miR-214 was obviously downregulated, whereas MED19 was upregulated in CRC clinical specimens compared with corresponding normal mucosa (Figures 7C and D). Spearman's correlation analyses showed a positive correlation between FOXD3 and miR-214 and negative correlations between miR-214 and MED19, FOXD3 and MED19 expressions (Figure 7E).
Figure 7.
Correlations of miR-214 with MED19, FOXD3 expression in CRC cell lines and tissues. (A) Western blot analysis of endogenous expression of FOXD3 and MED19 in six CRC cell lines. Expression levels were normalised to GAPDH. (B) Spearman's correlation analysis of the expression between FOXD3 and miR-214, miR-214 and MED19, FOXD3 and MED19 in six CRC cell lines. (C) Real-time PCR analysis of miR-214 expression in 18 paired primary CRC tissues (T) and matched adjacent normal mucosa (N). (D) Western blot analysis of FOXD3 and MED19 expression level in 18 paired primary CRC tissues (T) and corresponding normal mucosa (N). Expression levels were normalised to β-actin. (E) Spearman's correlation analysis of the expression between FOXD3 and miR-214, miR-214 and MED19, FOXD3 and MED19 in 18 paired CRC tissues. (F) Schematic diagram of FOXD3/miR-214/MED19 axis in the regulation of proliferation, migration and metastasis of human colorectal cancer.
Discussion
Increasing evidence has highlighted that miRNAs are aberrantly expressed or mutated in human cancer, indicating that they may function as a novel class of oncogenes or tumour suppressor genes (Calin et al, 2002; Ma et al, 2013); Chan and Wang, 2015. For instance, miR-204 suppressed self-renewal, stem-cell-associated phenotype, and migration of glioma cells by targeting for SOX4 and EphB2 (Ying et al, 2013); miR-21 was markedly elevated in human glioblastoma tumour and contribute to the maliganant phenotype by blocking expression of critical apoptosis-related genes (Chan et al, 2005). In our previous research, a list of deregulated miRNAs were screened out using miRNA microarray, including miR-137, miR-371-5p and miR-214. MiR-137 was able to suppress CRC invasion and metastasis by way of regulating FMNL2 (Liang et al, 2013); miR-371-5p inhibited EMT, stem cell properties and metastasis by targeting SOX2 in CRC (Li et al, 2015). MiR-214, located in the 14th intron of DNM3, was another significantly deregulated miRNA we found in CRC specimens. Emerging studies showed that miR-214 was implicated in many human physiological and pathological process. For instance, Li et al (2016) found that increased osteoclastic miR-214-3p reduced bone formation in elderly woman with fractures and in ovariectomised mice (Li et al, 2016); miR-214 was decreased significantly in Parkinson's disease (PD) patients and may represent a novel biomarker for the early detection of PD (Dong et al, 2016); miR-214 might promote Th17 cell differentiation by targeting mTOR signalling in purified CD4+ T cells of multiple sclerosis (Ahmadian-Elmi et al, 2016); Sun et al (2015) found that miR-214 mediated CF proliferation and collagen synthesis via inhibition of Mfn2 and activation of ERK1/2 MAPK signalling. In addition, miR-214 has been reported to be deregulated in many human tumours. Owing to ‘oncomir' or ‘tumour suppressor-mir' functions of miRNA in different tumour, it is still urged to investigate its potential regulatory molecular mechanisms for gaining better clinic diagnosis and treatment of CRC. In the present study, we set out to explore the role of miR-214 in vitro and in vivo and put forward a FOXD3/miR-214/MED19 axis involved in cell proliferation, migration and metastasis of CRC.
MiR-214 expression was apparently downregulated in CRC clinical specimens by real-time PCR, which was consistent with the microarray assay, and its downregulation was associated with CRC lymphatic metastasis. What is more, we found a hypermethylation status of promoter region in miR-214-encoding gene DNM3 by BSP, and this might explain the downregulation mechanism of miR-214 in CRC. Expression of miR-214 is deregulated in many tumours including pancreatic cancer, melanoma, and hepatocellular carcinoma (Zhang et al, 2010; Penna et al, 2013; Zhang et al, 2015), whereas upregulated in breast cancer by targeting p53 (Wang et al, 2015). The pleiotropic and tumour-specific of miR-214 contributes to various cancer formation and progression via its several target genes including p53, Bcl-2/Bax, TFAM, EZH2 (Wen et al, 2014; Yang et al, 2014; Tian et al, 2015). Recently, Chen et al (2014) reported that miR-214 negatively regulated liver metastasis in CRC. However, the molecular mechanism of miR-214 in CRC metastasis has still not been illustrated. Therefore, we explored potential miR-214-mediated mechanism in the progression of CRC. Generally, miRNAs showed to be regulated by the upstream transcription factors (O'Donnell et al, 2005). We analysed the promoter region of miR-214 and found FOXD3 transcriptionally regulated miR-214. FOXD3 has been reported to inhibit growth, invasion, metastasis and angiogenesis in several tumours (Li et al, 2013; Chu et al, 2014; Liu et al, 2014). Recently, van Roon et al (2013) found that FOXD3 gene expression was repressed due to hypermethylation of promoter in human CRC. We speculate that hypermethylation of FOXD3 gene could lead to its low expression in CRC, and this might subsequently cause low miR-214 expression. Combined with bioinformatics search and dual-luciferase assay, MED19 was found to be a major downstream effector of miR-214. Meanwhile, increasing evidence has revealed MED19 as functional target involved in tumour progression (Li et al, 2011). Our results revealed that FOXD3 inhibited the expression of MED19 by upregulating the expression of miR-214 in CRC cells. Thus, the FOXD3/miR-214/MED19 axis might act as a key pathway in CRC metastasis.
Next, we identify that miR-214, induced by its upstream transcription factor FOXD3, can suppress tumour growth and metastasis in CRC by targeting MED19. The in vitro and in vivo ‘MED19 rescue' experiments proved that miR-214 suppressed tumour growth and metastasis in CRC mainly by targeting MED19. Mediator complex subunit 19 (MED19) is a member of the mediator that has a key role in the activation and repression of signal transduction or the regulation of transcription in carcinomas (Casamassimi and Napoli, 2007; Sun et al, 2011). Accumulating evidence has shown that MED19 has important roles in cancer cell proliferation and tumorigenesis, and suppression of MED19 expression induces inhibition of cell proliferation and tumorigenesis in several different tumour types including lung cancer, pancreatic cancer, ovarian cancer and breast cancer (Li et al, 2011; Liu et al, 2012; Wei et al, 2015). Then we speculated that miR-214 played tumorigenic role by its target MED19 in CRC. Further research confirmed that FOXD3 stimulated the transcription activity of miR-214, subsequently significantly suppressed cell proliferation and metastasis by downregulation of MED19 in CRC. FOXD3 as a novel tumour suppressor has been reported highly connected with carcinogenesis (Chu et al, 2014). Hypermethylation of FOXD3 suppresses cell proliferation, invasion and metastasis in hepatocellular carcinoma (He et al, 2015). And recent study also suggested that decreased FOXD3 expression is associated with poor prognosis in patients with high-grade tumour (Du et al, 2015). Therefore, we provide evidence that the involvement of FOXD3/miR-214/MED19 axis in tumour growth and metastasis of CRC.
Eventually, we detected the correlations of miR-214, FOXD3 and MED19 in six CRC cell lines and 18 paired cases of human CRC tissues. Our results showed that there was a positive relationship between the expression level of miR-214 and FOXD3, and a negative relationship between miR-214 and MED19, FOXD3 and MED19. On the basis of these evidence, we clearly validate that FOXD3 induces miR-214 expression and consequently represses its target MED19 in CRC.
In summary, FOXD3/miR-214/MED19 axis has an important role in the regulation of CRC progression. Hypermethylation of DNM3 promoter leads to low expression of miR-214 in CRC. Function experiments demonstrate that miR-214 mediates the suppressive role of FOXD3 in proliferation, invasion and metastasis of CRC by targeting MED19 (Figure 7F). The FOXD3/miR-214/MED19 signalling axis might offer a promising therapeutic target for CRC treatment.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program, 2015CB554002), Key project of National Natural Science Fund (Guangdong Province NSFC- joint fund, U1201226), National Natural Science Foundation of China (81272759, 81172382, 81472313, 81401927); Natural Science Foundation of Guangdong Province (S2013010014544). We thank Professor Reddy for editing the English writing.
Author contributions
GYH, JLH, LZ, XHZ, SNX, DZ, GFL, WTL carried out experiments. YQD and LL conceived experiments and analysed the data. All authors were involved in writing the paper and had final approval of the submitted and published versions.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Ahmadian-Elmi M, Bidmeshki PA, Naghavian R, Ghaedi K, Tanhaei S, Izadi T, Nasr-Esfahani MH (2016) miR-27a and miR-214 exert opposite regulatory roles in Th17 differentiation via mediating different signaling pathways in peripheral blood CD4+ T lymphocytes of patients with relapsing-remitting multiple sclerosis. Immunogenetics 68: 43–54. [DOI] [PubMed] [Google Scholar]
- Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S, Xiang C, Poisson L, DeCarvalho AC, Slavin S, Jacoby E, Yalon M, Toren A, Mikkelsen T, Brodie C (2013) MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget 4: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Perez DCI, Malumbres M (2008) Control of cell proliferation pathways by microRNAs. Cell Cycle 7: 3143–3148. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamassimi A, Napoli C (2007) Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie 89: 1439–1446. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033. [DOI] [PubMed] [Google Scholar]
- Chan SH, Wang LH (2015) Regulation of cancer metastasis by microRNAs. J Biomed Sci 22: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, Wang FH, Chiao LJ, Pelicano H, Huang P, Li YH, Xu RH (2014) Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology 60: 598–609. [DOI] [PubMed] [Google Scholar]
- Chen X, Koh E, Yoder M, Gumbiner BM (2009) A protocadherin-cadherin-FLRT3 complex controls cell adhesion and morphogenesis. PLoS ONE 4: e8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu TL, Zhao HM, Li Y, Chen AX, Sun X, Ge J (2014) FoxD3 deficiency promotes breast cancer progression by induction of epithelial-mesenchymal transition. Biochem Biophys Res Commun 446: 580–584. [DOI] [PubMed] [Google Scholar]
- Derfoul A, Juan AH, Difilippantonio MJ, Palanisamy N, Ried T, Sartorelli V (2011) Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis 32: 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Wang C, Lu S, Yu C, Huang L, Feng W, Xu H, Chen X, Zen K, Yan Q, Liu W, Zhang C, Zhang CY (2016) A panel of four decreased serum microRNAs as a novel biomarker for early Parkinson's disease. Biomarkers 21: 129–137. [DOI] [PubMed] [Google Scholar]
- Du W, Pang C, Wang D, Zhang Q, Xue Y, Jiao H, Zhan L, Ma Q, Wei X (2015) Decreased FOXD3 expression is associated with poor prognosis in patients with high-grade gliomas. PLoS ONE 10: e0127976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Zhang Y, Deng M, Ye R, Zhao S, Wang Y, Li J, Zhao Z (2014) MicroRNA control of epithelial-mesenchymal transition in cancer stem cells. Int J Cancer 135: 1019–1027. [DOI] [PubMed] [Google Scholar]
- He G, Hu S, Zhang D, Wu P, Zhu X, Xin S, Lu G, Ding Y, Liang L (2015) Hypermethylation of FOXD3 suppresses cell proliferation, invasion and metastasis in hepatocellular carcinoma. Exp Mol Pathol 99: 374–382. [DOI] [PubMed] [Google Scholar]
- Hu L, Sham JS, Xie D, Wen JM, Wang WS, Wang Y, Guan XY (2007) Up-regulation of fibroblast growth factor 3 is associated with tumor metastasis and recurrence in human hepatocellular carcinoma. Cancer Lett 252: 36–42. [DOI] [PubMed] [Google Scholar]
- Huang HJ, Liu J, Hua H, Li SE, Zhao J, Yue S, Yu TT, Jin YC, Cheng SY (2014) MiR-214 and N-ras regulatory loop suppresses rhabdomyosarcoma cell growth and xenograft tumorigenesis. Oncotarget 5: 2161–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S (2010) MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Ioshikhes I, Roy S, Sen CK (2007) Algorithms for mapping of mRNA targets for microRNA. DNA Cell Biol 26: 265–272. [DOI] [PubMed] [Google Scholar]
- Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, Poisson LM, Mikkelsen T, Slavin S, Jacoby E, Yalon M, Toren A, Rempel SA, Brodie C (2013) MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS ONE 8: e54652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu J, Guo B, Liang C, Dang L, Lu C, He X, Cheung HY, Xu L, Lu C, He B, Liu B, Shaikh AB, Li F, Wang L, Yang Z, Au DW, Peng S, Zhang Z, Zhang BT, Pan X, Qian A, Shang P, Xiao L, Jiang B, Wong CK, Xu J, Bian Z, Liang Z, Guo DA, Zhu H, Tan W, Lu A, Zhang G (2016) Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun 7: 10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mei H, Qi M, Yang D, Zhao X, Xiang X, Pu J, Huang K, Zheng L, Tong Q (2013) FOXD3 is a novel tumor suppressor that affects growth, invasion, metastasis and angiogenesis of neuroblastoma. Oncotarget 4: 2021–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LH, He J, Hua D, Guo ZJ, Gao Q (2011) Lentivirus-mediated inhibition of Med19 suppresses growth of breast cancer cells in vitro. Cancer Chemother Pharmacol 68: 207–215. [DOI] [PubMed] [Google Scholar]
- Li XH, Fang DN, Zeng CM (2011) Knockdown of MED19 by short hairpin RNA-mediated gene silencing inhibits pancreatic cancer cell proliferation. Cancer Biother Radiopharm 26: 495–501. [DOI] [PubMed] [Google Scholar]
- Li Y, Lv Z, He G, Wang J, Zhang X, Lu G, Ren X, Wang F, Zhu X, Ding Y, Liao W, Ding Y, Liang L (2015) The SOX17/miR-371-5p/SOX2 axis inhibits EMT, stem cell properties and metastasis in colorectal cancer. Oncotarget 6: 9099–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Li X, Zhang X, Lv Z, He G, Zhao W, Ren X, Li Y, Bian X, Liao W, Liu W, Yang G, Ding Y (2013) MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology 144: 624–635.e4. [DOI] [PubMed] [Google Scholar]
- Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W, Yang YZ, Luo RZ, Zhang CZ, Yun JP (2014) FoxD3-regulated microRNA-137 suppresses tumour growth and metastasis in human hepatocellular carcinoma by targeting AKT2. Oncotarget 5: 5113–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tao X, Fan L, Jia L, Gu C, Feng Y (2012) Knockdown of mediator complex subunit 19 inhibits the growth of ovarian cancer. Mol Med Rep 6: 1050–1056. [DOI] [PubMed] [Google Scholar]
- Ma Y, Li W, Wang H (2013) Roles of miRNA in the initiation and development of colorectal carcinoma. Curr Pharm Des 19: 1253–1261. [DOI] [PubMed] [Google Scholar]
- Momose K, Minami A, Shimono Y, Mizutani K, Nobutani K, Azuma T, Takai Y (2013) miR-214 and hypoxia down-regulate Necl-2/CADM1 and enhance ErbB2/ErbB3 signaling. Genes Cells 18: 195–202. [DOI] [PubMed] [Google Scholar]
- Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA (2009) MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 9: 293–302. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843. [DOI] [PubMed] [Google Scholar]
- Penna E, Orso F, Cimino D, Vercellino I, Grassi E, Quaglino E, Turco E, Taverna D (2013) miR-214 coordinates melanoma progression by upregulating ALCAM through TFAP2 and miR-148b downmodulation. Cancer Res 73: 4098–4111. [DOI] [PubMed] [Google Scholar]
- Penna E, Orso F, Taverna D (2015) miR-214 as a key hub that controls cancer networks: small player, multiple functions. J Invest Dermatol 135: 960–969. [DOI] [PubMed] [Google Scholar]
- Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC (2006) Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res 66: 1277–1281. [DOI] [PubMed] [Google Scholar]
- Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC, Huang CH, Lee YS, Yen TC, Hsieh SY (2012) MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol 57: 584–591. [DOI] [PubMed] [Google Scholar]
- Sun M, Jiang R, Li JD, Luo SL, Gao HW, Jin CY, Shi DL, Wang CG, Wang B, Zhang XY (2011) MED19 promotes proliferation and tumorigenesis of lung cancer. Mol Cell Biochem 355: 27–33. [DOI] [PubMed] [Google Scholar]
- Sun M, Yu H, Zhang Y, Li Z, Gao W (2015) MicroRNA-214 mediates isoproterenol-induced proliferation and collagen synthesis in cardiac fibroblasts. Sci Rep 5: 18351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Maruyama R, Yamamoto E, Kai M (2013) Epigenetic alteration and microRNA dysregulation in cancer. Front Genet 4: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Zeng G, Li X, Wu Z, Wang L (2015) Cantharidin inhibits cell proliferation and promotes apoptosis in tongue squamous cell carcinoma through suppression of miR-214 and regulation of p53 and Bcl-2/Bax. Oncol Rep 33: 3061–3068. [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87–108. [DOI] [PubMed] [Google Scholar]
- van Roon EH, Boot A, Dihal AA, Ernst RF, van Wezel T, Morreau H, Boer JM (2013) BRAF mutation-specific promoter methylation of FOX genes in colorectal cancer. Clin Epigenetics 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM (2010) MicroRNAs: oncogenes, tumor suppressors or master regulators of cancer heterogeneity? Biochim Biophys Acta 1805: 72–86. [DOI] [PubMed] [Google Scholar]
- Wang F, Lv P, Liu X, Zhu M, Qiu X (2015) microRNA-214 enhances the invasion ability of breast cancer cells by targeting p53. Int J Mol Med 35: 1395–1402. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z, Jiang J (2012. a) MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of beta-catenin. Biochem Biophys Res Commun 428: 525–531. [DOI] [PubMed] [Google Scholar]
- Wang YS, Wang YH, Xia HP, Zhou SW, Schmid-Bindert G, Zhou CC (2012) MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac J Cancer Prev 13: 255–260. [DOI] [PubMed] [Google Scholar]
- Wei L, Wang XW, Sun JJ, Lv LY, Xie L, Song XR (2015) Knockdown of Med19 suppresses proliferation and enhances chemo-sensitivity to cisplatin in non-small cell lung cancer cells. Asian Pac J Cancer Prev 16: 875–880. [DOI] [PubMed] [Google Scholar]
- Wen H, Feng CC, Ding GX, Meng DL, Ding Q, Fang ZJ, Xia GW, Xu G, Jiang HW (2013) Med19 promotes bone metastasis and invasiveness of bladder urothelial carcinoma via bone morphogenetic protein 2. Ann Diagn Pathol 17: 259–264. [DOI] [PubMed] [Google Scholar]
- Wen Z, Lei Z, Jin-An M, Xue-Zhen L, Xing-Nan Z, Xiu-Wen D (2014) The inhibitory role of miR-214 in cervical cancer cells through directly targeting mitochondrial transcription factor A (TFAM). Eur J Gynaecol Oncol 35: 676–682. [PubMed] [Google Scholar]
- Yang T, Gu H, Chen X, Fu S, Wang C, Xu H, Feng Q, Ni Y (2014) Cardiac hypertrophy and dysfunction induced by overexpression of miR-214 in vivo. J Surg Res 192: 317–325. [DOI] [PubMed] [Google Scholar]
- Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M (2013) Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res 73: 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LL, Guo YJ, Zhao CN, Gao JY (2015) Effects and mechanism of miR-214 on hepatocellular carcinoma. Asian Pac J Trop Med 8: 392–398. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ (2010) Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol 3: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen C, Guo W, Zheng S, Sun Z, Geng X (2016) DNM3 attenuates hepatocellular carcinoma growth by activating P53. Med Sci Monit 22: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.