Abstract
Expansion of (CAG)•(CTG) repeats causes a number of familial neurodegenerative disorders. Although the underlying mechanism remains largely unknown, components involved in DNA mismatch repair, particularly mismatch recognition protein MutSβ (a MSH2-MSH3 heterodimer), are implicated in (CAG)•(CTG) repeat expansion. In addition to recognizing small insertion-deletion loop-outs, MutSβ also specifically binds DNA hairpin imperfect heteroduplexes formed within (CAG)n•(CTG)n sequences. However, whether or not and how MutSβ binding triggers expansion of (CAG)•(CTG) repeats remain unknown. We show here that purified recombinant MutSβ physically interacts with DNA polymerase β (Polβ) and stimulates Polβ-catalyzed (CAG)n or (CTG)n hairpin retention. Consistent with these in vitro observations, MutSβ and Polβ interact with each other in vivo, and colocalize at (CAG)•(CTG) repeats during DNA replication. Our data support a model for error-prone processing of (CAG)n or (CTG)n hairpins by MutSβ and Polβ during DNA replication and/or repair: MutSβ recognizes (CAG)n or (CTG)n hairpins formed in the nascent DNA strand, and recruits Polβ to the complex, which then utilizes the hairpin as a primer for extension, leading to (CAG)•(CTG) repeat expansion. This study provides a novel mechanism for trinucleotide repeat expansion in both dividing and non-dividing cells.
Keywords: trinucleotide repeat expansion, MutSβ, DNA polymerase β, (CAG)n•(CTG)n hairpin
Introduction
Trinucleotide repeat (TNR) expansion causes a number of familial neurological, neurodegenerative and neuromuscular disorders, including Huntington's disease, myotonic dystrophy type 1 and fragile X syndrome1,2,3,4. Despite extensive studies, the molecular mechanism that causes TNR expansion is not fully understood.
Several models have been proposed to interpret this process. Given that (CAG)n and (CTG)n sequences form stable hairpins in vitro5, a favored model is that hairpin formation in the nascent DNA strand during DNA replication and repair leads to TNR expansion. Indeed, Liu et al.6,7 showed that (CAG)•(CTG) repeat instability is associated with DNA replication, most likely through hairpin formation. We recently demonstrated that hairpin-mediated (CAG)n•(CTG)n expansion during DNA replication and repair requires both DNA polymerase δ (Polδ) and polymerase β (Polβ)8. Polδ alone removes the hairpin structure using its 3′-5′ proofreading activity, but in the presence of Polβ, Polδ preferentially facilitates hairpin retention. This is because Polβ can use the hairpin structure as a primer for DNA synthesis, and the resulting hairpin-containing structure is effectively extended by Polδ, leading to (CAG)•(CTG) repeat expansion8. However, how Polβ is preferentially recruited to the hairpin structure over Polδ is unknown.
DNA mismatch repair (MMR) is a critical genome maintenance system that corrects base-base and small insertion-deletion mispairs generated during DNA replication9,10,11. Surprisingly, key components of this well-known genome maintenance system, especially MutSβ (a heterodimer of MSH2 and MSH3) and MutLα (a heterodimer of MLH1 and PMS2), have been implicated in (CAG)•(CTG) repeat expansion12,13. As a mismatch recognition protein for MMR, MutSβ recognizes and participates in repair of small insertion-deletion mispairs11,14. Animal studies revealed that knockouts of the MutSβ subunits MSH2 or MSH3 efficiently block (CAG)n and (CTG)n expansion15,16,17. Similarly, siRNA knockdown of MutSβ in cell lines inhibits TNR expansions18. In vitro studies showed that MutSβ stimulates human cell extract-catalyzed instability of (CAG)•(CTG) repeats19. These findings imply a direct involvement of MutSβ in promoting expansion of (CAG)•(CTG) repeats. Consistent with this idea, purified MutSβ was found to specifically recognize DNA hairpins formed within (CAG)•(CTG) repeats20,21. However, the molecular mechanism by which MutSβ promotes expansion of (CAG)•(CTG) repeats is unclear. Given that human cells possess a strand-specific DNA hairpin repair pathway22,23, it has been postulated that binding of MutSβ to the unusual DNA structure traps the protein on the DNA so that it inhibits hairpin repair and/or MMR20,24; but this assumption is not in agreement with the fact that an excess amount of MutSβ does not inhibit DNA hairpin repair activity of HeLa extracts21, suggesting involvement of a different mechanism.
In this study, we report the discovery of a novel role for MutSβ to promote (CAG)•(CTG) repeat expansion. We show here that MutSβ physically interacts with Polβ and stimulates Polβ-catalyzed (CAG)n•(CTG)n hairpin retention in vitro. Consistent with these results, we find that MutSβ and Polβ colocalize at long (CAG)•(CTG) repeat regions in S phase in HeLa cells. These observations suggest that MutSβ mediates (CAG)•(CTG) repeat expansion during DNA synthesis, i.e., MutSβ recognizes DNA hairpins formed within (CAG)n•(CTG)n sequences at the site of DNA synthesis and recruits Polβ to the complex, where Polβ uses the newly formed hairpin as a primer for extension, fixing the hairpin structure and leading to (CAG)•(CTG) repeat expansion.
Results
MutSβ promotes (CAG)n or (CTG)n hairpin retention during in vitro DNA synthesis
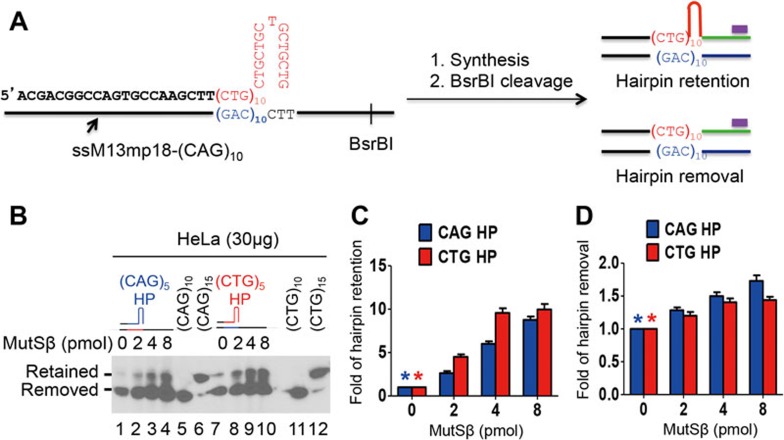
To determine whether MutSβ promotes (CAG)•(CTG) repeat expansion by enhancing (CAG)n•(CTG)n hairpin retention in the nascent strand, in vitro DNA synthesis was conducted in HeLa nuclear extracts using a (CAG)5 and (CTG)5 hairpin-containing primer extension system8 in the presence or absence of MutSβ. The reaction products were subjected to Southern blot analysis to determine hairpin retention or removal using a 32P-labeled probe specifically annealing to the downstream sequence of the newly synthesized strand as described8 (also see Figure 1A). As expected, incubation of (CAG)5 or (CTG)5 hairpin substrate with HeLa nuclear extracts in the absence of MutSβ yielded a major hairpin-removed band (lower band) and a minor hairpin-retained band (upper band) (Figure 1B). Interestingly, when purified MutSβ was added to the reaction, the amounts of the hairpin-retained product increased proportionally to the increasing amounts of MutSβ for both the CAG hairpin substrate (Figure 1B, reactions 2-4) and the CTG hairpin substrate (Figure 1B, reactions 8-10), with 8 pmol of MutSβ stimulating the hairpin retention activity by as much as 10-fold (Figure 1C). Given that MutSβ specifically binds (CAG)n and (CTG)n hairpins20,21, these results suggest that the binding of (CAG)n•(CTG)n hairpins formed in the nascent DNA strand by MutSβ induces (CAG)•(CTG) repeat expansion.
Figure 1.
MutSβ promotes (CAG)n or (CTG)n hairpin retention synthesis in HeLa nuclear extracts. (A) Diagram of hairpin removal/retention assay by Southern blot analysis. The purple bar shows the 32P-labeled oligonucleotide probe, which specifically anneals to the newly synthesized strand near the BsrBI site. The complete primer sequence of a CTG hairpin substrate used in this study is also shown. (B) Southern blot analysis showing the effect of MutSβ on (CAG)5 or (CTG)5 hairpin retention/removal during DNA synthesis in HeLa nuclear extracts. DNA hairpin substrate (0.15 pmol) was incubated with limited amount (30 μg) of HeLa nuclear extracts in the presence of increasing amounts of purified MutSβ. The resulting products were examined by Southern blot analysis. (C, D) Quantification of hairpin-retained products and hairpin-removed products shown in B, respectively. The data were from three independent experiments and the error bar represents SD.
It was noted that addition of exogenous MutSβ also led to an increase in hairpin removal activity in HeLa extracts, with almost a 2-fold stimulation in reactions containing 8 pmol of MutSβ (Figure 1B and 1D). This is likely due to repair of the retained hairpin by MutSβ-initiated MMR. We demonstrated previously that although MutSβ is not involved in removing DNA hairpins consisting of 10 or more CAG and CTG repeats21,25, it stimulates (CAG)5 or (CTG)5 hairpin repair in nuclear extracts derived from a MSH2-deficient cancer cell line25. This observation is consistent with the fact that MMR can process up to 16-nucleotide insertion-deletion loop-outs26. We therefore conclude that the observed enhancement of (CAG)5 or (CTG)5 hairpin removal is due to MutSβ-stimulated MMR in HeLa nuclear extracts.
MutSβ specifically stimulates DNA polymerase β-induced (CAG)n and (CTG)n hairpin retention during DNA synthesis
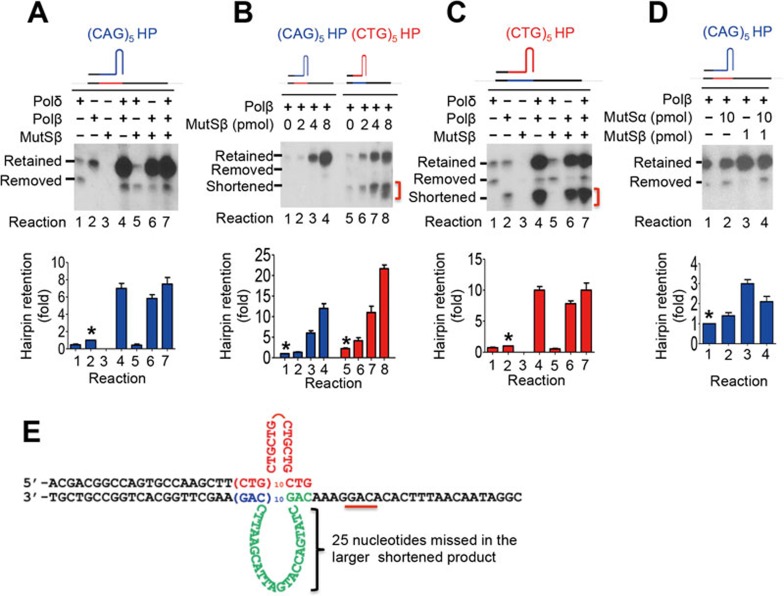
DNA polymerase β (Polβ) is capable of utilizing (CAG)n or (CTG)n hairpins as a primer for DNA synthesis, leading to (CAG)•(CTG) repeat expansion8. To examine potential role for MutSβ in Polβ-catalyzed (CAG)n or (CTG)n hairpin retention, purified MutSβ was included in the in vitro DNA synthesis reactions conducted by Polβ or Polδ (Figure 2). Consistent with our previous observations8, both hairpin removal and hairpin retention were detected when the CAG hairpin primer extension was carried out by Polδ alone (Figure 2A, reaction 1), but only hairpin retention products were obtained when the reaction was conducted by Polβ (Figure 2A, reaction 2). Addition of MutSβ did not change the product pattern in the Polδ-catalyzed reaction (Figure 2A, compare reaction 5 with reaction 1). However, addition of MutSβ dramatically stimulated the production of (CAG)5 hairpin retention in the Polβ-catalyzed reaction (Figure 2A, compare reaction 6 with reaction 2), and the stimulation is proportional to the amount of MutSβ in the reaction (Figure 2B, reactions 1-4). Similar results were also obtained when in vitro synthesis utilized a DNA substrate containing a (CTG)5 hairpin in the nascent DNA strand (Figure 2B, reactions 5-8; Figure 2C).
Figure 2.
MutSβ stimulates Polβ-induced (CAG)n and (CTG)n hairpin retention during DNA synthesis. Unless mentioned otherwise, hairpin retention/removal assays were performed in a 40-μL purified system containing 0.15 pmol (CAG)5 or (CTG)5 DNA hairpin substrate, 4 pmol MutSβ, 110 fmol RFC and 2 pmol PCNA in addition to the indicated polymerase (600 fmol Polδ, 260 fmol Polβ). Primer extension products were analyzed by Southern blot analysis as described in Figure 1 legends. (A) Effect of MutSβ on Polβ- and Polδ-catalyzed DNA synthesis using a (CAG)5 hairpin as primer. (B) Increased hairpin retention activity of Polβ is proportional to the increasing concerntrations of MutSβ. (C) Effect of MutSβ on Polβ- and Polδ-catalyzed DNA synthesis using a (CTG)5 hairpin as primer. (D) Effect of MutSα:MutSβ ratio on Polβ-catalyzed DNA synthesis using a (CAG)5 hairpin as primer. Relative hairpin retention activity in each group was calculated by using the hairpin retention activity conducted by Polβ alone as a reference, i.e., dividing hairpin retention level of each reaction with that catalyzed by Polβ alone (see *). The data in A-D were from three independent experiments and the error bar represents SD. (E) Proposed DNA structures for shortened DNA products.
The enhanced hairpin retention in the Polβ-catalyzed reaction in the presence of MutSβ is not due to the possibility that MutSβ preparation contains or is contaminated with a polymerase activity, as the protein by itself does not exhibit any polymerase activity (Figure 2A, reaction 3 and Figure 2C, reaction 3), nor does it enhance the Polδ-catalyzed reaction (Figure 2A, reaction 5 and Figure 2C, reaction 5). Thus, the simplest explanation is that MutSβ facilitates the Polβ-catalyzed (CAG) and (CTG) hairpin retention.
In addition to MutSβ, eukaryotic cells possess a second mismatch recognition protein designated MutSα, which consists of subunits MSH2 and MSH6. The cellular MutSα:MutSβ ratio in human cells is ∼10:127,28. To determine the impact of the cellular MutSα:MutSβ ratio on (CAG) or (CTG) hairpin retention, we performed the hairpin primer extension assay in the presence of MutSα alone or both proteins in their cellular ratio. The amounts of MutSα and MutSβ used in this experiment represent their relative concentrations in 100 μg HeLa nuclear extracts29. The result showed that MutSβ at 1 pmol stimulated the Polβ-catalyzed hairpin retention by ∼3-fold, as compared with the same reaction without MutSβ (Figure 2D, reactions 1 and 3); 10 pmol MutSα had little influence on the Polβ-catalyzed hairpin retention (Figure 2D, reactions 1 and 2), but slightly inhibited the MutSβ-stimulating activity (Figure 2D, reactions 3 and 4). These results indicate that MutSα is not involved in expansion of (CAG)•(CTG) repeats, consistent with previous observations in MSH3 and MSH6 knockout/knockdown organisms13,16,17,18.
It is worth noting that at least two new species, designated shortened products, were observed in Polβ-catalyzed (CTG)5 hairpin reactions (Figure 2B and 2C, red brackets). Previous sequencing analysis revealed that the larger band of these shortened species still contained the hairpin repeats (i.e., hairpin retained) but missed 25 nucleotides immediately 3′ to the CTG repeats in the primer strand or immediately 5′ to the CAG repeats in the template strand, because these 25-nucleotides in the template strand were looped out8 (also see Figure 2E). Although we did not sequence the smaller band of the shortened products, we speculate that it is probably missing 32 nucleotides as a result of pairing of the last CTG repeat in the hairpin primer with the underlined CAG sequence in the template strand (Figure 2E). Therefore, these shortened bands are actually hairpin-retained products.
We also note that Polδ stimulated the Polβ-catalyzed hairpin retention essentially as well as MutSβ, and that the combination of all three proteins yielded no additional stimulation (Figure 2A and 2C). These results may suggest that the rate-determining step in hairpin retention may be the elongation of the first product of Polβ-catalyzed primer extension, which is stimulated equally well by binding of MutSβ to the hairpin and Polβ or by adding Polδ. However, whether or not Polβ recruitment to a hairpin in vivo requires MutSβ remains to be investigated (see below).
MutSβ-stimulated DNA synthesis by Polβ requires a (CAG)n or (CTG)n hairpin
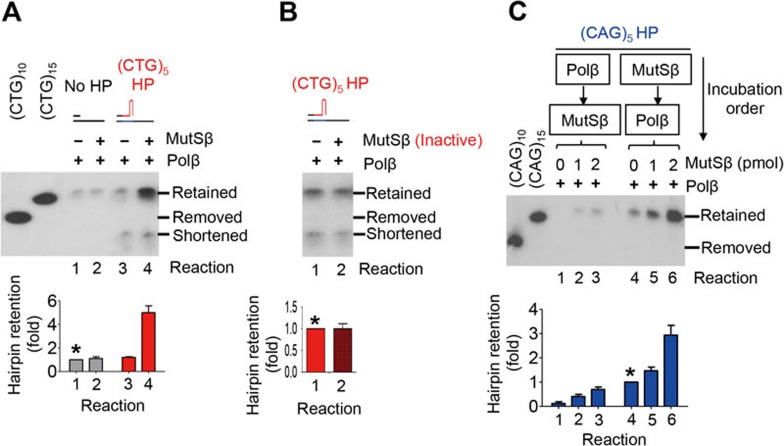
To determine whether a (CAG)n or (CTG)n hairpin is required for MutSβ-stimulated DNA synthesis by Polβ, we performed the following experiments. First, we compared DNA synthesis efficiency in reactions using a primer with or without a (CAG)n or (CTG)n hairpin right at the 3′ end. The result revealed that MutSβ had little influence on Polβ-catalyzed DNA synthesis when the primer did not contain a hairpin at the 3′ end (Figure 3A, reactions 1 and 2). However, when a (CTG)5 hairpin primer or a (CAG)5 hairpin primer (data not shown) was used in the reaction, MutSβ greatly stimulated Polβ-catalyzed DNA synthesis (Figure 3A, reactions 3 and 4). This stimulation was not due to the extra amount of protein in the reaction, as the same amount of heat-inactivated MutSβ failed to stimulate the Polβ-catalyzed synthesis (Figure 3B).
Figure 3.
MutSβ-stimulated DNA synthesis by Polβ requires a (CAG)n or (CTG)n hairpin. Unless mentioned otherwise, hairpin retention/removal assays were performed in a 40-μL purified system containing 0.15 pmol (CAG)5 or (CTG)5 DNA hairpin substrate, 4 pmol MutSβ, 110 fmol RFC, 2 pmol PCNA and 260 fmol Polβ. DNA synthesis products were analyzed by Southern blot analysis as described in Figure 1 legends. (A) Comparison of hairpin retention activity of Polβ in reactions with or without MutSβ. DNA substrate in non-hairpin reactions (reactions 1 and 2) used a ssM13mp18 derivative containing 15 CAG repeats to match the size of hairpin-retained products. (B) Hairpin retention activity in reaction with heat-inactivated MutSβ. (C) Dependence of hairpin retention activity on the incubation order of MutSβ and Polβ. Relative hairpin retention activity in each group was calculated by using the hairpin retention activity conducted by Polβ alone as a reference, i.e., dividing hairpin retention activity of each reaction with that catalyzed by Polβ alone (see *). The data were from three independent experiments and the error bar represents SD.
Second, we incubated hairpin primer extension substrates with one protein first for 5 min, followed by additional 5-min incubation after adding the other protein into the reaction. A slightly enhanced DNA synthesis was observed if Polβ was added first and MutSβ was added later (Figure 3C, reactions 1-3); however, a great stimulation was achieved when the order was reversed (Figure 3C, reactions 4-6). The simplest explanation of these results is that binding of a (CAG)n or (CTG)n hairpin by MutSβ recruits Polβ to the hairpin primer, allowing Polβ to add nucleotides to the hairpin primer.
MutSβ physically interacts with Polβ
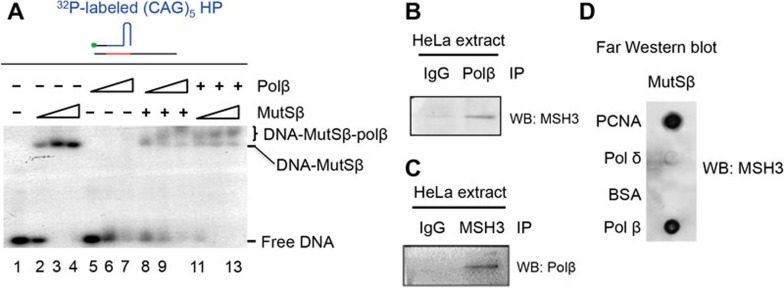
The results described above suggest that MutSβ interacts with Polβ. To test this hypothesis, we performed gel shift analysis using hairpin primer extension DNA substrates in the presence of MutSβ and/or Polβ. Consistent with previous studies showing that MutSβ specifically recognizes (CAG)n and (CTG)n hairpins20,21, a distinct MutSβ-DNA complex was observed when increasing amount of MutSβ was incubated with a (CAG)5 hairpin-containing DNA substrate (Figure 4A, lanes 2-4). Although Polβ by itself does not form any complex with the DNA substrate (Figure 4A, lanes 5-7), it induced a super-shifted complex together with MutSβ and DNA (Figure 4A, lanes 8-13), suggesting that Polβ participates in the big complex formation via its interaction with MutSβ. To explore this possibility, we performed co-immunoprecipitation in HeLa nuclear extracts. We found that an antibody against Polβ pulled down MSH3 (Figure 4B), a subunit of MutSβ and that an MSH3 antibody co-precipitated Polβ (Figure 4C). To confirm a direct interaction between these two proteins, we demonstrated that like proliferating cellular nuclear antigen (PCNA), which has been shown to specifically interact with MutSβ30,31,32, purified Polβ retained MutSβ on the far-western blot (Figure 4D). We therefore conclude that MutSβ physically interacts with Polβ.
Figure 4.
MutSβ physically interacts with Polβ. (A) Gel shift assay to determine the interaction of MutSβ, Polβ and a 32P-labeled primer extension substrate containing a (CAG)5 hairpin. (B, C) Co-immunoprecipitation (Co-IP) assay to determine interactions between MutSβ and Polβ in HeLa nuclear extract. Co-IP by IgG was used as a negative control. (D) Far-western blot analysis showing direct interaction between purified MutSβ and Polβ. PCNA and polδ were used as positive and negative controls, respectively.
The MutSβ-Polβ complex colocalizes with (CAG)•(CTG) repeats during DNA replication
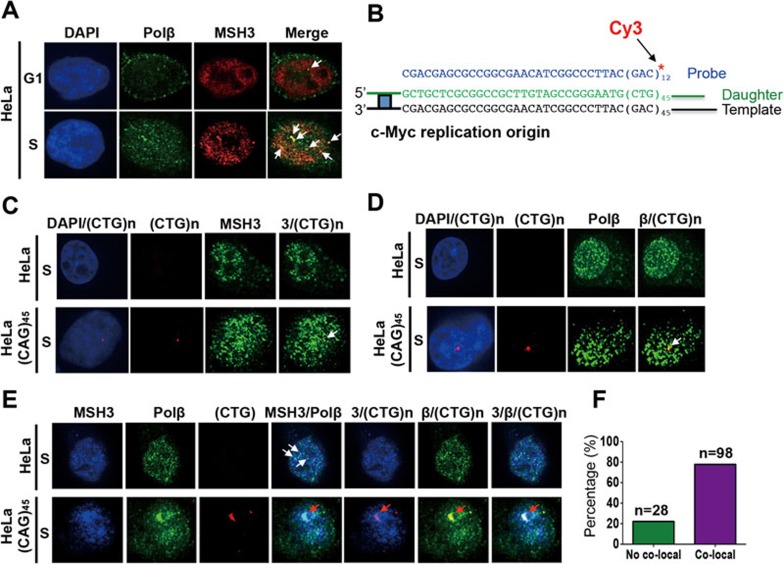
We reasoned that if MutSβ promotes (CAG)•(CTG) repeat expansion by recruiting Polβ to conduct DNA synthesis using a hairpin as primer, these two proteins should form a complex that colocalizes with (CAG)•(CTG) repetitive sequences in S phase. To test this hypothesis, we performed immunofluorescence analysis in HeLa cells. Cells were arrested in G1-S boundary by double thymidine blocks and released to S phase as described33, and the cellular distributions of MSH3 and Polβ were monitored by immunofluorescence using their corresponding antibodies. As shown in Figure 5A, despite that both MSH3 (i.e., MutSβ) and Polβ are rich in the nucleus in G1 and S phases, there seems to be only one detectable colocalizing focus between polβ and MutSβ in G1 phase, but there are at least 6 colocalizing foci between these two proteins in S phase, suggesting that MutSβ and Polβ interact with each other more frequently in S phase than in G1 phase. Since MutSβ binds to (CAG)n•(CTG)n hairpins, the number of the MutSβ-polβ colocalizing foci likely corresponds to the number of hairpins formed within the repetitive DNA sequences triggered by DNA synthesis. It is apparent that cells in S phase would form more (CAG)n and (CTG)n hairpins than cells in G1 phase, as all repetitive DNA sequences undergo replication in S phase, but only limited repair and synthesis occur in damaged (CAG)n•(CTG)n sequences in G1 phase.
Figure 5.
MutSβ-Polβ complex colocalizes with (CAG)•(CTG) repeats during DNA synthesis. (A) Confocal immunofluorescence analysis showing MSH3 and Polβ foci and their colocalization in HeLa cells in G1 phase (top) and S phase (bottom). (B) DNA sequence of 5 end Cy3-labeled oligonucleotide used in FISH analysis and its mapping site at the ectopic c-Myc replication origin in HeLa-(CAG)45 cells. (C, D) Confocal immunofluorescence analysis showing MSH3-(CTG)45 and Polβ-(CTG)45 colocalizations in HeLa-(CAG)45 cells in S phase, respectively. (E) Confocal immunofluorescence analysis showing colocalization of MSH3, Polβ and (CTG)45 in HeLa-(CAG)45 cells in S phase (bottom), with HeLa cells without the ectopic (CAG)45 sequence as a negative control (top). β and 3 represent Polβ and MSH3, respectively. (F) Percentage of cells showing colocalization of MSH3, Polβ and (CTG)45 repeats.
Does the MutSβ-Polβ interaction occur at the site of the (CAG)•(CTG) repeats? To answer this question, we combined immunofluorescence analysis with fluorescence in situ hybridization (FISH) in HeLa cells with or without 45 (CAG)•(CTG) repeats alongside a single ectopic copy of the wild-type 2.4 kb c-Myc replication origin, with a (CAG)45 repeat sequence in the leading strand template for replication from the proximal c-Myc origin6. A Cy3-labeled 66-mer oligonucleotide that contains 12 CAG repeats and 30 additional nucleotides complementing the c-Myc replication origin was used to map the location of the ectopic (CAG)•(CTG) repeats and c-Myc replication origin (Figure 5B). Thus, MutSβ foci, Polβ foci and the (CAG)45•(CTG)45 location could be detected simultaneously. Indeed, the results show that both MutSβ and polβ colocalized with (CTG)45 in S phase (Figure 5C and 5D, arrows); and all these three components were found in the same location in S phase (Figure 5E, bottom, red arrows). In contrast, we did not see a (CTG)n repeat picked up by the Cy3-labeled probe in HeLa cells without the ectopic c-Myc replication origin containing the (CAG)45 repeat sequence, although we did observe some colocalizations between MSH3 and Polβ (Figure 5E, top), which is likely due to their interactions at endogenous (CAG)•(CTG) repeats in the genome, as shown in Figure 5A. These results strongly suggest that MutSβ and polβ interact with each other at the (CAG)•(CTG) repeats and promote expansion of the latter during DNA synthesis.
Discussion
MutSβ is an important MMR protein involved in recognizing and removing small insertion-deletion mispairs generated during DNA replication. Given that MMR is well known for its role in maintaining replication fidelity, it was surprising that MutSβ was found to promote (CAG)•(CTG) repeat expansion15. However, the mechanism by which this genome safeguard protein promotes TNR instability has been a long-standing puzzle in the field. The data presented in this study provides novel insights into the mechanism of MutSβ-mediated (CAG)•(CTG) repeat expansion, i.e., MutSβ collaborates with Polβ to conduct error-prone DNA synthesis using a (CAG)n or (CTG)n hairpin formed in the nascent DNA strand right at the replication fork as a primer, which leads to the hairpin retention and (CAG)•(CTG) repeat expansion.
In addition to recognizing small insertion-deletion mispairs, MutSβ was also found to specifically recognize (CAG)n and (CTG)n hairpins20,21. The binding of the unusual hairpin structures by MutSβ was thought to trap the protein on the DNA structure so that it prevents hairpin removal as well as impairs MutSβ's activity in heteroduplex repair20,24. However, later studies showed that MutSβ at excess amounts did not inhibit DNA hairpin repair activity in HeLa nuclear extracts and that ATP can release MutSβ from (CAG)n•(CTG)n hairpins21, a well-known “sliding activity” of all MutS family proteins in response to ATP34,35,36. These observations suggest that other mechanism(s) are involved in MutSβ-mediated (CAG)•(CTG) repeat expansion.
It is logical to think that (CAG)•(CTG) repeat expansion is associated with DNA synthesis. Indeed, we show here that MutSβ stimulates retention of (CAG)n and (CTG)n hairpins during DNA synthesis conducted in HeLa nuclear extracts (Figure 1). Our previous studies have identified translesion polymerase Polβ37 as the enzyme catalyzing the hairpin retention synthesis by efficiently using (CAG)n and (CTG)n hairpins as a primer8. Does MutSβ promote expansion of (CAG)•(CTG) repeats via Polβ-catalyzed hairpin-retaining DNA synthesis? A positive answer is provided in this study. First, MutSβ stimulates Polβ-catalyzed primer extension only when the primer contains a (CAG)n or (CTG)n hairpin (Figure 3A), and this stimulation is Polβ-specific, as MutSβ has little influence on Polδ-catalyzed hairpin primer synthesis (Figure 2A and 2C). Second, MutSβ binds to (CAG)n and (CTG)n hairpin substrates used for primer extension, and this binding appears to be responsible for recruiting Polβ to the MutSβ-DNA complex. This is because Polβ by itself does not bind to the DNA substrate, but it forms a super-shifted complex together with MutSβ and DNA substrates (Figure 4A). Co-immunoprecipitation and far-western blot experiments confirmed that these two proteins interact with each other (Figure 4B–4D). Finally, we show that MutSβ and Polβ colocalize right at the (CAG)45•(CTG)45 sequence of the ectopic c-Myc replication origin in HeLa cells (Figure 5), and the colocalization of MutSβ, Polβ and (CAG)n•(CTG)n repeats appears to occur more frequently in S phase than in G1 phase in HeLa cells (Figure 5), suggesting that MutSβ-mediated expansion of (CAG)•(CTG) repeats is via DNA replication. We previously showed that although Polδ alone does not conduct hairpin retention synthesis, it promotes hairpin retention in the presence of Polβ8, which is also confirmed in this study (Figure 2). This is because Polβ uses (CAG)n or (CTG)n hairpin as a primer for extension; once the hairpin primer carries a complementary 3′ sequence with 2- or more nucleotides that fix the hairpin structure, Polδ can continue to add nucleotides to the Polβ-generated hairpin-containing product, leading to hairpin retention8.
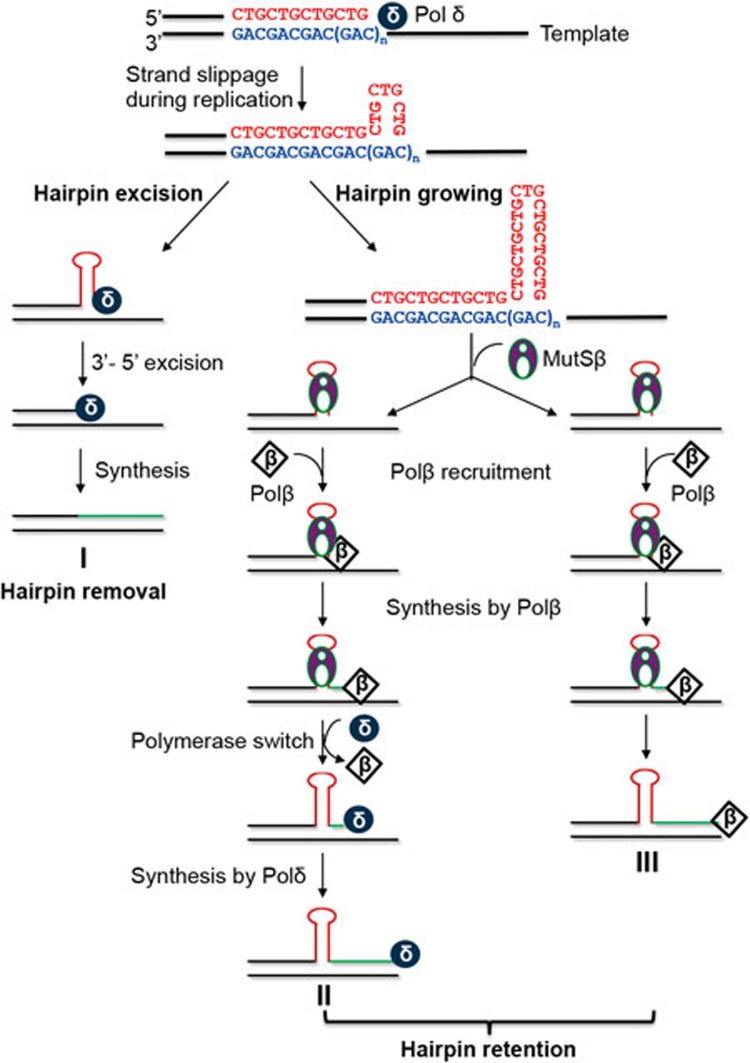
Taken together, these studies support a model by which MutSβ promotes (CAG)n•(CTG)n expansion during DNA replication (Figure 6). First, a newly replicated (CAG)n or (CTG)n sequence forms a hairpin structure at the 3′ end of the primer strand via strand slippage and the size of the hairpin can grow as the repetitive sequence is being further replicated. Despite that Polδ is capable of removing the hairpin structure8 (also see Figure 6, I), binding of the hairpin by MutSβ recruits Polβ to the complex through physical interactions between these two proteins. Polβ then uses the hairpin as a primer to add several nucleotides to the 3′-end of the hairpin, which fixes the hairpin structure in the nascent DNA strand. Polδ is then recruited to the site for the high-fidelity and highly processive DNA synthesis via polymerase switch, a not fully understood process yet. It is the collaborative efforts by MutSβ, Polβ and Polδ that render the hairpin remained in the newly synthesized strand, leading to (CAG)n•(CTG)n expansion (Figure 6, II).
Figure 6.
Proposed model for MutSβ's role in promoting (CAG)•(CTG) repeat expansion. During DNA synthesis (i.e., DNA replication or DNA repair), (CAG)•(CTG) repeats form a hairpin via strand slippage. The hairpin can be removed by Polδ (I). However, hairpin binding by MutSβ recruits Polβ to the complex, where Polβ uses the hairpin as a primer to add several nucleotides to the 3′ end of hairpin. The resulting hairpin product can be utilized for the high-fidelity and highly processive DNA synthesis by Polδ, leading to hairpin retention (II). Alternatively, Polβ can carry out the hairpin retention DNA synthesis independent of Polδ (III).
Given the fact that in the presence of MutSβ, Polβ can efficiently carry out hairpin retention DNA synthesis independent of Polδ (Figure 2), the MutSβ-Polβ-facilitated (CAG)•(CTG) repeat expansion can apply to DNA repair-mediated repeat expansion in non-dividing cells38. Various DNA lesions, e.g., oxidative DNA damage, can occur within a (CAG)n•(CTG)n sequence39, and repair of these lesions by various DNA repair pathways, including base excision repair, nucleotide excision repair and non-canonical DNA MMR, involves DNA synthesis. As in DNA replication, a (CAG)n•(CTG)n hairpin can form via strand slippage during the repair DNA synthesis step, which provides MutSβ and Polβ the opportunity to conduct error-prone DNA synthesis to expand the repetitive DNA sequence (Figure 6, III). This may explain why non-replicating neuronal cells undergo TNR expansions.
It is worth mentioning that MutSβ has been shown to be involved in both TNR expansions and contractions40, which may depend on if a hairpin is formed in the newly synthesized/repaired strand or in the template strand, respectively. However, our model suggests that in the absence of MutSβ, a (CAG)n or (CTG)n hairpin in the primer strand will be preferentially removed by Polδ, which will greatly reduce the probability of TNR expansion, but such hairpins in the template strand could still induce TNR contractions, as Polδ cannot process TNR hairpins in the template strand, consistent with a recent study41. Although we demonstrated colocalization of MutSβ, Polβ and (CAG)45•(CTG)45 sequence in HeLa cells during DNA replication (Figure 5), we did not directly show the presence of a (CAG)n or (CTG)n hairpin in these cells. However, it is in these (CAG)n•(CTG)n-containing HeLa cells that in vivo hairpin formation was demonstrated within (CAG)n•(CTG)n sequences as the basis of (CAG)•(CTG) repeat instability during DNA replication6. Based on the fact that MutSβ does not bind to a perfectly paired (CAG)n•(CTG)n DNA20,21, colocalization of MutSβ with (CAG)45•(CTG)45 sequence indicates the presence of an imperfect heteroduplex (i.e., slip-out or hairpin) in the repetitive DNA sequence. This model also explains recent observations showing enrichment of MutSβ at (CAG)•(CTG) repeats in human somatic cells18 and Polβ in the striatum of Huntington disease mice42. Interestingly, MutSβ enrichment was also observed in long GAA repeat tracts in Friedreich's ataxia induced pluripotent stem cells43, implying that expansion of GAA repeats might be through the MutSβ-Polβ collaborative mechanism as well.
A fundamental but critical question for the model is how a (CAG)n•(CTG)n hairpin is formed at the 3′ end of the primer strand in vivo? Several lines of in vitro evidence have shown that “naked” (CAG) or (CTG) repeats can form a hairpin structure spontaneously2,5,38,44,45. Despite that DNA in vivo is bound with many proteins, it is relatively “naked” at the replication fork, and has an “open” 3′ end. This open end provides the repetitive DNA sequence freedom to form secondary structures, including hairpins and/or loop-outs at the replication fork, where mismatch recognition proteins MutSα and MutSβ are on “duty” detecting replication-generated base-base mismatches and insertion-deletion loop-outs/hairpins, respectively. Upon formation, the (CAG)n or (CTG)n hairpin will be bound and stabilized by MutSβ. The latter then recruits Polβ for error-prone DNA synthesis, as described in Figure 6. Another fundamental question related to the model is how MutSβ is recruited to nucleosomes carrying the repetitive sequence before replication? Based on the fact that MutSα recruitment to chromatin is through H3K36me333, an unidentified histone mark may be responsible for loading MutSβ to chromatin prior to its interactions with (CAG)n and (CTG)n hairpins at the replication fork. Recent studies by Lahue and colleagues have linked a role for MutSβ in (CAG)•(CTG) repeat expansion with histone deacetylase complexes18, indicating that histone acetylation is a potential candidate for MutSβ recruitment. However, thorough investigations are required to elucidate the relationship between these molecules, as well as the molecular mechanism of primer hairpin formation.
Materials and Methods
Cell culture and nuclear extract preparation
HeLa and HeLa-(CAG)45•(CTG)45 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn bovine serum (NBS) and 4 mM L-glutamine at 37 °C in a humidified atmosphere with 5% CO2. The nuclear extract was prepared as described46.
Recombinant protein expression and preparation
MutSα, MutSβ, Polδ, replication factor C (RFC) and PCNA were overexpressed and purified as described29. MutSα, MutSβ, Polδ and RFC were expressed in High Five insect cells using baculovirus system, and PCNA was expressed in Escherichia coli. cDNA of Polβ were cloned into the pFastBac-HTb vector and expressed in insect High Five cells using Bac-to-Bac expression system. The recombinant proteins were purified to near homogeneity. Protein concentration was determined by the Coomassie (Bradford) Protein Assay Reagent (Pierce). The purified proteins were stored in aliquot at −80 °C.
(CAG)n or (CTG)n hairpin removal/retention assay
DNA hairpin substrates were prepared as described previously8. Hairpin removal/retention assay was performed by incubating DNA substrates (0.15 pmol) with either HeLa nuclear extracts (30 μg) or purified proteins (260 fmol Polβ, 1-8 pmol MutSβ, 600 fmol Polδ, 110 fmol RFC and 2 pmol PCNA) in a 40-μL reaction containing 110 mM KCl, 20 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 1 mM glutathione, 1.5 mM ATP, 0.1 mM of each dNTP, and 0.05 mg/mL BSA at 37 °C for 30 min. The reaction was terminated by incubating with 60 μL of proteinase K solution containing 0.67% (w/v) of SDS, 2.5 mM EDTA and 20 mg/mL proteinase K. After phenol extraction and ethanol precipitation, the DNA sample was digested with 0.3 units of BsrBI (New England Biolab) and resolved in a 6% polyacrylamide denaturing gel, followed by Southern blotting analysis using a 32P-end-labeled probe specifically annealing to the newly synthesized strand near the BsrBI site as described23.
Gel shift analysis
32P-labeled oligonucleotides containing 15 CAG and 15 CTG repeats were annealed with oligonucleotides containing 10 CTG and 10 CAG repeat to form (CAG)5 and (CTG)5 hairpin substrates (Figure 1A), respectively. The first 5′ bases of oligonucleotide are phosphothioated to prevent nuclease degradation. Gel shift assays were performed in 20-μL reactions containing 10 mM HEPES-KOH (pH 7.5), 110 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 32P-labeled oligonucleotide duplexes, and MutSβ in the presence of 10-fold excess amount of unlabeled oligonucleotide homoduplex. The reactions were incubated on ice for 20 min, followed by the addition of 5 μL of 50% (w/v) sucrose. The products were then loaded on and resolved in 6% non-denaturing polyacrylamide gel in buffer containing 50 mM Tris borate (pH 7.5) and 1 mM EDTA. The buffer was recirculated during electrophoresis. The gel was dried and analyzed by a Storm PhosphorImager (GE Healthcare).
Co-immunoprecipitation, western blot and far-western blot analyses
Nuclear extract (1 mg) was incubated with 5 μg of primary antibody overnight at 4 °C with rotation. The antibodies used were control mouse IgG (Santa Cruz Biotechnology), and those against MSH3 (BD Pharmingen), Polβ (Santa Cruz Biotechnology). Fifty microliter of agarose-protein G beads (50% slurry; Invitrogen) pre-equilibrated with washing buffer (25 mm Hepes-KOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 50 μg/ml bovine serum albumin (BSA), 0.05% Triton X-100) were then added, and the reactions were incubated by rotation at 4 °C for 3 h. The beads were washed three times with the washing buffer containing 600 mM NaCl and resuspended in 30 μl of 2× Laemmli SDS buffer. After SDS-PAGE, the proteins were then transferred onto nitrocellulose (0.45 micron, GE Water and Process Technologies), followed by western blot analysis using chemiluminescence.
Far-western blot analysis was performed as described previously47. Purified proteins (1.0 μg of PCNA, Polδ, Polβ and BSA were spotted onto a nitrocellulose membrane and incubated with 1.0 μg of purified MutSβ. The membrane was immunoblotted with antibody against MSH3. The protein was detected by western blot analysis as described above.
Cell synchronization, DNA FISH and immunofluorescence analysis
Cell synchronization was performed as described previously33. Cells were arrested at G1/S by culturing for 18 h in complete medium with 2 mM thymidine, thymidine-free medium for 10 h, and then thymidine-containing medium for an additional 15 h before release into complete medium. Cells were harvested at 0 h (G1 phase), 3 h (S phase), and 8 h (G2/M). Cell cycle status was confirmed by flow cytometry.
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in Phosphate-Buffered Saline (PBS, 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4). After incubation with 10 U/ml RNase A (Sigma) in PBS for 3 h at 37 °C, the fixed cells were blocked by 5% (w/v) BSA for 1 h at room temperature, followed by incubation with primary antibody overnight at 4 °C. After washing with PBS for 3 times (5 min each), the cells were incubated with a secondary antibody for 2 h at room temperature and washed for three times with PBS. Hybridization was performed in a humidified chamber for 3 h at room temperature with a Cy3-labeled DNA oligonucleotide probe (10 ng per hybridization) containing 12 CAG repeats and 30 additional nucleotides complementary to the ectopic c-Myc replication origin (see Figure 5B), 1 mg/mL BSA, 10 U/mL RNase A in PBS. Cells were then washed three times with PBS and mounted in ProLong GOLD (Invitrogen) and left in the dark room overnight. Immunofluorescence images were obtained using Zeiss Observer Z1 confocal scanner laser microscopy system.
Author Contributions
JG performed the experiments; LG developed the in vitro hairpin retention/removal assay; ML generated the HeLa-(CAG)45 cell line; G-ML contributed to the overall experimental design and interpretation of experimental results; JG, LG, ML and G-ML wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Guangshuo Ou for making his confocal scanner laser microscopy system available for this study, and Guogen Mao for technical help and discussions. This work was supported in part by the National Institutes of Health of the United States (GM089684 to GML; GM099874 to ML) and the National Natural Science Foundation of China (31370766), by the Joint NSFC-ISF Research Program, jointly funded by the National Natural Science Foundation of China and the Israel Science Foundation (31461143005), by Tsinghua-Peking Joint Center for Life Sciences, and by the University of Southern California Norris Comprehensive Cancer Center. JG is supported by a postdoctoral grant (2015M570079) from the Ministry of Human Resources and Social Security of the People's Republic of China.
References
- Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 2010; 11:165–170. [DOI] [PubMed] [Google Scholar]
- McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet 2010; 11:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet 2005; 6:729–742. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature 2007; 447:932–940. [DOI] [PubMed] [Google Scholar]
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 1995; 81:533–540. [DOI] [PubMed] [Google Scholar]
- Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) × (CAG) repeat hairpins in human cells. Nat Chem Biol 2010; 6:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chen X, Leffak M. Oligodeoxynucleotide binding to (CTG)•(CAG) microsatellite repeats inhibits replication fork stalling, hairpin formation, and genome instability. Mol Cell Biol 2013; 33:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NL, Guo J, Zhang T, et al. Coordinated processing of 3′ slipped (CAG)n/(CTG)n hairpins by DNA polymerases beta and delta preferentially induces repeat expansions. J Biol Chem 2013; 288:15015–15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Ann Rev Biochem 1996; 65:101–133. [DOI] [PubMed] [Google Scholar]
- Kolodner RD. A personal historical view of DNA mismatch repair with an emphasis on eukaryotic DNA mismatch repair. DNA Repair 2016; 38:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. Eukaryotic mismatch repair in relation to DNA replication. Ann Rev Genet 2015; 49:291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Napierala M, Wells RD. DNA triplet repeat expansion and mismatch repair. Ann Rev Biochem 2015; 84:199–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Pearson CE. Disease-associated repeat instability and mismatch repair. DNA Repair 2016; 38:117–126. [DOI] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res 2008; 18:85–98. [DOI] [PubMed] [Google Scholar]
- Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet 1999; 23:471–473. [DOI] [PubMed] [Google Scholar]
- van den Broek WJ, Nelen MR, Wansink DG, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet 2002; 11:191–198. [DOI] [PubMed] [Google Scholar]
- Foiry L, Dong L, Savouret C, et al. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet 2006; 119:520–526. [DOI] [PubMed] [Google Scholar]
- Gannon AM, Frizzell A, Healy E, Lahue RS. MutSβ and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res 2012; 40:10324–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JR, Lahue EE, Li GM, Lahue RS. Trinucleotide repeat expansions catalyzed by human cell-free extracts. Cell Res 2013; 23:565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen BA, Yang Z, Lai M, et al. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol 2005; 12:663–670. [DOI] [PubMed] [Google Scholar]
- Tian L, Hou C, Tian K, Holcomb NC, Gu L, Li GM. Mismatch recognition protein MutSβ does not hijack (CAG)n hairpin repair in vitro. J Biol Chem 2009; 284:20452–20456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol 2005; 12:654–662. [DOI] [PubMed] [Google Scholar]
- Hou C, Chan NL, Gu L, Li GM. Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts. Nat Struct Mol Biol 2009; 16:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang WH, Coats JE, Majka J, et al. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc Natl Acad Sci USA 2011; 108:E837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Huang J, Gu L, Li GM. In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats. DNA Repair 2012; 11:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Gu L, Li GM. Bi-directional processing of DNA loops by mismatch repair-dependent and -independent pathways in human cells. J Biol Chem 2003; 278:3891–3896. [DOI] [PubMed] [Google Scholar]
- Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutS/hMutSβ ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci USA 1997; 94:10144–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA 1998; 95:8568–8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 2005; 122:693–705. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet 2000; 26:375–378. [DOI] [PubMed] [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem 2000; 275:36498–36501. [DOI] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev 2001; 15:724–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, et al. The Histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 2013; 153:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science 1995; 268:1909–1912. [DOI] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R. The role of mismatched nucleotides in activating the hMSH2-hMSH6 molecular switch. J Biol Chem 2000; 275:3922–3930. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem 2005; 280:22245–22257. [DOI] [PubMed] [Google Scholar]
- Yamtich J, Sweasy JB. DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta 2010; 1804:1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K, House NC, Freudenreich CH. Repeat instability during DNA repair: Insights from model systems. Crit Rev Biochem Mol Biol 2015; 50:142–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 2007; 447:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XN, Kumari D, Gupta S, et al. Mutsβ generates both expansions and contractions in a mouse model of the Fragile X-associated disorders. Hum Mol Genet 2015; 24:7087–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slean MM, Panigrahi GB, Castel AL, Pearson AB, Tomkinson AE, Pearson CE. Absence of MutSβ leads to the formation of slipped-DNA for CTG/CAG contractions at primate replication forks. DNA Repair (Amst) 2016. 42:107– 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goula AV, Berquist BR, Wilson DM 3rd, Wheeler VC, Trottier Y, Merienne K. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington's disease transgenic mice. PLoS Genet 2009; 5:e1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S, Soragni E, Campau E, et al. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell 2010; 7:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GK, Jie J, Fox GE, Gao X. DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res 1995; 23:4303–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res 1996; 24:1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JJr, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA 1990; 87:5837–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J, Li JY, Lee S, Tong D, Gu L, Li GM. Phosphorylation of PCNA by EGFR inhibits mismatch repair and promotes misincorporation during DNA synthesis. Pro Natl Acad Sci USA 2015; 112:5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]