Abstract
Breast cancer is the most common female cancer, affecting approximately one in eight women during their life-time. Besides environmental triggers and hormones, inherited mutations in the breast cancer 1 (BRCA1) or BRCA2 genes markedly increase the risk for the development of breast cancer. Here, using two different mouse models, we show that genetic inactivation of the key osteoclast differentiation factor RANK in the mammary epithelium markedly delayed onset, reduced incidence, and attenuated progression of Brca1;p53 mutation-driven mammary cancer. Long-term pharmacological inhibition of the RANK ligand RANKL in mice abolished the occurrence of Brca1 mutation-driven pre-neoplastic lesions. Mechanistically, genetic inactivation of Rank or RANKL/RANK blockade impaired proliferation and expansion of both murine Brca1;p53 mutant mammary stem cells and mammary progenitors from human BRCA1 mutation carriers. In addition, genome variations within the RANK locus were significantly associated with risk of developing breast cancer in women with BRCA1 mutations. Thus, RANKL/RANK control progenitor cell expansion and tumorigenesis in inherited breast cancer. These results present a viable strategy for the possible prevention of breast cancer in BRCA1 mutant patients.
Keywords: BRCA1, RANK, RANKL, inherited breast cancer, mammary progenitor cells
Introduction
Risk factors for breast cancer development include exposure to environmental factors such as synthetic sex steroid hormones, endogenous hormones, or genetic predisposition. In particular, germline mutations in breast cancer 1 (BRCA1) and BRCA2 account for 2%-10% of breast cancer cases depending on the ethnic population1. Although BRCA1/2 are involved in the repair of double-strand breaks in DNA, human evidence suggests a relationship between BRCA1/2 mutations, sex hormone levels, and cancer risk2. In addition, progesterone has been shown to play a role in mammary tumorigenesis of Brca1/p53 mutant mice3. However, the molecular mediators of BRCA1/2-associated risk and sex steroid exposure have not been identified.
RANKL (receptor activator of NF-κB ligand), its receptor RANK, and the natural inhibitor osteoprotegerin (OPG) are essential for the development and activation of osteoclasts4,5. Based on these findings, RANKL inhibition with a monoclonal antibody has been successfully developed as a rational therapy against osteoporosis and skeletal-related events in cancer patients6,7,8,9. In addition, RANKL/RANK are essential for the formation of a lactating mammary gland during pregnancy10. RANKL is secreted by estrogen receptor (ER)- and progesterone receptor (PR)-positive mammary epithelial cells in response to progesterone, and subsequently acts in a paracrine fashion on ER/PR-negative epithelial progenitor cells, promoting proliferation and expansion of mammary epithelial cells9,11,12. Using genetic mouse models, we and others have previously shown that RANKL/RANK control progestin-driven mammary cancer13,14. Interestingly, RANK signaling acts on progenitor cells, which are also believed to be “seed cells” for triple-negative breast cancer in carriers with BRCA1/2 mutations15. We therefore speculated that RANKL/RANK might have a role in the etiology of BRCA1/2 mutation-driven breast cancer.
Results
Genetic inactivation of Rank protects from Brca1 deletion-driven tumorigenesis
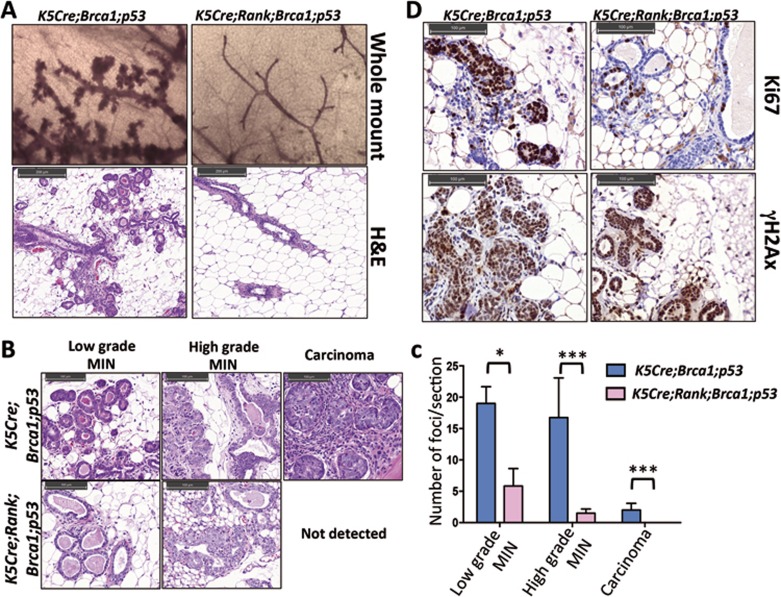
In mice the incidence of mammary tumors in the presence of only Brca1 mutation is low. Therefore, to directly assess the role of RANKL/RANK in Brca1 mutation-mediated tumorigenesis in vivo, we first deleted Brca1 and p53 in basal mammary epithelial cells and mammary progenitor cells using K5Cre mice16 to induce mammary cancer as previously reported17,18. This line was then crossed into Rankflox/flox mice to examine Brca1 deletion-induced tumorigenesis in the presence or absence of RANK expression (Supplementary information, Figures S1A, S1B and S2). All mouse lines examined appeared to develop normal mammary glands at puberty. In 4-month-old control K5Cre;Brca1;p53 double-mutant mice, we observed widespread epithelial hyperplasia (Figure 1A and Supplementary information, Figure S3) as well as low and high-grade mammary intraepithelial neoplasias (MINs) and invasive carcinomas (Figure 1B and 1C). The mammary glands of age-matched females with concomitant ablation of Rank appeared largely normal, displaying a significantly lower number of MINs and no detectable carcinomas (Figure 1A-1C and Supplementary information, Figure S3). Quantification of branching points in whole mount stainings from female littermates further showed that loss of RANK significantly decreased proliferation and pre-neoplasia observed in the absence of Brca1 and p53 (Supplementary information, Figure S1C). Enhanced proliferation of mammary epithelial cells in K5Cre;Brca1;p53 double-mutant mice was confirmed using Ki67 immunostaining (Figure 1D and Supplementary information, Figure S1D). Importantly, we observed marked DNA damage in both double- and triple-mutant mice as determined by γH2AX immunostaining (Figure 1D and Supplementary information, Figure S1E). DNA damage was confirmed using a second marker, p53BP1 (Supplementary information, Figure S4A). Moreover, low- and high-grade MINs that developed in 4-month-old double- and triple-mutant mice expressed Cytokeratin5 (KRT5/CK5) and β-catenin, confirming the basal epithelial origin of these tumors (Supplementary information, Figure S4B and S4C). These data show that genetic deletion of Rank in basal mammary epithelial cells markedly abrogates the development of intraepithelial neoplasms and invasive carcinomas as a consequence of Brca1;p53 mutations.
Figure 1.
Ablation of Rank in mammary epithelial cells markedly decreases tumor formation in Brca1/p53 mutant female mice. (A) Representative whole mount images (haematoxylin staining, magnification 52×) and paraffin sections (H&E staining, scale bar, 200 μm) of mammary glands from 4-month-old K5Cre;Brca1;p53 double- and K5Cre;Rank;Brca1;p53 triple-knockout littermate mice. (B) Representative images (H&E staining, scale bar, 100 μm) and (C) quantification of low-grade MINs, high-grade MINs and adenocarcinomas in mammary glands from 4-month-old K5Cre;Brca1;p53 and K5Cre;Rank;Brca1;p53 mutant littermates. Data are shown as average number of foci/section of 1 inguinal and 2 thoracic mammary glands per mouse +/− SEM. n ≥ 4 mice/group. *P < 0.05, ***P < 0.001 (2-way ANOVA). (D) Representative images of Ki67 and γH2AX immunostaining of mammary glands from 4-month-old K5Cre;Brca1;p53 double- and K5Cre;Rank;Brca1;p53 triple-knockout littermates. Scale bar, 100 μm.
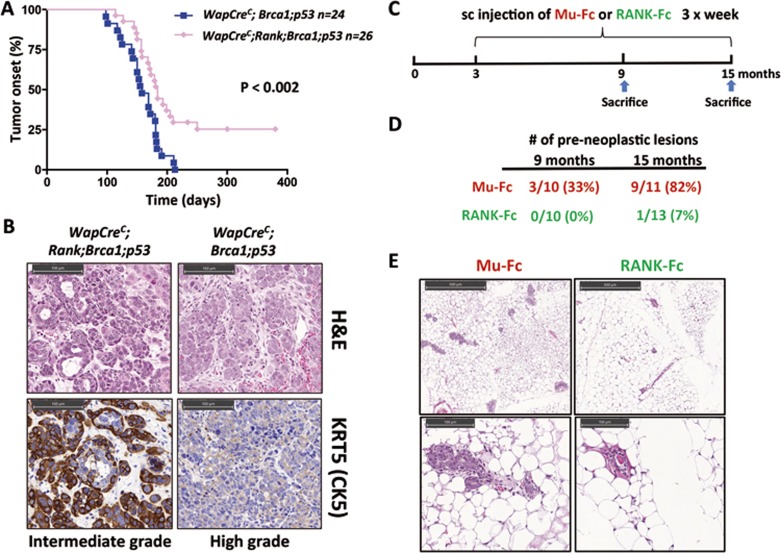
The occurrence of skin cancer commonly observed in the K5Cre;Brca1;p53 double-17,18 as well as K5Cre;Rank;Brca1;p53 triple-mutant mice precluded further analysis of mammary tumorigenesis beyond the 4 month time point. We therefore switched the Cre deleter line and introduced all three conditional alleles onto a WapCreC mouse background19 (Supplementary information, Figure S5A). Of note, in this mouse line, the whey-acid protein (Wap) activity is independent of doxycycline and pregnancy, and the Cre activity is present in both luminal and basal epithelial cells in the mammary gland19 (Supplementary information, Figure S2). As expected from our previous work using whole-body Rank mutants or MMTVCre- and K5Cre-driven Rank deletion, WapCreC-mediated Rank deletion had no apparent effect on formation of the mammary gland during puberty (Supplementary information, Figure S5B). While all WapCreC;Brca1;p53 mutant females developed palpable tumors starting around day 100 after birth, concomitant Rank deletion in the mammary epithelium significantly delayed tumor onset (Figure 2A). The median tumor onset for WapCreC;Brca1;p53 mice was 158 days, whereas the median onset for WapCreC;Rank;Brca1;p53 triple-mutant mice was 184 days. Importantly, while all WapCreC;Brca1;p53 double-mutant females developed mammary carcinomas, 25% of WapCreC;Rank;Brca1;p53 triple-mutant littermates never developed any tumors (Figure 2A). This was also reflected by the overall survival rates (Supplementary information, Figure S5C), even when we followed these females up to 2 years of age. Once mammary tumors developed in both double- and triple-mutant mice, their growth curves appeared to be similar (Supplementary information, Figure S5D). Thus, WapCreC-driven deletion of Rank delays the onset and in 25% of cases even completely prevents the development of Brca1;p53 mutation-driven mammary cancer.
Figure 2.
Genetic deletion or pharmacological inhibition of RANK reduces the incidence of Brca1 deletion-driven mammary tumorigenesis. (A) Onset of palpable mammary tumors in WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 mice. Data are shown as percentage of tumor-free mice. (B) Representative histological sections of mammary tumors isolated from WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 mice showing different histological grades (H&E) and altered, grade-dependent KRT5/CK5 immunostainings. Scale bar, 100 μm. (C) Schematic of the regimen used to treat MMTV-CreBrca1flox11/flox11 mice with control Mu-Fc or RANK-Fc to block RANKL/RANK in vivo. (D) Numbers and percentages of MMTV-Cre Brca1flox11/flox11 mice with pre-neoplastic lesions that received subcutaneous (sc) injection of Mu-Fc or RANK-Fc for 6 or 12 months (3 times/week) and were sacrificed for analysis at 9 and 15 month of age. (E) Representative H&E-stained sections of mammary tissue of MMTV-CreBrca1flox11 mice that received Mu-Fc or RANK-Fc and were sacrificed at 15 month of age. Magnification, 400×.
Loss of RANK impairs tumor progression to high-grade malignancies
Histopathologic analysis revealed that double-knockout WapCreC;Brca1;p53 mice feature high-grade tumors (60%) as well as intermediate-grade tumors (40%). These high-grade tumors typically exhibit more foci with higher degrees of anisocytosis and anisokaryosis, higher mitotic rates and fewer regions of glandular differentiation (Figure 2B and Supplementary information, Figure S6A). By contrast, triple-knockout mice never developed high-grade tumors but displayed intermediate-grade tumors characterized by low to intermediate anisocytosis and anisokaryosis, low to moderate mitotic rate and some foci of glandular differentiation (Figure 2B and Supplementary information, Figure S6A). In these tumors we observed effective deletion of Rank, Brca1 and p53 (Supplementary information, Figure S6B and S6C). All intermediate-grade tumors that developed in double- and triple-mutant mice expressed the basal epithelial marker CK5; however, in high-grade tumors from WapCreC;Brca1;p53 double-mutant females we observed marked downregulation of CK5 expression (Figure 2B, Supplementary information, Figure S7A and S7B), supporting the notion of manifest epithelial dedifferentiation of these high-grade mammary cancers. Similar to CK5, we detected high expression of the basal myoepithelial marker p63 in Rank mutant tumors, whereas p63 expression was largely lost in the high-grade tumors from WapCreC;Brca1;p53 double-mutant females (Supplementary information, Figure S7C). Tumor cells of both cohorts still expressed β-catenin, confirming the epithelial lineage, whereas ERα and PRs were not detectable (Supplementary information, Figure S7D-S7F), confirming that these tumors are hormone receptor negative. RNAseq profiling of mammary carcinomas from WapCreC;Brca1;p53 and littermate WapCreC;Rank;Brca1;p53 females showed differences in their molecular signatures (Supplementary information, Figure S8A and S8B; all primary data have been deposited to Gene Expression Omnibus reference GSE71362); hierarchical clustering of the Spearman's correlation showed similarity of our mouse tumors to previously defined basal-like mammary cancer20 (Supplementary information, Figure S8C). Thus, loss of RANK not only delays tumor onset, but also offsets progression to higher grades of malignancy.
Ki67 immunostaining of mammary tumors from WapCreC;Brca1;p53 double-mutant and WapCreC;Rank;Brca1;p53 triple-mutant females showed comparable proliferation of tumor cells in both genotypes (Supplementary information, Figure S9A and S9B), a finding that might explain that established tumors grow at a comparable “speed” (Supplementary information, Figure S5D). Immunostaining for γH2AX as well as p53BP1 showed that loss of Brca1 and p53 resulted in massive DNA damage in intermediate as well as high-grade tumors, irrespective of the presence or absence of RANK (Supplementary information, Figure S9C-S9F). Importantly, we also observed marked DNA damage in the mammary epithelium of a 2-year-old WapCreC;Rank;Brca1;p53 triple-mutant female that had never developed any tumor (Supplementary information, Figure S10A). Although there was substantial DNA damage, the mammary glands displayed normal histology with sparse proliferative foci as determined by low Ki67 positivity (Supplementary information, Figure S10A and S10B). Analysis of mammary epithelial cells (MECs) isolated from the tumor-free 2-year-old WapCre;Rank;Brca1;p53 female confirmed efficient deletion of Brca1, p53 and Rank (Supplementary information, Figure S10C). These data show that loss of Rank protects mice from mammary tumorigenesis and tumor progression despite the presence of DNA damage due to the inactivation of Brca1.
Pharmacological inhibition of RANKL prevents the development of Brca1 mutation-driven pre-neoplastic mammary lesions
We next assessed whether, in addition to our genetic experiments, preventive pharmacological RANKL inhibition could also be used to mitigate tumor development in a Brca1 mutant mouse background. To test this, we treated MMTV-Cre;Brca1flox11/flox11 mice with RANK-Fc to block RANKL/RANK in vivo and as a control with Mu-Fc and followed the cohorts for up to 15 months (Figure 2C). We switched to this mouse model because these animals still express p53 and develop pre-neoplastic mammary lesions, which are thus driven solely by the loss of Brca1. Therefore, they better mimic the genetics of the human situation prior to the development of overt breast tumors, which is when a prophylactic would be used. 3/10 (33%) MMTV-Cre;Brca1flox11/flox11 females developed pre-neoplastic lesions in the control group (Mu-Fc-treated). Intriguingly, not a single (0/10) MMTV-Cre;Brca1flox11/flox11 female treated for 6 months with RANK-Fc developed pre-neoplastic lesions in the mammary gland. All tissues analyzed retained morphological characteristics resembling age-matched wild-type (WT) control females (Figure 2D and Supplementary information, Figure S11A). In mice treated for 12 month with control Mu-Fc, 9/11 (82%) females developed mammary lesions, while such lesions were only detectable in 1/13 (7%) females treated with RANK-Fc (Figure 2D and 2E). Further characterization of the mammary lesions by immunohistochemistry verified that these lesions were of epithelial origin (Supplementary information, Figure S11B). Moreover, we found that the pre-neoplastic lesions in Mu-Fc-treated mice did not express ERα or PRs, indicative of pre-neoplastic expansion of ERα- and PR-negative epithelial progenitors (Supplementary information, Figure S11C). These data show that pharmacological inhibition of RANKL can prevent the development of Brca1 mutation-driven pre-neoplastic mammary lesions.
RANK controls the expansion of Brca1-mutated mouse and human mammary progenitor cells
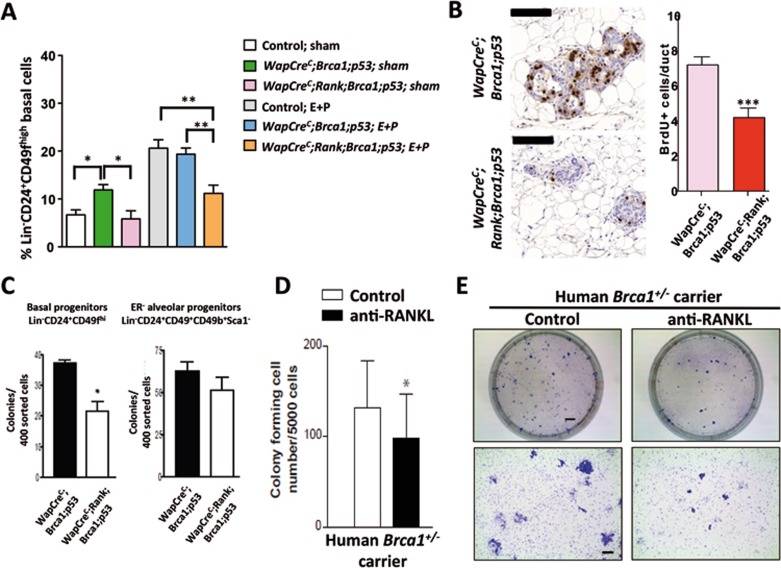
Since genetic deletion or pharmacological inhibition of RANK delays the onset and even prevents Brca1 and Brca1;p53 mutation-driven mammary tumors, we speculated that RANKL/RANK might affect proliferation and expansion of mammary progenitor cells. To assess this hypothesis, we performed ovariectomy (ovx) on female mice followed by sham treatment (no hormones) or reconstitution with estrogen and progesterone (E+P). As previously reported11,12, sex hormone treatment resulted in a marked expansion of Lin−CD24+CD49fhi basal mammary progenitor cells in both control as well as WapCreC;Brca1;p53 double-mutant mice; however, this expansion was markedly reduced in the absence of RANK expression (Figure 3A and Supplementary information, Figure S12A). We also observed expansion of the basal Lin-CD24+CD49fhi progenitor compartment in sham operated WapCre;Brca1;p53 double-mutant mice, a phenotype that was again dependent on the expression of RANK (Figure 3A and Supplementary information, Figure S12A). Sex hormone-driven proliferation in ovx females was further determined by in vivo BrdU labelling. WapCreC-driven deletion of Brca1 and p53 resulted in markedly increased numbers of cycling mammary epithelial cells; this sex hormone-induced proliferation of mammary epithelial cells was significantly reduced in Rank mutant females (Figure 3B). Importantly, deletion of Rank significantly abrogated the in vitro colony formation capacity of basal Lin−CD24+CD49fhi mammary progenitors from WapCreC;Brca1;p53 mutant mice; the colony formation capacity within the ER−PR−Sca1− alveolar progenitor compartment was also lower in the absence of RANK, albeit less pronounced (Figure 3C). Similar to our data using WapCreC deleter mice, sex hormone treatment resulted in expansion of the basal Lin−CD24+CD49fhi progenitor compartment in ovarectomized K5Cre;Brca1;p53 double-knockout mice as compared with sham operated mice, while this expansion was not observed in the absence of RANK (Supplementary information, Figure S12B). Thus, RANKL/RANK control sex hormone-induced expansion and activity of Brca1;p53 mutant basal mammary progenitor cells. To determine the relevance of these results for the human disease, we isolated mammary progenitor cells from three women carriers of heterozygous BRCA1 mutations who underwent prophylactic mastectomy, which is the standard of care. Importantly, in vitro colony-forming cell (CFC) assays performed on these human breast epithelial cells showed that RANKL inhibition using the clinically approved RANKL blocking antibody Denosumab significantly decreased the frequency of colony-forming cells (Figure 3D, 3E and Supplementary information, Figure S12C), indicating suppression of human mammary progenitor activity. These data reveal the capacity of RANKL inhibition to reduce the activity of mammary progenitor cells from women that carry germline BRCA1 mutations.
Figure 3.
RANK mediates growth and expansion of Brca1 mutant murine and BRCA1 mutant human mammary progenitor cells. (A) Quantification of Lin−CD24+CD49fhi basal mammary epithelial cells in ovariectomized WapCre-negative control, WapCreC;Brca1;p53 double- and WapCreC;Rank;Brca1;p53 triple-knockout mice treated with E+P or without hormone treatment (sham). Data are from 10-12-week-old mice, at which age we never observed any evident tumors, and represent mean +/− SEM (n ≥ 3 mice/group). *P < 0.05, **P < 0.005 (Student's t-test). (B) Representative histologic images and quantification of BrdU immunostaining of mammary glands of ovariectomized WapCreC;Brca1;p53 double-knockout and WapCreC;Rank;Brca1;p53 triple-knockout female mice treated with E+P showing reduced mammary epithelial proliferation (reduced BrdU positivity). Quantification data are shown as average numbers of BrdU+ cells per duct +/− SEM (n = 4 mice/cohort). ***P < 0.0001 (Student's t-test). Scale bar, 100 μm. (C) Colony forming capacity of basal progenitors and ER-negative alveolar mammary epithelial progenitor cells from WapCreC;Rank;Brca1;p53 and WapCreC;Brca1;p53 knockout mice. Data are shown as number of colonies per 400 sorted cells +/− SEM (n = 3 mice/group). *P < 0.05, ns, not significant (Student's t-test). (D, E) Colony forming capacity (CFC) of human mammary progenitor cells. Human mammary progenitor epithelial cells were isolated from three women carrying heterozygous BRCA1 mutations. Single mammary cell preparations were generated from organoids for CFC assays, plated, and were either untreated or treated with the anti-RANKL blocking Ab Denosumab (1 μg/ml). The quantification is shown in D. Data are shown as number of colonies per 5 000 plated cells +/− SEM (n = 3 different BRCA1 carriers per group). *P < 0.01 (Paired Student's t-test). Representative images of paired untreated control and anti-RANKL (Denosumab)-treated (1 μg/ml) mammary progenitor colonies are shown in E. Scale bars: upper panels, 5 mm; lower panels, 1mm.
RANKL and RANK are highly expressed in pre-malignant lesions and breast cancer from human BRCA mutation carriers
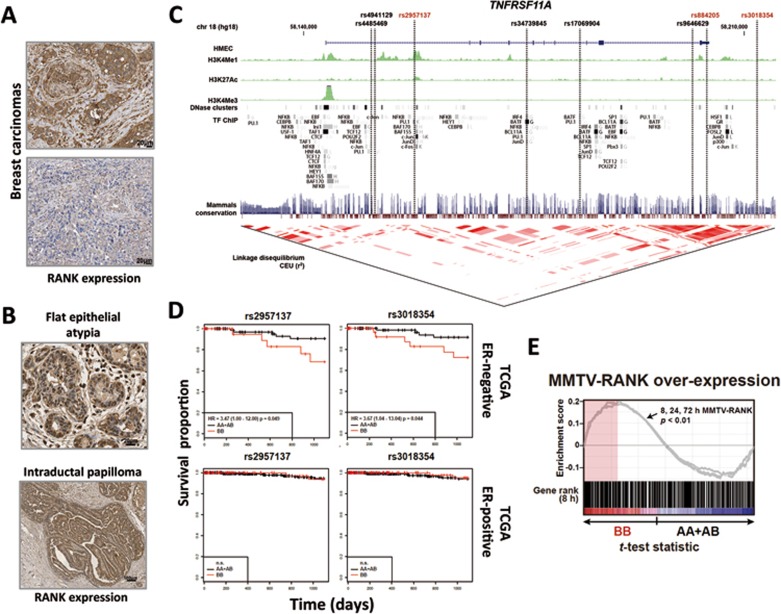
Following our observation that RANKL/RANK control Brca1 mutation-driven tumorigenesis in mice and affect expansion of human BRCA1+/mut mammary progenitor cells, we analyzed RANK protein expression in human breast tumors that developed in BRCA1 or BRCA2 mutation carriers, and in non-BRCA1/2 mutated individuals. RANK protein expression was absent or low (0 and 1+) in 74.5% of malignant breast tumors from BRCA1/2 WT patients, with intermediate (2+) expression detectable in 25.5% of cases (Table 1). Notably, high RANK protein expression (3+) was not detected in any BRCA1/2 WT patients. Conversely, intermediate or high levels of RANK were observed in 70.4% of BRCA1- and 80.7% of BRCA2-mutated tumors (Figure 4A and Table 1). A similar pattern was observed for RANKL protein expression which was detectable in 40.8% of tumors from BRCA WT patients, with intermediate or high expression in only 5.1% of breast cancer samples, but which was found in 59.1% of malignant tumors from BRCA1 germline mutation carriers with intermediate or high expression in 14.8% samples. Similarly, 69.6% of tumors from BRCA2 mutation carriers expressed RANKL with intermediate or high expression in 12.5% samples (Table 1). There was also a highly significant correlation between tumor grade and RANK protein expression in the overall series. While low-grade (G1) tumors expressed intermediate or high RANK levels in 28% of cases, 50.8% of grade 2 (G2) and 73.3% of all grade 3 (G3) tumors exhibited intermediate or high RANK protein expression (P < 0.001) (Supplementary information, Figure S12D and Table S1). RANKL protein staining was detected in 41.7% of G1 tumors, 56.3% of G2 tumors, and 51.2% of G3 tumors and there was no significant difference in respect to grading (P = 0.47) (Supplementary information, Figure S12D and Table S1). We next analyzed whether RANK expression was already present in the earliest tumor lesions detectable in the breast tissue of BRCA1 mutation carriers. In all cases of such early lesions analyzed (n = 11 patients) including flat epithelial atypia and intraductal papillomas we detected high RANK expression (Figure 4B). Thus, RANK is highly expressed in pre-malignant lesions as well as in breast cancer that has developed in BRCA1 and BRCA2 mutation carriers.
Table 1. RANKL and RANK protein expression in BRCA1 mutant, BRCA2 mutant, and BRCA1/2 WT human breast tumors.
| Intensity | RANK1 |
RANKL1 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT |
BRCA12 |
BRCA23 |
WT |
BRCA12 |
BRCA23 |
|||||||
| (n = 98) | (n = 88) | (n = 62) | (n = 98) | (n = 88) | (n = 56) | |||||||
| N | % | N | % | N | % | N | % | N | % | N | % | |
| 0 | 0 | 0 | 2 | 2.3 | 0 | 0 | 58 | 59.2 | 36 | 40.9 | 17 | 30.4 |
| 1+ | 73 | 74.5 | 24 | 27.3 | 12 | 19.4 | 35 | 35.7 | 39 | 44.3 | 32 | 57.1 |
| 2+ | 25 | 25.5 | 34 | 38.6 | 30 | 48.4 | 5 | 5.1 | 13 | 14.8 | 7 | 12.5 |
| 3+ | 0 | 0 | 28 | 31.8 | 20 | 32.3 | 0 | 0 | 0 | 0 | 0 | 0 |
1Column percentages presented.
2Includes obligate carriers: BRCA1, n = 15.
3Includes obligate carriers: BRCA2, n = 2.
RANK and RANKL expression levels were determined by immunohistochemistry by board certified pathologists. In all cases, the BRCA1 and BRCA2 mutation status was determined by sequencing. P values (χ2 or Fisher's Exact test): RANK (0/1+ vs 2+/3+): WT vs BRCA1/2 mutations, P < 0.001; WT vs BRCA1, P < 0.001; WT vs BRCA2, P < 0.001; BRCA1 vs BRCA2, P = 0.158. RANKL (0 vs 1+/2+/3+): WT vs BRCA1/2 mutations, P < 0.004; WT vs BRCA1, P = 0.013; WT vs BRCA2, P = 0.001; BRCA1 vs BRCA2, P = 0.201.
Figure 4.
RANK expression in tumor tissues and TNFRSF11A variations linked to BRCA1-mutated status, risk of breast cancer, and cancer progression. (A) Representative images showing high and low RANK expression in invasive breast cancer tissues from BRCA1 mutation carriers. (B) Representative examples of RANK expression in early epithelial pre-neoplastic lesions in women with BRCA1 mutations. Representative images of a flat epithelial atypia and a typical intraductal papilloma are shown. Scale bars are shown. (C) Graph depicting the TNFRSF11A locus including the variants associated with breast cancer risk in BRCA1 mutation carriers, and regulatory evidence from the ENCODE project and human mammary epithelial cells (HMECs). The rs884205 variant and the two in linkage disequilibrium and analyzed in TCGA dataset are marked in red. The exons are marked by blue-filled rectangles and the direction of transcription is marked by arrows in the gene structure. The chromosome 18 positions (base pairs (bp)) and linkage disequilibrium (r2) from a HapMap CEU panel are also shown. (D) Kaplan-Meier survival plots for the indicated TNFRSF11A genotypes (A and B represent the major and minor allele, respectively) in ER-negative and ER-positive breast tumors using TCGA data. (E) Gene Set Expression Analysis (GSEA) graphical output showing the positive correlation between the gene expression in tumors with the minor genotype (associated with poorer prognosis in ER-negative human breast cancer) and gene sets that characterize RANK overexpression in mammary epithelial cells of mice at 8, 24 or 72 h (GSE66174). All three sets were found to be significantly associated (P < 0.01); the score distribution curves overlap. The gene rank corresponds to the 8 h time point and the GSEA enrichment scores are shown26.
Common variants in RANK are associated with increased breast cancer risk in human BRCA mutation carriers
Since RANK/RANKL critically influence Brca1 mutation-driven breast carcinogenesis in mice, we assessed the role of genetic modifiers of breast cancer risk in women with inherited BRCA1 mutations at the corresponding locus encoding for human RANK, TNFRSF11A. This analysis was performed using data from the Collaborative Oncological Gene-environment Study (iCOGS) that included 51 TNFRSF11A single-nucleotide polymorphisms (SNPs) genotyped in ∼15 200 BRCA1 and ∼8 200 BRCA2 mutation carriers21,22 (Supplementary information, Table S2). Using a retrospective likelihood approach, we identified six SNPs that were significantly associated with breast cancer risk in the overall series of BRCA1 mutation carriers and/or in ER-negative or triple-negative subtypes (Figure 4C, Table 2 and Supplementary information, Table S2). In addition, we found two SNPs in TNFRSF11A significantly associated (P < 0.05) with breast cancer risk in BRCA2 mutation carriers (Supplementary information, Table S3). Interestingly, the rs884205 change (C→A) introduces an ATTAAA motif in the 3′-untranslated (3′-UTR) region of TNFRSF11A, providing a poly(A) signal for the nearby alternative polyA site (PolyA_DB Hs.204044.1.8; PolyA-seq GSE30198). Of note, the minor A allele, which we find predicts increased risk of breast cancer, correlates with enhanced RANK expression in various tissues using GTEX analysis, and the same allele has been previously associated with decreased bone density23,24, consistent with an over-activation of RANK-mediated signaling. Moreover, the G allele of rs4485469 was found to be associated with a reduced risk of breast cancer in the BRCA1 mutation carriers (Figure 4C, Supplementary information, Tables S2 and S3), and we again find increased RANK expression linked to the complementary allele (A) in GTEX data. To assess these observations further, we analyzed data from The Cancer Genome Atlas (TCGA)25. While rs884205 was not present in TCGA, it was partially correlated (r2 ≈ 0.45) with two included variants, rs2957137 and rs3018354; a survival analysis using the recessive model for these variants revealed a significant association in ER-negative, but not in ER-positive breast cancer (Figure 4D). These two SNPs were not genotyped in iCOGS, but their imputed results (r2 > 0.94) again revealed significant associations (P = 0.021) with breast cancer risk in BRCA1 mutation carriers, with effect estimations similar to rs884205: rs2957137 HR = 1.079, rs3018354 HR = 1.052. Moreover, expression analyses revealed that the ER-negative tumors with poorer prognosis have activated RANK signaling, as shown by their positive expression correlation with time series signatures of RANK overexpression26 (Figure 4E). These data indicate that common variations in TNFRSF11A modify the risk of developing breast cancer in BRCA mutation carriers.
Table 2. TNFRSF11A genotyped iCOGS variants associated (P < 0.05) with breast cancer risk in BRCA1 mutation carriers and/or ER-negative or overall triple-negative subtypes.
| SNP ID | All BRCA1-mutated |
ER-negative |
Triple-negative |
rs884205 (r2) | |||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||
| rs9646629 | 1.052 (1.006-1.096) | 2.15E-02 | 1.061 (1.008-1.116) | 2.20E-02 | 1.072 (1.008-1.140) | 2.60E-02 | 0.54 |
| rs4485469 | 0.956 (0.918-0.996) | 3.55E-02 | 0.944 (0.899-0.992) | 2.10E-02 | 0.943 (0.889-1.001) | 5.40E-02 | 0.06 |
| rs34739845 | 0.933 (0.876-0.999) | 4.07E-02 | 0.9154 (0.847-0.990) | 2.70E-02 | 0.907 (0.826-0.997) | 4.30E-02 | 0.003 |
| rs4941129 | 1.048 (1.002-1.098) | 4.50E-02 | 1.059 (1.003-1.117) | 3.70E-02 | 1.057 (0.990-1.129) | 9.40E-02 | 0.12 |
| rs17069904 | 0.935 (0.878-1.001) | 5.22E-02 | 0.918 (0.849-0.993) | 3.30E-02 | 0.887 (0.805-0.979) | 1.70E-02 | 0.002 |
| rs884205 | 1.048 (0.995-1.098) | 6.23E-02 | 1.063 (1.004-1.125) | 3.60E-02 | 1.066 (0.996-1.141) | 6.70E-02 | 1 |
Discussion
RANKL acts in a paracrine fashion on the membranous RANK of ER/PR-negative epithelial cells of the breast to expand the stem and progenitor cell populations27,28. Our data show that RANKL/RANK also regulate the expansion and functional capacity of progenitor cells on a Brca1/p53 mutant background and, most importantly, on mammary stem cells from women with heterozygous germline BRCA1 mutations. Since RANKL is induced by progesterone, these data could explain why sex hormones, in particular progesterone, play an important role in BRCA1 mutation-driven tumorigenesis in mouse models and humans. Studies have also demonstrated that Brca1 deficiency increases the activity of E+P signaling pathways during mammary gland development and promotes mammary epithelial cell growth29,30,31. In particular, loss of full-length Brca1 exaggerates the mammary growth in response to exogenous progesterone in virgin mice30. The importance of PR signaling in mutant Brca1 tumorigenesis was further demonstrated by Poole et al.3 who showed that tumor development in Brca1/p53 deficient mice could be prevented by the PR antagonist RU-486. Together, these findings highlight the potential value for RANKL inhibition in BRCA1-associated cancers at the early stages of tumorigenesis. Whether RANKL inhibition has an advantage over Tamoxifen and/or oophorectomy needs to be demonstrated in future experiments, especially in careful, clinical trials.
We also report, analyzing iCOGS data for more than 23 000 women with germline BRCA1/2 mutations, that common variation in TNFRSF11A, linked to altered RANK expression, may be associated with breast cancer risk in BRCA1 mutation carriers and, furthermore, the survival of patients with ER-negative tumors. Some of these identified polymorphisms are directly associated with altered gene expression and function of RANK. Further fine-scale mapping genetic analyses and studies on the regulation of RANK expression, and also expression of RANKL and the natural RANK inhibitor OPG, are warranted, since the 3′-UTR of RANK can be additionally regulated by various microRNAs, some of which have been directly associated with human breast cancer32. Since BRCA1 mutation carriers are at high risk of ovarian cancer, it will be important to also explore the role of RANKL/RANK blockade in ovarian carcinogenesis. Most importantly, our work here shows that pharmacological inhibition of RANKL almost completely prevents the development of pre-neoplastic lesions due to a Brca1 mutation. Our findings therefore raise the possibility that inhibition of RANKL, for which there is an already approved drug with a good safety record, could offer a novel, targeted approach for breast cancer prevention in BRCA1 mutation carriers.
Materials and Methods
Mice
Rankfloxed (Tnfrsf11tm1.1Pngr) and WapCreC;Brca1floxed;p53floxed (Tg-WapCreC, Brca1tm2Cxd, and Trp53tm1Elee) knockout mice have been previously generated in our laboratories3,16. K5Cre (Tg-KRT5-Cre) mice were purchased from the Jackson Laboratory. Of note, in our cohorts, the control mice examined were either Cre-negative but carried the Brca1flox;p53flox, and/or Rankflox alleles or were K5Cre positive in the absence of dual floxed alleles; none of these controls examined showed any epithelial hyperplasia in the mammary glands. Brca1 conditional knockout mice with two exon 11 floxed Brca1 alleles (Brca1flox11/flox11) carrying the mouse mammary tumor virus (MMTV)-Cre recombinase gene (MMTVCre;Brca1flox11/flox11) were maintained on C57Bl/6 genetic background. The Rosa26eYPF reporter mouse line has been previously reported33. Animals were genotyped by PCR analysis of genomic DNA. Mice were maintained in temperature-controlled conditions. All animal experiments were carried out in agreement with the ethical animal license protocol in accordance with the current laws of Austria and in accordance with institutional guidelines approved by the University of Maryland, Baltimore Animal Care and Use Committee.
Histology and immunohistochemistry
Mouse tissue samples were fixed in 4% paraformaldehyde (PFA) overnight at 4°C and embedded in paraffin after dehydration in ascending concentrations of ethanol. For histological analysis, 2-4 μm-thick paraffin sections were prepared and stained with haematoxylin and eosin. For immunohistochemistry, after routine processing and antigen retrieval procedures, the following antibodies were used. Paraffin sections were stained using previously described protocols34 with antibodies against: KRT/CK5 (1:1 000, Rabbit anti-human KRT5, Sigma Aldrich, SAB4501651), γH2AX (1:500, Rabbit anti-human H2AFX, ph-Ser139, Novus Biologicals, NB100-79967), TP53BP1 (1:200, Rabbit anti-human TP53BP1 (E247), Bethyl Laboratories, IHC-00001), Ki67 (1:1 000, Rabbit anti-human Ki67, Novocastra, NCL-Ki67p), Ki67 (1:500, Rat anti-mouse Ki67, Affymetrix E-bioscience, 14-5698), BrdU (1:200, Rat anti-BrdU (BU1/75(ICR1)), Abcam, ab6326), CTNNB1 (1:250, Rabbit anti-CTNNB1 (E247), Abcam, ab32572). CASP3, Cleaved (1:200, Rabbit anti-human Cleaved Caspase 3, Cell Signalling, 9661), ERα (1:100, Rabbit anti-mouse ERα, Santa Cruz, sc542), PR (1:100, Rabbit anti-human PR, Santa Cruz, sc538), TP63 (1:150, Mouse anti-human TP63, Santa Cruz, sc8341), CD265/RANK-patient samples (1:1 000, Mouse anti-human CD265 (9A725), Acris Antibodies, TA336373), and CD254/RANKL ― patient samples (1:1 000, anti-human CD254, Acris Antibodies, ID8600).
Histomorphometric indices were calculated as the number of positive epithelial cells divided by the total number of epithelial cells, with no fewer than 1 000 counted nuclei for Ki67 and γH2AX staining. Mammary epithelial neoplasias (MINs) and adenocarcinomas in histologic sections were designated and evaluated in accordance with predefined criteria proposed by the Mouse Models of Human Cancers Consortium and INHAND35,36. Briefly, low-grade MINs were defined by the presence of ducts with intact basement membrane, at least one layer of atypical cells, hyperchromatic nuclei, luminal and/or myoepithelial cells with little cytoplasm, at least one layer and an increased mitotic rate. High-grade MIN lesions were defined by less organized glandular patterns, increased layers of epithelium, pleomorphism of nuclei and/or epithelial cells and/or an increase in mitotic figures with an intact basement membrane. Carcinomas were defined by an invasive growth pattern, increased cellular and nuclear pleomorphism and transgression of the basement membrane. The extent of KRT5 and TP53BP1 immunopositivity was quantified in mouse mammary lesions with Definiens Tissue StudioTM software. The software algorithm was trained by manual delineation and classification of representative areas in representative slides to identify positive targets and exclude non-targets and to create a quantification solution. Target regions were identified on the basis of morphology and positive immunohistochemical (IHC) staining. Subsequently the solution was applied to the digital slide sets to obtain automated quantification results. The validity of the analysis was confirmed by histopathologic verification of representative slides from both groups. For imaging and digital quantification, stained slides were scanned using a Pannoramic slide scanner (3D Histech).
For murine Rank immunostainings on frozen samples, tissue samples were snap frozen in OCT and 5-10-μm-thick frozen sections were prepared. After drying at room temperature, sections were fixed in Aceton at −20 °C for 5-10 min. Samples were first blocked for 45 min in freshly prepared H2O2 (0.3%), followed by blocking in TNB buffer for 30 min (Perkin Elmer, TSA fluorescein amplification kit) and Avidin and Biotin block for 15 min respectively (Avidin/Biotin Blocking kit, Vector Lab). Samples were incubated with a primary biotinylated anti-mRANK antibody (BAF692, R&D, 1:50) overnight at 4 °C followed by incubation with Streptavidin-HRP (1:100) for 30 min at room temperature and incubation with the working solution for 5-10 min. Samples were mounted with DAPI. For analysis of mammary glands after RANK-Fc treatment, the right inguinal mammary gland from each mouse was fixed in 10% buffered formalin (Fisher Scientific, Pittsburgh, PA) overnight at 4 °C and embedded in paraffin using standard techniques for H&E staining and IHC analysis. ERα IHC was performed on formalin-fixed, paraffin-embedded tissue sections using mouse on mouse (M.O.M) peroxidase kit (PK-2200, Vector Laboratories Inc., Burlingame, CA, USA) as previously described37 using a 1:25 dilution of the ERα antibody (M7047, Dako, Carpinteria, CA, USA) for 30 min. Polyclonal Rabbit Anti-Human Progesterone Receptor, Dako, A0098). Digital photographs were taken using a Nikon 50i Upright Microscope System with a high Resolution 5 Megapixel Color Digital Camera system (Nikon Instruments Inc., Melville, NY, USA). All histomorphology and immunohistochemistry of the subsets of tissues were qualitatively or semiquantitatively evaluated by board certified pathologists (AK, HP, LK, ZBH).
Whole mount stainings
Mammary glands were dissected, placed on a microscope slide and fixed in 4% PFA overnight. To remove the adipose tissue, mammary glands were incubated in acetone for 3 h followed by rehydration in 100% ethanol and 95% ethanol for 1 h, respectively. Mammary glands were stained with hematoxylin for 3 h and afterwards incubated in slightly basic tap water for 2 h. Excessive hematoxylin was removed by destaining the glands in 50% ethanol acidified with 25 ml 1.0 M HCl/liter. After dehydration in 70%, 95%, and 100% ethanol, respectively, mammary glands were stored in xylene.
Ovariectomy and hormone treatments
Six to eight-week-old female mice were anesthetized by intraperitoneal injections of Ketasol (5 mg/ml) and Xylasol (0.8 mg/ml). Bilateral small incisions were made under sterile conditions to open the peritoneal cavity. Ovaries, including the surrounding adipose tissue, were excised. Mice were allowed to recover for at least 2 weeks before further experiments were performed. For hormone supplementation mice were implanted subcutaneously on the right flank with slow release pellets containing a combination of progesterone and 17β-estradiol (Innovative Research of America, 21 day release, 0.14 mg 17β-estradiol + 14 mg progesterone).
qRT-PCR
Total RNA of tumors and isolated mammary epithelial cells was prepared using the RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions. cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad). For qRT-PCR the following primers were used:
β-actin forward primer: 5′-GCTCATAGCTCTTCTCCAGGG-3′
β-actin reverse primer: 5′-CCTGAACCCTAAGGCCAACCG-3′.
Rank forward primer: 5′-CCCAGGAGAGGCATTATGAG-3′
Rank reverse primer: 5′-CAGCACTCGCAGTCTGAGTT-3′
Brca1 forward primer: 5′-TAAGCCAGGTGATTGCAGTG-3′
Brca1 reverse primer: 5′-TGCCCTCAGAAAACTCACAA-3′
P53 forward primer: 5′-TGGAAGACAGGCAGACTTTTC-3′
P53 reverse primer 5′-CCCCATGCAGGAGCTATTAC-3′
Mammary epithelial cell isolation
Mammary epithelial cells were isolated as previously described13. Briefly, mice were sacrificed and mammary fat pads were removed using sterile dissection instruments. Mammary glands were transferred in 50 ml Falcon tubes and incubated in complete EpiCult medium (EpiCult-B basal medium, EpiCult-B proliferation supplements, 10 ng/ml rh bFGF, 10 ng/ml rh EGF, 4 μg/ml Heparin and 5% FCS) and 2.5× collagenase/hyaluronidase at 37 °C in a shaking incubator for 2.5 h. Red blood cell lysis was performed using ammonium chloride. Pellets were consecutively treated with 0.25% trypsin-EDTA and prewarmed dispase containing 200 μl of 1 mg/ml DNase. After filtering single mammary epithelial cells were prepared for FACS analysis or lysed for RNA purification.
Flow cytometry
Multiparameter flow cytometry analyses were performed on mammary epithelial single-cell suspensions by immunostaining for 20 min at 4 °C in FACS buffer (PBS, 2% FCS, 2 mM EDTA). Before adding the fluorescently labeled antibodies, Fc-receptors were blocked with anti-CD16/CD32 (Pharmingen) antibodies was performed. Mammary epithelial cells were labeled with antibodies directed against CD24 (Pharmingen #553261), CD49f (Pharmingen #551129) and CD61 (eBioscience #11-0622-82). Non-epithelial cells were excluded using antibodies directed against CD45 (BD #553078), CD31 (BioLegend #102404) and Ter119 (BioLegend #116204). Flow cytometry data were acquired using a Fortessa LSRII flow cytometer (FACSFortessa™, BD), equipped with FacsDiva™ software (BD). Data analysis was performed using FlowJo™ software (Tree Star).
Bromodesoxyuridin (BrdU) labeling
For BrdU pulse labeling, ovariectomized mice were stimulated with 17β-estradiol and progesterone for 2 weeks. BrdU (0.4 mg/g body weight) was injected intraperitoneally. Mice were sacrificed 2 h later and tissues were harvested.
Human breast epithelial preparations
Human breast tissue samples were obtained from disease-free pre-menopausal women undergoing reduction mammoplasty or prophylactic mastectomy (BRCA1 mutation carriers) with informed patient consent and Institutional Research Ethics Board approval from St. Michael's Hospital and from the University Health Network. Single-cell suspensions were prepared from previously isolated and cryopreserved organoids as previously described38.
Murine and human mammary colony-formation assay
Mouse mammary epithelial subsets were FACS-sorted from individual mice and plated with irradiated fibroblasts in DMEM:F12 (3:1) medium containing 10% FBS, insulin (Life Technologies), cholera toxin (Sigma), adenine (Sigma), hydrocortisone (STEMCELL Technologies), and Rock inhibitor (Reagents Direct) and cultured in 5% oxygen conditions. Colonies were scored after 7-10 days. For human CFC assay, mammary epithelial progenitors were isolated from Brca1 heterozygous mutation carriers undergoing pre-emptive surgery, and assays were performed in dishes precoated with a thin layer of collagen (STEMCELL technologies) in complete Epicult-B medium (STEMCELL Technologies) with irradiated feeders as previously documented39 and cultured in 5% oxygen conditions.
Bioinformatics
PolyA-mRNA of WapCreC;Brca1;p53, and WapCreC;Rank;Brca1;p53 mammary tumors was isolated and the generated libraries were sequenced by 50-bp single-end Illumina mRNA sequencing. Reads were aligned using Tophat v2.0.10 and bowtie v0.12.9, FPKM estimation was performed with Cufflinks v2.1.1, aligned reads were counted with HTSeq v0.6.1p1, and differential expression analysis was performed with DESeq2 v1.6.2. Significantly differentially expressed (DE) genes were selected with a false discovery rate (FDR) of 5%. Pathway enrichment was evaluated using GSEA against the Molecular Signature Database MSigDB v5 gene-set collection. For GSEA, human to mouse conversion of official gene symbols was performed using HomoloGene ortholog assignment. Gene sets with significant enrichment were selected based on a FDR q-value cut-off of 1%. RNAseq data and GSEA analysis data have been deposited to the NCBI Expression Omnibus and all data are accessible through the GEO accession number GSE71362 (NCBI tracking system #17490198). Briefly, gene set enrichment analysis (GSEA) using KEGG comparing mammary tumors growing in WapCreC;Brca1;p53 double- vs WapCreC;Rank;Brca1;p53 triple-mutant mice showed significantly enhanced expression of genes annotated to Basal Cell Carcinoma, Notch signaling, Hedgehog signaling, and metabolism of xenobiotics by cytochrome P450 in the triple-mutant tumors, whereas in double-mutant tumors genes annotated to aminoacyl tRNA biosynthesis, RNA degradation, and Citrate/TCA cycle were higher expressed. Using Hallmark GSEA analysis, we found enhanced expression of genes annotated to Wnt-catenin signaling, Notch signaling, the p53 pathway, late estrogen response, and Ras signaling in Rank-deficient tumors, whereas genes annotated to Myc and E2F targets, mTORC1 signaling, G2M checkpoint and mitotic spindle, inflammation and allograft rejection, as well as the unfolded protein response (UPR) were significantly upregulated in the double-knockout tumors. Of note, the UPR has recently been associated with triple-negative breast cancer40, aminoacyl-tRNA synthetases have been linked to tumorigenesis41, and multiple cross-talks have been established between the Wnt and RANKL/RANK pathways, including molecular cross-talks that control mammary progenitor cells11,42. Thus, Rank inactivation in a WapCreC;Brca1;p53 mutant background results in mammary tumors that exhibit expression differences in genes annotated to oncogenesis, basic metabolism, RNA metabolism, or the regulation of mammary stem cells. The GSEA tool was run using default values for all parameters and using the t-statistic of the difference of gene expression between ER-negative tumors with the AA+AB and BB genotypes. We used transcriptome data to compare our mouse model with murine data previously assigned to mouse breast-cancer subtypes and linked to human subtypes20,43. Log expression estimates of our and published mouse models (NCBI GEO GSE3165) were combined by adjusting for the batch effect using Combat44. Hierarchical clustering of the Spearman correlation of samples shows similarity of our mouse model to basal-like samples. No consistent classification could be obtained using TNBCtype, a tool designed to identify potential Triple-negative breast cancer (TNBC) subtype membership45,46 (applied to log2FPKM expression data from our mouse model after mapping the data to human orthologous identifiers using HomoloGene).
RANK-Fc treatment
Three-month-old MMTV-Cre;Brca1flox11/flox11 mutant were treated with RANK-Fc (10 mg/kg three times/week; n = 26, RANK-Fc was provided by Amgen Pharmaceuticals, Inc.) subcutaneous injection (s.c.) or mouse Fc-fragment (Mu-Fc; 10 mg/kg three times/week, s.c. as a control; n = 22). The animal weights were monitored weekly. The treatment regime was well tolerated by all the mice and the weights were stably maintained throughout the length of treatment. Only one mouse in the Mu-Fc 15 months cohort (control) group was found dead in the cage due to unknown causes. Mammary tumor formation was determined by palpation and confirmed by histologic examination. Cohorts of mice from each experimental group were euthanized at 9 and 15 months of age. At necropsy, all ten mammary glands were exposed following a midline incision and glands were inspected visually, palpated, and digital photographs taken. Mammary glands were collected and processed for whole mount, hematoxylin and eosin (H&E) and IHC analyses, or snap frozen in liquid nitrogen and stored at −80 °C for gene and/or protein expression.
Human breast cancer samples
Tumor samples were provided as formalin fixed, paraffin embedded tissues by the Center for Familial Breast and Ovarian Cancer at Medical University of Vienna and by the Kathleen Cunningham Foundation Consortium for research into Familial Breast cancer. The analysis was approved by the local Institutional Review Board-IRB: the Ethikkommissionsbescheid (IRB) number which covers the analyses is EK 056/2005, Amendment Vers. 1.0 of 4.12.2014 “Modifiers des Krebsrisikos in BRCA1/2 Mutationsträgerinnen”. Data were analyzed using SPSS Ver21.0 (Chicago, IL, USA). Women diagnosed with breast cancer who had undergone surgeries and gave consent to germline mutation testing were included in this analysis. All samples, including the BRCA1/2 WT samples, were confirmed by sequencing. χ2-test and Fisher's Exact tests were used to compare the difference within RANK and RANKL categories. Samples classified as unknown for any variables were excluded from analyses. Two-sided P values ≤ 0.05 were considered to be statistically significant.
Human cohort studies
This analysis was performed using data from the Collaborative Oncological Gene-environment Study (iCOGS) that included 51 TNFRSF11A (encoding for RANK) SNPs genotyped in ∼15 200 BRCA1 and ∼8 200 BRCA2 mutation carriers. The iCOGS design, quality controls, and statistical analyses have been previously described21,22. The TCGA genotype data from primary breast tumors were obtained following an approved request and through the data portal (https://tcga-data.nci.nih.gov/tcga). The survival analysis as a function of the genotypes was performed using the Cox proportional hazards regression and the significance of the association assessed using the log-rank test.
Statistics
All values in the paper are given as means ± sem. Comparisons between groups were made by Student's t-test or 2-way ANOVA using GraphPad Prism (GraphPad Software, San Diego, CA, USA) or R statistical software. For the Kaplan–Meier analysis of tumor onset and survival a log-rank test was performed. P < 0.05 was accepted as statistically significant.
Author Contributions
VS performed most of the experiments in RANK mutant mice with help from IK, DS, RS, MT, IU, LT and SR. GW performed lineage tracing experiments. KOB performed majority of the RANK-Fc treatment experiments and data analysis. JH and AS administered RANK-Fc and Mu-Fc treatments and data analysis. NE and AOA contributed to the ER and PR IHC experiments and data analysis. PAJ and RK analyzed and FACS-sorted mammary epithelial populations for colony forming assays and performed the human CFC assays. JC and HB provided human mammary gland epithelial cells. MN performed all bioinformatics analysis. AK, ZBH, and LK are board certified pathologists and performed histology and immunohistology assessments. MAP, EV and HH provided and analyzed TCGA data, CL, EGS and MAP analyzed and provided human data for CIMBA. HP performed immunostainings on human breast cancer samples provided by YT and CS. EYHPL provided BRCA1 and p53 mutant mice. LPJ directed and designed the RANK-Fc treatment experiments. JMP coordinated the project and wrote the manuscript.
Competing Financial Interests
IMBA has applied for a patent on using RANKL inhibition to block breast cancer. Amgen, Inc. provided RANK-Fc and partial financial support (LPJ) for this study.
Acknowledgments
We thank all members of our laboratories for helpful discussions. We thank Tobias Müller, Pawel Pasierbek, Gabriele Petri, Thomas Lendl and Gerald Schmauss for BioOptics support, Andrea Güthlin (Histology Laboratory, Clinical Institute of Pathology, Medical University of Vienna), Michaela Schlederer (Ludwig Boltzmann Institute), Agnieszka Piszczek (VBCF Histopathology) and Mihaela Zeba (VBCF Histopathology) for histology and immunohistochemistry and Dr Sousan Alaei for retrieval of archived material. We thank Daniel Branstetter and William C Dougall (Amgen, Inc.) for data analysis. LPJ is supported by National Cancer Institute, NIH grant CA167268. Additional support was also provided by NIH Award Number T32GM007753 (KOB) and T34GM08663 (KOB and NE) from the National Institute of General Medical Sciences (KOB) and CA186872 from NCI (AOA). The content is solely the responsibility of the authors and does not necessarily represent the official views of Amgen, Inc. or the National Institutes of Health. RK is supported by grants from the Canadian Cancer Society Research Institute (CCSRI). PAJ holds a postdoctoral fellowship from the Canadian Breast Cancer Foundation (CBCF). LK is supported by FWF, P26011 and a Marie-Sklodowska-Curie-Initial Training Networks-European Training Networks (ITN-ETN) ALKATRAS from the H2020. EYHPL is supported by BCRF-35127 and NIH NCI CA137102. MAP is supported by AGAUR SGR 2014-364, ISCIII Spanish Ministry of Economy and Competitiveness “Fondo Europeo de Desarrollo Regional (FEDER) una manera de hacer Europa” PI15/00854 and RTICC RD12/0036/0008. JMP is supported by grants funded by the Austrian Ministry of Sciences, the Austrian Academy of Sciences, an ERC Advanced Grant, and an Era of Hope/DoD Innovator award. DS is recipient of the HFSP Career Development Award.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Generation of K5Cre;Brca1;p53 double and K5Cre;Rank;Brca1;p53 triple knockout mice.
Cre-mediated deletion efficiency in K5Cre and WapCreC mice using a Rosa26eYFP reporter line.
Rank deletion in basal mammary epithelial cells reduces Brca1;p53 mutation-driven hyperplastic proliferation.
DNA damage and epithelial origin of WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 tumors.
Generation of WapCreC;Brca1;p53 double and WapCreC;Rank;Brca1;p53 triple knockout mice.
Histopathologic assessment.
Histological assessment of tumors from WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 mutant mice.
Gene expression profiling of WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 tumors.
Detection of proliferation and DNA damage in WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 mammary tumors.
Analysis of a tumor free, 2 year old WapCreC;Rank;Brca1;p53 triple mutant female.
Pharmacological RANKL inhibition prevents the development of pre-neoplastic regions.
Analysis of murine mammary progenitor cells.
RANKL and RANK protein expression in human breast tumors irrespective of BRCA status.
Genotyped iCOGS variants and breast cancer association in BRCA1 mutation carriers.
Genotyped iCOGS variants and breast cancer association in BRCA2 mutation carriers.
References
- Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266:66–71. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Rosenthal AN, Philpott S, et al. The sex hormone system in carriers of BRCA1/2 mutations: a case-control study. Lancet Oncol 2013; 14:1226–1232. [DOI] [PubMed] [Google Scholar]
- Poole AJ, Li Y, Kim Y, Lin SC, Lee WH, Lee EY. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 2006; 314:1467–1470. [DOI] [PubMed] [Google Scholar]
- Dougall WC, Glaccum M, Charrier K, et al. RANK is essential for osteoclast and lymph node development. Genes Dev 1999; 13:2412–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YY, Yoshida H, Sarosi I, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 1999; 397:315–323. [DOI] [PubMed] [Google Scholar]
- Cummings SR, San Martin J, McClung MR, et al. Denosumab for prevention of fractures in postmenopausal women with osteoporosis. N Engl J Med 2009; 361:756–765. [DOI] [PubMed] [Google Scholar]
- McClung MR, Lewiecki EM, Cohen SB, et al. Denosumab in postmenopausal women with low bone mineral density. N Engl J Med 2006; 354:821–831. [DOI] [PubMed] [Google Scholar]
- Smith MR, Egerdie B, Hernandez Toriz N, et al. Denosumab in men receiving androgen-deprivation therapy for prostate cancer. N Engl J Med 2009; 361:745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D, Sigl V, Penninger JM. RANKL and RANK in sex hormone-induced breast cancer and breast cancer metastasis. Trends Endocrinol Metab 2011; 22:188–194. [DOI] [PubMed] [Google Scholar]
- Fata JE, Kong YY, Li J, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 2000; 103:41–50. [DOI] [PubMed] [Google Scholar]
- Joshi PA, Jackson HW, Beristain AG, et al. Progesterone induces adult mammary stem cell expansion. Nature 2010; 465:803–807. [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Vaillant F, Sheridan JM, et al. Control of mammary stem cell function by steroid hormone signalling. Nature 2010; 465:798–802. [DOI] [PubMed] [Google Scholar]
- Schramek D, Leibbrandt A, Sigl V, et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature 2010; 468:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Jacob AP, Jones J, et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature 2010; 468:103–107. [DOI] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci USA 2008; 105:1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada R, Leibbrandt A, Hanada T, et al. Central control of fever and female body temperature by RANKL/RANK. Nature 2009; 462:505–509. [DOI] [PubMed] [Google Scholar]
- Berton TR, Matsumoto T, Page A, et al. Tumor formation in mice with conditional inactivation of Brca1 in epithelial tissues. Oncogene 2003; 22:5415–5426. [DOI] [PubMed] [Google Scholar]
- Liu X, Holstege H, van der Gulden H, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA 2007; 104:12111–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lee KF, Nikitin AY, et al. Somatic mutation of p53 leads to estrogen receptor alpha-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer Res 2004; 64:3525–3532. [DOI] [PubMed] [Google Scholar]
- Pfefferle AD, Herschkowitz JI, Usary J, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol 2013; 14:R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FJ, Wang X, McGuffog L, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet 2013; 9:e1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet MM, Kuchenbaecker KB, Vijai J, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet 2013; 9:e1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 2012; 44:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira F, Styrkarsdottir U, Estrada K, et al. Genetic Factors for Osteoporosis (GEFOS) Consortium, Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet 2009; 41:1199–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero A, Pellegrini P, Sanz-Moreno A, et al. Rankl impairs lactogenic differentiation through inhibition of the prolactin/Stat5 pathway at midgestation. Stem Cells 2016; 34:1027–1039. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Gallego-Ortega D, Ledger A, et al. Progesterone drives mammary secretory differentiation via RankL-mediated induction of Elf5 in luminal progenitor cells. Development 2013; 140:1397–1401. [DOI] [PubMed] [Google Scholar]
- Obr AE, Grimm SL, Bishop KA, Pike JW, Lydon JP, Edwards DP. Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol Endocrinol 2013; 27:1808–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Wang J, Yuan R, Rosen EM, et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science 1999; 284:1354–1356. [DOI] [PubMed] [Google Scholar]
- Ma Y, Katiyar P, Jones LP, et al. The breast cancer susceptibility gene BRCA1 regulates progesterone receptor signaling in mammary epithelial cells. Mol Endocrinol 2006; 20:14–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LP, Li M, Halama ED, et al. Promotion of mammary cancer development by tamoxifen in a mouse model of Brca1-mutation-related breast cancer. Oncogene 2005; 24:3554–3562. [DOI] [PubMed] [Google Scholar]
- Dvinge H, Git A, Graf S, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013; 497:378–382. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 2001; 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencik J, Schlederer M, Gruber W, et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun 2015; 6:7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudmann D, Cardiff R, Chouinard L, et al. Proliferative and nonproliferative lesions of the rat and mouse mammary, Zymbal's, preputial, and clitoral glands. Toxicol Pathol 2012; 40:7S–39S. [DOI] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 2000; 19:968–988. [DOI] [PubMed] [Google Scholar]
- Frech MS, Halama ED, Tilli MT, et al. Deregulated estrogen receptor a expression in mammary epithelial cells of transgenic mice results in the development of ductal carcinoma in situ. Cancer Res 2005; 65:681–685. [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Stingl J, Raouf A, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med 2008; 14:1384–1389. [DOI] [PubMed] [Google Scholar]
- Stingl J, Emerman JT, Eaves CJ. Enzymatic dissociation and culture of normal human mammary tissue to detect progenitor activity. Methods Mol Biol 2005; 290:249–263. [DOI] [PubMed] [Google Scholar]
- Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature 2014; 508:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer 2011; 11:708–718. [DOI] [PubMed] [Google Scholar]
- Joshi PA, Waterhouse PD, Kannan N, et al. RANK signaling amplifies WNT-responsive mammary progenitors through R-SPONDIN1. Stem Cell Reports 2015; 5:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007; 8:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012; 28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li J, Gray WH, et al. TNBCtype: a subtyping tool for triple-negative breast cancer. Cancer Inform 2012; 11:147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121:2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of K5Cre;Brca1;p53 double and K5Cre;Rank;Brca1;p53 triple knockout mice.
Cre-mediated deletion efficiency in K5Cre and WapCreC mice using a Rosa26eYFP reporter line.
Rank deletion in basal mammary epithelial cells reduces Brca1;p53 mutation-driven hyperplastic proliferation.
DNA damage and epithelial origin of WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 tumors.
Generation of WapCreC;Brca1;p53 double and WapCreC;Rank;Brca1;p53 triple knockout mice.
Histopathologic assessment.
Histological assessment of tumors from WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 mutant mice.
Gene expression profiling of WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 tumors.
Detection of proliferation and DNA damage in WapCreC;Brca1;p53 and WapCreC;Rank;Brca1;p53 mammary tumors.
Analysis of a tumor free, 2 year old WapCreC;Rank;Brca1;p53 triple mutant female.
Pharmacological RANKL inhibition prevents the development of pre-neoplastic regions.
Analysis of murine mammary progenitor cells.
RANKL and RANK protein expression in human breast tumors irrespective of BRCA status.
Genotyped iCOGS variants and breast cancer association in BRCA1 mutation carriers.
Genotyped iCOGS variants and breast cancer association in BRCA2 mutation carriers.