ABSTRACT
The Rags represent a unique family of evolutionarily conserved, heterodimeric, lysosome-localized small GTPases that play an indispensible role in regulating cellular metabolism in response to various amino acid signaling mechanisms. Rapid progress in the field has begun to unveil a picture in which Rags act as central players in translating information regarding cellular amino acid levels by modulating their nucleotide binding status through an ensemble of support proteins localized in and around the lysosomes. By cooperating with other signaling pathways that converge on the lysosomes, Rags promote anabolic processes through positively affecting mTORC1 signaling in the presence of abundant amino acids. Conversely, Rag inactivation plays an indispensible role in switching cellular metabolism into a catabolic paradigm by promoting the activity of the master lysosomal/autophagic transcription factors TFEB and TFE3. Precise control of Rag signaling is necessary for cells to adapt to constantly changing cellular demands and emerging evidence has highlighted their importance in a wide variety of developmental and pathological conditions.
KEYWORDS: autophagy, lysosomes, mTOR, Rags, TFE3, TFEB
Introduction
Recent advances in the field of lysosome biology have established a growing consensus that in addition to their well-characterized roles in cellular cargo degradation, lysosomes act as signaling platforms for coordinating a host of critical functions governing cellular metabolism. Specifically, lysosomes are the site of activation of mechanistic target of rapamycin (mTOR), a central regulator of cellular growth, proliferation, and accumulation of energy stores. mTOR is an evolutionarily conserved atypical serine/threonine kinase in the phosphoinositide 3-kinase-related kinase (PIKK) family. It comprises the core of 2 functionally distinct subcomplexes termed mTORC1 and mTORC2 that are defined by their unique constituents such as Raptor (mTORC1) and Rictor (mTORC2).1
Recent evidence has shown that 2 different types of small GTPases, Rags and Rheb, cooperate to accurately regulate mTOR activity. The Rags operate on the lysosome surface where they sense amino acid levels from the cytosol and lysosomal lumen. When amino acid levels are high, the Rags adopt an active conformation and promote the recruitment of mTORC1 from the cytosol to the lysosome surface (Fig. 1).1 Full mTORC1 activation requires a second small GTPase, Rheb, which is activated under conditions of high cellular ATP and upstream growth factor signals. Together, these GTPases act as coincidence detectors, which integrate signals relating to nutrient, energy, and growth factor status. Active mTORC1 coordinates global cellular metabolism to promote cell growth and proliferation while actively suppressing catabolic processes such as autophagy (Fig. 1).
Figure 1.
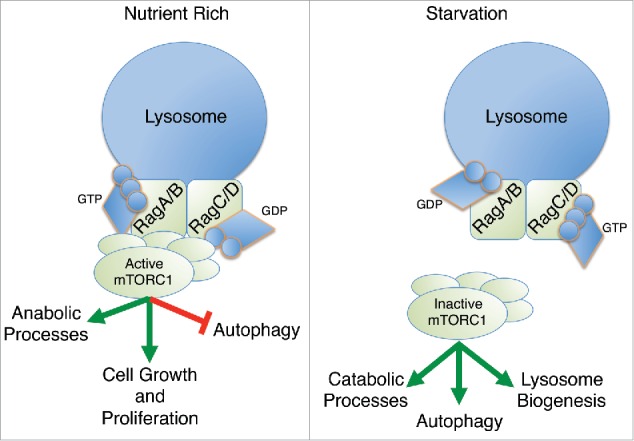
Rags localize to the lysosome surface and serve as a signaling scaffold for mTORC1 activation in response to cellular nutrient levels. Under nutrient rich conditions, Rags promote mTORC1 lysosomal localization where it is activated and promotes an anabolic signaling environment to promote cellular growth and proliferation. Under conditions of low nutrient availability, mTORC1 dissociates from the Rags at the lysosome surface and the cell switches to a catabolic metabolic program featuring increased autophagy and lysosome biogenesis, which serves to restore cellular nutrient pools.
The Rag proteins operate in a complex of obligate heterodimers of functionally redundant small GTPases composed of either RagA or RagB and RagC or RagD. They are considered “active” when RagA/B is in the GTP bound state (RagA/BGTP) and RagC/D is in the GDP bound state (RagC/DGDP).1 Many recent studies have provided insight into the complex machinery implicated in the regulation of Rag activity that allows them to function as nutrient sensors. While multiple amino acid-dependent pathways activate Rag signaling, a common feature among them is the control of Rag nucleotide status, particularly through activation of guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). Here, we review the major mechanisms governing amino acid dependent Rag GTPase signaling, with a particular emphasis on metazoan and mammalian mechanisms and their relevance to human health and disease.
Mechanisms of Rag activation
Extensive work has revealed a number of mechanisms that activate Rag signaling on lysosomal surfaces in response to amino acids (Fig. 2). Unlike most Ras superfamily GTPases, the Rags are not bound to membranes through direct lipidation modifications, but instead localize to lysosome membranes through their interaction with Ragulator, a pentameric protein complex, discovered by mass spectrometric analysis of Rag immunoprecipitates.2 The Ragulator complex is itself anchored to the lysosome membrane through palmitoylation and myristoylation of its p18 subunit.3 After the discovery of Ragulator, an RNAi screen in Drosophila cells revealed the vacuolar H(+)-adenosine triphosphatase (V-ATPase) as critical for amino acid-dependent mTORC1 signaling.4 The V-ATPase senses amino acids located within the lumen of the lysosome and different subunits of the V-ATPase directly interact with the Ragulator complex, RagA/B, or both. Later, it was shown that the Ragulator complex acts as a GEF toward RagA and RagB in response to signals of amino acid sufficiency through the V-ATPase.5
Figure 2.
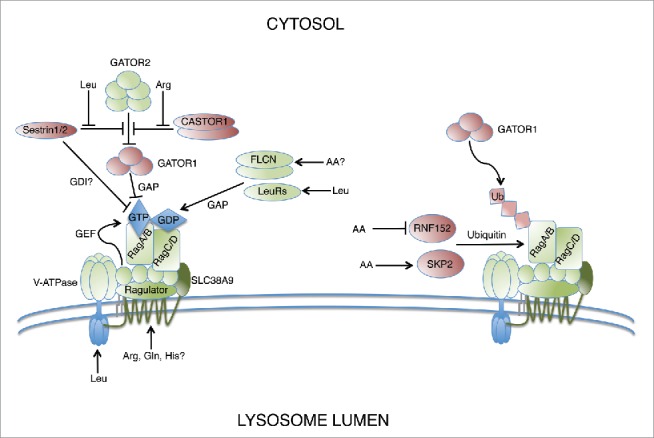
Rags signal amino acid availability through a variety of nutrient sensors that modulate nucleotide binding status. Positive regulators of Rag signaling are shown in green while negative regulators are shown in red.
Recently, SLC38A9, a lysosomal amino acid transporter has been independently identified using mass spectrometric techniques by 3 different groups, as being another critical component of the V-ATPase-Ragulator-RagA/B machinery.6-8 SLC38A9 binds preferentially to Rag mutants mimicking RagA/BGDP and RagC/DGTP, consistent with its proposed role in coupling amino acid sensing to Ragulator mediated RagA/B GEF activity. There remains some uncertainty as to the specific amino acids sensed by SLC38A9 under physiological conditions. In vitro studies suggest a broad substrate specificity with Rebsamen et al. suggesting glutamine as the major substrate required for Rag signaling, while Wang et al. propose that arginine is the dominant amino acid sensed by SLC38A9, with leucine also contributing to its activity.7,8 Further, SLC38A9 overexpression could partially rescue mTORC1 signaling in V-ATPase inhibited cells, suggesting that parallel amino acid sensing pathways may exist to activate the Rags.8 Taken together, these studies suggest a mechanistic model in which both the V-ATPase and SLC38A9 sense intralysosomal amino acids and convert those signals to the Ragulator complex which in turn changes the nucleotide binding of RagA/B to the GTP-bound state, ultimately promoting productive Rag signaling.
Since RagA/BGTP is associated with RagC/DGDP in active Rag signaling complexes, GAP activity toward RagC and RagD is another mechanism that contributes to Rag signaling. To date, 2 different proteins, leucyl-tRNA synthetase (LeuRS) and folliculin (FLCN), have been proposed to have GAP activity toward either RagC or RagD in response to amino acid stimulation (Fig. 2). LeuRS is best characterized for its role in charging leucine to its cognate tRNA for protein translation. LeuRS is reported to localize to lysosomes in a leucine dependent manner and bind with high affinity to RagDGTP. Additional biochemical evidence suggests that LeuRS acts as a GAP against RagD, but not RagC.9 The yeast homolog of LeuRS, Cdc60, was also shown to activate mTORC1, albeit through a different mechanism which involves GTP-loading of Gtr1 via the EGO complex, the yeast homologues of RagA/B and Ragulator, respectively.10 LeuRS represents a highly conserved regulator of Rag function, however the mechanism by which this occurs has likely diverged throughout evolution and some controversy remains as to whether its reported GAP activity occurs under physiological conditions in mammalian cells.11 FLCN has also been implicated in the amino acid dependent regulation of Rag signaling. FLCN is recruited to lysosomes through interaction with Rag A/BGDP/RagC/DGTP heterodimers where, in concert with the FLCN-interacting protein 1 (FNIP1), it exhibits GAP activity against RagC and RagD.11-13 Together, these studies suggest a model in which FLCN-FNIP remains bound to inactive RagA/BGDP, primed to exert its GAP activity toward RagC/DGTP upon stimulation by the appropriate amino acid signals.
Other mechanisms of Rag activation have been proposed that act in concert with the previously described mechanisms by modulating intracellular amino acid and/or amino acid metabolite pools, thereby indirectly affecting GEF and GAP activity toward the Rags. The importance of glutamine and leucine in Rag activation are highlighted by a number of studies. Leucine has been shown to promote glutaminolysis through allosteric binding of glutamate dehydrogenase, promoting the formation of α-ketoglutarate which promotes GTP-loading of RagB through a mechanism dependent on proteins in the prolyl hydroxylase (PHD) family.14,15 Aside from its direct roles in activation of Rag signaling, glutamine also plays an essential role in the intracellular transport of leucine, which is directly involved in Rag activation. In contrast to the Rag-dependent glutaminolysis model, Nicklin, et al. propose a model in which glutamine drives intracellular accumulation of leucine through the heterodimeric antiporter LAT1-4F2hc (SLC7A5-SLC3A2).16 The multipass lysosome membrane protein LAPTM4b, was further shown to be essential for localizing LAT1-4F2hc to the lysosome, allowing lumenal accumulation of leucine, which in turn activates the V-ATPase-Ragulator-Rag pathway.17 In addition, the proton-assisted amino acid transporter, PAT1, has been shown to interact directly with the Rag complex and drive mTORC1 activation through an as yet unknown mechanism.18,19 PAT1 causes efflux of small neutral amino acids from the lysosome and sufficient overexpression can inhibit mTORC1 signaling.4 Milkereit and colleagues speculate that PAT1 may be a candidate to provide a counter-transport mechanism to allow intralysosomal accumulation of leucine.17
Finally, mechanisms that do not directly depend on modulation of Rag GEF or GAP activity per se have also been reported. Additional cytosolic proteins identified as positive regulators of Rag signaling include MAP4K3 and p62.20-23 MAP4K3 is a Ste20 family kinase identified in a cell culture screen for protein kinases affecting mTORC1 signaling.22 A subsequent study showed that it is involved in regulating body size and metabolism in Drosophila and that it preferentially binds RagCGDP.20 Additionally, MAP4K3 autophosphorylates in the presence of amino acids and this modification is essential for promoting mTORC1 signaling.23 These roles of MAP4K3 require its kinase activity as well as the presence of its Citron homology (CNH) domain, however the exact mechanism by which it modulates Rag signaling is unclear at present.22 Finally, the autophagy adaptor protein, p62, was shown to have a non-canonical role in Rag signaling by binding and stabilizing active, RagC/DGDP in a leucine-dependent manner.21
Mechanisms of Rag inactivation
Major mechanistic insights into amino acid dependent Rag signaling have been gained since the identification of the GATOR complex by protein cross-linking coupled mass spectrometry (Fig. 2). The GATOR complex consists of 2 subcomplexes; the trimeric GATOR1, which exhibits GAP activity toward RagA/B, and the pentameric GATOR2, which binds and inhibits GATOR1 in the presence of amino acids. Thus, GATOR2 acts as a negative regulator of GATOR1 and subsequent mTORC1 inactivation.24
Recently, the sestrin proteins have emerged as key players in GATOR mediated Rag inactivation. Sestrins are a group of highly conserved stress-inducible metazoan proteins capable of suppressing mTORC1 signaling.25,26 Sestrin1, Sestrin2, and Sestrin3 have recently been shown to interact with GATOR2 in the absence of amino acids.27,28 In addition, Sestrin2 inhibits mTORC1 activity in response to stressors such as inhibitors of mitochondrial respiration or induction of ER stress, in a GATOR-dependent manner.28 However, the mechanism of sestrin-mediated mTORC1 inactivation is not fully understood. Co-immunoprecipitation experiments by Parmigiani et al. showed that increasing levels of Sestrin2 fail to inhibit the GATOR2-GATOR1 interaction and that Sestrin2 overexpression fails to enhance GATOR1 activity.28 Furthermore, Sestrin2 was capable of co-immunoprecipitating subunits of both GATOR2 and GATOR1.27 Together, these studies suggest a novel, yet to be identified mechanism by which Sestrins suppresses Rag signaling through the GATOR complex.27,28 In contrast, an alternative mechanism for Sestrin2s inhibitory effect on Rag signaling has been proposed. Reciprocal co-immunoprecipitations between components of the GATOR2 and GATOR1 complex in the presence or absence of Sestrin2 revealed that Sestrin2 inhibits GATOR1 and GATOR2 binding and potentiates GATOR1s GAP activity against RagB, arguing for a model in which Sestrin2 inhibits Rag signaling through de-repression of GATOR1.29 Recently, Sestrin1 and Sestrin2 were revealed to be intracellular leucine sensors that specifically dissociate from GATOR2 in the presence of sufficient intracellular leucine. Sestrin3 binding to GATOR2 was insensitive at physiological leucine concentrations, suggesting the possibility that other cellular stressors regulate its binding to GATOR2 or that it is constitutively bound when expressed at sufficient levels.30 A crystal structure of Sestrin2 revealed that the side-chain of intracellular leucine binds a surface exposed hydrophobic pocket, while leucine's amine and carboxyl groups are stabilized by salt bridges on either side of the pocket.31 Sestrins may also inhibit Rag signaling by functioning as guanine nucleotide dissociation inhibitors (GDIs) for RagA/B, locking them in their GDP-bound state.32 This mode of regulation is supported by the presence of a highly conserved GDI motif in the C-terminus of the Sestrins which, when mutated abolishes the ability to suppress Rag signaling. Saxton and colleagues offer an alternative model in which the GDI activity plays a less prominent role owing to the fact that the critical GDI residues are buried in their crystal structure and the lack of structural similarity to known GDIs.31 While it is clear that Sestrins inhibit Rag signaling in vivo, it remains to be determined how they specifically affect GATOR complex function and if they represent a unique class of bona fide GDIs. It is conceivable that both mechanisms are relevant during different physiological conditions and determining the precise regulatory mechanisms between the different models will require ongoing work.
Another recent study analyzed a high-throughput affinity-purification mass spectrometry dataset to identify a protein that acts as an arginine sensor to regulate GATOR activity in a way analogous to the aforementioned Sestrins.33 In the absence of sufficient intracellular arginine, the CASTOR complex, composed of a homodimer of CASTOR1 or a heterodimer of CASTOR1 and CASTOR2; binds to GATOR2, thus inhibiting Rag signaling.33 Arginine binds to CASTOR through its aspartate kinase, chorismate mutase and TyrA (ACT) domain, which allows dissociation of the CASTOR complex from GATOR2.33 Interestingly, CASTOR2 itself remains insensitive to arginine, much like Sestrin3 is insensitive to leucine, suggesting the possibility that the presence of CASTOR2 in the complex may modulate intracellular sensitivity to signals of arginine sufficiency.33 Questions still remain as to the exact effect the CASTOR complex has on GATOR2 and ultimately GATOR1 function and if these mechanisms act in parallel with Sestrins on GATOR2 or if they are regulated under different physiological or developmental conditions.
Ubiquitination has recently emerged as another key post-translational modification capable of inhibiting Rag signaling through the recruitment of GATOR1 to RagA. Two different groups independently screened panels of ubiquitin E3 ligases and identified unique ligases that catalyze K63-linked polyubiquitination of RagA on distinct residues under differing nutrient conditions.34,35 RNF152 is a RING family ubiquitin E3 ligase that preferentially polyubiquitinates RagAGDP at residues K142, K220, K230, and K244 in the absence of amino acids, thereby providing a mechanism for maintaining Rags in an inactivate state under nutrient poor conditions.34 Skp2, a member of the F-box family forms a Skp1-Cul1-F box (SCF) ubiquitin E3 ligase complex; was found to mediate polyubiquitination of RagA at K15 under amino acid replete conditions, suggesting negative feedback regulation of Rag signaling to prevent hyperactivation of mTORC1.35 Both RNF152 and Skp2 regulate RagA (and likely RagB) in similar ways by bridging their ubiquitination function to GATOR1 GAP activity, however they do so under completely different nutrient conditions further highlighting the intricate fine-tuning of Rag signaling that occurs in vivo.
The cytosolic proteins SH3BP4 and C17orf59 were both identified as negative regulators of Rag signaling through their abilities to disrupt the Ragulator-Rag-mTORC1 complex on the surface of lysosomes.36,37 SH3BP4 preferentially binds the inactive Rag GTPase complex, thereby inhibiting its ability to recruit mTORC1.36 C17orf59 directly binds the Ragulator complex, preventing Rags from associating on the lysosomal surface regardless of the nucleotide-bound state of the individual Rags.37 This may represent a mode of Rag inhibition that can override other nutrient signals by upregulation of C17orf59 levels. Alternatively, C17orf59 may have its own set of unique effectors, independent of the amino acid activated Rag complex.
Finally, the Rag GTPases have been shown to exhibit direct crosstalk with Rheb, the other major regulatory GTPase required for mTORC1 activation. In the absence of amino acids, the Rags were shown to recruit TSC2, the GAP component of the TSC complex, which inactivates Rheb (Fig. 3).38 Thus, Rags are required not only for stimulating mTORC1 activity in the presence of amino acids, but also play an active role in terminating its signaling in response to amino acid depletion. It bears mentioning that a pair of recent studies have begun to identify Rag-independent roles of amino acid signaling in the activation of mTORC1. In one study, glutamine was shown to activate mTORC1 in RagA/B knockout cells through an Arf1-dependent mechanism.39 Echoing this finding, another study demonstrated a Rag-independent role of arginine on mTORC1 activation in which arginine inhibited TSC2 binding and GAP activity toward Rheb.40 Collectively, these studies have begun to reveal hitherto unappreciated redundancies and cross-talk with other pathways involved in amino acid dependent mTORC1 signaling.
Figure 3.
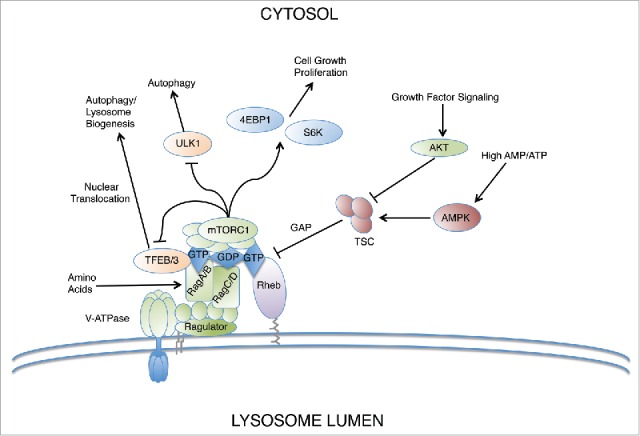
Rags coordinate lysosome biogenesis and autophagy induction at the lysosome surface. The Rag GTPases sense amino acid availability and serve as a signal integration hub for mTORC1 activation. Another small GTPase, Rheb, coordinates signaling in response to growth factor signals and cellular energy level cues via AKT and AMPK, which positively and negatively regulate Rheb through TSC. Under anabolic signaling conditions, mTORC1 activates protein synthesis through phosphorylation of intermediaries such as 4EBP1 and S6K. Under catabolic conditions, autophagy and lysosomal biogenesis is induced via de-repression of TFEB and TFE3 at the lysosome surface.
Regulation of lysosomal biogenesis and autophagy by Rags
In situations of cellular stress, such as nutrient deprivation, autophagy is induced and targets material to lysosomes, thereby providing energy supply. To ensure efficient autophagic flux under stress conditions, autophagosome synthesis is linked to autophagosome-lysosome fusion and lysosomal degradative activity. Importantly, Rag GTPases' role in mediating mTOR recruitment to lysosomes is critical in linking autophagy induction to the nutrient status in the cell. In fully fed cells, RagA/BGTP promotes the binding of Raptor and subsequent assembly and activation of mTORC1.5,41 While Rags do not directly stimulate the kinase activity of mTORC1, they are required for its localization to the lysosomal surface where the small GTPase Rheb stimulates it. Active mTORC1 then inhibits autophagy by phosphorylating and subsequently inactivating a few pathways. Under nutrient sufficiency, mTORC1 phosphorylates and represses the kinase activity of unc-51-like kinase 1 (ULK1), an autophagy regulator.42 It also phosphorylates TFEB and TFE3, members of the MiTF/TFE family of basic helix-loop-helix leucine zipper transcription factors that are critical for inducing autophagy and lysosomal biogenesis (Fig. 3).12,43,44 This phosphorylation requires recruitment of the transcription factors to lysosomes, which is mediated by direct interaction between TFEB/TFE3 and active Rag GTPases.12,45 Inactive or individual Rags fail to interact with TFEB/TFE3 suggesting the nucleotide binding state and dimerization of Rags are important for binding.12,45 Once phosphorylated at lysosomes, TFEB/TFE3 bind to 14-3-3 proteins, which mask their nuclear localization signals and keep them sequestered in the cytosol.12,44,46,47
Reduction in intracellular amino acid levels leads to Rag inactivation, thus preventing mTORC1 recruitment to the lysosome and subsequent activation; and, results in autophagy induction. Furthermore, depletion or inactivation of Rags or overexpression of inactive Rags prevents TFEB and TFE3 redistribution to lysosomes thereby preventing their inhibition by mTORC1 and resulting in their nuclear accumulation.45,47 Here, TFEB and TFE3 bind directly to a 10-base pair modified E-box, termed the Coordinated Lysosomal Expression and Regulation (CLEAR) element.48,49 These CLEAR motifs have been found in the promoters of multiple autophagic and lysosomal genes, in most cased located in close proximity to the transcription start site.48-51 Through expansion of autophagic and lysosomal compartments, this transcriptional network increases the degradative and recycling capabilities of the cell, thus sustaining energy levels and generating new cellular components in response to nutritional cues from the cell (Fig. 3). Furthermore, the regulation of metabolic genes by TFEB is as an important contributor to cell survival during nutrient deprivation.
Rags in disease
Given the prevalence of cancer- or lysosomal storage disorder-related mutations in pathways that regulate autophagy and lysosomal function (Guertin 2007, Lieberman 2012), it is not surprising that mTOR function is dysregulated in several diseases.52,53 Rag GTPases are critical for neonatal autophagy and survival and signal both amino acid and glucose sufficiency.54 Mice with constitutively active RagA were unable to trigger autophagy and produce glucose; and, in low glucose conditions, mTORC1 was diffusely localized in the cytosol rather than clustered at lysosomes.
Aberrant Rag GTPases function may have deleterious effects in cancer. Mutations in the RagA/B GAP, GATOR1, have been identified in several human cancers, which disrupt Rag function and made mTORC1 hyperactive and insensitive to nutrient starvation.24 Furthermore, some patients with colon cancers show low levels of Sestrin2, suggesting that dysregulation of Rag GTPases and hyperactive mTORC1 may contribute to cancer development.55 Rag signaling dysfunction has also been implicated in Birt-Hogg Dubé (BHD) syndrome, an inherited renal cancer syndrome that arises as a result of germline mutations in FLCN.56 Conflicting data from Flcn-deficient Caenorhabditis elegans, mouse, and cell line models and tumors of BHD-affected patients support a role of FLCN as an activator and inhibitor of mTORC1 and autophagy, suggesting the effect of FLCN deficiency on mTOR may depend on the cell type or circumstance in which the deficiency occurs.57-61 It is perplexing that a tumor suppressor such as FLCN activates mTORC1 activity since FLCN appears to facilitate mTORC1 recruitment to the lysosome by activating Rags in an amino-acid dependent manner.11-13 Furthermore, FLCN mutations have been shown to cause constitutive nuclear localization of TFE3 in BHD cancer cell lines.62 Given the sensitivity of the MiTF/TFE family and mTORC1 function to Rag GTPase activity, it is possible that dysregulation of TFEB/TFE3 and its contribution to BHD pathology may arise from reduced RagA/B activation.
It is also possible that Rags contribute to the pathology of some lysosomal storage disorders (LSDs). At this point it is unknown as to what extent the accumulation of undigested material and lysosomal dysfunction observed in LSDs may affect Rag/mTORC1 signaling, but it could potentially have important consequences on energy homeostasis, cellular clearance, and protein synthesis. Several examples support a general framework in which lysosomal dysfunction and subsequent impairment of mTORC1 signaling acts as a primary disease mechanism. For instance, mutations in the ion channel TRPML1 cause the LSD mucolipidosis type IV. Loss of the Drosophila homolog, TRPML, results in late endosomes that fail to fuse with lysosomes, which then exhibit impaired TORC1 signaling.63
The heart is a major organ affected in many LSDs; for example, childhood onset Pompe disease is generally lethal due to heart failure.64 Mouse models lacking RagA/B in cardiomyocytes exhibit hypertrophic cardiomyopathy which phenocopied LSDs.65 Furthermore, lysosomal acidification defects arose due to decreased V-ATPase expression in RagA/B KO cells, implicating Rags as key mediators of cardioprotective lysosomal functions.65 LSDs are also characterized by neurodevelopmental deficits and several neurological disorders display lysosomal dysfunction. Recent work utilizing zebrafish showed that the Rag-Ragulator complex is an essential regulator of lysosomes in microglia.66 Zebrafish lacking RagA function were unable to digest apoptotic neuronal debris suggesting an essential role for the Rag-Ragulator complex in lysosomal function and phagocytic flux in microglia.66 Drosophila models have also revealed that Rag GTPases are important in lysosome-mediated growth of neuromuscular junctions (NMJ).67 Larvae lacking RagC showed fewer NMJ and expression of constitutively active RagA in fly LSD models rescued synaptic growth defects. Altogether, Rag GTPases play an essential role in mediating lysosomal function and autophagy; and, when dysregulated can contribute to numerous pathologies.
Concluding remarks
Rapid advances in the understanding of Rag signaling mechanisms have underscored their importance in bridging cellular nutrient sensing mechanisms with reprograming of cell metabolism. In addition, the lysosome has emerged as the critical signaling platform where multiple pathways converge and ultimately regulate Rag activity. Comprehensive studies have implicated Rag signaling dysfunction in a wide array of pathological conditions, including cancer and lysosomal storage disorders, underscoring their potential as therapeutic targets. Further studies on the integration of different stress signals to better understand Rag biology and improved disease models may allow for the design of specific modulators of Rag function with wide reaching implications.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute.
References
- [1].Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141:290-303; PMID:20381137; http://dx.doi.org/ 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. Embo J 2009; 28:477-89; PMID:19177150; http://dx.doi.org/ 10.1038/emboj.2008.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 2011; 334:678-83; PMID:22053050; http://dx.doi.org/ 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator Is a GEF for the Rag GTPases that signal amino acid levels tomTORC1. Cell 2012; 150:1196-208; PMID:22980980; http://dx.doi.org/ 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol 2015; 35:2479-94; PMID:25963655; http://dx.doi.org/ 10.1128/MCB.00125-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rebsamen M, Pochini L, Stasyk T, de Araujo MEG, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al.. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015; 519:477-+; PMID:25561175; http://dx.doi.org/ 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang S, Tsun Z-Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al.. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015; 347:188-94; PMID:25567906; http://dx.doi.org/ 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell 2012; 149:410-24; PMID:22424946; http://dx.doi.org/ 10.1016/j.cell.2012.02.044 [DOI] [PubMed] [Google Scholar]
- [10].Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Mol Cell 2012; 46:105-10; PMID:22424774; http://dx.doi.org/ 10.1016/j.molcel.2012.02.009 [DOI] [PubMed] [Google Scholar]
- [11].Tsun Z-Y, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013; 52:495-505; PMID:24095279; http://dx.doi.org/ 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Martina JA, Diab HI, Lishu L, Jeong-A L, Patange S, Raben N, Puertollano R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 2014; 7:ra9; PMID:24448649; http://dx.doi.org/ 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 2013; 202:1107-22; PMID:24081491; http://dx.doi.org/ 10.1083/jcb.201307084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Duran RV, MacKenzie ED, Boulahbel H, Frezza C, Heiserich L, Tardito S, Bussolati O, Rocha S, Hall MN, Gottlieb E. HIF-independent role of prolyl hydroxylases in the cellular response to amino acids. Oncogene 2013; 32:4549-56; PMID:23085753; http://dx.doi.org/ 10.1038/onc.2012.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis Activates Rag-mTORC1 Signaling. Mol Cell 2012; 47:349-58; PMID:22749528; http://dx.doi.org/ 10.1016/j.molcel.2012.05.043 [DOI] [PubMed] [Google Scholar]
- [16].Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al.. Bidirectional Transport of Amino Acids Regulates mTOR and Autophagy. Cell 2009; 136:521-34; PMID:19203585; http://dx.doi.org/ 10.1016/j.cell.2008.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Milkereit R, Persaud A, Vanoaica L, Guetg A, Verrey F, Rotin D. LAPTM4b recruits the LAT1-4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat Commun 2015; 6:7250; PMID:25998567; http://dx.doi.org/ 10.1038/ncomms8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Heublein S, Kazi S, Oegmundsdottir MH, Attwood EV, Kala S, Boyd CAR, Wilson C, Goberdhan DCI. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene 2010; 29:4068-79; PMID:20498635; http://dx.doi.org/ 10.1038/onc.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Oegmundsdottir MH, Heublein S, Kazi S, Reynolds B, Visvalingam SM, Shaw MK, Goberdhan DCI. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. Plos One 2012; 7:e36616; PMID:22574197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bryk B, Hahn K, Cohen SM, Teleman AA. MAP4K3 regulates body size and metabolism in Drosophila. Dev Biol 2010; 344:150-7; PMID:20457147; http://dx.doi.org/ 10.1016/j.ydbio.2010.04.027 [DOI] [PubMed] [Google Scholar]
- [21].Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 Is a Key Regulator of Nutrient Sensing in the mTORC1 Pathway. Mol Cell 2011; 44:134-46; PMID:21981924; http://dx.doi.org/ 10.1016/j.molcel.2011.06.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J 2007; 403:13-20; PMID:17253963; http://dx.doi.org/ 10.1042/BJ20061881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF. PP2A(T61 epsilon) is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell 2010; 37:633-42; PMID:20227368; http://dx.doi.org/ 10.1016/j.molcel.2010.01.031 [DOI] [PubMed] [Google Scholar]
- [24].Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013; 340:1100-6; PMID:23723238; http://dx.doi.org/ 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Budanov AV, Karin M. p53 target genes Sestrin1 and Sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008; 134:451-60; PMID:18692468; http://dx.doi.org/ 10.1016/j.cell.2008.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, et al.. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 2010; 327:1223-8; PMID:20203043; http://dx.doi.org/ 10.1126/science.1182228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. The sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 2014; 9:1-8; PMID:25263562; http://dx.doi.org/ 10.1016/j.celrep.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, Guan K-L, Karin M, Budanov AV. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep 2014; 9:1281-91; PMID:25457612; http://dx.doi.org/ 10.1016/j.celrep.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim JS, Ro S-H, Kim M, Park H-W, Semple IA, Park H, Cho U-S, Wang W, Guan K-L, Karin M, et al.. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes (vol 5, 9502, 2015). Sci Rep 2015; 5:14029; PMID:26390293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. METABOLISM Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2016; 351:43-8; PMID:26449471; http://dx.doi.org/ 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, Sabatini DM. METABOLISM Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2016; 351:53-8; PMID:26586190; http://dx.doi.org/ 10.1126/science.aad2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 2014; 159:122-33; PMID:25259925; http://dx.doi.org/ 10.1016/j.cell.2014.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 2016; 165:153-64; PMID:26972053; http://dx.doi.org/ 10.1016/j.cell.2016.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Deng L, Jiang C, Chen L, Jin J, Wei J, Zhao L, Chen M, Pan W, Xu Y, Chu H, et al.. The ubiquitination of RagA GTPase by RNF152 negatively regulates mTORC1 activation. Mol Cell 2015; 58:804-18; PMID:25936802; http://dx.doi.org/ 10.1016/j.molcel.2015.03.033 [DOI] [PubMed] [Google Scholar]
- [35].Jin G, Lee S-W, Zhang X, Cai Z, Gao Y, Chou P-C, Rezaeian AH, Han F, Wang C-Y, Yao J-C, et al.. Skp2-mediated RagA ubiquitination elicits a negative feedback to prevent amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1. Mol Cell 2015; 58:989-1000; PMID:26051179; http://dx.doi.org/ 10.1016/j.molcel.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim Y-M, Stone M, Hwang TH, Kim Y-G, Dunlevy JR, Griffin TJ, Kim D-H. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol Cell 2012; 46:833-46; PMID:22575674; http://dx.doi.org/ 10.1016/j.molcel.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schweitzer LD, Comb WC, Bar-Peled L, Sabatini DM. Disruption of the Rag-ragulator complex by c17orf59 inhibits mTORC1. Cell Rep 2015; 12:1445-55; PMID:26299971; http://dx.doi.org/ 10.1016/j.celrep.2015.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 2014; 156:786-99; PMID:24529380; http://dx.doi.org/ 10.1016/j.cell.2014.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jewell JL, Kim YC, Russell RC, Yu F-X, Park HW, Plouffe SW, Tagliabracci VS, Guan K-L. Differential regulation of mTORC1 by leucine and glutamine. Science 2015; 347:194-8; PMID:25567907; http://dx.doi.org/ 10.1126/science.1259472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Carroll B, Maetzel D, Maddocks OD, Otten G, Ratcliff M, Smith GR, Dunlop EA, Passos JF, Davies OR, Jaenisch R, et al.. Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife 2016; 5:e11058; PMID:26742086; http://dx.doi.org/ 10.7554/eLife.11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/ 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biology 2011; 13:132-U71; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Settembre C, Di Malta C, Polito VA, Garcia-Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Tuong H, Ferron M, Karsenty G, Vellard MC, et al.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/ 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Martina JA, Puertollano R. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes. J Cell Biol 2013; 200:475-91; PMID:23401004; http://dx.doi.org/ 10.1083/jcb.201209135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/ 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/ 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Martina JA, Diab HI, Brady OA, Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. Embo J 2016; 35:479-95; PMID:26813791; http://dx.doi.org/ 10.15252/embj.201593428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 2011; 20:3852-66; PMID:21752829; http://dx.doi.org/ 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- [50].Pastore N, Brady O, Diab H, Martina J, Sun L, Huynh T, Lim J, Zare H, Raben N, Ballabio A, et al.. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 2016; 12:1240-1258; PMID:27171064; http://dx.doi.org/ 10.1080/15548627.2016.1179405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al.. A Gene Network Regulating Lysosomal Biogenesis and Function. Science 2009; 325:473-7; PMID:19556463 [DOI] [PubMed] [Google Scholar]
- [52].Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell 2007; 12:9-22; PMID:17613433; http://dx.doi.org/ 10.1016/j.ccr.2007.05.008 [DOI] [PubMed] [Google Scholar]
- [53].Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy 2012; 8:719-30; PMID:22647656; http://dx.doi.org/ 10.4161/auto.19469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 2013; 493:679-+; PMID:23263183; http://dx.doi.org/ 10.1038/nature11745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ro S, Xue X, Ramakrishnan S, Cho C, Namkoong S, Jang I, Semple I, Ho A, Park H, Shah Y, et al.. Tumor suppressive role of sestrin2 during colitis and colon carcinogenesis. eLife 2016; 5:e12204; PMID:26913956; http://dx.doi.org/ 10.7554/eLife.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, et al.. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell 2002; 2:157-64; PMID:12204536; http://dx.doi.org/ 10.1016/S1535-6108(02)00104-6 [DOI] [PubMed] [Google Scholar]
- [57].Baba M, Furihata M, Hong S-B, Tessarollo L, Haines DC, Southon E, Patel V, Igarashi P, Alvord WG, Leighty R, et al.. Kidney-targeted Birt-Hogg-Dube gene inactivation in a mouse model: Erk1/2 and Akt-mTOR activation, cell hyperproliferation, and polycystic kidneys. J Natl Cancer Inst 2008; 100:140-54; PMID:18182616; http://dx.doi.org/ 10.1093/jnci/djm288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dunlop EA, Seifan S, Claessens T, Behrends C, Kamps MAF, Rozycka E, Kemp AJ, Nookala RK, Blenis J, Coull BJ, et al.. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy 2014; 10:1749-60; PMID:25126726; http://dx.doi.org/ 10.4161/auto.29640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong S-B, Yamaguchi TP, et al.. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci U S A 2009; 106:18722-7; PMID:19850877; http://dx.doi.org/ 10.1073/pnas.0908853106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hudon V, Sabourin S, Dydensborg AB, Kottis V, Ghazi A, Paquet M, Crosby K, Pomerleau V, Uetani N, Pause A. Renal tumour suppressor function of the Birt-Hogg-Dube syndrome gene product folliculin. J Med Genet 2010; 47:182-9; PMID:19843504; http://dx.doi.org/ 10.1136/jmg.2009.072009 [DOI] [PubMed] [Google Scholar]
- [61].Stamatakis L, Metwalli AR, Middelton LA, Linehan WM. Diagnosis and management of BHD-associated kidney cancer. Fam Cancer 2013; 12:397-402; PMID:23703644; http://dx.doi.org/ 10.1007/s10689-013-9657-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM. Inactivation of the FLCN Tumor Suppressor Gene Induces TFE3 Transcriptional Activity by Increasing Its Nuclear Localization. Plos One 2010; 5(12):e15793; PMID:21209915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wong CO, Li RX, Montell C, Venkatachalam K. Drosophila TRPML is required for TORC1 activation. Curr Biol 2012; 22:1616-21; PMID:22863314; http://dx.doi.org/ 10.1016/j.cub.2012.06.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Linhart A, Elliott PM. Heart in Anderson-Fabry disease and other lysosomal storage disorders. Heart 2007; 93:528-35; PMID:17401074; http://dx.doi.org/ 10.1136/hrt.2005.063818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kim YC, Park HW, Sciarretta S, Mo J-S, Jewell JL, Russell RC, Wu X, Sadoshima J, Guan K-L. Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun 2014; 5:4241; PMID:24980141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shen K, Sidik H, Talbot WS. The Rag-ragulator complex regulates lysosome function and phagocytic flux in microglia. Cell Rep 2016; 14:547-59; PMID:26774477; http://dx.doi.org/ 10.1016/j.celrep.2015.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wong C-O, Palmieri M, Li J, Akhmedov D, Chao Y, Broadhead GT, Zhu MX, Berdeaux R, Collins CA, Sardiello M, et al.. Diminished MTORC1-dependent JNK activation underlies the neurodevelopmental defects associated with lysosomal dysfunction. Cell Rep 2015; 12:2009-20; PMID:26387958; http://dx.doi.org/ 10.1016/j.celrep.2015.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]


