ABSTRACT
ARL2 is among the most highly conserved proteins, predicted to be present in the last eukaryotic common ancestor, and ubiquitously expressed. Genetic screens in multiple model organisms identified ARL2, and its cytosolic binding partner cofactor D (TBCD), as important in tubulin folding and microtubule dynamics. Both ARL2 and TBCD also localize to centrosomes, making it difficult to dissect these effects. A growing body of evidence also has found roles for ARL2 inside mitochondria, as a regulator of mitochondrial fusion. Other studies have revealed roles for ARL2, in concert with its closest paralog ARL3, in the traffic of farnesylated cargos between membranes and specifically to cilia and photoreceptor cells. Details of each of these signaling processes continue to emerge. We summarize those data here and speculate about the potential for cross-talk or coordination of cell regulation, termed higher order signaling, based upon the use of a common GTPase in disparate cell functions.
KEYWORDS: ARF family GTPases, ARL2, ARL3, BART, cofactor D, microtubule dynamics, mitochondrial fusion, PDE6D, TBCD
Introduction
With the discovery of the RAS,1,2 ARF,3,4 RHO,5,6 and RAB 7,8 families as large collections of homologous regulators of an incredibly diverse array of cellular functions, it became increasingly evident how challenging it was, and will be, to describe the mechanisms of action for each family member. This typically involves identifying at least 2 other components in the pathway regulated by the GTPase: (i) the guanine nucleotide exchange factor(s) (GEFs) that act immediately upstream of the GTPase to promote the release of GDP and allow formation of the active, GTP-liganded regulator, and (ii) the effector that acts immediately downstream of the GTPase and is the obligate next step leading to the biological response. The GEF:GTPase:effector triad is a paradigm of GTPase biology, first worked out for the heterotrimeric G proteins using hormone-stimulated adenylyl cyclase activation as the model.9 However, it quickly became apparent that any one GTPase can be activated by several different GEFs, typically sharing a common GEF domain (e.g., the SEC7 domains for ARF GEFs).10-12 Similarly, most activated GTPases have been found capable of directly binding to numerous effectors, some of which have GTPase activating (GAP) activity and thus are capable of providing temporal limits on the activated state.13 And if this isn't complex enough, researchers consistently find that the key protein-protein interactions occur optimally only after solid-phasing (i.e., after association of components to a lipid bilayer). Thus, regulated translocation of GEF, GTPase, and effector onto a membrane surface is often fundamental to the process. Finally, specific lipids (notably the phosphatidylinositol phosphates) in the bilayer can, and often do, provide specificity to the site of action and serve as allosteric activators of the GEFs, GTPases, effectors, or GAPs. As this system has evolved in eukaryotes there is ample evidence of redundancies in signaling between closely related GTPases or their regulators. This is perhaps most evident in the yeast S. cerevisiae in which the genome was thought to have largely duplicated, resulting in 2 paralogous ARFs (ARF1 and ARF2) that are 97% identical in primary sequence and differ only in the levels of expression, with ARF1 expressed ∼10X the levels of ARF2.14 Thus, while ARFs are essential in this yeast, deletion of either one results in only rather subtle phenotypes. In this article, we consider the opposite scenario: What are the ramifications or implications when cells employ one GTPase for 2 or more essential functions?
We first ask, why would such a scenario have arisen? With expansion of GTPase families being the norm, why haven't 2 GTPases evolved into completely distinct pathways without shared components, allowing clean separation of functions and independent regulation of each? The simple answer is that there may be selective advantage to one GTPase regulating 2 or more pathways. We propose that the use of shared components in 2 different, essential pathways provides a ready means of “cross-talk” between pathways and consequently a higher level of cell organization than is possible with only linear pathways. A related question is how could such a system have arisen? Almost certainly one of the essential functions of the GTPase in question was in place before it acquired a new and second function. It seems unlikely that the GTPase co-opted for the second pathway was chosen at random. Rather, we propose that the 2 pathways are linked in some non-linear fashion and in doing so, this linkage provides selective advantage(s) to the cells. To test these ideas, we sought a regulatory GTPase with 2 essential functions. This article summarizes our studies of the roles of ARL2 in mitochondrial fusion and microtubule dynamics. Though we have not yet described how the common use of ARL2 as a key regulator integrates or connects the 2 systems, our studies have laid the groundwork for testing several of the ideas associated with higher order signaling.
Early history of ARL2 and roles in microtubule dynamics
ARL2 is among the most highly conserved proteins known, sharing >50% identity between human and S. pombe proteins. It is ubiquitous in every eukaryote investigated and predicted to have been present in the last eukaryotic common ancestor.4 The name ARF-like 2 (ARL2) results from it being the second in a series of cDNAs amplified by redundant primer PCR from a human cDNA library and with open reading frames encoding proteins displaying high homology to the ARFs. Most of these proteins, including ARL2, have been shown to lack the canonical ARF activity, co-factor in the ADP-ribosylation of Gs catalyzed by cholera toxin, and were thus named ARF-like.15 But even before that, the ortholog had been identified in 2 different genetic screens in S. cerevisiae, with mutations resulting in changes in microtubules.16,17 Very similar results were later repeated in genetic screens using A. thaliana (TTN5),18 S. pombe (Alp41),19 C. elegans, (evl-20)20 T. brucei (TbArl2),21 and D. melanogaster (ARL2).22 Therefore, the evidence that ARL2 plays important role(s) in tubulin and microtubule biologies, with links to cell division, is abundantly clear. The ARL2 ortholog is essential in each of these organisms, save S. cerevisiae. Mitochondria from mammals and yeast are known to have a number of important differences in composition and regulation (e.g., see van der Bliek, et al.)23 making it difficult to ascribe this difference to any one factor.
While genomic mutations in ARL2 can clearly result in compromised microtubule arrays and later cell death, decreased expression of ARL2 resulting from its regulated expression has also been observed. ARL2 has been found to be a target of microRNAs, with links to heart disease,24 G0/G1 cell cycle check point arrest,25 and apoptosis in neural progenitor cells.26 Thus, as is often the case with essential genes, it may well turn out that alterations in the level of expression of ARL2, resulting from transcriptional responses or miRNA targeting, play important roles in disease processes.
In a number of the same genetic screens that linked ARL2 to microtubules and cell division, orthologs of the tubulin co-chaperone cofactor D (TBCD) were also found (CIN1 in S. cerevisiae;16,17,27 Alp1 in S. pombe;19 and TTN1 in A. thaliana28). The close genetic linkage between ARL2 and TBCD became understood as a physical one when endogenous ARL2 was first purified from mammalian tissues and found to co-purify with TBCD in an apparent 1:1 complex.29
TBCD is one of 5 tubulin co-chaperones, TBCA-E, identified in the Cowan lab using an assay involving in vitro transcribed and translated β-tubulin and native gels to monitor its binding to added components.30,31 This powerful approach clearly succeeded in identifying many of the key components required to assemble native tubulin heterodimers in cells. Yet surprisingly, it failed to identify ARL2 as a required component in this process, perhaps because it was present in sufficient amounts in the reticulocyte lysate used to generate the labeled β-tubulin. Instead, the binding of ARL2 and TBCD was first shown using a combination of in vitro transcribed and translated proteins.32 The cellular interaction between ARL2 and TBCD was demonstrated after expression of tagged proteins in mammalian cell culture.32 Over-expression of TBCD was first shown to disrupt microtubules in S. pombe.33 It was later shown that overexpression of bovine TBCD in HeLa cells resulted in the loss of microtubules.32,34 Co-expression of ARL2 with the TBCD, particularly the dominant, inactivating mutant ([T30N]ARL2), reversed this effect, which was termed the tubulin destruction pathway.32,35 Whether such a pathway exists in mammalian cells or is a consequence of protein over-expression is uncertain. Studies in our lab add further confusion to this question as we found that human and bovine TBCD differ in their ability to reduce microtubule densities upon overexpression, though this difference is likely only in sensitivity of the response.35,36 Further, we showed that expression of either dominant activating or inactivating point mutants of human ARL2 caused the loss of microtubules.37 Such a result is typically interpreted as evidence for the required cycling of the GTPase between its active and inactive states. Thus, rather than ARL2 displacing β-tubulin from TBCD in a destructive pathway, we propose that the stable scaffold on which tubulins are folded consists of the trimer of ARL2/TBCD/β-tubulin (J.W. Francis and R.A. Kahn, manuscript in preparation) and that the ARL2 in this trimer can bind and hydrolyze GTP to regulate the activity of the complex. This is also in contrast to results in Nithianantham, et al. in which they argue, based upon results from reconstitution of yeast orthologs expressed in bacteria, that the basic scaffold for tubulin folding consists of a different trimer, TBCD/ARL2/TBCE.38 While it is likely that one or more of these apparent discrepancies results simply from different rates of dissociation of subunits between orthologs or cell/tissue sources during isolation, clearly more work is needed before we can understand the tubulin folding process, the biological importance of the tubulin destruction pathway, and the strongly suggested additional roles of ARL2 and TBCD in regulating tubulin polymerization.
In addition to its presence in the cytosol, ARL2 is also found in mitochondria, at centrosomes and in the nucleus in every mammalian cell line or tissue examined. Because of this diversity in locations, and for technical reasons, it has proven to be difficult to get absolute values for the distribution of ARL2 in each of these organelles. Despite the majority of ARL2 being cytoplasmic, and in contrast to other members of the ARF family, we find scant evidence for a pool of monomeric ARL2 in cytosol. Using gel filtration or native gel electrophoresis of cell or tissue homogenates we estimate that a clear majority, perhaps as much as 90%, of total cellular ARL2 is in the cytosol and almost all of this is tightly bound to TBCD. By immunofluorescence it is also evident that pools of ARL2 specifically localize to centrosomes, mitochondria, and the nucleus.36,39-41 TBCD is also present at centrosomes, likely in complex with ARL2 though this has not been shown directly.36,42 In contrast, TBCD is absent from mitochondria. Thus, ARL2 is clearly found in multiple compartments in cells and while most is in a stable, functional complex with TBCD, it also clearly functions independently of it.
The specific localization of both ARL2 and TBCD to centrosomes strongly suggests roles for each protein in microtubule dynamics over and above their role in tubulin folding. Although it is technically very difficult to clearly resolve effects on pools of polymerizable tubulins (i.e., folding) from the polymerization process itself, the presence of ARL2 and TBCD in the cytosol and at centrosomes suggests 2 related functions. The fact that ARL2 and TBCD are present in much lower amounts than are the tubulins implies that their actions in each process are catalytic. Thus, it does not appear likely that a tubulin destruction pathway can exist if it requires TBCD to directly bind sufficient β-tubulin to prevent polymerization. Rather, we currently model ARL2 and TBCD as having actions at centrosomes distinct from tubulin biogenesis, to regulate microtubule dynamics. This model is supported by studies showing the effects of TBCD on centriologenesis, centrosome integrity, ciliogenesis, mitotic spindle formation, and midbody abscission.42 TBCD is also required for microtubule stability and dendrite formation in Drosophila progenitor neurons, and it may provide a link between cell surface receptors and microtubules.43 Another recent study suggests that ARL2 and TBCD are involved in microtubule growth and asymmetric division of Drosophila neural stem cells.22 There are also data suggesting a role for ARL2 and TBCD in the regulation of cell-cell adhesion through the apical junctional complex, providing further evidence of non-tubulin-folding activities for these proteins.44
Thus, ARL2 and TBCD are very closely linked in function, likely both in the cytosol and at centrosomes. Molecular models for these actions are incomplete, though the large size of TBCD and lack of defined domains suggest a scaffolding role. And given the nature of ARL2 as a GTPase, we believe it likely to act as a regulator of TBCD in either tubulin biogenesis, polymerization, or both. Because tubulins are targets of some clinically important chemotherapeutics, and changes in ARL2 activities have already been found capable of influencing those actions,45-47 future studies offer promise of the design of better and more selective therapies.
Roles of ARL2 in mitochondrial fusion
Despite the strong evidence that ARL2 regulates one or more aspects of microtubule dynamics, most of the early studies in our lab employed biochemical approaches and suggested ARL2 also functions at mitochondria. Characterization of the first antibody to ARL2 found specific staining of mitochondria by immunofluorescence in a number of cell lines and by cell fractionation.39 The first effector of ARL2, termed Binder of ARL2 (BART; though arbitrarily re-named by the human genome curators as ARL2BP) was also found to partially localize to mitochondria.39,48 We then purified the adenine nucleotide translocase (ANT1), which resides in the inner mitochondrial membrane, as a specific interactor with the ARL2(GTP)-BART complex.39 In this same study we noted that upon deletion of ANT1 in mouse muscle tissues specifically, the fraction of mitochondrial ARL2 (but not total cellular/tissue ARL2) was markedly increased. We have more recently found that a variety of energetic stressors increase ARL2 immunofluorescence at mitochondria (Newman, Schiavon, Zhou, and Kahn, manuscript in preparation). These results suggest that the mitochondrial pool of ARL2 is regulated in response to stressors.
ARL2 siRNA results in no evident changes to the microtubule array yet has profound effects on mitochondria; including fragmentation, perinuclear clustering, and loss of ∼50% of cellular ATP, with later cell death.37 Expression of the dominant inactivating mutant ARL2[T30N] mimics the knockdown with regard to fragmentation and clustering phenotypes but has no effect on ATP levels. Thus, we propose that ARL2 is acting in 2 distinct pathways in mitochondria – one resulting in the regulation of ATP production and the other influencing mitochondrial morphology and motility (see Fig. 1).
Figure 1.
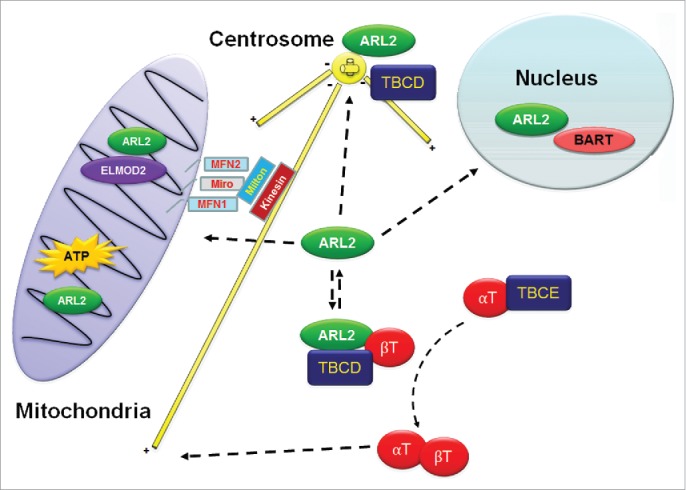
Model of the actions of ARL2 at distinct sites in cells. ARL2 (green) and TBCD (dark blue) are shown at centrosomes at the top, and free in the cytosol (toward the bottom of the figure), in complex with β-tubulin. The small pool of monomeric ARL2 is in the middle and is the presumptive source of ARL2 that is either imported into mitochondria (left side) or the nucleus (upper right). The role of ARL2 in the folding of the αβ-tubulin heterodimer (red) is depicted, showing α-tubulin bound initially to TBCE, omitting the earlier steps in folding, including the roles of the other tubulin co-chaperones. Also shown here are the proposed 2 distinct pathways inside mitochondria; one involving the ARL2 GAP ELMOD2 and leading to mitochondrial fusion and the other to changes in ATP production through unknown mechanism(s). The yellow bars represent microtubules that may be both regulated by ARL2/TBCD at the centrosome and along which mitochondria traffic, in yet another process influenced by ARL2. See text for details.
The model of 2 different ARL2-sensitive pathways in mitochondria is supported by the observation that one of the 3 members of the ELMOD family of ARL2 GAPs, ELMOD2, also localizes to mitochondria.37,49-51 When ELMOD2 is depleted by siRNA in cultured cells they also display fragmented mitochondria with perinuclear clustering but no changes in ATP levels (or in microtubule arrays).37 We conclude from such data that ELMOD2 acts as an effector for ARL2 in mitochondria, leading to changes in morphology and motility. But because the lack of ARL2 has such profound effects on ATP levels, while knockdown of ELMOD2 lacks such effects, we propose the existence of a distinct ARL2 pathway in mitochondria that regulates ATP production independently of ELMOD2 (see Fig. 1).
In order for ARL2 to regulate the putative effector ANT1, a protein spanning the inner mitochondrial membrane, it has to be able to enter mitochondria. Fractionation of mitochondria using well-established methods, yielded evidence that ARL2 was indeed inside, though perhaps not limited to a single compartment. We have also used differential solubilization of the outer membrane using digitonin or tBid, in each case followed by immunofluorescence, and found that there is always a pool of ARL2 inside mitochondria and that it may reside in both the inter-membrane space (IMS) as well as the matrix.37,39
ARL2 is highly unusual both in being predominantly found outside of mitochondria with only a small pool inside, but also in being present in more than one compartment of mitochondria. Thus, in order to develop a rigorous test of its site of action in regulating mitochondrial morphology, we generated a collection of plasmids directing expression of ARL2 (or dominant mutants) with strong N-terminal leader sequences that drive it either to the matrix, to the IMS, or prevents mitochondrial import. We found that only when directed to the IMS is ARL2[T30N] capable of influencing mitochondrial morphology. This study also provides strong supporting evidence that it acts to promote mitochondrial fusion, and not fission. Thus, ARL2 is the first regulatory GTPase and first soluble factor to regulate fusion, and motility, from the IMS. In doing so, it joins a growing list of nuclear encoded genes with well-defined functions outside of mitochondria but with important additional roles inside mitochondria; including STAT3,52-54 MEF2,55 fumarase,56 and some DNA repair proteins.57 ARL2 is among the list of GTPases with essential regulatory functions acting in or on mitochondria, that currently includes the well-established regulators OPA1, MFNs, MIROs, and DRP1, and recent addition of ARL4D.58-60
The findings that ARL2 regulates 2 such fundamental and ancient aspects of eukaryotic biology is quite striking but consistent with its predicted presence at the origin of eukaryotes, its ubiquity, and high degree of sequence conservation. The stable (at least through several days of purification in vitro) association of ARL2 with TBCD and low levels of monomer in the cytosol is expected to limit the availability of ARL2 for import into mitochondria. The observation that deletion of ANT1 in mice results in increased mitochondrial import of ARL2 in affected tissues,39 suggests that import of ARL2 into mitochondria is a regulated process. Though controversial, it is currently thought that proteins imported into the IMS cannot later be exported, with the exception of during apoptosis. Thus, in its simplest form, we propose that metabolic stress inside mitochondria may be capable of sending a signal to the cytosol that results in increased dissociation of ARL2 from TBCD, allowing increased mitochondrial import, fusion, and motility. This model requires extensive testing but is an example of the type of higher order signaling alluded to in the title of this article.
ARL2 shares with ARL3 several binding partners and related roles in traffic of farnesylated cargos
In addition to the technical challenges in dealing with a regulatory GTPase that acts in multiple sites and systems in the same cell, there are additional challenges when 2 or more GTPase family members share a common effector. Although this functional redundancy may have arisen to protect the cell/organism against loss of an essential function, it can also provide different inputs and inter-dependent regulation of a common output. ARL2 provides an interesting example of this phenomenon. Human ARL2 shares 53% primary sequence identity with ARL3, making them the closest paralogs. Activated (GTP-bound) ARL2 and ARL3 bind to some of the same GAPs and effectors, including ELMODs, BART, HRG4, and PDEδ (PDE6D).51,61-64 Despite these similarities, ARL2 and ARL3 generally perform distinct cellular functions.40 One exception is that both ARL2 and ARL3 regulate the traffic of farnesylated cargoes between membranes, particularly important in cilia and in rods and cones of the eye.63-68 Yeast-2-hybrid screening revealed that ARL2 and ARL3 both bind to phosphodiesterase delta (PDEδ). Activated ARL2 and ARL3 have been found to allosterically modulate PDEδ by inducing a conformational change in the farnesyl-binding pocket to release lipid-modified RHEB and RAS GTPases.62,63,67,69 Upon dissociation from PDEδ, RHEB and RAS bind to membranes via their farnesylated C-termini. Thus, the PDEδ/ARL2/ARL3 system is thought to regulate the traffic of farnesylated cargos between membranes, including specific targeting to cilia, with release of signaling proteins in a temporally and spatially-regulated manner. The PDEδ/ARL2/ARL3 system has been implicated in both endomembrane transport of farnesylated cargo to the plasma membrane as well as the recruitment of factors to the transition zone of primary cilia.70,71 As the complexity of signaling by any one GTPase has increased throughout eukaryotic evolution, it has increased the costs to the cell of mutations that alter central features of the GTPase. Perhaps for this reason we often find that it is mutations in the downstream effectors that are commonly found linked to diseases in humans; examples include BART, HRG4, and PDEδ.72-76
Summary
We summarize here work from a number of laboratories that focus on different aspects of ARL2 biology. While cellular roles in regulating microtubules, mitochondria, and traffic of farnesylated protein cargos are well-established, many of the details are not. For example, one might notice the lack of mention of any guanine nucleotide exchange factor(s) for ARL2. This may be because ARL2 displays unusually low affinity for guanine nucleotides and high stability in the nucleotide-free form. While it is possible that the activation of ARL2 is independent of any GEF, this seems unlikely given the precedence for all other GTPases. Similarly, the biochemical characterization of ARL2 GAP activities, specifically the ELMOD proteins, reveals a broader substrate specificity and range of specific activities than most GAPs. Thus, we expect that we are still missing some fundamental information on the regulation of ARL2 and its interactors. As the number of ARL2 interactors climbs, it increasingly resembles the situation with the ARFs, which have well over 20 known effectors. In seeking a rationale for the shared use of one or a few GTPases for several different biological purposes, we propose that this allows coordination between otherwise disparate functions in cells, termed higher order signaling. With cell-based assays growing increasingly robust and specific for ARL2 functions, we look forward to performing critical tests of this model in the near future.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the many colleagues whose discussions and ideas so often contribute to our own research and writing and apologies to the many researchers whose work in this field were not cited due to limited space.
Funding
The research in our lab was supported by several grants from the National Institute of General Medical Sciences, including most recently R01-GM090158.
References
- [1].Cox AD, Der CJ. Ras history: The saga continues. Small GTPases 2010; 1:2-27; PMID:21686117; http://dx.doi.org/ 10.4161/sgtp.1.1.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE 2004; 2004:RE13; PMID:15367757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kahn RA. ARF Family GTPases. Dordrecht: Kluwer Academic Publishers, 2004. [Google Scholar]
- [4].Li Y, Kelly WG, Logsdon JM Jr., Schurko AM, Harfe BD, Hill-Harfe KL, Kahn RA. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J 2004; 18:1834-50; PMID:15576487; http://dx.doi.org/ 10.1096/fj.04-2273com [DOI] [PubMed] [Google Scholar]
- [5].Wennerberg K, Der CJ. Rho-family GTPases: it's not only Rac and Rho (and I like it). J Cell Sci 2004; 117:1301-12; PMID:15020670; http://dx.doi.org/ 10.1242/jcs.01118 [DOI] [PubMed] [Google Scholar]
- [6].Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol 2001; 11:471-7; PMID: 11719051; http://dx.doi.org/ 10.1016/S0962-8924(01)02153-5 [DOI] [PubMed] [Google Scholar]
- [7].Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol 2001; 2; PMID:11387043; http://dx.doi.org/ 10.1186/gb-2001-2-5-reviews3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elias M, Brighouse A, Gabernet-Castello C, Field MC, Dacks JB. Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J Cell Sci 2012; 125:2500-8; PMID:22366452; http://dx.doi.org/ 10.1242/jcs.101378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gilman AG. The Albert Lasker Medical Awards. G proteins and regulation of adenylyl cyclase. JAMA 1989; 262:1819-25; PMID:2506367; http://dx.doi.org/ 10.1001/jama.1989.03430130095041 [DOI] [PubMed] [Google Scholar]
- [10].Wright J, Kahn RA, Sztul E. Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell Mol Life Sci 2014; 71:3419-38; PMID:24728583; http://dx.doi.org/ 10.1007/s00018-014-1602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cherfils J, Menetrey J, Mathieu M, Le Bras G, Robineau S, Beraud-Dufour S, Antonny B, Chardin P. Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature 1998; 392:101-5; PMID:9510256; http://dx.doi.org/ 10.1038/32210 [DOI] [PubMed] [Google Scholar]
- [12].Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 2007; 8:1476-85; PMID:17850229; http://dx.doi.org/ 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- [13].East MP, Kahn RA. Models for the functions of Arf GAPs. Semin Cell Dev Biol 2011; 22:3-9; PMID:20637885; http://dx.doi.org/ 10.1016/j.semcdb.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol 1990; 10:6690-9; PMID:2123295; http://dx.doi.org/ 10.1128/MCB.10.12.6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Clark J, Moore L, Krasinskas A, Way J, Battey J, Tamkun J, Kahn RA. Selective amplification of additional members of the ADP-ribosylation factor (ARF) family: cloning of additional human and Drosophila ARF-like genes. Proc Natl Acad Sci U S A 1993; 90:8952-6; PMID:8415637; http://dx.doi.org/ 10.1073/pnas.90.19.8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol 1990; 10:223-34; PMID:2403635; http://dx.doi.org/ 10.1128/MCB.10.1.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stearns T, Hoyt MA, Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics 1990; 124:251-62; PMID:2407611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McElver J, Patton D, Rumbaugh M, Liu C, Yang LJ, Meinke D. The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. The Plant cell 2000; 12:1379-92; PMID:10948257; http://dx.doi.org/ 10.1105/tpc.12.8.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Radcliffe PA, Vardy L, Toda T. A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett 2000; 468:84-8; PMID:10683446; http://dx.doi.org/ 10.1016/S0014-5793(00)01202-3 [DOI] [PubMed] [Google Scholar]
- [20].Antoshechkin I, Han M. The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev Cell 2002; 2:579-91; PMID:12015966; http://dx.doi.org/ 10.1016/S1534-5807(02)00146-6 [DOI] [PubMed] [Google Scholar]
- [21].Price HP, Peltan A, Stark M, Smith DF. The small GTPase ARL2 is required for cytokinesis in Trypanosoma brucei. Mol Biochem Parasitol 2010; 173:123-31; PMID:20653091; http://dx.doi.org/ 10.1016/j.molbiopara.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen K, Koe CT, Xing ZB, Tian X, Rossi F, Wang C, Tang Q, Zong W, Hong WJ, Taneja R, et al.. Arl2- and Msps-dependent microtubule growth governs asymmetric division. J Cell Biol 2016; 212:661-76; PMID:26953351; http://dx.doi.org/ 10.1083/jcb.201503047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harbor perspectives in biology 2013; 5; PMID:23732471; http://dx.doi.org/ 10.1101/cshperspect.a011072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, Takemura G, Kinoshita M, Kuwabara Y, Mori RT, Hasegawa K, et al.. MicroRNA-15b modulates cellular ATP levels and degenerates mitochondria via Arl2 in neonatal rat cardiac myocytes. J Biol Chem 2010; 285:4920-30; PMID:20007690; http://dx.doi.org/ 10.1074/jbc.M109.082610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang K, Li P, Dong Y, Cai X, Hou D, Guo J, Yin Y, Zhang Y, Li J, Liang H, et al.. A microarray-based approach identifies ADP ribosylation factor-like protein 2 as a target of microRNA-16. J Biol Chem 2011; 286:9468-76; PMID:21199864; http://dx.doi.org/ 10.1074/jbc.M110.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhou Y, Jiang H, Gu J, Tang Y, Shen N, Jin Y. MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis 2013; 4:e695; PMID:23807224; http://dx.doi.org/ 10.1038/cddis.2013.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hoyt MA, Macke JP, Roberts BT, Geiser JR. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics 1997; 146:849-57; PMID:9215891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu CM, Meinke DW. The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 1998; 16:21-31; PMID:9807824; http://dx.doi.org/ 10.1046/j.1365-313x.1998.00268.x [DOI] [PubMed] [Google Scholar]
- [29].Shern JF, Sharer JD, Pallas DC, Bartolini F, Cowan NJ, Reed MS, Pohl J, Kahn RA. Cytosolic Arl2 is complexed with cofactor D and protein phosphatase 2A. J Biol Chem 2003; 278:40829-36; PMID:12912990; http://dx.doi.org/ 10.1074/jbc.M308678200 [DOI] [PubMed] [Google Scholar]
- [30].Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded beta-tubulin. Cell 1996; 86:287-96; PMID:8706133; http://dx.doi.org/ 10.1016/S0092-8674(00)80100-2 [DOI] [PubMed] [Google Scholar]
- [31].Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol 1997; 138:821-32; PMID:9265649; http://dx.doi.org/ 10.1083/jcb.138.4.821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bhamidipati A, Lewis SA, Cowan NJ. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol 2000; 149:1087-96; PMID:10831612; http://dx.doi.org/ 10.1083/jcb.149.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J 1998; 17:658-66; PMID:9450991; http://dx.doi.org/ 10.1093/emboj/17.3.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Martin L, Fanarraga ML, Aloria K, Zabala JC. Tubulin folding cofactor D is a microtubule destabilizing protein. FEBS Lett 2000; 470:93-5; PMID:10722852; http://dx.doi.org/ 10.1016/S0014-5793(00)01293-X [DOI] [PubMed] [Google Scholar]
- [35].Tian G, Thomas S, Cowan NJ. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton (Hoboken) 2010; 67:706-14; PMID:20740604; http://dx.doi.org/ 10.1002/cm.20480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cunningham LA, Kahn RA. Cofactor D functions as a centrosomal protein and is required for the recruitment of the gamma-tubulin ring complex at centrosomes and organization of the mitotic spindle. J Biol Chem 2008; 283:7155-65; PMID:18171676; http://dx.doi.org/ 10.1074/jbc.M706753200 [DOI] [PubMed] [Google Scholar]
- [37].Newman LE, Zhou CJ, Mudigonda S, Mattheyses AL, Paradies E, Marobbio CM, Kahn RA. The ARL2 GTPase is required for mitochondrial morphology, motility, and maintenance of ATP levels. PloS One 2014; 9:e99270; PMID:24911211; http://dx.doi.org/ 10.1371/journal.pone.0099270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nithianantham S, Le S, Seto E, Jia W, Leary J, Corbett KD, Moore JK, Al-Bassam J. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alphabeta-tubulin pool for microtubule dynamics. eLife 2015; 4; PMID:26208336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sharer JD, Shern JF, Van Valkenburgh H, Wallace DC, Kahn RA. ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol Biol Cell 2002; 13:71-83; PMID:11809823; http://dx.doi.org/ 10.1091/mbc.01-05-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell 2006; 17:2476-87; PMID:16525022; http://dx.doi.org/ 10.1091/mbc.E05-10-0929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Muromoto R, Sekine Y, Imoto S, Ikeda O, Okayama T, Sato N, Matsuda T. BART is essential for nuclear retention of STAT3. International immunology 2008; 20:395-403; PMID:18234692; http://dx.doi.org/ 10.1093/intimm/dxm154 [DOI] [PubMed] [Google Scholar]
- [42].Fanarraga ML, Bellido J, Jaen C, Villegas JC, Zabala JC. TBCD links centriologenesis, spindle microtubule dynamics, and midbody abscission in human cells. PloS One 2010; 5:e8846; PMID:20107510; http://dx.doi.org/ 10.1371/journal.pone.0008846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Okumura M, Sakuma C, Miura M, Chihara T. Linking cell surface receptors to microtubules: tubulin folding cofactor D mediates Dscam functions during neuronal morphogenesis. J Neurosci 2015; 35:1979-90; PMID:25653356; http://dx.doi.org/ 10.1523/JNEUROSCI.0973-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shultz T, Shmuel M, Hyman T, Altschuler Y. Beta-tubulin cofactor D and ARL2 take part in apical junctional complex disassembly and abrogate epithelial structure. FASEB J 2008; 22:168-82; PMID:17704193; http://dx.doi.org/ 10.1096/fj.06-7786com [DOI] [PubMed] [Google Scholar]
- [45].Beghin A, Matera EL, Brunet-Manquat S, Dumontet C. Expression of Arl2 is associated with p53 localization and chemosensitivity in a breast cancer cell line. Cell Cycle 2008; 7:3074-82; PMID:18818514; http://dx.doi.org/ 10.4161/cc.7.19.6777 [DOI] [PubMed] [Google Scholar]
- [46].Beghin A, Honore S, Messana C, Matera EL, Aim J, Burlinchon S, Braguer D, Dumontet C. ADP ribosylation factor like 2 (Arl2) protein influences microtubule dynamics in breast cancer cells. Exp Cell Res 2007; 313:473-85; PMID:17188265; http://dx.doi.org/ 10.1016/j.yexcr.2006.10.024 [DOI] [PubMed] [Google Scholar]
- [47].Beghin A, Belin S, Hage-Sleiman R, Brunet Manquat S, Goddard S, Tabone E, Jordheim LP, Treilleux I, Poupon MF, Diaz JJ, et al.. ADP ribosylation factor like 2 (Arl2) regulates breast tumor aggressivity in immunodeficient mice. PloS One 2009; 4:e7478; PMID:19829707; http://dx.doi.org/ 10.1371/journal.pone.0007478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sharer JD, Kahn RA. The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J Biol Chem 1999; 274:27553-61; PMID:10488091; http://dx.doi.org/ 10.1074/jbc.274.39.27553 [DOI] [PubMed] [Google Scholar]
- [49].Bowzard JB, Cheng D, Peng J, Kahn RA. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J Biol Chem 2007; 282:17568-80; PMID:17452337; http://dx.doi.org/ 10.1074/jbc.M701347200 [DOI] [PubMed] [Google Scholar]
- [50].East MP, Bowzard JB, Dacks JB, Kahn RA. ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for arf protein family GTPases. J Biol Chem 2012; 287:39538-53; PMID:23014990; http://dx.doi.org/ 10.1074/jbc.M112.417477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ivanova AA, East MP, Yi SL, Kahn RA. Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J Biol Chem 2014; 289:11111-21; PMID:24616099; http://dx.doi.org/ 10.1074/jbc.M114.548529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NB. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J Biol Chem 2013; 288:4723-32; PMID:23271731; http://dx.doi.org/ 10.1074/jbc.M112.378984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al.. Function of mitochondrial Stat3 in cellular respiration. Science 2009; 323:793-7; PMID:19131594; http://dx.doi.org/ 10.1126/science.1164551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 2009; 324:1713-6; PMID:19556508; http://dx.doi.org/ 10.1126/science.1171721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].She H, Yang Q, Shepherd K, Smith Y, Miller G, Testa C, Mao Z. Direct regulation of complex I by mitochondrial MEF2D is disrupted in a mouse model of Parkinson disease and in human patients. J Clin Invest 2011; 121:930-40; PMID:21393861; http://dx.doi.org/ 10.1172/JCI43871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yogev O, Naamati A, Pines O. Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J 2011; 278:4230-42; PMID:21929734; http://dx.doi.org/ 10.1111/j.1742-4658.2011.08359.x [DOI] [PubMed] [Google Scholar]
- [57].Bauer NC, Doetsch PW, Corbett AH. Mechanisms Regulating Protein Localization. Traffic 2015; 16:1039-61; PMID:26172624; http://dx.doi.org/ 10.1111/tra.12310 [DOI] [PubMed] [Google Scholar]
- [58].Li CC, Wu TS, Huang CF, Jang LT, Liu YT, You ST, Liou GG, Lee FJ. GTP-binding-defective ARL4D alters mitochondrial morphology and membrane potential. PloS One 2012; 7:e43552; PMID:22927989; http://dx.doi.org/ 10.1371/journal.pone.0043552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Reis K, Fransson A, Aspenstrom P. The Miro GTPases: at the heart of the mitochondrial transport machinery. FEBS Lett 2009; 583:1391-8; PMID:19376118; http://dx.doi.org/ 10.1016/j.febslet.2009.04.015 [DOI] [PubMed] [Google Scholar]
- [60].Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell 2009; 20:3525-32; PMID:19477917; http://dx.doi.org/ 10.1091/mbc.E09-03-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Van Valkenburgh H, Shern JF, Sharer JD, Zhu X, Kahn RA. ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J Biol Chem 2001; 276:22826-37; PMID:11303027; http://dx.doi.org/ 10.1074/jbc.M102359200 [DOI] [PubMed] [Google Scholar]
- [62].Hanzal-Bayer M, Linari M, Wittinghofer A. Properties of the interaction of Arf-like protein 2 with PDEdelta. J Mol Biol 2005; 350:1074-82; PMID:15979089; http://dx.doi.org/ 10.1016/j.jmb.2005.05.036 [DOI] [PubMed] [Google Scholar]
- [63].Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol 2011; 7:942-9; PMID:22002721; http://dx.doi.org/ 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- [64].Watzlich D, Vetter I, Gotthardt K, Miertzschke M, Chen YX, Wittinghofer A, Ismail S. The interplay between RPGR, PDEdelta and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep 2013; 14:465-72; PMID:23559067; http://dx.doi.org/ 10.1038/embor.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fansa EK, Kosling SK, Zent E, Wittinghofer A, Ismail S. PDE6delta-mediated sorting of INPP5E into the cilium is determined by cargo-carrier affinity. Nat Commun 2016; 7:11366; PMID:27063844; http://dx.doi.org/ 10.1038/ncomms11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hanke-Gogokhia C, Zhang H, Frederick JM, Baehr W. The function of arf-like proteins ARL2 and ARL3 in photoreceptors. Adv Exp Med Biol 2016; 854:655-61; PMID:26427472; http://dx.doi.org/ 10.1007/978-3-319-17121-0_87 [DOI] [PubMed] [Google Scholar]
- [67].Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J 2002; 21:2095-106; PMID:11980706; http://dx.doi.org/ 10.1093/emboj/21.9.2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ismail SA, Chen YX, Miertzschke M, Vetter IR, Koerner C, Wittinghofer A. Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J 2012; 31:4085-94; PMID:22960633; http://dx.doi.org/ 10.1038/emboj.2012.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Renault L, Hanzal-Bayer M, Hillig RC. Coexpression, copurification, crystallization and preliminary X-ray analysis of a complex of ARL2-GTP and PDE delta. Acta Crystallogr D Biol Crystallogr 2001; 57:1167-70; PMID:11468408; http://dx.doi.org/ 10.1107/S0907444901009556 [DOI] [PubMed] [Google Scholar]
- [70].Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, Bastiaens PI. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 2014; 157:459-71; PMID:24725411; http://dx.doi.org/ 10.1016/j.cell.2014.02.051 [DOI] [PubMed] [Google Scholar]
- [71].Wätzlich D, Vetter I, Gotthardt K, Miertzschke M, Chen YX, Wittinghofer A, Ismail S. The interplay between RPGR, PDEδ and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep 2013; 14:465-72; PMID:Can't; http://dx.doi.org/ 10.1038/embor.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Thomas S, Wright KJ, Le Corre S, Micalizzi A, Romani M, Abhyankar A, Saada J, Perrault I, Amiel J, Litzler J, et al.. A homozygous PDE6D mutation in Joubert syndrome impairs targeting of farnesylated INPP5E protein to the primary cilium. Human mutation 2014; 35:137-46; PMID:24166846; http://dx.doi.org/ 10.1002/humu.22470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Davidson AE, Schwarz N, Zelinger L, Stern-Schneider G, Shoemark A, Spitzbarth B, Gross M, Laxer U, Sosna J, Sergouniotis PI, et al.. Mutations in ARL2BP, encoding ADP-ribosylation-factor-like 2 binding protein, cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet 2013; 93:321-9; PMID:23849777; http://dx.doi.org/ 10.1016/j.ajhg.2013.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mori N, Ishiba Y, Kubota S, Kobayashi A, Higashide T, McLaren MJ, Inana G. Truncation mutation in HRG4 (UNC119) leads to mitochondrial ANT-1-mediated photoreceptor synaptic and retinal degeneration by apoptosis. Invest Ophthalmol Vis Sci 2006; 47:1281-92; PMID:16565359; http://dx.doi.org/ 10.1167/iovs.05-0493 [DOI] [PubMed] [Google Scholar]
- [75].Ishiba Y, Higashide T, Mori N, Kobayashi A, Kubota S, McLaren MJ, Satoh H, Wong F, Inana G. Targeted inactivation of synaptic HRG4 (UNC119) causes dysfunction in the distal photoreceptor and slow retinal degeneration, revealing a new function. Exp Eye Res 2007; 84:473-85; PMID:17174953; http://dx.doi.org/ 10.1016/j.exer.2006.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, Fujimaki T, McLaren MJ, Weleber RG, Inana G. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol Vis Sci 2000; 41:3268-77; PMID:11006213 [PubMed] [Google Scholar]


