ABSTRACT
Arf GTPases assemble protein complexes on membranes to carry out major functions in cellular traffic. An essential step is their activation by guanine nucleotide exchange factors (GEFs), whose Sec7 domain stimulates GDP/GTP exchange. ArfGEFs form 2 major families: ArfGEFs with DCB, HUS and HDS domains (GBF1 and BIG1/BIG2 in humans), which act at the Golgi; and ArfGEFs with a C-terminal PH domain (cytohesin, EFA6 and BRAG), which function at the plasma membrane and endosomes. In addition, pathogenic bacteria encode an ArfGEF with a unique membrane-binding domain. Here we review the allosteric regulation of Arf GTPases and their GEFs at the membrane interface. Membranes contribute several regulatory layers: at the GTPase level, where activation by GTP is coupled to membrane recruitment by a built-in structural device; at the Sec7 domain, which manipulates this device to ensure that Arf-GTP is attached to membranes; and at the level of non-catalytic ArfGEF domains, which form direct or GTPase-mediated interactions with membranes that enable a spectacular diversity of regulatory regimes. Notably, we show here that membranes increase the efficiency of a large ArfGEF (human BIG1) by 32-fold by interacting directly with its N-terminal DCB and HUS domains. The diversity of allosteric regulatory regimes suggests that ArfGEFs can function in cascades and circuits to modulate the shape, amplitude and duration of Arf signals in cells. Because Arf-like GTPases feature autoinhibitory elements similar to those of Arf GTPases, we propose that their activation also requires allosteric interactions of these elements with membranes or other proteins.
KEYWORDS: allostery, Arf, guanine nucleotide exchange factors, membrane, small GTPase, structure
Introduction
Small GTPases of the Arf family control many aspects of lipid and membrane traffic, such as the formation of vesicles, the assembly of membrane contact sites or the regulation of lipid-modifying enzymes, and they are also subverted by several pathogens to manipulate host traffic (reviewed in ref. 1). In their membrane-attached and GTP-bound form, Arf GTPases interact with a variety of effectors with unrelated structures, which they assemble into multivalent membrane-binding platforms (reviewed in ref. 2). It is accepted that guanine nucleotide exchange factors (GEFs), which activate small GTPases by stimulating exchange of GDP for GTP, also have important roles in specifying the subcellular localization and downstream effectors of GTP-bound GTPases (reviewed in ref. 3). Arf GTPases are activated by GEFs characterized by a conserved Sec7 domain which is responsible for stimulating GDP/GTP exchange.4,5 Two major families of ArfGEFs in eukaryotes and one bacterial family can be defined based on their related domain organizations6-8 (Fig. 1). ArfGEFs with a dimerization and cyclophilin-binding (DCB) and homology upstream Sec7 (HUS) domain and 3 or 4 homology downstream Sec7 (HDS) domains (refered to as large ArfGEFs hereafter) are found in all eukaryotes. In humans, this family comprises 2 subfamilies: GBF1, which functions primarily at the cis-Golgi, and BIG1 and BIG2, which activate Arf GTPases at the trans Golgi network (TGN). ArfGEFs with a phosphoinositide-binding pleckstrin homology (PH) domain in C-terminus (PH domain-containing ArfGEFs hereafter) are found only in animals, where they activate Arf GTPases at the cell periphery. This family comprises 3 subfamilies, cytohesins, Brefeldin A-Resistant ArfGEF (BRAG) and Exchange Factor for Arf6 (EFA6), which diverge in the regions located upstream of their Sec7 domain. The intracellular pathogens Legionella pneumophila, which is responsible for Legionnaire's disease and Rickettsia prowazekii, which causes epidemic typhus, encode a type IV effector called RalF with a domain homologous to the eukaryotic Sec7 domain and a domain unrelated to any eukaryotic domain. RalF is a bacterial ArfGEF, which is secreted to activate host Arf GTPases during infection.
Figure 1.
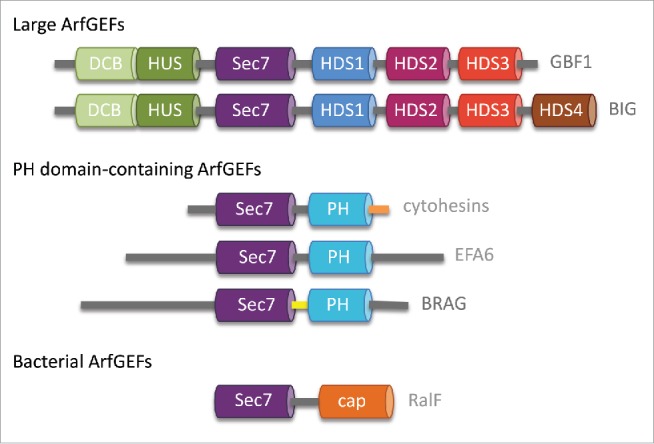
Conserved domains in human large ArfGEFs and PH domain-containing ArfGEFs and in bacterial ArfGEFs. Alternative names for PH domain-containing ArfGEFs subfamily members are: cytohesin-1/GRP1/ARNO2; cytohesin-2/ARNO/ARNO1; cytohesin-2/ARNO3; cytohesin-4/ARNO4; EFA6A/PSD1; EFA6B/PSD4,EFA6C/PSD2; EFA6D/PSD3; BRAG1/IQSEC2; BRAG2/IQSEC1/GEP100, BRAG3/IQSEC3.
Here we discuss the current knowledge in the molecular and structural mechanisms for the regulation of Arf GTPases and their GEFs at the interface with membranes, and how these processes can be described under the different facets of allostery.
Built-in allostery in the structural mechanism of Arf activation at the membrane interface
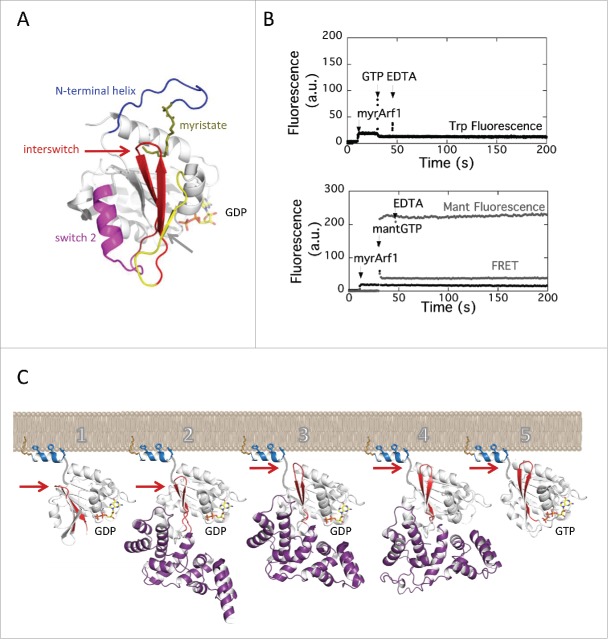
The core mechanism of activation of Arf GTPases at the membrane interface has been extensively investigated by biochemical and structural studies and its general features are now well understood. Arf GTPases differ from “classical” small GTPases of the Ras superfamily in that, in addition to the nucleotide-binding switch 1 and switch 2 regions, they have 2 unique switch regions that undergo large remodeling upon GDP/GTP exchange: a N-terminal myristoylated amphipathic helix located opposite to the nucleotide-binding site9-13 and the interswitch, a β-hairpin that connects switch 1 and switch 2 and runs across the GTPase10,11,14 (Fig. 2A). The myristoylated N-terminal helix is critical for the association of Arf-GTP with membranes; biochemical and NMR studies showed that it interacts with the membrane bilayer by both the myristate and aromatic/aliphatic residues.13,15-17 In contrast to Arf-GTP, in Arf-GDP the N-terminal helix forms intramolecular interactions with the core of the GTPase that shield its myristate and membrane-binding residues, and thus block its association with membranes.10,18 Remarkably, the autoinhibited conformation of the N-terminal helix also inhibits the binding of GTP: structures of Arf-GDP showed that it stabilizes the interswitch in a retracted conformation that obstructs the binding site of the γ-phosphate of GTP.10,11 The priming event for the activation by GTP is thus the displacement of the myristoylated N-terminal helix by membranes to release this first layer of autoinhibition. The second layer of autoinhibition is released by the toggle of the interswitch β-hairpin, a 2-residue shift that converts the GTPase to the canonical conformation observed in other GTPases and remodels the nucleotide-binding site to bind GTP. Remarkably, the interswitch toggle obstructs the intramolecular binding site for the N-terminal helix, thereby ensuring that Arf-GTP is securely bound to membranes by its N-terminus. Because of these autoinhibitory elements, myristoylated Arf-GDP is strictly resistant to activation by GTP in the absence of membranes, whether by an ArfGEF5,19 or by chelating Mg2+ with EDTA to facilitate GDP dissociation (Fig. 2B). Using tryptophan fluorescence to monitor the conformational change or mant-nucleotides and FRET fluorescence to monitor nucleotide exchange directly, we show that EDTA is unable to promote the conformational change or the loading of GTP. Thus, caution is advised that it does not appear to be possible to produce myrArf-GTP in solution, which should be taken into account when devising experimental protocols.
Figure 2.
Coupled activation by GTP and membrane recruitment of Arf GTPases. (A) The switch elements of Arf GTPases. The switch 1 and switch 2 are found in all small GTPases; the myristoylated N-terminal helix and the interswitch are switch elements unique to Arf and Arf-like small GTPases. The NMR structure of myristoylated yeast Arf1-GDP is from ref. 18; PDB entry 2K5U. (B) EDTA does not promote GTP binding on myrArf1 in solution. No nucleotide exchange was measured upon addition of EDTA, whether by tryptophan fluorescence (top) or by monitoring mant-GTP fluorescence (bottom) (see Methods for wavelengths). Analysis of mant-GTP fluorescence in the absence of protein showed that the large initial increase is due to its intrinsic fluorescence, and that the slow increase following EDTA addition is due to an effect of EDTA on the interactions between mant-GTP and Mg2+, as it was not observed in a Mg2+-free buffer (data not shown). (C) Schematic representation of the stimulation of GDP/GTP exchange by the Sec7 domain on membranes. One: the N-terminal helix, which locks Arf-GDP in an autoinhibited conformation in solution, is displaced by membranes; 2: membrane-attached Arf-GDP is recognized by the Sec7 domain (here, stabilized by the drug Brefeldin A); 3: the Sec7 domain promotes the toggle of the interswitch, which releases autoinhibition of the GTP-binding site and secures Arf to the membranes (here, stabilized by mutation of the catalytic glutamate to lysine); 4: the Sec7 domain dissociates GTP from Arf to form a nucleotide-free complex; 5: GTP binds to membrane-associated Arf. Arf-GDP is from ref. 11, PDB entry 1RRG; the Arf-GDP-Sec7 complexes trapped by Brefeldin A and by mutation of the catalytic glutamate are from ref. 26, PDB entries 1SD9 and 1R8S; nucleotide-free Arf-Sec7 is from ref. 14; Arf1-GTP is from ref. 86, PDB entry 1O3Y. The structure of the N-terminal helix and the position of the complexes with respect to membranes are dicative. The toggle of the interswitch is indicated by an arrow.
Altogether, the N-terminal helix and the interswitch couple the recruitment of Arf to membranes to its activation by GTP such that Arf is autoinhibited in the cytosol and Arf-GTP is entirely membrane-associated. With crystal structures depicting the GDP/GTP cycle of Arf110,11, 14 and Arf6,20,21 we realized that this spectacular conformational switch is a common feature that defines Arf and Arf-like GTPases as a distinct GTPase family and that it is encoded in specific sequence signatures.22 This mechanism has the hallmarks of allostery: it allows interactions on the side of the GTPase where it interacts with membranes to propagate information to the opposite side of the protein where it binds guanine nucleotides.
Important questions regarding the energetics and specificity of this built-in allosteric mechanism still remain only partially understood. Deletion of the N-terminal helix resulted in hydrogen bonds in the interswitch of Arf1-GDP to exchange protons more rapidly, as seen in NMR H/D exchange experiments,23 and in Arf6-GDP it resulted in the partial unfolding of the interswitch in the crystal.24 These experiments suggest that displacement of the N-terminal helix by membranes, which is mimicked in solution by its deletion, increases the internal dynamics to facilitate the allosteric transition. Another important question is the identification of the structural and physico-chemical determinants that allow Arf GTPases, notably Arf1, to function on membranes as diverse as those of the Golgi, plasma membrane or vesicles.25 It is possible that the N-terminus of Arf1 has sequence or structural features that allow it to recognize different types of membrane, and that other Arf family members, which diverge from Arf1 mostly in their N-terminus, recognize membranes in a more specific manner. Also, it cannot be excluded that subtle effects are brought about by interaction with GEFs.
The allosteric contribution of membranes to the activation of Arf GTPases by the Sec7 domain
It was observed early that the Sec7 domain, which suffices to stimulate GDP dissociation in solution using N-terminally truncated Arf GTPases, cannot activate an intact Arf GTPase in the absence of a membrane.5,19 Thus, the Sec7 domain contributes little, if at all, to the displacement of the autoinhibitory N-terminal helix implying that myrArf-GDP must have been primed for activation by membranes prior to its activation by an ArfGEF. This probably occurs spontaneously through the equilibrium between soluble myrArf-GDP and a myrArf-GDP intermediate that is weakly bound to membranes (Fig. 2C). Crystallographic analysis of Arf/ArfGEF intermediates captured by the drug BFA,26,27 by a mutation of catalytic glutamate that occupies the Mg2+ binding site26,28,29 and by removal of GDP14 revealed that ArfGEFs activate membrane-associated Arf-GDP in 2 discrete steps (Fig. 2C). First, they promote the toggle of the interswitch, which secures Arf to membranes before dissociation of GDP;26 then GDP is displaced, allowing for the entry of GTP.14 In other words, activation of Arf GTPases results from the combined actions of a membrane, which unlocks the N-terminal helix, and of a GEF, which promotes the subsequent structural remodeling. Communication between the membrane and the GEF domain involves little direct contact, if any, and can thus be described in the framework of allostery. It is well established that Sec7 domains have little conformational flexibility,26,27,29-33 which makes it unlikely that the GEF activity is regulated by conformational changes at the level of the Sec7 domain. In the following sections, we discuss our current knowledge on the regulatory roles of non-catalytic domains of eukaryotic and bacterial ArfGEFs at the membrane interface.
Regulation of large ArfGEFs by direct and small GTPase-mediated interactions with membranes
The large ArfGEFs family includes 2 subfamilies, GBF1 (the yeast orthologs of which are Gea1 and Gea2) and BIG1/BIG2 (Sec7 in yeast), which in humans activate Arf GTPases at the cis Golgi and the trans-Golgi network (TGN), respectively.8 Conserved domains outside the Sec7 domain in these ArfGEFs were first identified based on sequence analysis.34 They include the DCB and HUS domains upstream of the Sec7 domain and the HDS1 to HDS3 domains downstream of the Sec7 domain, none of them has homology to any conventional membrane-binding domain (Fig. 1). The role of non-catalytic domains in regulation and membrane localization of large ArfGEFs has begun to be unraveled, highlighting multiple layers of interactions.
The DCB domain was originally described in the plant homolog GNOM as supporting dimerization35 and the HUS domain was named after a conserved motif, the HUS,box 36 whose mutation in yeast Gea2 impairs anterograde Golgi transport.37 The DCB and HUS domains of yeast Sec7 form a compact helical structure in which the conserved HUS motif is unstructured and they have no structural resemblance to known membrane-binding domains.38 We demonstrate here that the DCB-HUS tandem is a membrane-binding element and that it contributes to the efficiency of Arf activation. A purified human BIG1 construct spanning the DCB-HUS-Sec7 domains associated directly with membranes in a liposome co-sedimentation assay (Fig. 3A), and membranes increased its activation of Arf1 by 32 fold (kcat/Km determined using a range of GEF concentrations: 0.11 × 105 M−1s−1 in solution and 3.5 × 105 M−1s−1 on membranes) (Fig. 3B). The fact that this construct had significant GEF activity in solution suggests that the DCB-HUS domains do not form autoinhibitory interactions with the Sec7 domain. Consistent with our study, the DCB-HUS domains are required for the association of mammalian BIG1 and BIG2 to the TGN39 and of human GBF1, which belongs to the other subfamily of large ArfGEFs, to the Golgi.40 In a contrasting set of experiments, a yeast Sec7 construct carrying the DCB-HUS-Sec7 domains did not bind to membranes autonomously and was 3-4-fold less active on membranes than in solution.38,41 This construct was however about 4-fold more active on membranes than the Sec7 domain and this led to a model in which it was proposed to assist in the displacement of the N-terminal helix of Arf.38 Together, these studies point to a conserved function of the DCB-HUS domains in the association of large ArfGEFs with membranes and in the regulation of their activity, possibly with different structural implementations between subfamilies and/or species that remain to be established.
Figure 3.
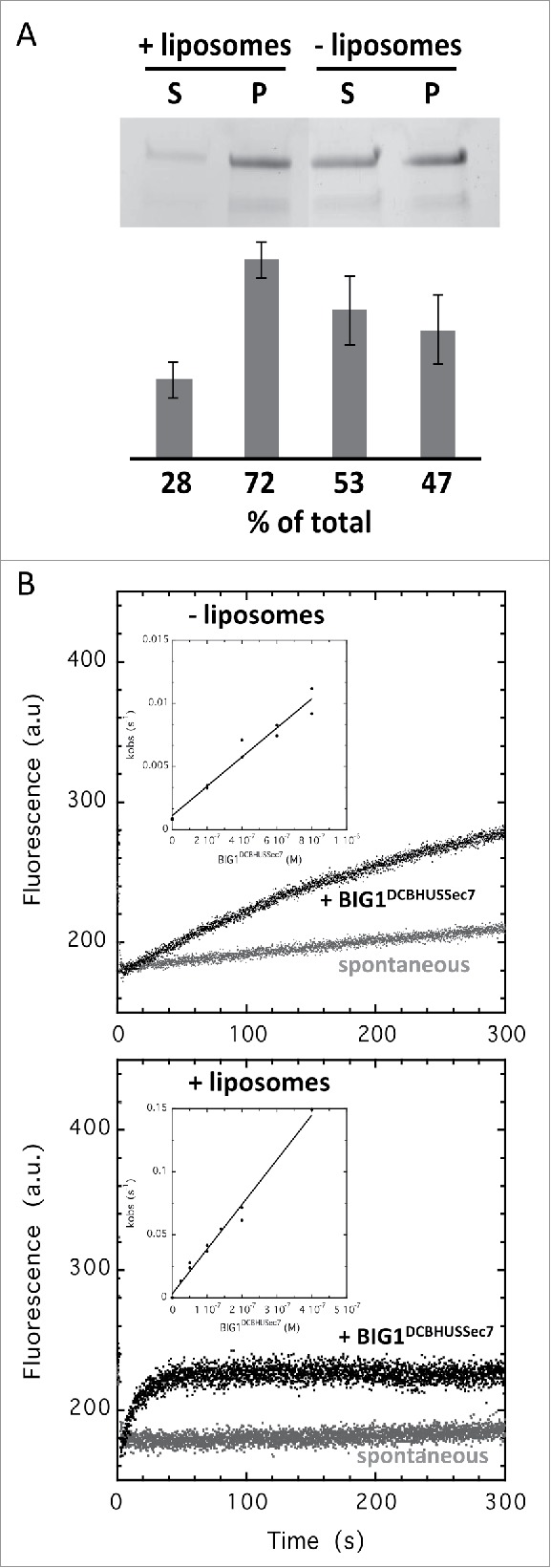
The DCB-HUS domains potentiates BIG1 activity on membranes. (A) BIG1DCBHUSSec7 binds to liposomes containing 40% PC, 35% DOPC, 10% PE, 10% DOPE and 5% PI(4)P in a co-sedimentation assay; S = supernatant, P = pellet (n = 2, error bars = SD). (B) BIG1DCBHUSSec7 is 32 times more active on membranes. Representative nucleotide exchange kinetics traces for BIG1DCBHUSSec7 (200 nM) activation of Δ17Arf1 in solution (top) or of myrArf1 in the presence of liposomes containing 38% PC, 10% DOPC, 10% PE, 10% DOPE, 5% PS, 2% PI(4)P, 10% PI and 15% cholesterol (bottom). Nucleotide exchange was monitored by tryptophan fluorescence. Insets show kcat/Km determination of BIG1DCBHUSSec7 for each substrate (n = 2). Values are given in Table 1.
The HDS1 domain, which is located immediately downstream to the Sec7 domain, is another region that has been associated with the regulation of large ArfGEFs on membranes. The HDS1 domain of human GBF1 bound directly to “Golgi-mix” liposomes and to artificial lipid droplets, and this interaction was inhibited in a construct comprising both the Sec7 and HDS1 domains.40 Consistent with the inhibition of membrane-binding elements in the HDS1 domain by the Sec7 domain, a yeast Sec7 construct encompassing the DCB-HUS-Sec7-HDS1 domains did not bind autonomously to membranes, yet it was about 5-fold more active on membranes than the shorter DCB-HUS-Sec7 construct.41 These studies are consistent with a mechanism in which membrane recruitment and activation are coupled through an autoinhibitory mechanism involving the HDS1 domain, possibly with differences between the 2 subfamilies the details of which are currently unknown.
Binding of Arf and other small GTPases provides a second layer of regulation of large ArfGEFs on membranes. Arf1-GTP bound to the yeast Sec7 construct encompassing the DCB-HUS-Sec7-HDS1 domains and recruited it to membranes, and it enhanced its GEF activity up to 3-fold on membranes.41 BIG1 and BIG2 (the mammalian orthologs of Sec7) also interacted with Arf-GTP in cells, but the region critical for interaction was mapped to the N-terminal DCB-HUS domains.39 Interaction with Arf-GTP was not observed for yeast Gea1, which belongs to the other large ArfGEF subfamily 41 or for its human ortholog GBF1,40 suggesting that it is specific to the Sec7/BIG subfamily. Other GTP-bound small GTPases have been reported to interact with large ArfGEFs and to regulate their localization and/or activities. The Arf1-related GTPase Arl1 was shown to be necessary for the recruitment of BIG1 to the Golgi42,43 and this is mediated by a direct interaction of Arl1-GTP with the DCB domain.43 Another trafficking small GTPase, Rab1b, was required for the localization of GBF1 to the Golgi and was shown to interact with the DCB-HUS N-terminal domains.44 Ypt1-GTP, the yeast ortholog of Rab1, Arl1-GTP and Ypt31-GTP, the yeast ortholog of Rab11, also bound to yeast Sec7.45 Overexpression of Ypt1 suppressed a growth phenotype defect and mislocalization of a Sec7 carrying a mutation in the DCB domain, suggesting that it interacts with the N-terminal region as observed in GBF1.45 This study showed that while Arf1-GTP and Arl1-GTP did not increase the GEF activity of full-length yeast Sec7 and Ypt1-GTP increased it only 2-fold, Ypt31-GTP increased its activity 20-fold, suggesting that it is a major regulator of Sec7 activity on membranes.45
This ensemble of observations suggests that the 2 subfamilies of large ArfGEFs are recruited to membranes by multivalent interactions, both direct and mediated by trafficking GTPases, some of them may also regulate autoinhibitory and feedback effects. Understanding the structural organization, membrane preference and subfamily specificity of these elaborate patterns of autoinhibitory, GTPase and membrane interactions will be an important next step toward understanding the regulation of large ArfGEFs. Reconstitutions using purified proteins and artificial membranes have already provided important insight into the molecular mechanisms of large ArfGEFs and should continue to be pivotal, together with structural studies, for deciphering the full “instruction manual” of large ArfGEF regulation. Since individual regulatory interactions may only have modest effects, stimulatory effects should be measured as accurately as possible to detect small differences with confidence. For that reason, we recommend that ArfGEF activities are determined by kcat/Km measured by kinetics experiments over a range of GEF concentrations (see Methods).
Regulation of PH domain-containing ArfGEFs by membranes
ArfGEFs with a PH domain are found only in animals, where they function at the plasma membrane and in endosomal processes.6 They are subdivided in 3 subfamilies which diverge in the regions located upstream of their Sec7 domain (Fig. 1). While the function of N-terminal regions has remained poorly understood, the regulatory mechanisms of PH domains have been extensively studied and this revealed an unexpected diversity of regulatory regimes. Although they display minimal homology to each other, the PH domains of cytohesins,5 BRAG29,46,47 and EFA648,49 all bind PI(4,5)P2 phosphoinositides. Cytohesins also interact with PI(3,4,5)P3, with splice variants with a di-glycine instead of a tri-glycine in the β1-β2 loop of the PH domain showing a marked preference for this rare phosphoinositide.50-52 The polybasic region in the C-terminus of the PH domain of cytohesins makes additional contributions to their interaction with acidic membranes.53 Biophysical and molecular dynamics simulations suggest that the PH domain and the polybasic region bind additional acidic lipids at non-canonical binding sites.54,55 Whereas the activities and specificities of these 3 subfamilies differ broadly in solution, all are highly potent at activating both Arf1 and Arf6 in the presence of membranes, including EFA6 which is not active on Arf1 in solution.29,46, 49,56-58 These stark differences highlight the importance to assess specificity using membranes and myristoylated Arf GTPases, as this is often the starting point for building models of the biology of the proteins being studied. kcat/Km toward Arf1 in solution and on membranes determined using a range of GEF concentrations are given in Table 1 for Sec-PH constructs of representative members of the cytohesin (ref. 33, this study), BRAG29 and EFA649 subfamilies. In contrast to the similarities of these 3 subfamilies in their lipid and Arf specificities, their regulation is unexpectedly different.
Table 1.
Catalytic efficiencies of ArfGEFs toward Arf1 are highly enhanced by membranes.
| GEFs | myrArf1 (kcat/Kmx 105 M−1s−1) | Δ17Arf1 (kcat/Kmx 105 M−1s−1) | fold increase myrArf1/ Δ17Arf1 |
|---|---|---|---|
| ARNOSec7PH | 165 ± 9# | 0.015 ± 0.0004 33 | 11000 |
| BRAG2Sec7PH | 346 ± 46 29 | 2.5 ± 0.06 29 | 138 |
| EFA6Sec7PH | 56 ± 1 49 | 0.011 ± 0.005 49 | 5090 |
| BIG1DCBHUSSec7 | 3.53 ± 0.04# | 0.11 ± 0.01# | 32 |
#this study.
Cytohesins have a short region in the N-terminus of the Sec7 domain which promotes homotypic and heterotypic dimerization, a conserved linker between the Sec7 domain and the PH domain, and a positively charged extension in the C-terminus. Elements proximal to the PH domain (Sec7-PH linker and C-terminal helix/polybasic region) shut down the GEF activity by obstructing the active site of the Sec7 domain.33 Autoinhibition is released by Arf1-GTP and Arf6-GTP, which bind to the PH domain as effectors33,59,60 and enable a positive feedback mechanism whereby the activated Arfs modulate the GEF activity of cytohesins58,61 and enhance their recruitment to membranes.58 Another activated small GTPase, Arl4-GTP, binds to the PH domain of cytohesins and recruits them to the plasma membrane, although whether this is associated with autoinhibition release is not known.62 The phosphoinositide-binding site of cytohesins is fully accessible in their autoinhibited conformation,33 which suggests that phosphoinositides can recruit cytohesins to membranes regardless of whether or not they are autoinhibited or bound to Arf-GTP. Thus, the PH domain of cytohesins has a dual regulatory role on membranes: it responds to PIP2 or PIP3 phosphoinositides signals by recruiting cytohesins to membranes through its canonical phosphoinositide-binding site; and it modulates the shape of the Arf-GTP signal by autoinhibition of the Arf-binding site and its release by membrane-bound Arf-GTP. Depending on whether the substrate and effector Arf isoforms are the same or distinct, this mechanisms has the ability to implement a positive feedback loop or a signaling cascade, in both cases resulting in non-linear amplification of Arf-GTP generation.
BRAG ArfGEFs have a ≈500-residue N-terminal region of essentially unknown function, and a linker between the Sec7 and PH domains which is longer than in cytohesins and extends the structure of the PH domain.29,36 Unlike in cytohesins, this extended PH domain is not autoinhibitory.29,46 The crystal structure of a human BRAG2 construct comprising the Sec7 and PH domains in complex with Arf showed that the PH domain forms extensive intramolecular interactions with the Sec7 domain.29 It also contacts Arf in a manner that positions the GTPase and the canonical PIP2-binding site in register to interact with membranes. Remarkably, although BRAG2Sec7PH is highly active in solution on Arf1 and Arf6, its activity is further increased by more than 2 orders of magnitude by membranes (Table 1),29,46 approaching the theoretical maximal value of about 108 M−1s−1, corresponding to a rate which is limited only by the diffusion of the proteins.63 Arf-GTP did not modify the activity of BRAG2, indicating that the PH domain is not involved in a feedback mechanism.29 This major difference between BRAG and cytohesins is consistent with the observation that the side of the PH domain that binds Arf-GTP in cytohesins33 is masked by the linker in BRAG2.29 The origin of the allosteric effect of membranes on BRAG efficiency is best explained by multiple discrete intramolecular, protein-protein and protein-lipid interactions that result in an optimized conformation and orientation of the Arf/BRAG complex at the membrane interface. Mutations in the related ArfGEF BRAG1 are found in patients with nonsyndromic intellectual disability.64 Remarkably, several mutations are located at the Sec7-PH interface where they could impair the coupling between the ArfGEF and membrane-binding functions, pointing to a defect in allosteric regulation on membranes as a possible component of the disease.
EFA6 members have a poorly conserved region in the N-terminus of their Sec7 domain whose function is currently poorly understood, and a positively charged domain of ≈150 residues in the C-terminus of their PH domain (Ct hereafter).56 Neither the PH nor the PH-Ct domains are autoinhibitory; yet, PIP2-containing membranes increase activity by 2 orders of magnitude48,49 (Table 1). Remarkably, although EFA6 is strictly specific for Arf6 in solution, it activates both Arf1 and Arf6 efficiently on membranes, highlighting the fact that membranes can modulate the specificity of GEFs for their small GTPase substrates.49 The regulatory regime of EFA6 is however strikingly different from that of the other 2 subfamilies. EFA6 activity decreases as more Arf-GTP is produced on membranes, suggesting that it might be regulated by negative feedback.49 Although the structural details of this effect remain to be investigated, titration by the PH-Ct domains suggests that Arf-GTP regulates EFA6 by interacting with this domain.
In summary, each subfamily uses its PH domain for a different allosteric usage on membranes: autoinhibition and positive feedback in cytohesins, negative feedback in EFA6, and constitutive activity without feedback in BRAG.
Regulation of bacterial ArfGEFs by membranes
Membrane traffic pathways are often subverted by pathogens, and manipulation of small GTPases regulation is among their primary targets. The genomes of Legionella and of Rickettsia encode a type IV effector with a Sec7 domain homologous to that of eukaryotic ArfGEFs which functions as a GEF to activate host Arf GTPases.65 The structure of Legionella RalF showed that the Sec7 domain is strongly autoinhibited by a C-terminal domain that blocks access to the Arf-binding site,66 and a similar autoinhibited conformation was observed in the Rickettsia RalF homolog.67 Both bacterial ArfGEFs are strongly activated by membranes, and the membrane-binding site is identical to the elements in the autoinhibitory domain that blocks the Arf-binding site.68,69 Thus, activation of Arf GTPases by RalF strictly depends on its recruitment to membranes. Legionella RalF produces Arf-GTP at the surface of the Legionella-containing vacuole where the pathogen hides and replicates;65 however Rickettsia replicates in the host cytosol and therefore should use RalF for different functions. In that regard, it is remarkable that Legionella and Rickettsia RalF respond to different membrane characteristics for their activation.69 Legionella RalF is activated equally well by membranes that contain acidic lipids or are enriched in unsaturated lipids, suggesting that it is tailored to remain active at the surface of the phagosome-derived vacuole as it matures by incorporating ER-derived vesicles. It is also a unique situation in which a membrane-binding domain recognizes 2 membranes with different characteristics. In contrast, Rickettsia RalF has a strict requirement for anionic membranes, probably reflecting the cytosolic lifestyle of the pathogen. These differences in membrane specificities are explained by the ratio between positively charged residues, which bind to acidic lipids, and aromatic residues, which wedge in lipid packing defects, which differs between Legionella and Rickettsia RalF.69 Thus, both Legionella and Rickettsia have evolved a simplified ArfGEF version to subvert host Arf GTPases, in which the same structural element is responsible for autoinhibition and for membrane binding to support allosteric regulation by membranes.
Discussion
50 years after the celebrated Monod-Wyman-Changeux thermodynamic model was first formulated,70 the concept of allostery experiences a spectacular renaissance. The original concept, which was underlined by structural communication between subunits in oligomeric proteins, has undergone a major change in trademark to encompass any situation in which distant sites in a protein communicate in response to cellular clues to trigger a change in activity.71 Recently, the realization that co-localization on membranes is an important source at the origin of allosteric interactions72 and that allosteric interactions can establish through changes in dynamics even in the absence of conformational changes,73 further broadened the scope of the concept. The regulation of Arf family activation by GEFs on membranes can be described in this general framework at multiple levels: at the structural level of the Arf GTPase itself, through changes in conformation and dynamics that propagate information between the guanine nucleotide-binding site and the membrane-binding site; at the level of the Sec7 domains of ArfGEFs, which stabilize the interaction of Arf GTPase with membranes prior to nucleotide exchange by interacting with Arf at the opposite of its membrane-binding site; and at the level of non-catalytic domains of the ArfGEFs, which modulate the activity of the GEF domain by interacting with membranes and Arf-GTP and other small GTPases using sites remote from the Arf-binding site. It is important to point out however that not all increase in catalytic efficiency resulting from addition of membranes is due to allostery, since co-association of ArfGEFs with Arf GTPases on a membrane reduces the diffusion volume, thereby decreasing the apparent Km and hence increases catalytic efficiency.
As predicted from sequence signatures,22 structural studies of Arf-like and Arf-related GTPases, including Golgi Arl1,74,75 cilium Arl376,77,78 and Arl679 and endoplasmic reticulum Sar180,81 have now shown that autoinhibition by the N-terminal extension and the interswitch takes place in these GTPases and is released by the displacement of the N-terminus and the toggle of the interswitch. Thus, the allosteric mechanism that uses the displacement of the N-terminal extension as a priming event to autoinhibition release and the remodeling of the interswitch as the means whereby information is propagated to the nucleotide-binding site is probably general to the entire Arf-like family. The N-termini of Arf-related GTPases vary in length, sequence and post-translational modifications, and there is therefore ample reasons to believe that they are displaced in different ways to release autoinhibition. For instance, the N-terminus of Arl3-GTP interacts with another protein,77,78,82 and it is conceivable that there exists Arf-like GTPases in which autoinhibition release involves interactions of the N-terminus with both a protein and a membrane. Understanding this process will be a major theme for future investigations of the molecular mechanisms that regulate the activity of Arf-like GTPases.
A major advance over the last few years has been the discovery that ArfGEfs have an unexpected diversity of allosteric regulatory regimes (Fig. 4). This diversity evokes important directions for future research. At the biochemical and structural levels, our understanding of the full repertoire and hierarchy of regulatory interactions, and of the structures and conformational changes that support them, still remains fragmentary. Notably, structures of the N-terminal domains of PH-domain containing ArfGEFs and of the C-terminal domains in large ArfGEFs are still missing, and no structure of a fully active, membrane-associated ArfGEFs, is currently known. We anticipate that analysis of the structure and biochemistry of full-length ArfGEFs will reveal aspects of their regulation and functions that cannot be identified by working on isolated domains or truncated proteins. There are also fascinating issues to be addressed at the functional level, which depend in large part on an accurate understanding of the “instruction manual” of active ArfGEFs. The observation that the localization and regulation of large ArfGEFs depend on their product and/or on other trafficking GTPases suggests that they may be interconnected in cascades and that this should contribute to defining trafficking checkpoints and enhancing the fidelity of anterograde cargo transport at the Golgi.45,61,83 Likewise, the diversity of regulatory regimes of PH-containing ArfGEFs raises exciting prospects relevant to Arf functions at the cell periphery. Notably, the realization that PH domain-containing ArfGEFs have little if any autonomous discrimination between Arf1 and Arf6 and they all recognize PIP2 phosphoinositides, suggests that they have the ability to activate both Arf isoforms at the same time and on the same membrane. These diverging regulatory regimes could then allow ArfGEFs to function in cascades, in which an Arf GTPase activated by a first ArfGEF binds to a second ArfGEF to activate another Arf GTPase, or in circuits, in which an activated Arf GTPase modulates the activity of an upstream ArfGEF by positive or negative feedback effects. This could expand the shape, duration and amplitude of Arf GTPases activity and match them to specific cellular situations. The observation that both EFA6 and BRAG2 were required to usurp Arf GTPases during Salmonella invasion of host cells may be an example of such coordinated activity.84 As for large ArfGEFs, determination of the structure, interactions and regulatory functions of N-terminal elements in PH domain-containing ArfGEFs and the analysis of the shape, duration and efficiency of ArfGEFs activity in comprehensively reconstituted assays will be important steps in deciphering how Arf GTPases functions are orchestrated by ArfGEFs in cells.
Figure 4.
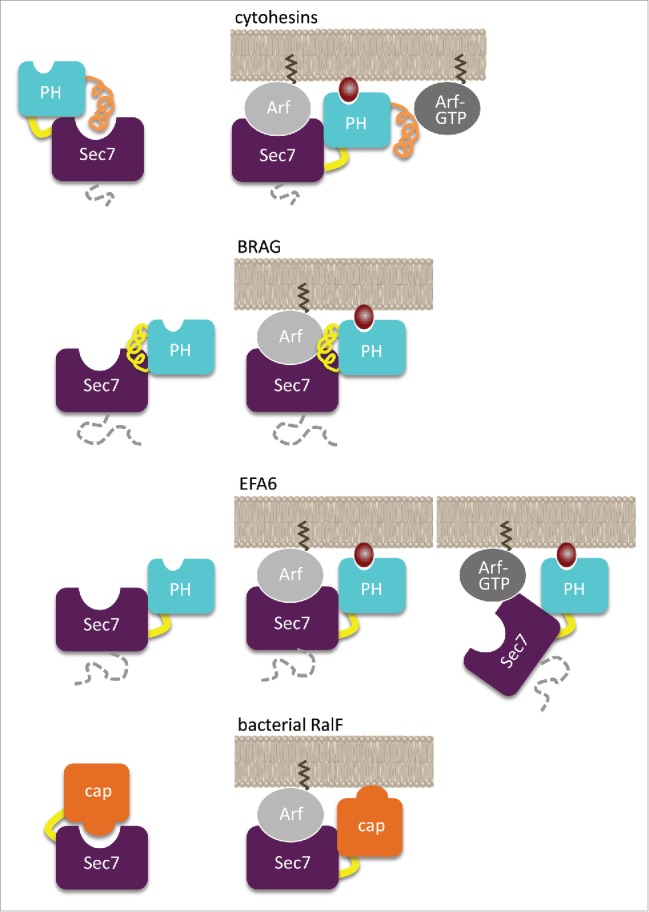
Models of allosteric activation of PH domain-containing ArfGEFs and of bacterial ArfGEFs by membranes. Note that each subfamily regulatory regime is a unique combination of membrane-binding site autoinhibition and/or Arf-binding site autoinhibition and/or feedback effect.
Material and methods
Proteins
Human Δ17Arf1 (residues 18-181) and myristoylated Arf1 were obtained as described previously.49 ARNOSec7PH (residues 50-399) constructs cloned into the pET-8c vector is a kind gift of Bruno Antonny (CNRS, France). ARNOSec7PH was purified on NiNTA affinity column followed by size exclusion chromatography. Human BIG1DCBHUSSec7 (residues 2-888) was expressed in insect cells using the baculovirus system and purified as described in ref. 85.
Liposomes binding assay
All lipids were obtained from Avanti Polar Lipids, except the NBD- phosphatidylethanolamine (NBD-PE), which is from Invitrogen. Sucrose-loaded liposomes that contained 40% phosphatidylcholine (PC), 35% DOPC, 10% phosphatidylethanolamine (PE), 10% DOPE and 5% phosphatidylinositol-4-phosphate (PI(4)P) were freshly extruded through a 0.2 μm filter (Whatman) and used within 2 days. Co-sedimentation experiments were carried out as described previously69 after incubation of 1 mM of liposomes with 3 µM of BIG1DCBHUSSec7 for 15 min at 20°C. Proteins were analyzed by SDS-PAGE on a 4–20% TGX-stain free gel (Bio-Rad) and detected by fluorescence after 2.5 min activation using a ChemiDoc imaging system (Bio-Rad). Quantifications were done with the Bio-Rad Image Lab™ software (v5.2).
Nucleotide exchange kinetics
Analysis of nucleotide exchange stimulation of myrArf1 by EDTA in solution was monitored by tryptophan fluorescence (emission/excitation wavelengths of 292/340 nm), mant-GTP fluorescence (emission/excitation wavelengths of 350/440 nm) or FRET between protein tryptophans and mant-GTP (emission/excitation wavelengths of 292/440 nm), in the absence of any detergent. Briefly, myrArf1 was incubated in 50 mM Hepes pH 7.4 and 120 mM potassium acetate, 1mM MgCl2 buffer then 100µM GTP or 5µM mant-GTP was added. The reaction was initiated by addition of 10mM EDTA. ArfGEFs catalytic efficiencies (kcat/Km) were determined in solution using Δ17Arf1 at 3 μM and/or in the presence of 100 μM liposomes using myrArf1 at 0,4 μM as described.28,29 Liposomes contained 38% PC, 10% DOPC, 10% PE, 10% DOPE, 5% PS, 2% PI(4)P, 10% PI and 15% cholesterol (BIG1DCBHUSSec7) or 38% PC, 20% PE, 20% PS, 2% PI(3,4,5)P3 and 20% cholesterol (ARNOSec7PH). The concentration range for ARNOSec7PH was 1nM-4nM; under these conditions, the lag phase corresponding to inhibition release by Arf-GTP was negligible and kinetic traces could be fit with a mono-exponential function. For all constructs, values are mean ± SD of 2 independent series of measurements performed with different batches of liposomes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We greatly thank François Peurois and Lurlène Akendengue in our lab for the purification of ARNOSec7PH and BIG1DCBHUSSec7.
Funding
This work was supported by grants from the Fondation pour la Recherche Médicale, the Institut National du Cancer and the CNRS to JC.
References
- [1].Jackson CL, Bouvet S. Arfs at a glance. Journal of cell science 2014; 127:4103-9; PMID:25146395; http://dx.doi.org/ 10.1242/jcs.144899 [DOI] [PubMed] [Google Scholar]
- [2].Cherfils J. Arf GTPases and their effectors: assembling multivalent membrane-binding platforms. Curr Opin Struct Biol 2014; 29:67-76; PMID:25460270; http://dx.doi.org/ 10.1016/j.sbi.2014.09.007 [DOI] [PubMed] [Google Scholar]
- [3].Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; http://dx.doi.org/ 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- [4].Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 1996; 384:479-81; PMID:8945477; http://dx.doi.org/ 10.1038/384479a0 [DOI] [PubMed] [Google Scholar]
- [5].Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 1996; 384:481-4; PMID:8945478; http://dx.doi.org/ 10.1038/384481a0 [DOI] [PubMed] [Google Scholar]
- [6].Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 2007; 8:1476-85; PMID:17850229; http://dx.doi.org/ 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- [7].Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 2011; 12:362-75; PMID:21587297; http://dx.doi.org/ 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wright J, Kahn RA, Sztul E. Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell Mol Life Sci 2014; 71:3419-38; PMID:24728583; http://dx.doi.org/ 10.1007/s00018-014-1602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kahn RA, Goddard C, Newkirk M. Chemical and immunological characterization of the 21-kDa ADP-ribosylation factor of adenylate cyclase. J Biol Chem 1988; 263:8282-7; PMID:3131341 [PubMed] [Google Scholar]
- [10].Amor JC, Harrison DH, Kahn RA, Ringe D. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature 1994; 372:704-8; PMID:7990966; http://dx.doi.org/ 10.1038/372704a0 [DOI] [PubMed] [Google Scholar]
- [11].Greasley SE, Jhoti H, Teahan C, Solari R, Fensome A, Thomas GM, Cockcroft S, Bax B. The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms. Nature structural biology 1995; 2:797-806; PMID:7552752; http://dx.doi.org/ 10.1038/nsb0995-797 [DOI] [PubMed] [Google Scholar]
- [12].Franco M, Chardin P, Chabre M, Paris S. Myristoylation of ADP-ribosylation factor 1 facilitates nucleotide exchange at physiological Mg2+ levels. J Biol Chem 1995; 270:1337-41; PMID:7836400; http://dx.doi.org/ 10.1074/jbc.270.3.1337 [DOI] [PubMed] [Google Scholar]
- [13].Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 1997; 36:4675-84; PMID:9109679; http://dx.doi.org/ 10.1021/bi962252b [DOI] [PubMed] [Google Scholar]
- [14].Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 1998; 95:237-48; PMID:9790530; http://dx.doi.org/ 10.1016/S0092-8674(00)81754-7 [DOI] [PubMed] [Google Scholar]
- [15].Losonczi JA, Prestegard JH. Nuclear magnetic resonance characterization of the myristoylated, N-terminal fragment of ADP-ribosylation factor 1 in a magnetically oriented membrane array. Biochemistry 1998; 37:706-16; PMID:9425095; http://dx.doi.org/ 10.1021/bi9717791 [DOI] [PubMed] [Google Scholar]
- [16].Gizachew D, Oswald R. NMR structural studies of the myristoylated N-terminus of ADP ribosylation factor 6 (Arf6). FEBS Lett 2006; 580:4296-301; PMID:16839550; http://dx.doi.org/ 10.1016/j.febslet.2006.06.086 [DOI] [PubMed] [Google Scholar]
- [17].Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf*GTP. Nat Struct Mol Biol 2010; 17:876-81; PMID:20601958; http://dx.doi.org/ 10.1038/nsmb.1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu Y, Kahn RA, Prestegard JH. Structure and membrane interaction of myristoylated ARF1. Structure 2009; 17:79-87; PMID:19141284; http://dx.doi.org/ 10.1016/j.str.2008.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Paris S, Beraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. Role of protein-phospholipid interactions in the activation of ARF1 by the guanine nucleotide exchange factor Arno. J Biol Chem 1997; 272:22221-6; PMID:9268368; http://dx.doi.org/ 10.1074/jbc.272.35.22221 [DOI] [PubMed] [Google Scholar]
- [20].Menetrey J, Macia E, Pasqualato S, Franco M, Cherfils J. Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nature structural biology 2000; 7:466-9; PMID:10881192; http://dx.doi.org/ 10.1038/75863 [DOI] [PubMed] [Google Scholar]
- [21].Pasqualato S, Menetrey J, Franco M, Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep 2001; 2:234-8; PMID:11266366; http://dx.doi.org/ 10.1093/embo-reports/kve043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pasqualato S, Renault L, Cherfils J. Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO Rep 2002; 3:1035-41; PMID:12429613; http://dx.doi.org/ 10.1093/embo-reports/kvf221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Buosi V, Placial JP, Leroy JL, Cherfils J, Guittet E, van Heijenoort C. Insight into the role of dynamics in the conformational switch of the small GTP-binding protein Arf1. J Biol Chem 2010; 285:37987-94; PMID:20861011; http://dx.doi.org/ 10.1074/jbc.M110.134445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Biou V, Aizel K, Roblin P, Thureau A, Jacquet E, Hansson S, Guibert B, Guittet E, van Heijenoort C, Zeghouf M, et al.. SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6. J Mol Biol 2010; 402:696-707; PMID:20709080; http://dx.doi.org/ 10.1016/j.jmb.2010.08.002 [DOI] [PubMed] [Google Scholar]
- [25].D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 2006; 7:347-58; PMID:16633337; http://dx.doi.org/ 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- [26].Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 2003; 426:525-30; PMID:14654833; http://dx.doi.org/ 10.1038/nature02197 [DOI] [PubMed] [Google Scholar]
- [27].Mossessova E, Corpina RA, Goldberg J. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell 2003; 12:1403-11; PMID:14690595; http://dx.doi.org/ 10.1016/S1097-2765(03)00475-1 [DOI] [PubMed] [Google Scholar]
- [28].Beraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J 1998; 17:3651-9; PMID:9649435; http://dx.doi.org/ 10.1093/emboj/17.13.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aizel K, Biou V, Navaza J, Duarte LV, Campanacci V, Cherfils J, Zeghouf M. Integrated conformational and lipid-sensing regulation of endosomal ArfGEF BRAG2. PLoS Biol 2013; 11:e1001652; PMID:24058294; http://dx.doi.org/ 10.1371/journal.pbio.1001652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cherfils J, Menetrey J, Mathieu M, Le Bras G, Robineau S, Beraud-Dufour S, Antonny B, Chardin P. Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature 1998; 392:101-5; PMID:9510256; http://dx.doi.org/ 10.1038/32210 [DOI] [PubMed] [Google Scholar]
- [31].Mossessova E, Gulbis JM, Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human arno and analysis of the interaction with ARF GTPase. Cell 1998; 92:415-23; PMID:9476900; http://dx.doi.org/ 10.1016/S0092-8674(00)80933-2 [DOI] [PubMed] [Google Scholar]
- [32].Cronin TC, DiNitto JP, Czech MP, Lambright DG. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J 2004; 23:3711-20; PMID:15359279; http://dx.doi.org/ 10.1038/sj.emboj.7600388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].DiNitto JP, Delprato A, Gabe Lee MT, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol Cell 2007; 28:569-83; PMID:18042453; http://dx.doi.org/ 10.1016/j.molcel.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, Jürgens G, Melançon P, Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC genomics 2005; 6:20; PMID:15717927; http://dx.doi.org/ 10.1186/1471-2164-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jürgens G. A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. The Plant cell 2000; 12:343-56; PMID:10715321; http://dx.doi.org/ 10.1105/tpc.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mansour SJ, Skaug J, Zhao XH, Giordano J, Scherer SW, Melancon P. p200 ARF-GEP1: a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci U S A 1999; 96:7968-73; PMID:10393931; http://dx.doi.org/ 10.1073/pnas.96.14.7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Park SK, Hartnell LM, Jackson CL. Mutations in a highly conserved region of the Arf1p activator GEA2 block anterograde Golgi transport but not COPI recruitment to membranes. Mol Biol Cell 2005; 16:3786-99; PMID:15930122; http://dx.doi.org/ 10.1091/mbc.E05-04-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Richardson BC, Halaby SL, Gustafson MA, Fromme JC. The Sec7 N-terminal regulatory domains facilitate membrane-proximal activation of the Arf1 GTPase. Elife 2016; 5; pii: e12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lowery J, Szul T, Styers M, Holloway Z, Oorschot V, Klumperman J, Sztul E. The Sec7 guanine nucleotide exchange factor GBF1 regulates membrane recruitment of BIG1 and BIG2 guanine nucleotide exchange factors to the trans-Golgi network (TGN). J Biol Chem 2013; 288:11532-45; PMID:23386609; http://dx.doi.org/ 10.1074/jbc.M112.438481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bouvet S, Golinelli-Cohen MP, Contremoulins V, Jackson CL. Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. Journal of cell science 2013; 126:4794-805; PMID:23943872; http://dx.doi.org/ 10.1242/jcs.134254 [DOI] [PubMed] [Google Scholar]
- [41].Richardson BC, McDonold CM, Fromme JC. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell 2012; 22:799-810; PMID:22516198; http://dx.doi.org/ 10.1016/j.devcel.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Christis C, Munro S. The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J Cell Biol 2012; 196:327-35; PMID:22291037; http://dx.doi.org/ 10.1083/jcb.201107115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Galindo A, Soler N, McLaughlin SH, Yu M, Williams RL, Munro S. Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi. Cell Rep 2016; 16(3):839-50; PMID:27373159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell 2007; 18:2400-10; PMID:17429068; http://dx.doi.org/ 10.1091/mbc.E06-11-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].McDonold CM, Fromme JC. Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-golgi network. Dev Cell 2014; 30:759-67; PMID:25220393; http://dx.doi.org/ 10.1016/j.devcel.2014.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jian X, Gruschus JM, Sztul E, Randazzo PA. The pleckstrin homology (PH) domain of the Arf exchange factor Brag2 is an allosteric binding site. J Biol Chem 2012; 287:24273-83; PMID:22613714; http://dx.doi.org/ 10.1074/jbc.M112.368084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Roy NS, Yohe ME, Randazzo PA, Gruschus JM. Allosteric properties of PH domains in Arf regulatory proteins. Cell Logist 2016; 6:e1181700; PMID:27294009; http://dx.doi.org/ 10.1080/21592799.2016.1181700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Macia E, Partisani M, Favard C, Mortier E, Zimmermann P, Carlier MF, Gounon P, Luton F, Franco M. The pleckstrin homology domain of the Arf6-specific exchange factor EFA6 localizes to the plasma membrane by interacting with phosphatidylinositol 4,5-bisphosphate and F-actin. J Biol Chem 2008; 283:19836-44; PMID:18490450; http://dx.doi.org/ 10.1074/jbc.M800781200 [DOI] [PubMed] [Google Scholar]
- [49].Padovani D, Folly-Klan M, Labarde A, Boulakirba S, Campanacci V, Franco M, Zeghouf M, Cherfils J. EFA6 controls Arf1 and Arf6 activation through a negative feedback loop. Proc Natl Acad Sci U S A 2014; 111:12378-83; PMID:25114232; http://dx.doi.org/ 10.1073/pnas.1409832111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 1997; 275:1927-30; PMID:9072969; http://dx.doi.org/ 10.1126/science.275.5308.1927 [DOI] [PubMed] [Google Scholar]
- [51].Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem 2000; 275:32816-21; PMID:10913124; http://dx.doi.org/ 10.1074/jbc.M002435200 [DOI] [PubMed] [Google Scholar]
- [52].Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell 2000; 6:385-94; PMID:10983985; http://dx.doi.org/ 10.1016/S1097-2765(00)00038-1 [DOI] [PubMed] [Google Scholar]
- [53].Macia E, Paris S, Chabre M. Binding of the PH and polybasic C-terminal domains of ARNO to phosphoinositides and to acidic lipids. Biochemistry 2000; 39:5893-901; PMID:10801341; http://dx.doi.org/ 10.1021/bi992795w [DOI] [PubMed] [Google Scholar]
- [54].Lai CL, Srivastava A, Pilling C, Chase AR, Falke JJ, Voth GA. Molecular mechanism of membrane binding of the GRP1 PH domain. J Mol Biol 2013; 425:3073-90; PMID:23747485; http://dx.doi.org/ 10.1016/j.jmb.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Naughton FB, Kalli AC, Sansom MS. Association of Peripheral Membrane Proteins with Membranes: Free Energy of Binding of GRP1 PH Domain with Phosphatidylinositol Phosphate-Containing Model Bilayers. J Phys Chem Lett 2016; 7:1219-24; PMID:26977543; http://dx.doi.org/ 10.1021/acs.jpclett.6b00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J 1999; 18:1480-91; PMID:10075920; http://dx.doi.org/ 10.1093/emboj/18.6.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Macia E, Chabre M, Franco M. Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J Biol Chem 2001; 276:24925-30; PMID:11342560; http://dx.doi.org/ 10.1074/jbc.M103284200 [DOI] [PubMed] [Google Scholar]
- [58].Stalder D, Barelli H, Gautier R, Macia E, Jackson CL, Antonny B. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J Biol Chem 2011; 286:3873-83; PMID:21118813; http://dx.doi.org/ 10.1074/jbc.M110.145532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell 2007; 18:2244-53; PMID:17409355; http://dx.doi.org/ 10.1091/mbc.E06-11-0998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Malaby AW, van den Berg B, Lambright DG. Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors. Proc Natl Acad Sci U S A 2013; 110:14213-8; PMID:23940353; http://dx.doi.org/ 10.1073/pnas.1301883110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Stalder D, Antonny B. Arf GTPase regulation through cascade mechanisms and positive feedback loops. FEBS Lett 2013; 587:2028-35; PMID:23684643; http://dx.doi.org/ 10.1016/j.febslet.2013.05.015 [DOI] [PubMed] [Google Scholar]
- [62].Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr Biol 2007; 17:711-6; PMID:17398095; http://dx.doi.org/ 10.1016/j.cub.2007.03.007 [DOI] [PubMed] [Google Scholar]
- [63].Itzen A, Rak A, Goody RS. Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J Mol Biol 2007; 365:1359-67; PMID:17134721; http://dx.doi.org/ 10.1016/j.jmb.2006.10.096 [DOI] [PubMed] [Google Scholar]
- [64].Shoubridge C, Tarpey PS, Abidi F, Ramsden SL, Rujirabanjerd S, Murphy JA, Boyle J, Shaw M, Gardner A, Proos A, et al.. Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet 2010; 42:486-8; PMID:20473311; http://dx.doi.org/ 10.1038/ng.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 2002; 295:679-82; PMID:11809974; http://dx.doi.org/ 10.1126/science.1067025 [DOI] [PubMed] [Google Scholar]
- [66].Amor JC, Swails J, Zhu X, Roy CR, Nagai H, Ingmundson A, Cheng X, Kahn RA. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem 2005; 280:1392-400; PMID:15520000; http://dx.doi.org/ 10.1074/jbc.M410820200 [DOI] [PubMed] [Google Scholar]
- [67].Folly-Klan M, Sancerne B, Alix E, Roy CR, Cherfils J, Campanacci V. On the use of Legionella/Rickettsia chimeras to investigate the structure and regulation of Rickettsia effector RalF. J Struct Biol 2015; 189:98-104; PMID:25498244; http://dx.doi.org/ 10.1016/j.jsb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- [68].Alix E, Chesnel L, Bowzard BJ, Tucker AM, Delprato A, Cherfils J, Wood DO, Kahn RA, Roy CR. The capping domain in RalF regulates effector functions. PLoS Pathog 2012; 8:e1003012; PMID:23166491; http://dx.doi.org/ 10.1371/journal.ppat.1003012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Folly-Klan M, Alix E, Stalder D, Ray P, Duarte LV, Delprato A, Zeghouf M, Antonny B, Campanacci V, Roy CR, et al.. A novel membrane sensor controls the localization and ArfGEF activity of bacterial RalF. PLoS Pathog 2013; 9:e1003747; PMID:24244168; http://dx.doi.org/ 10.1371/journal.ppat.1003747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol 1965; 12:88-118; PMID:14343300; http://dx.doi.org/ 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- [71].Changeux JP. 50 years of allosteric interactions: the twists and turns of the models. Nat Rev Mol Cell Biol 2013; 14:819-29; PMID:24150612; http://dx.doi.org/ 10.1038/nrm3695 [DOI] [PubMed] [Google Scholar]
- [72].Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature 2007; 450:983-90; PMID:18075577; http://dx.doi.org/ 10.1038/nature06524 [DOI] [PubMed] [Google Scholar]
- [73].Nussinov R, Tsai CJ. Allostery without a conformational change? Revisiting the paradigm. Curr Opin Struct Biol 2015; 30:17-24; PMID:25500675; http://dx.doi.org/ 10.1016/j.sbi.2014.11.005 [DOI] [PubMed] [Google Scholar]
- [74].Amor JC, Horton JR, Zhu X, Wang Y, Sullards C, Ringe D, Cheng X, Kahn RA. Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases. J Biol Chem 2001; 276:42477-84; PMID:11535602; http://dx.doi.org/ 10.1074/jbc.M106660200 [DOI] [PubMed] [Google Scholar]
- [75].Panic B, Perisic O, Veprintsev DB, Williams RL, Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol Cell 2003; 12:863-74; PMID:14580338; http://dx.doi.org/ 10.1016/S1097-2765(03)00356-3 [DOI] [PubMed] [Google Scholar]
- [76].Hillig RC, Hanzal-Bayer M, Linari M, Becker J, Wittinghofer A, Renault L. Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein. Structure 2000; 8:1239-45; PMID:11188688; http://dx.doi.org/ 10.1016/S0969-2126(00)00531-1 [DOI] [PubMed] [Google Scholar]
- [77].Ismail SA, Chen YX, Miertzschke M, Vetter IR, Koerner C, Wittinghofer A. Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J 2012; 31:4085-94; PMID:22960633; http://dx.doi.org/ 10.1038/emboj.2012.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lokaj M, Kosling SK, Koerner C, Lange SM, van Beersum SE, van Reeuwijk J, Roepman R, Horn N, Ueffing M, Boldt K, et al.. The Interaction of CCDC104/BARTL1 with Arl3 and Implications for Ciliary Function. Structure 2015; 23:2122-32; PMID:26455799; http://dx.doi.org/ 10.1016/j.str.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mourao A, Nager AR, Nachury MV, Lorentzen E. Structural basis for membrane targeting of the BBSome by ARL6. Nat Struct Mol Biol 2014; 21:1035-41; PMID:25402481; http://dx.doi.org/ 10.1038/nsmb.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Huang M, Weissman JT, Beraud-Dufour S, Luan P, Wang C, Chen W, Aridor M, Wilson IA, Balch WE. Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J Cell Biol 2001; 155:937-48; PMID:11739406; http://dx.doi.org/ 10.1083/jcb.200106039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 2002; 419:271-7; PMID:12239560; http://dx.doi.org/ 10.1038/nature01040 [DOI] [PubMed] [Google Scholar]
- [82].Kapoor S, Fansa EK, Mobitz S, Ismail SA, Winter R, Wittinghofer A, Weise K. Effect of the N-Terminal Helix and Nucleotide Loading on the Membrane and Effector Binding of Arl2/3. Biophys J 2015; 109:1619-29; PMID:26488653; http://dx.doi.org/ 10.1016/j.bpj.2015.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Richardson BC, Fromme JC. Autoregulation of Sec7 Arf-GEF activity and localization by positive feedback. Small GTPases 2012; 3:240-3; PMID:22996016; http://dx.doi.org/ 10.4161/sgtp.21828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc Natl Acad Sci U S A 2013; 110:16880-5; PMID:24085844; http://dx.doi.org/ 10.1073/pnas.1311680110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ramaen O, Joubert A, Simister P, Belgareh-Touze N, Olivares-Sanchez MC, Zeeh JC, Chantalat S, Golinelli-Cohen MP, Jackson CL, Biou V, et al.. Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J Biol Chem 2007; 282:28834-42; PMID:17640864; http://dx.doi.org/ 10.1074/jbc.M705525200 [DOI] [PubMed] [Google Scholar]
- [86].Shiba T, Kawasaki M, Takatsu H, Nogi T, Matsugaki N, Igarashi N, Suzuki M, Kato R, Nakayama K, Wakatsuki S. Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport. Nat Struct Biol 2003; 10:386-93; PMID:12679809; http://dx.doi.org/ 10.1038/nsb920 [DOI] [PubMed] [Google Scholar]