ABSTRACT
Formation of autophagosomes requires vesicular trafficking from virtually every subcellular compartment to the formation site. This traffic must be tightly regulated but also adaptable as different membrane compartments will contribute varying amounts of membrane, lipids and proteins to the forming autophagosome depending on the stimulus. In mammalian cells, efforts to understand how autophagosomes form have been focused on the role of Rab proteins in autophagy. Rab proteins provide specificity through their interaction with coat proteins, vesicle tethers and SNAREs.
Recent data emerging from these studies have defined a subset of Rab proteins and their regulators, the RabGAPS (GTPase activating proteins) in both autophagosome formation and maturation. This review will focus on the role of a set of RabGAPs shown to regulate autophagy, in particular TBC1D14, and its interactors, RAB11 and TRAPPIII. Through our studies on TBC1D14, we have gained an understanding of the contribution of membrane from the recycling endosome, and the role of TRAPPIII in maintaining ATG (Autophagy protein) 9 trafficking in autophagosome formation.
KEYWORDS: ATG9, autophagosome, autophagy, Rab proteins, TBC domains, TRAPPIII
Introduction
Macroautophagy, usually called autophagy, is a lysosome-mediated catabolic pathway that is conserved in eukaryotes (for review see ref. 1). During the process of autophagy, organelles, macromolecular complexes, lipids, proteins and cytosol are sequestered by double membrane vesicles, called autophagosomes, which fuse with the lysosome for degradation (Fig. 1). Autophagy was traditionally considered to be a non-selective process. Selective autophagy pathways, for example mitophagy, use cargo receptors to recruit cargo (for example mitochondria) into forming autophagosomes. Cargo receptors bind Atg8 proteins (Atg8 in yeast, LC3 and GABARAP proteins in mammals) on the autophagosome membrane via a LC3-interacting region (LIR) motif and selectively target cytoplasmic components (eg mitochondria) for degradation. Selective autophagy pathways which use cargo receptors with high avidity for the Atg (autophagy) proteins, Atg8, LC3 and GABARAP, have been proposed2 to be able to exclude untargeted soluble cytosolic components and thus potentially spare these from degradation. Cargo adaptors are a second type of Atg8-binding LIR-domain containing proteins, which recruit cytoplasmic cargo not for degradation but to add functionality to the autophagosome (for example recruitment of trafficking molecules) (for review see ce 3).
Figure 1.
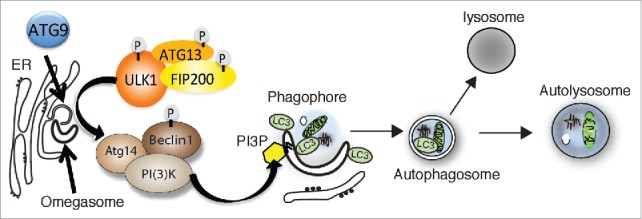
Membrane compartments (omegasome, phagophore, autophagosome) and the early stage autophagy proteins ATG9, the protein kinase ULK complex, and the VPS34 PI3K (phosphatidylinositol 3-kinase) complex (called complex I) involved in autophagosome formation. Not shown is WIPI2 the PI3P (phosphatidylinositol 3-phosphate) effector, the ubiquitin-like conjugation complexes. LC3 is depicted on the phagophore but this could also be a GABARAP family member. Autophagosomes are depicted fusing with lysosomes to create an autolysosome but other endocytic compartments are also involved in the autophagosome maturation prior to fusion with the lysosome, include the recycling endosome.
The origin of autophagosomal membranes in mammalian cells is not entirely clear despite intense investigation. The earliest structure identified in yeast is the PAS (pre-autophagosome structure), defined as the site where the Atg proteins accumulate. The mammalian equivalent has been called the phagophore or isolation membrane. While there is some evidence that the phagophore may be derived from Golgi, mitochondria, or plasma membrane-derived vesicles,3 it is widely accepted that during amino acid starvation the omegasome is the precursor membrane of the phagophore (Fig. 1). The omegasome is a platform formed on the endoplasmic reticulum (ER) that is rich in PI3P (Phosphatidylinositol 3-phosphate) and is marked by the presence of DFCP-1, a double FYVE domain containing protein.4
The omegasome and phagophore expand rapidly upon amino acid starvation, initiated primarily due to the rapid inactivation of the negative regulator of autophagy, mTORC1 (Target of Rapamycin complex 1). Loss of mTORC1 activity results in the activation and membrane association of the ULK (Unc-51 like kinase) kinase complex, and of other early Atg protein complexes to the omegasome and phagophore (Fig. 1). The ULK complex contains a serine/threonine kinase and 3 additional proteins, FIP200, ATG13 and ATG101.5,6 In mammalian cells, the hierarchy of recruitment of the Atg protein complexes to the phagophore has been described.7,8 ATG9, the only membrane spanning Atg protein, is the first protein to translocate to phagophore structures,9 followed by the VPS34 complex I and the ULK complex.
While the function of ATG9 is not known it transiently interacts with the omegasome, the phagophore and the autophagosome.9 The PI3P contained on the omegasome is produced by the Vps34 complex I, the autophagy specific PI3P complex, containing BECLIN1, VPS34, p150, and ATG14L1.10,11 The autophagy specific effector of the PI3P on the omegasome is WIPI2, and is required to recruit the LC3 and GABARAP lipidation machinery (not shown in Fig. 1).12 Both the ULK complex and WIPI2 are required for ATG9 trafficking to and possibly from the phagophore and omegasome.9
The growth of the phagophore, and its closure to become an autophagosome, requires the lipidated Atg8s, LC3 and GABARAPs.13 The Atg8's are cytosolic proteins which become membrane associated by the covalent attachment of PE (phosphatidlyethanolamine) at their C-terminal glycine. Similar to ubiquitination, the Atg8's are modified the activity of the E1-like ATG7 and the E2-like ATG3. ATG7 and ATG3 are downstream of the ATG12-5-16 conjugate produced in a second ubiquitin-like cascade using ATG7 and ATG10.1
Growing the phagophore with Rab proteins
The rapid expansion of the omegasome and phagophore suggests that additional membrane must be rapidly incorporated into these structures. There are 2 likely sources of membrane, de novo synthesis of lipids facilitated by the lipid biosynthetic machinery on the ER, and vesicular trafficking from cellular compartments. While the former events are largely uncharacterized, there is evidence which supports vesicular delivery from several compartments to aid formation and expansion of the autophagosome (for review see ref. 3). Vesicular delivery or transport requires vesicle formation at the donor site and accurate delivery and fusion to the acceptor membrane. The machinery that performs these steps are classified as: coat protein complexes (to form the vesicle), Rab GTPases (to direct the vesicle to the correct compartment), tethers (to anchor the vesicle at the acceptor membrane) and SNARE proteins (to catalyze fusion of the vesicle with the acceptor membrane).
COPII (coat protein II) coated vesicles and the ERGIC (ER-Golgi intermediate compartment) facilitate LC3 lipidation and likely contribute membrane to the expanding phagophore.14 In addition, several Rabs are involved in early and late autophagosome formation steps.15 However, the autophagy-specific function of most of these Rab proteins are not known. An advance in understanding the role of Rab proteins in the formation of autophagosomes came with the identification of a group of TBC (tre2-bub2-cdc16)-domain containing RabGAPS, and non-TBC RabGAPs (for review see ref. 16) which regulate autophagosome formation. RabGAPs were screened for colocalization, or direct association with LC3, occurring most probably through LC3-interacting regions (LIR motifs).17,20 RAB33B interacts with ATG16L1,17 and subsequently a RabGAP for RAB33B, OATL1 (TBC1D25) was shown to regulate autophagosome maturation.18 Fourteen RabGAPs were identified in a screen for binding to the Atg8 family,19 and TBC1D5, a RabGAP for RAB7 association with the retromer complex,19 was shown to directly bind Atg8's through a LIR motif. 20 Furthermore, in an overexpression screen, 11 TBC domain containing RabGAPs were identified that inhibited autophagy.21 Importantly, one of these, TBC1D14, which does not contain a LIR motif, co-localizes with and binds the ULK complex on the recycling endosome.21
Effectors of the Rabs and RabGAPs
Additional insight into how the RabGAPs function in autophagosome formation came from the identification of novel RabGAP interactors, not including their cognate Rabs. During autophagy, TBC1D5 binding to LC3 via the LIR motif is responsible for the transfer of TBC1D5 from the retromer to LC3 present on the autophagosome.22 TBC1D5 also binds AP-2, and this binding during autophagy regulates ATG9 trafficking through the AP-2 coat complex, facilitating formation of ATG9-positive vesicles from the plasma membrane and retromer-positive endosomes which contribute to the formation of autophagosomes.
TBC1D14 is an inactive GAP which however binds RAB11 (Fig. 2).21 While active RAB11 is required for autophagosome formation, acting at the recycling endosome, TBC1D14 is likely to act independently of RAB11. TBC1D14 binds the mammalian TRAPPIII complex,23 which is functionally equivalent to yeast TRAPPIII complex (Fig. 2). Yeast TRAPPIII, required for autophagy in yeast24,25 acts as a GEF for Ypt1 (homologue of RAB1). During nutrient deprivation, yeast TRAPPIII delivers Atg9-positive vesicles to the PAS. Atg9 is retrieved from the vacuole once the yeast autophagosome has fused with the vacuole.26 In mammals, the TRAPPIII subunit TRAPPC8, equivalent to yeast Trs85, binds TBC1D14.23 TRAPPIII and TBC1D14 coordinate cycling of ATG9 from a RAB11-positive recycling endosome through a RAB1-positive ER-Golgi intermediate compartment to the Golgi (Fig. 3). Interestingly, TBC1D14 together with TRAPPIII is responsible for maintaining active RAB1B-GTP levels. This coordinated trafficking of ATG9 may utilize a “Rab conversion”27 or a GEF-GAP cascade,28 between RAB11 and RAB1 to maintain a pool of ATG9 in the Golgi complex and the ATG9-compartment. Furthermore, unlike yeast, the cycling of ATG9 through these compartments is required to provide a pool of ATG9 vesicles to the omegasome, phagophore and autophagosome during autophagy.
Figure 2.
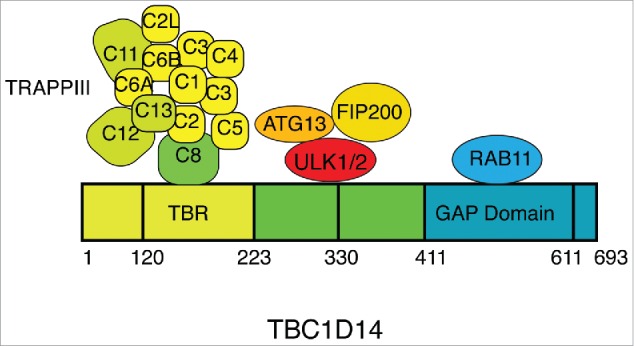
Scheme of human TBC1D14 and interactors. The TRAPPIII complexes which are specific to TRAPPII are colored green.
Figure 3.
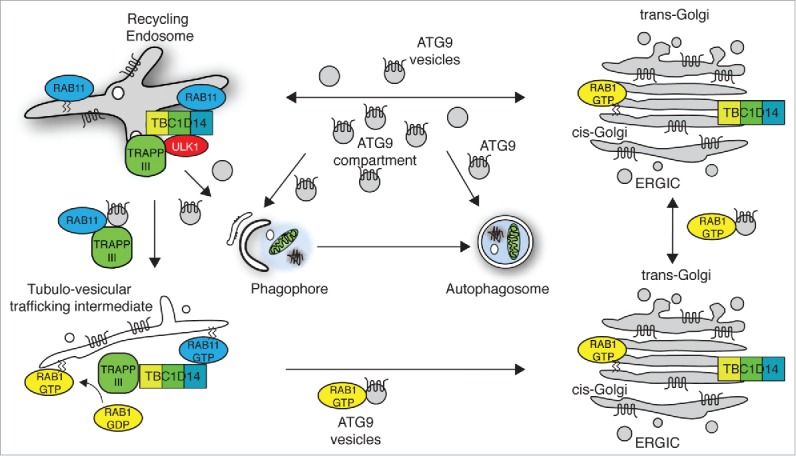
Model for function of TBC1D14 and TRAPPIII in controlling ATG9 trafficking. 23
Conclusions
Through the study of RabGaps, and Rab proteins, we have advanced our knowledge of aspects of vesicular trafficking required for autophagosome formation. In particular, we now understand the requirement of TBC1D14 interacting with TRAPPIII to maintain a reservoir of ATG9 in the Golgi and a tubular intermediate compartment which we believe is immediately mobilized upon induction of autophagy.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors wish to acknowledge the contributions of the members of the MCBA lab to the work described in the commentary.
Funding
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001187), the UK Medical Research Council (FC001187), and the Wellcome Trust (FC001187).
References
- [1].Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Ann Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- [2].Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol 2016; 428:1714-24; PMID:26876603; http://dx.doi.org/ 10.1016/j.jmb.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 2013; 14:759-74; PMID:24201109; http://dx.doi.org/ 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- [4].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/ 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hosokawa N, Sasaki T, Iemura SI, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009; 5:973-9; PMID:19597335; http://dx.doi.org/ 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- [6].Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009; 5:649-62; PMID:19287211; http://dx.doi.org/ 10.4161/auto.5.5.8249 [DOI] [PubMed] [Google Scholar]
- [7].Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, Ktistakis NT. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci 2013; 126:5224-38; PMID:24013547; http://dx.doi.org/ 10.1242/jcs.132415 [DOI] [PubMed] [Google Scholar]
- [8].Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 2013; 9:1491-9; PMID:23884233; http://dx.doi.org/ 10.4161/auto.25529 [DOI] [PubMed] [Google Scholar]
- [9].Orsi A, Razi M, Dooley H, Robinson D, Weston A, Collinson L, Tooze S. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Mol Biol Cell 2012; 23:1860-73; PMID:22456507; http://dx.doi.org/ 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 Forms Two Distinct Phosphatidylinositol 3-Kinase Complexes with Mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360-72; PMID:18843052; http://dx.doi.org/ 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 2008; 105(49):19211-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell 2014; 55:238-52; PMID:24954904; http://dx.doi.org/ 10.1016/j.molcel.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol 2011; 12:226; PMID:21867568; http://dx.doi.org/ 10.1186/gb-2011-12-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2013; 2:e00947; PMID:23930225; http://dx.doi.org/ 10.7554/eLife.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ 2014; 21:348-58; PMID:24440914; http://dx.doi.org/ 10.1038/cdd.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kern A, Dikic I, Behl C. The integration of autophagy and cellular trafficking pathways via RAB GAPs. Autophagy 2015; 11:2393-7; PMID:26565612; http://dx.doi.org/ 10.1080/15548627.2015.1110668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident Small GTPase Rab33B Interacts with Atg16L and Modulates Autophagosome Formation. Mol Biol Cell 2008; 19:2916-25; PMID:18448665; http://dx.doi.org/ 10.1091/mbc.E07-12-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Itoh T, Kanno E, Uemura T, Waguri S, Fukuda M. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J Cell Biol 2011; 192:839-53; PMID:21383079; http://dx.doi.org/ 10.1083/jcb.201008107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci 2009; 122:2371-82; PMID:19531583; http://dx.doi.org/ 10.1242/jcs.048686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Popovic D, Akutsu M, Novak I, Harper JW, Behrends C, Dikic I. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol 2012; 32:1733-44; PMID:22354992; http://dx.doi.org/ 10.1128/MCB.06717-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Longatti A, Lamb CA, Razi M, Yoshimura S-I, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11 and recycling endosomes. J Cell Biol 2012; 197:659-75; PMID:22613832; http://dx.doi.org/ 10.1083/jcb.201111079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Popovic D, Dikic I. TBC1D5 and the AP2 complex regulate ATG9 trafficking and initiation of autophagy. EMBO Rep 2014; 15:392-401; PMID:24603492; http://dx.doi.org/ 10.1002/embr.201337995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J 2016; 35:281-301; PMID:26711178; http://dx.doi.org/ 10.15252/embj.201592695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y. Atg9 vesicles recruit vesicle-tethering proteins, Trs85 and Ypt1, to the autophagosome formation site. J Biol Chem 2012; 287(53):44261-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A 2010; 107:7811-6; PMID:20375281; http://dx.doi.org/ 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012; 198:219-33; PMID:22826123; http://dx.doi.org/ 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005; 122:735-49; PMID:16143105; http://dx.doi.org/ 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- [28].Hutagalung AH, Novick PJ. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiological reviews 2011; 91:119-49; PMID:21248164; http://dx.doi.org/ 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]


