Abstract
Our previous genome-wide association studies showed that DNA methyltransferase 1 (DNMT1) is associated with increased susceptibility to type 2 diabetes (T2D) in Han Chinese individuals. Here, we aimed to further evaluate the role of DNMT1 in T2D. We performed a genome-wide DNA methylation array and found that the nuclear receptor subfamily 4 group A member 1 (NR4A1) promoter was hypermethylated in patients with T2D and in a mouse model of T2D. Moreover, DNA hypermethylation of the NR4A1 promoter reduced NR4A1 mRNA expression. Transient transfection of human NR4A1 into RIN-m5F and 293T cells caused DNMT1 inhibition and induced insulin receptor activation. NR4A1knockdown by shRNA resulted in overexpression of DNMT1 and inhibition of insulin receptor, suggesting that the NR4A1 gene is involved in the epigenetics pathway. Furthermore, T2D model mice treated with the DNMT1 inhibitor aurintricarboxylic acid (ATA) showed reduced activation of DNMT1 in pancreatic β cells; this effect reversed the changes in NR4A1 expression and decreased blood glucose in T2D model mice. Thus, our results showed for the first time that DNMT1 caused NR4A1 DNA hypermethylation and blocked insulin signaling in patients with T2D. Importantly, ATA therapy may be useful for decreasing blood glucose levels by reversing NR4A1-dependent insulin signaling. These findings improve our understanding of the crucial roles of these regulatory elements in human T2D.
Keywords: type 2 diabetes, DNA methylation, NR4A1, DNMT1, epigenetics, Gerotarget
INTRODUCTION
Type 2 diabetes (T2D) is a complex disease that is characterized by hyperglycemia, pancreatic β-cell dysfunction, decreased insulin signaling, and increased hepatic glucose production [1–4]. We previously identified the rs78789647 T allele in the DNA methyltransferase 1 (DNMT1) gene as the allele having the strongest association with T2D in Han Chinese individuals by epigenetic regulation of the protein tyrosine phosphatase receptor type delta (PTPRD) gene in the insulin signaling pathway [5–7]. Additionally, in the pancreas, DNA methylation is involved in regulating de novo β-cell formation [8, 9]. Recent studies have shown that these epigenetic factors contribute to the pathogenesis of T2D. However, the specific mechanisms through which DNMT1 regulates T2D are unclear, and the roles of DNMT1 in T2D onset and development have not been elucidated.
Nuclear receptor subfamily 4 group A (NR4A) family receptors are expressed as early response genes of physiological and pathological stimuli, including fatty acids, stress, prostaglandins, growth factors, calcium and inflammatory cytokines, peptide hormones, and neurotransmitters [10], and are implicated in important biological processes, including carcinogenesis, apoptosis control, inflammation, vascular disease, dopaminergic neuron development, and metabolism [11]. The NR4A member 1 (NR4A1) gene, which has been shown to be associated with insulin in cell models [9], encodes is a transcriptional regulator of glucose metabolism in liver and skeletal muscle [12–15].
Genome-wide DNA methylation arrays have become important in evaluation of epigenetic changes associated with disease progression. Here, we performed a pilot study using a DNA methylation array with samples from patients with T2D in order to identify biomarkers associated with the onset and development of T2D in Taiwan. Moreover, to gain insights into the functions and mechanisms through which DNMT1 affects the insulin signaling pathway, we examined epigenetic changes in patients with T2D and in a mouse model of T2D.
RESULTS
NR4A1 promoter hypermethylation in patients with T2D
First, we performed a genome-wide DNA methylation array of samples from patients with T2D. DNA methylation array data are accessible via Gene Expression Omnibus database (GEO), accession number GSE81868 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81868). Analysis of DNA methylation status, as indicated by model-based analysis of tiling-arrays (MAT) scores (range: 5.03684-8.45898), showed that the following genes had the highest methylation scores: ABR, ERICHI, WDFY2, GUSBP1, NR4A1, AFF2, PIPK1B, LOC100288637, APBA1, and PTPRN2 (Table 1). Further analysis of blood samples from patients with T2D and healthy controls showed that relative NR4A1 mRNA levels were lower in patients with T2D than in controls (1 versus 0.356, respectively; P < 0.05; Figure 1).
Table 1. Top 10 significant differentially hypermethylated genes in human T2D versis CTL.
| Transcript ID | Gene Symbol | MAT-scorea | p-value |
|---|---|---|---|
| NM_002847 | PTPRN2 | 8.45898 | 8.79E-06 |
| NM_001163 | APBA1 | 5.5031 | 8.79E-06 |
| NR_038253 | LOC100288637 | 5.40479 | 8.79E-06 |
| NM_001278253 | PIP5K1B | 5.39986 | 8.79E-06 |
| NM_001169122 | AFF2 | 5.22154 | 8.79E-06 |
| NM_001202233 | NR4A1 | 5.14054 | 8.79E-06 |
| NR_027026 | GUSBP1 | 5.11848 | 8.79E-06 |
| NM_052950 | WDFY2 | 5.08292 | 8.79E-06 |
| NR_073397 | ERICH1-AS1 | 5.0794 | 8.79E-06 |
| NM_001159746.1 | ABR | 5.03684 | 8.79E-06 |
MAT-score: Model-based Analysis of Tiling-arrays-score
Figure 1. NR4A1 mRNA was downregulated in patients with T2D.
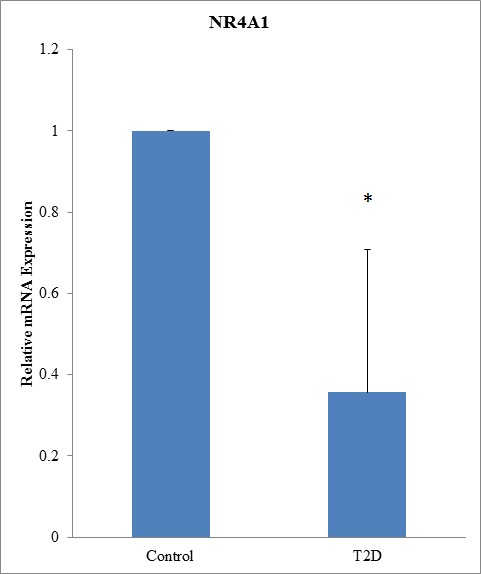
mRNA from 94 patients with T2D and 98 normal controls was used for qRT-PCR. Expression levels are shown relative to that in control patients (P < 0.05).
NR4A1 was involved in the insulin signaling pathway
The role of NR4A1 in the insulin signaling pathway is still unclear. Therefore, we used an in vitro model to analyze the effects of NR4A1 expression on insulin signaling. A plasmid containing a fragment of human NR4A1 (pcDNA-NR4A1) was constructed and transiently transfected into 293T and RIN-m5F cells. The results indicated that expression of human NR4A1 inhibited the activity of DNMT1, but induced insulin receptor overexpression in cells (Figure 2A, 2B), and DNMT1 and NR4A1 affected glucose-stimulated insulin secretion (GSIS; Figure S1), suggesting that NR4A1 was involved in the insulin signaling pathway and affected by DNMT1.
Figure 2. The NR4A1 gene was epigenetically regulated in the insulin signaling pathway.
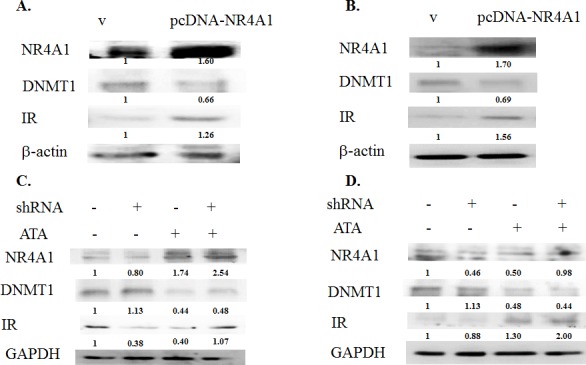
A. 293T cells were transfected with the pcDNA vector or pcDNA-NR4A1 for 48 h, and the effects of NR4A1 overexpression on DNMT1 inhibition and induction of insulin receptor (IR) overexpression were analyzed. B. RIN-m5F cells were transfected with NR4A1 for 48 h, and the effects of NR4A1 overexpression on DNMT1 inhibition and induction of IR expression were analyzed. C. Effects of NR4A1 knockdown by shRNA and treatment with ATA in 293T cells. D. Effects of NR4A1 knockdown by shRNA and treatment with ATA in RIN-m5F cells.
Effects of DNMT1 inhibition on NR4A1 expression
Interestingly, knockdown of NR4A1 expression by shRNA in 293T and RIN-m5F cells resulted in simultaneous downregulation of the insulin receptor and induction of DNMT1 in RIN-m5F cells. These results further supported that NR4A1 was involved in the insulin signaling pathway and affected by DNMT1. Therefore, we treated cells with the DNMT1 inhibitor aurintricarboxylic acid (ATA). The results showed that ATA induced NR4A1 expression in not only RIN-m5F cells and 293T cells but also NR4A1-knockdown cells. In addition, the insulin receptor was induced in RIN-m5F and 293T cells (Figure 2C, 2D).
ATA decreased blood glucose and induced changes in b-cells
Next, we used a mouse model of T2D to further elucidate the role of DNMT1 in diabetes. Sixteen-week-old mice showing insulin resistance were treated with ATA daily for 2 weeks. The results showed that blood glucose was significantly lower in ATA-treated T2D mice than in control mice (149.3 versus 526.7 mg/dL, respectively; P < 0.05; Figure 3). In yKK mice, which were used in this study, insulin resistance is associated with hypertrophy of pancreatic islets and degranulation of β-cells. After ATA treatment, pancreas islets showed decreased mass (Figure 4) and DNMT1 inhibition (Figure 5). Moreover, NR4A1 DNA hypermethylation was reduced by ATA (Figure S2), insulin signaling was triggered by insulin receptor activation, PTPRD induction, and NR4A1 overexpression (Figure 6).
Figure 3. ATA decreased blood glucose levels in T2D model mice.
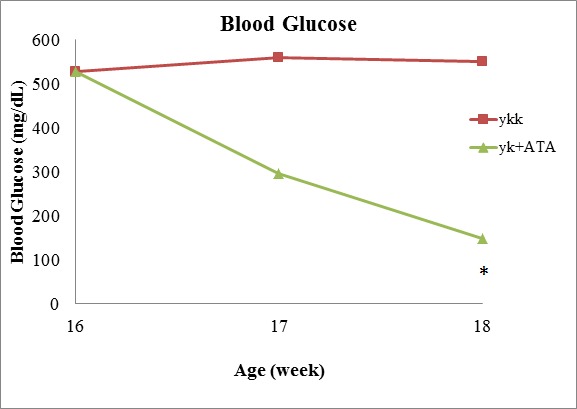
Sixteen-week-old mice were treated with ATA daily, and blood glucose was measured after 2 weeks.
Figure 4. Effects of ATA on pancreatic islet mass.
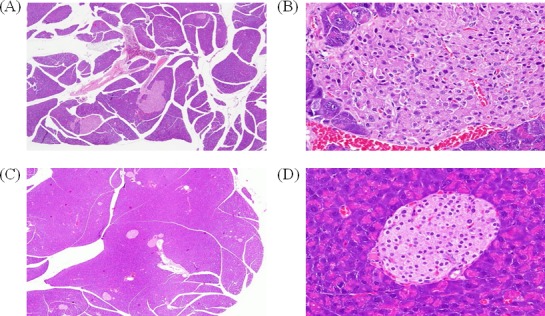
H&E staining of pancreas islet mass after 2 weeks of ATA treatment. A. Pancreas samples from yKK mice, 100×. B. Pancreas samples from yKK mice, 400×. C. Pancreas samples from ATA-treated yKK mice, 100×. D. Pancreas samples from ATA-treated yKK mice, 400×.
Figure 5. DNMT1 expression was inhibited by ATA in the pancreas.
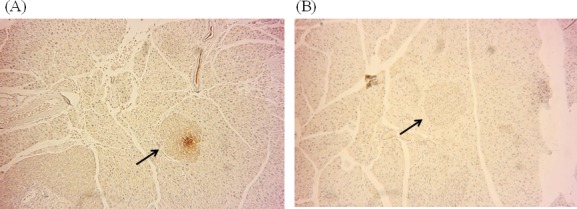
A. Immunocytochemical staining of pancreas tissues showing DNMT1 protein expression in yKK mice. B. Expression of DNMT1 in the pancreas after ATA treatment for 2 weeks.
Figure 6. Effects of ATA in the insulin signaling pathway.

Expression levels of DNMT1, NR4A1, PTPRD, and insulin receptor (IR) in livers from T2D model mice.
DISCUSSION
Genome-wide DNA methylation profiling data can reveal markers that may explain clinical and pathological specifics of T2D in a given population. DNA methylation may also have applications as a diagnostic and therapeutic target in patients with T2D [16]. In this study, we found that NR4A1 DNA was hypermethylated, resulting in downregulation of NR4A1 mRNA in patients with T2D. A recent microarray analysis indicated that NR4A1 does not induce genes associated with cell stress or apoptosis, suggesting that this protein may function mainly to stimulate proliferation when activated in the context of islet β-cells [17, 18]. Genetic deletion of NR4A1 increases susceptibility to diet-induced obesity and insulin resistance, and loss of NR4A1 expression in skeletal muscle impairs insulin signaling, markedly reduces GLUT4 protein expression, and increases triglyceride content [19]. Thus, our report presents the first findings to support the mechanism of NR4A1 function in T2D.
NR4A1 has been shown to be induced by insulin and thiazolidinedione drugs in 3T3-L1 adipocytes, and NR4A1 gene expression is enhanced in skeletal muscles and adipose tissues in multiple rodent models of insulin resistance [9]. Consistent with these findings, our results showed that the NR4A1 gene was epigenetically regulated by DNMT1 and was involved in the insulin signaling pathway, providing support for the participation of NR4A1 in pancreatic insulin receptor signaling.
In a previous study, we found that the DNMT1 gene was associated with susceptibility to T2D in Han Chinese individuals. Importantly, DNMT1 expression is significantly upregulated in mice and humans with T2D compared with that in controls [7]. Moreover, a single nucleotide polymorphism in DNMT1 (rs78789647) has been shown to be correlated with susceptibility to T2D, and DNMT1 has a preference for hemimethylated DNA [20–22].
KK-Ay mice carry the lethal yellow obese (Ay) mutation and develop diabetes of polygenic origin, showing severe obesity, hypertriglyceridemia, hyperglycemia, hyperinsulinemia, and insulin resistance by 16 weeks of age [23, 24]. In the current study, we treated 16-week-old yKK mice with ATA, which has been shown to inhibit DNMT1 [25]. Importantly, ATA treatment caused decreased blood glucose, increased NR4A1 expression, PTPRD recovery, and insulin receptor activation, providing further insights into the effects of ATA on T2D and DNMT1 function.
Hepatic expression of NR4A1 is induced by the cAMP axis in response to glucagon and fasting in vivo and is increased in diabetic mice exhibiting elevated gluconeogenesis [14]. Our previous report showed that PTPRD was inhibited by DNMT1 overexpression in patients with T2D and T2D model mice [7]. Moreover, inhibition of DNMT1 activity with 5-aza-2′-deocycytidine may facilitate the reprogramming of mouse embryonic fibroblasts into pluripotent stem cells [26], and drugs that seem to be inefficient in solid tumors could be applicable to particular subgroups of patients with pancreatic ductal adenocarcinoma [27]. Another group suggested that genomic hypomethylation caused by disrupted DNMT1 activity is correlated with a greater capability to form de novo beta cells in response to ablation [8]. Thus, our results showed, for the first time, that NR4A1 was induced by DNMT1 inhibition, resulting in insulin receptor activation and decreased blood glucose.
In summary, we found that NR4A1 expression was significantly lower in patients with T2D. Additionally, NR4A1 was involved in the insulin signaling pathway and could be epigenetically silenced by DNA methylation; epigenetic recovery of NR4A1 was observed following DNMT1 inhibition. We also found that the DNMT1 inhibitor ATA functioned to regulate blood glucose in T2D. These findings improve our understanding of the crucial roles of various regulatory elements in human T2D susceptibility.
PATIENTS AND METHODS
Isolation of methylated DNA by MBDCap
A MethylMiner Methylated DNA Enrichment Kit (Invitrogen, Carlsbad, CA, USA) was used to isolate methylated DNA. Genomic DNA was extracted from peripheral blood leukocytes using a Genomic DNA extraction kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions. One microgram of genomic DNA was sonicated to produce fragments ranging from 100 to 500 bp. Then, 3.5 mg (7 mL) of MBD-Biotin Protein was added to 10 mL Dynabeads M-280 Streptavidin according to the manufacturer's instructions. The MBD-magnetic bead conjugates were washed three times and suspended in 1 volume of 13 bind/wash buffer. The capture reaction was performed by adding 1 mg sonicated DNA to the MBD-magnetic beads on a rotating mixer for 1 h at room temperature. All capture reactions were carried out in duplicate. The beads were washed three times with 13 bind/wash buffer. The methylated DNA was eluted in one of the following two ways: (1) as a single fraction with a high-salt elution buffer (2000 mM NaCl), denoted as MBD-SF; or (2) as distinct subpopulations based on the degree of methylation using an increasing NaCl concentration of the elution buffer, from 200 to 2000 mM in a stepwise gradient (elution 1, 200 mM; elution 2, 350 mM; elution 3, 450 mM; elution 4, 600 mM; elution 5, 1000 mM; and elution 6, 2000 mM). Each fraction was concentrated by ethanol precipitation using 1 mL glycogen (20 mg/mL), 1/10th volume of 3 M sodium acetate (pH 5.2), and two sample volumes of 100% ethanol and was subsequently resuspended in 60 mL H20.
Promoter array analyses
Immunoprecipitated DNA and input DNA from MeDIP immunoprecipitations and MBD-Capture reactions were amplified with a GenomePlex Single Cell Whole Genome Amplification Kit (cat. no. WGA4; Sigma, St. Louis, MO, USA), according to the manufacturer's instructions. Fifty nanograms of DNA was used in each amplification reaction. The reactions were cleaned up using cDNA cleanup columns (cat. no. 900371; Affymetrix), and 7.5 μg of amplified DNA was fragmented and labeled according to Affymetrix Chromatin Immunoprecipitation Assay Protocol P/N 702238 Rev. 3. Affymetrix GeneChip Human Promoter 1.0R arrays (P/N 900777) were hybridized using a GeneChip Hybridization wash and stain kit (P/N 900720).
Hybridization was performed with an Affymetrix GeneChip Promoter 1.0R array. The arrays were hybridized for 17 h at 45°C and 60 rpm. Arrays were subsequently washed (Affymetrix Fluidics Station 450), stained with streptavidin-phycoerythrin (GeneChip Hybridization, Wash, and Stain Kit; cat. no. 900720), and scanned on an Affymetrix GeneChip Scanner 3000.
Patients and sample collection for quantitative real-time polymerase chain reaction (qRT-PCR) analysis
A total of 192 blood samples (98 from healthy controls and 94 from patients with T2D) were collected. All individuals attended the China Medical University Hospital in Taichung, and patients fulfilled the diagnostic criteria for T2D. Total RNA was isolated from human blood using a High Pure RNA Isolation Kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. cDNA was synthesized from 1 μg total RNA using a High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA), in a 20-μL reaction volume, according to the manufacturer's instructions. cDNA was diluted to 10 ng/L, and 1 μL cDNA was used for each qRT-PCR assay in a final reaction volume of 10 μL. For quantification of gene expression with the ABI ViiA 7 Real-Time PCR System (Applied Biosystems), FastStart Universal SYBR Green Master mix (Roche) was used. Primer sequences were as follows: NR4A1 sense: 5′-TCTATGTCCTCGCCTTGGTT-3′, antisense: 5′-ATTATCCCGTCTGCCTTCAG-3′; GAPDH sense: 5′-CAGCCTCAAGATC ATCAGCA-3′, antisense: 5′-TGTGGTCATGAGTCCTTCCA-3′. This study was approved by the Human Studies Committee of China Medical University Hospital, and informed consent was obtained from either the participants or their parents.
Transfection
RIN-m5F rat pancreatic β-cells and 293T cells were purchased from Food Industry Research and Development Institute (Hsinchu, Taiwan). Cells were seeded at 150,000 cells per well in six-well culture plates and incubated until the culture reached 50-80% confluence. Cells were then transfected with either the empty pcDNA3.0 vector, pcDNA3.0-NR4A1 construct, or lentiviral expression system for NR4A1 shRNA (provided by the National RNAi Core Facility, Academia Sinica, Taiwan) using Xfect Transfection Reagent (Clontech, Palo Alto, CA, USA), according to the manufacturer's instructions. At 48 h after transfection, total protein was isolated from the cells.
Protein extraction and western blotting
The cells were homogenized in ice-cold RIPA lysis buffer (Millipore, Temecula, CA, USA) with freshly added protease inhibitor and phosphatase inhibitor (FIVEphoton, San Diego, CA, USA). The homogenate was incubated on ice for 30 min and centrifuged at 13,000 × g for 30 min at 4°C. The supernatant was used for western blotting. Proteins in a 40-μg sample of the supernatant fraction were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a 10% acrylamide resolving gels and transferred to polyvinylidene difluoride membranes. Membranes were incubated in blocking solution containing 5% nonfat dry milk and 0.1% Tween-20 in Tris-buffered saline, followed by incubation with rabbit anti-NR4A1 (1:1,000; LifeSpan BioSciences, Seattle, WA, USA), anti-DNMT1 (1:1,000; LifeSpan BioSciences), anti-insulin receptor (1:1,000; GeneTex, Irvine, CA, USA), anti-PTPRD (1:1,000; GeneTex), and anti-actin (1:5,000; GeneTex) polyclonal primary antibodies. Membranes were then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000; Jackson ImmunoResearch, West Grove, PA, USA) secondary antibodies. Proteins were visualized using SuperSignal West Pico Chemiluminescent Substrate or SuperSignal West Femto Chemiluminescent Substrate (Thermo, Rockford, IL, USA).
Animals
Four-week-old male KK and KK.Cg-Ay/J mice were obtained from Jackson Laboratories (Bar Harbor, ME, USA). Sixteen-week-old mice were treated intraperitoneally with ATA (50 mg/kg/day) for 2 weeks. Animals were housed in individual cages and given lab chow ad libitum (LabDiet 5k52; St. Louis, MO, USA). The animals were housed in a room with a constant temperature (22-25°C), relative humidity (50-70%), and photoperiod (12-h light/12-h dark). This study was approved by the Institutional Animal Care and Use Committee (IACUC) of China Medical University (IACUC: 102-217).
Immunohistochemistry
DNMT1 was performed using rabbit anti-DNMT1 antibodies (LifeSpan BioSciences) and a streptavidin-biotin-peroxidase complex for immunohistochemistry staining. Formalin-fixed paraffin-embedded tissue sections were incubated in 3% H2O2 in distilled water for 30 min at room temperature, followed by antigen retrieval by boiling the sections in 0.01 M citrate buffer for 20 min for deparaffinization and rehydration. Sections were then washed in 50 mM Tris-HCl (pH 7.6) with 0.05% Tween for 2 min. To block nonspecific binding, all sections were incubated with 5% nonfat dry milk for 30 min at room temperature. The slides were then incubated with anti-DNMT1 antibodies (1:250) for 1 h at room temperature. The reaction was stopped by rinsing the sections with 0.01 M PBS. The sections were then incubated with biotinylated anti-mouse/rabbit IgG serum (secondary antibody), followed by treatment with a peroxidase-labeled streptavidin-biotin complex and diaminobenzidine substrate to visualize the positive cells. Finally, sections were counterstained with hematoxylin, prior to being mounted for examination by light microscopy. DNMT1-positive cells per area in the mouse pancreas were counted using a Q500MC Image Analysis System (Leica, Nussloch, Germany). DNMT1-positive cells were quantified in 20 fields from the cortex and 15 fields from the medulla at a magnification of 200×.
Statistical analysis
Data were organized using Microsoft Excel software and analyzed using SPSS 15.0 (SPSS, Chicago, IL, USA) or GraphPad Prism version 3 (GraphPad Software, San Diego, CA, USA). All values were expressed as means ± standard deviations. Normality of the data was tested using the Kolmogorov-Smirnov test. Hierarchical gene analysis and heat maps were determined using a Pearson correlation matrix. Depending on the probability distribution pattern and the total number of subjects, parametric (normal distribution and ≥ 50 subjects) or nonparametric tests (skewed distribution or < 50 subjects) were used. The level of significance was set at P < 0.05 (two-tailed).
SUPPLEMENTARY MATERIALS FIGURES
Acknowledgments
This work is supported by China Medical University Hospital (DMR-105-092) in Taiwan. This study is supported by research grants from Biosignature project, Academia Sinica, Taiwan.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interests associated with this article.
Authors' contributions
YTC and FJT designed the study. YTC, JWL, and YCT collected the subjects, performed experiments, and analyzed the data. YTC and FJT wrote the first draft of the manuscript. All authors contributed to the interpretation of the data and critical revision of the manuscript. All authors read and approved the final manuscript.
REFERENCES
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Liao WL, Tsai FJ. Personalized medicine in Type 2 Diabetes. BioMedicine. 2014;4:1–8. doi: 10.7603/s40681-014-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunasekaran U, Gannon M. Type 2 diabetes and the aging pancreatic beta cell. Aging (Albany NY) 2011;3:565–575. doi: 10.18632/aging.100350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozanne SE, Sandovivi I, Constancia M. Maternal diet, aging and diabetes meet at a chromatin loop. Aging (Albany NY) 2011;3:548–554. doi: 10.18632/aging.100330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao WL, Tsai FJ. Personalized medicine in Type 2 Diabetes. BioMedicine. 2014;4:1–8. doi: 10.7603/s40681-014-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, Chang CC, Chen P, Chen CH, et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen YT, Lin WD, Liao WL, Lin YJ, Chang JG, Tsai FJ. PTPRD silencing by DNA hypermethylation decreases insulin receptor signaling and leads to type 2 diabetes. Oncotarget. 2015;6:12997–13005. doi: 10.18632/oncotarget.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PDS, Shin D, Chi NC, Shin CH, Schlegel A, Halpern M, Stainier SYR. Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Developmental Biology. 2009;334:213–223. doi: 10.1016/j.ydbio.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Luo L, Luo N, Zhu X, Garvey WT. NR4A orphan nuclear receptors modelate insulin action and the glucose transport system: potential role in insulin resistance. J Biol Chem. 2007;282:31525–31533. doi: 10.1074/jbc.M701132200. [DOI] [PubMed] [Google Scholar]
- 10.McMorrow JP, Murphy EP. Inflammation: a role for NR4A orphan nuclear receptors? Biochem Soc Trans. 2011;39:688–693. doi: 10.1042/BST0390688. [DOI] [PubMed] [Google Scholar]
- 11.Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GE. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the β-adrenergic and an orphan nuclear hormone receptor pathway. J Biol Chem. 2005;280:12573–12584. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- 13.Pearen MA, Ryall JG, Maxwell MA, Ohkura N, Lynch GS, Muscat GE. The orphan nuclear receptor, NOR-1, is a target of β-adrenergic signaling in skeletal muscle. Endocrinology. 2006;147:5217–5227. doi: 10.1210/en.2006-0447. [DOI] [PubMed] [Google Scholar]
- 14.Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 15.Chao LC, Zhang Z, Pei L, Saito T, Tontonoz P, Pilch PF. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol Endocrinol. 2007;21:2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronn T, Ling C. DNA methylation as a diagnostic and therapeutic target in the battle against type 2 diabetes. Epigenomics. 2015;7:451–160. doi: 10.2217/epi.15.7. [DOI] [PubMed] [Google Scholar]
- 17.Li QX, Ke N, Sundaram R, Wong-Staal F. NR4A1, 2, 3—an orphan nuclear hormone receptor family involved in cell apoptosis and carcinogenesis. Histol Histopathol. 2006;21:533–540. doi: 10.14670/HH-21.533. [DOI] [PubMed] [Google Scholar]
- 18.Tessem JS, Moss LG, Chao LC, Arlotto M, Lu D, Jensen MV, Stephens SB, Tontonoz P, Hohmeier HE, Newgard CB. Nkx6. 1 regulates islet β-cell proliferation via Nr4a1 and Nr4a3 nuclear receptors. Proc Natl Acad Sci U S A. 2014 Apr 8;111:5242–7. doi: 10.1073/pnas.1320953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao LC, Worblewski K, Zhang Z, Pei L, Vergnes L, Ilkayeva OR, Ding SY, Reue K, Watt MJ, Newgard CB, Pilch PF, Hevener AL, Tontonoz P. Insulin resistance and altered systemic glucose metabolism in mice lacking nur77. Diabetes. 2009;58:2788–2796. doi: 10.2337/db09-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bestor TH, Ingram VM. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 22.Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 23.Iwatsuka H, Shino A, Suzuoki Z. General survey of diabetic features of yellow KK mice. Endocrinol Jpn. 1970;17:23–35. doi: 10.1507/endocrj1954.17.23. [DOI] [PubMed] [Google Scholar]
- 24.Castle CK, Colca JR, Melchior GW. Lipoprotein profile characterization of the KKA(y) mouse, a rodent model of type II diabetes, before and after treatment with the insulin-sensitizing agent pioglitazone. Arterioscler Thromb. 1993;13:302–309. doi: 10.1161/01.atv.13.2.302. [DOI] [PubMed] [Google Scholar]
- 25.Yoo J, Kim JH, Robertson KD, Medina-Franco JL. Molecular modeling of inhibitors of human DNA methyltransferase with a crystal structure: discovery of a novel DNMT1 inhibitor. Adv Protein Chem Struct Biol. 2012;87:219–247. doi: 10.1016/B978-0-12-398312-1.00008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gayet O, Loncle C, Duconseil P, Gilabert M, Lopez MB, Moutardier V, Turrini O, Valvo E, Ewald J, Giovannini M, Gasmi M, Bories E, Barthet M, Ouaissi M, et al. A subgroup of pancreatic adenocarcinoma is sensitive to the 5-aza-dC DNA methyltransferase inhibitor. Oncotarget. 2015;6:746–754. doi: 10.18632/oncotarget.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


