Abstract
Matrix metalloproteinase 14 (MMP-14), a membrane-anchored MMP that promotes the tumorigenesis and aggressiveness, is highly expressed in gastric cancer. However, the transcriptional regulators of MMP-14 expression in gastric cancer still remain largely unknown. In this study, through mining computational algorithm programs and chromatin immunoprecipitation datasets, we identified adjacent binding sites of myeloid zinc finger 1 (MZF1) and miRNA-337-3p (miR-337-3p) within the MMP-14 promoter. We demonstrated that MZF1 directly bound to the MMP-14 promoter to facilitate its nascent transcription and expression in gastric cancer cell lines. In contrast, endogenous miR-337-3p suppressed the MMP-14 expression through recognizing its binding site within MMP-14 promoter. Mechanistically, miR-337-3p repressed the binding of MZF1 to MMP-14 promoter via recruiting Argonaute 2 and inducing repressive chromatin remodeling. Gain- and loss-of-function studies demonstrated that miR-337-3p suppressed the growth, invasion, metastasis, and angiogenesis of gastric cancer cells in vitro and in vivo through repressing MZF1-facilitated MMP-14 expression. In clinical specimens and cell lines of gastric cancer, MZF1 was highly expressed and positively correlated with MMP-14 expression. Meanwhile, miR-337-3p was under-expressed and inversely correlated with MMP-14 levels. miR-337-3p was an independent prognostic factor for favorable outcome of gastric cancer, and patients with high MZF1 or MMP-14 expression had lower survival probability. Taken together, these data indicate that miR-337-3p directly binds to the MMP-14 promoter to repress MZF1-facilitatd MMP-14 expression, thus suppressing the progression of gastric cancer.
Keywords: gastric cancer, microRNA-337-3p, myeloid zinc finger 1, matrix metalloproteinase 14, transcriptional regulation
INTRODUCTION
Gastric cancer is one of the most common cancers worldwide, with approximately 1 million new cases being diagnosed annually [1]. The prognosis of advanced gastric cancer remains poor, mainly due to tumor recurrence, invasion and metastasis, with a 5-year survival rate below 30% [1]. Better elucidating the mechanisms of tumorigenesis and aggressiveness is important for improving the therapeutic efficiency of gastric cancer [2]. Matrix metalloproteinase 14 (MMP-14), a membrane-anchored MMP, is not only able to degrade structural extracellular matrix (ECM) components such as fibronectin, vitronectin, laminin-1, laminin-5, fibrin, and collagen, but also capable of activating MMP-2, thereby promoting tumor invasion and metastasis [3]. In addition, MMP-14 contributes to pro-angiogenic responses through facilitating the expression of vascular epidermal growth factor (VEGF) and releasing bioactive ECM products [4]. High MMP-14 expression has been documented in most types of human malignancies, including colon cancer [5], breast cancer [6], ovary cancer [7], and skin cancer [8], and its elevated expression is associated with tumor invasion and metastasis [9]. Previous studies have shown that MMP-14 is highly expressed in gastric cancer, and is correlated with poor outcome of patients [10], suggesting the importance of MMP-14 in the progression of gastric cancer.
Human MMP-14 gene is localized at chromosome 14q11 and mainly regulated at the transcription level [11]. Transcription factors specificity protein 1, hypoxia-inducible factor 2 alpha, and Krüppel-like factor 8 have been identified as potent regulators of MMP-14 expression in prostate cancer, renal cell carcinoma, and breast cancer [11–13]. In ovarian cancer cells, polyomavirus enhancer activator 3 (PEA3) is able to induce MMP-14 expression via direct binding to its promoter, and knockdown of PEA3 reduces the MMP-14 levels [14]. In addition, hepatocyte nuclear factor 4 alpha exhibits oncogenic activity through directly binding to the MMP-14 promoter and facilitating its transcription in neuroblastoma [15]. However, the transcriptional regulators and underlying mechanisms essential for MMP-14 expression in gastric cancer are limitedly identified.
In the current study, through mining computational algorithm programs and chromatin immunoprecipitation (ChIP) datasets, we identified adjacent binding sites of myeloid zinc finger 1 (MZF1) and miRNA-337-3p (miR-337-3p) within the MMP-14 promoter. We demonstrate, for the first time, that MZF1 is highly expressed and facilitates the transcription of MMP-14 in gastric cancer. Meanwhile, miR-337-3p is under-expressed and anti-correlated with MMP-14 expression in clinical gastric cancer specimens. In addition, miR-337-3p directly binds to the MMP-14 promoter to suppress its transcription via inducing chromatin remodeling and repressing MZF1 enrichment, thus inhibiting the growth, invasion, metastasis, and angiogenesis of gastric cancer cells in vitro and in vivo, suggesting the tumor suppressive functions of miR-337-3p in the progression of gastric cancer.
RESULTS
MZF1 facilitates the expression of MMP-14 in gastric cancer cell lines
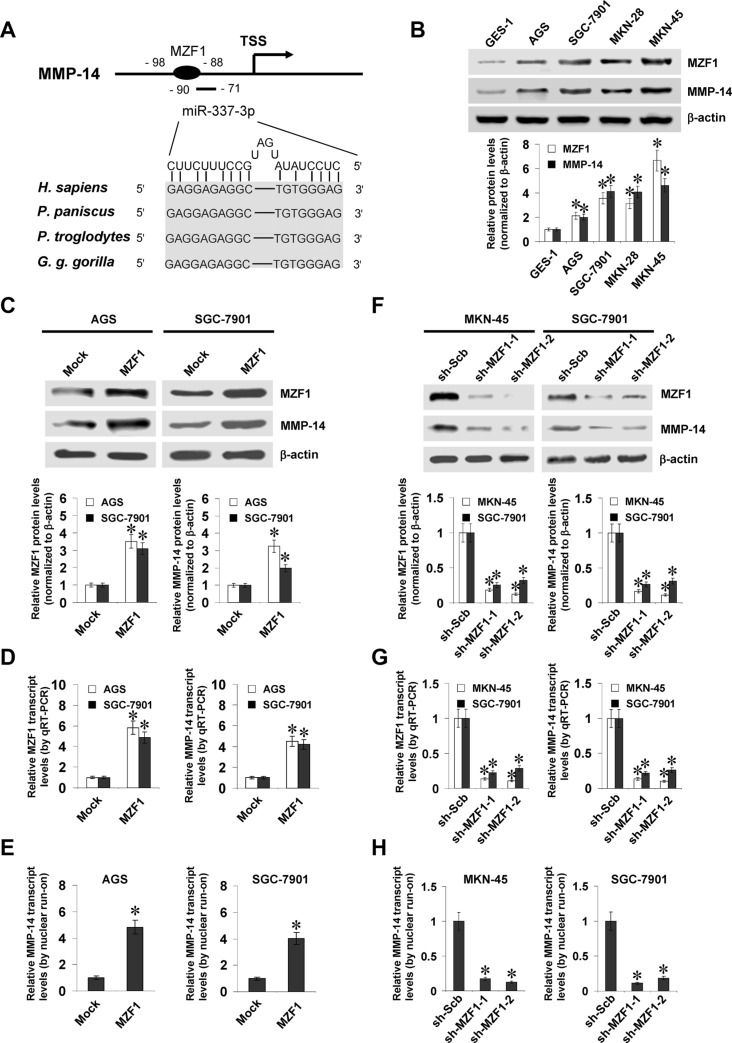
To investigate the mechanisms crucial for MMP-14 expression in gastric cancer, we first analyzed the activity of a series of MMP-14 promoter fragments. Dual-luciferase assay indicated that −384/−95 bp relative to the transcription start site (TSS) was essential for the MMP-14 promoter activity in cultured MKN-45 and SGC-7901 cells (Supplementary Figure S1A). Over-lapping analysis of computational algorithm programs Genomatrix [16], TFBIND [17], and PROMO [18] revealed the potential binding sites of MZF1, nuclear factor Y (NFY), and nuclear factor erythroid-2 related factor 2 (NRF2) within this region (chr14:23305444-23305733; Supplementary Figure S1B), locating at bases −98/−88, −158/−144, and −179/−159 relative to MMP-14 TSS, respectively. Mining of publicly available ChIP-seq dataset [19] indicated the enrichment of MZF1, but not of NFY or NRF2, on MMP-14 promoter region (Supplementary Figure S1B). Further analysis of Gene Expression Omnibus (GEO) datasets indicated the positive correlation between MZF1 and MMP-14 levels in different gastric cancer cohorts (Supplementary Figure S1B and S1C). In addition, the binding site of miR-337-3p with high complementarity was noted at −90/−71 bp region adjacent to that of MZF1 (Figure 1A). Higher MZF1 and MMP-14 levels were observed in gastric cancer cell lines than those in normal gastric epithelial cells (Figure 1B).
Figure 1. MZF1 facilitates the expression of MMP-14 in gastric cancer cells.
(A), scheme of potential binding sites of MZF1 and miR-337-3p within the MMP-14 promoter, locating at bases −98/−88 and −90/−71 relative to TSS. (B), western blot showing the expression of MZF1 and MMP-14 in normal gastric epithelial GES-1 cells and gastric cancer cell lines (AGS, SGC-7901, MKN-28, and MKN-45). (C and D), western blot and real-time quantitative RT-PCR indicating the expression of MZF1 and MMP-14 in AGS and SGC-7901 cells stably transfected with empty vector (mock) or MZF1. (E) nuclear run-on assay showing the nascent MMP-14 transcript levels in gastric cancer cells stably transfected with mock or MZF1. (F and G), western blot and real-time quantitative RT-PCR indicating the MZF1 and MMP-14 levels in MKN-45 and SGC-7901 cells stably transfected with scramble shRNA (sh-Scb) or MZF1 shRNA (sh-MZF1). (H), nuclear run-on assay showing the nascent MMP-14 transcript levels in gastric cancer cells transfected with sh-Scb or sh-MZF1. *P < 0.01 vs. GES-1, mock, or sh-Scb.
To address the hypothesis that MZF1 may influence the MMP-14 expression in gastric cancer cell lines, we performed the MZF1 over-expression and knockdown experiments. Western blot and real-time quantitative RT-PCR demonstrated that stable transfection of MZF1 into AGS and SGC-7901 cells obviously increased the protein and transcript levels of MZF1 and MMP-14, than those stably transfected with empty vector (mock; Figure 1C and 1D). Nuclear run-on assay demonstrated that stable over-expression of MZF1 increased the nascent transcript levels of MMP-14 in gastric cancer cells, than those in mock cells (Figure 1E). On the other hand, short hairpin RNA (shRNA) specific for MZF1 (sh-MZF1) was stably transfected into MKN-45 and SGC-7901 cells, resulting in decreased protein and transcript levels of MZF1 and MMP-14, when compared to those stably transfected with scramble shRNA (sh-Scb) (Figure 1F, 1G, and 1H). These results demonstrated that MZF1 considerably facilitated the MMP-14 expression at transcriptional levels in gastric cancer cells.
MZF1 increases the transcription of MMP-14 through direct binding to its promoter
To determine whether MZF1 could bind to the MMP-14 promoter to increase its transcription, the MMP-14 promoter luciferase reporter and its mutation vectors were transfected into gastric cancer cells (Figure 2A). Dual-luciferase assay indicated that ectopic expression or knockdown of MZF1 enhanced and attenuated the promoter activity of MMP-14 in gastric cancer cells, respectively (Figure 2B and 2C), and mutation of MZF1 binding site abolished these effects (Figure 2B and 2C). In addition, ChIP and real-time quantitative PCR (qPCR) were applied to measure the enrichment of MZF1 on MMP-14 promoter with primer sets spanning its binding sites. In cultured AGS and SGC-7901 cells, enrichment of MZF1 was observed around its binding site (−122/+69 bp relative to TSS) (Figure 2D). As controls, no MMP-14 promoter regions were immunoprecipitated with unspecific antibody (isotype IgG) or detected by qPCR with primer set (−326/−157 bp) distal to the binding site of MZF1 (Figure 2D). Stable transfection of MZF1 into gastric cancer cells resulted in enrichment of MZF1 on the MMP-14 promoter (Figure 2D). Meanwhile, stable knockdown of MZF1 with shRNA construct decreased the binding of MZF1 to MMP-14 promoter in MKN-45 and SGC-7901 cells (Figure 2D). These results indicated that MZF1 directly bound to the promoter of MMP-14 to increase its transcription in gastric cancer cells.
Figure 2. MZF1 increases the transcription of MMP-14 through direct binding to its promoter.
(A), scheme and sequence of the intact MZF1 binding site (WT) and its mutation (Mut) within the MMP-14 promoter-luciferase reporter vectors. (B), dual-luciferase assay showing the activity of MMP-14 promoter and its mutant in AGS and SGC-7901 cells stably transfected with empty vector (mock) or MZF1. (C), dual-luciferase assay indicating the activity of MMP-14 promoter and its mutant in MKN-45 and SGC-7901 cells stably transfected with scramble shRNA (sh-Scb) or MZF1 shRNA (sh-MZF1). (D), ChIP and qPCR assay (normalized to input DNA) showing the enrichment of MZF1 on the MMP-14 promoter in AGS, SGC-7901, and MKN-45 cells transfected with mock, MZF1, sh-Scb, or sh-MZF1. *P < 0.01 vs. mock, sh-Scb, or IgG.
miR-337-3p recruits AGO2 to repress the transcription of MMP-14
Since our previous studies have shown the functions of miR-337-3p in repressing MMP-14 transcription [20], we observed the effects of miR-337-3p on MMP-14 expression in gastric cancer cells. Lower miR-337-3p expression was observed in gastric cancer cell lines than those in normal gastric epithelial cells (Figure 3A). Stable transfection of miR-337-3p precursor into MKN-45 and SGC-7901 cells increased the miR-337-3p levels (Figure 3B). Meanwhile, transfection of anti-miR-337-3p inhibitor obviously decreased the miR-337-3p levels in AGS and SGC-7901 cells (Figure 3C). Western blot, real-time quantitative RT-PCR, and dual-luciferase assays demonstrated that over-expression or knockdown of miR-337-3p decreased and increased the protein and transcript levels of MMP-14 in gastric cancer cells, than those transfected with empty vector (mock) or negative control (anti-NC) inhibitor, respectively (Figure 3D–3G, Supplementary Figure S2A and S2B). The expression of VEGF, the MMP-14 downstream gene in gastric cancer [21], was significantly decreased or increased in miR-337-3p over-expressing and knockdown gastric cancer cells, consistent with the MMP-14 levels (Figure 3D–3F, Supplementary Figure S2A and S2B). The analysis of microPIR database revealed no potential binding site of miR-337-3p within the VEGF promoter, ruling out the possibility that miR-337-3p may directly suppress the transcription of VEGF. Since previous studies have revealed the potential involvement of AGO2 in miR-337-3p-induced transcriptional repression [20], shRNAs against AGO2 were transfected into MKN-45 and SGC-7901 cells. Western blot, real-time quantitative RT-PCR, and dual-luciferase assays demonstrated that knockdown of AGO2 abolished the miR-337-3p-induced transcriptional repression of MMP-14 in gastric cancer cells (Figure 3H, 3I, Supplementary Figure S2C, and S2D). The MMP-14 promoter region was detected for enrichment of AGO2 by ChIP and real-time qPCR. In cultured AGS and SGC-7901 cells, enrichment of AGO2 was observed around the binding site of miR-337-3p (Figure 3J). As controls, no MMP-14 promoter regions were immunoprecipitated with unspecific antibody (isotype IgG) (Figure 3J). In addition, RNase H treatment, but not RNase A treatment, abolished the enrichment of AGO2 on the MMP-14 promoter in gastric cancer cells (Figure 3J). Stable transfection of miR-337-3p precursor into gastric cancer cells resulted in increased binding of AGO2 and epigenetic markers histone H3 lysine 9 dimethylation (H3K9me2) and histone H3 lysine 27 trimethylation (H3K27me3), and decreased binding of histone H3 lysine 4 trimethylation (H3K4me3) and MZF1 on MMP-14 promoter (Figure 3K), which were abolished by knockdown of AGO2 (Figure 3K). Collectively, these results indicated that miR-337-3p recognized the binding site and recruited AGO2 to repress the MMP-14 transcription in gastric cancer cells.
Figure 3. miR-337-3p recruits AGO2 to repress the transcription of MMP-14.
(A), real-time quantitative RT-PCR assay showing the miR-337-3p levels in normal gastric epithelial GES-1 cells and gastric cancer cell lines (AGS, SGC-7901, MKN-28, and MKN-45). (B and C), real-time quantitative RT-PCR assay indicating the miR-337-3p levels in gastric cancer cells transfected with empty vector (mock), miR-337-3p precursor, negative control inhibitor (anti-NC, 100 nmol/L), or anti-miR-337-3p inhibitor (100 nmol/L). (D and E), western blot and dual-luciferase assays showing the expression of MMP-14 and VEGF and promoter activity of MMP-14 in MKN-45 and SGC-7901 cells stably transfected with mock or miR-337-3p precursor. (F and G), western blot and dual-luciferase assays indicating the expression of MMP-14 and VEGF and promoter activity of MMP-14 in AGS and SGC-7901 cells transfected with anti-NC or anti-miR-337-3p inhibitor (100 nmol/L). (H and I), western blot and dual-luciferase assays showing the expression of AGO2 and MMP-14 and promoter activity of MMP-14 in MKN-45 and SGC-7901 cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with scramble shRNA (sh-Scb) or AGO2 shRNA (sh-AGO2). (J) ChIP and qPCR assay indicating the binding of AGO2 to MMP-14 promoter in gastric cancer cells treated with RNase H or RNase A. (K) ChIP and qPCR assay showing the enrichment of AGO2, H3K9me2, H3K27me3, H3K4me3, and MZF1 on MMP-14 promoter in MKN-45 and SGC-7901 cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with sh-Scb or sh-AGO2. *P < 0.01 vs. GES-1, mock, anti-NC, mock + sh-Scb, or IgG.
miR-337-3p suppresses the growth, invasion, and angiogenesis of gastric cancer cells through repressing MZF1-facilitated MMP-14 expression in vitro
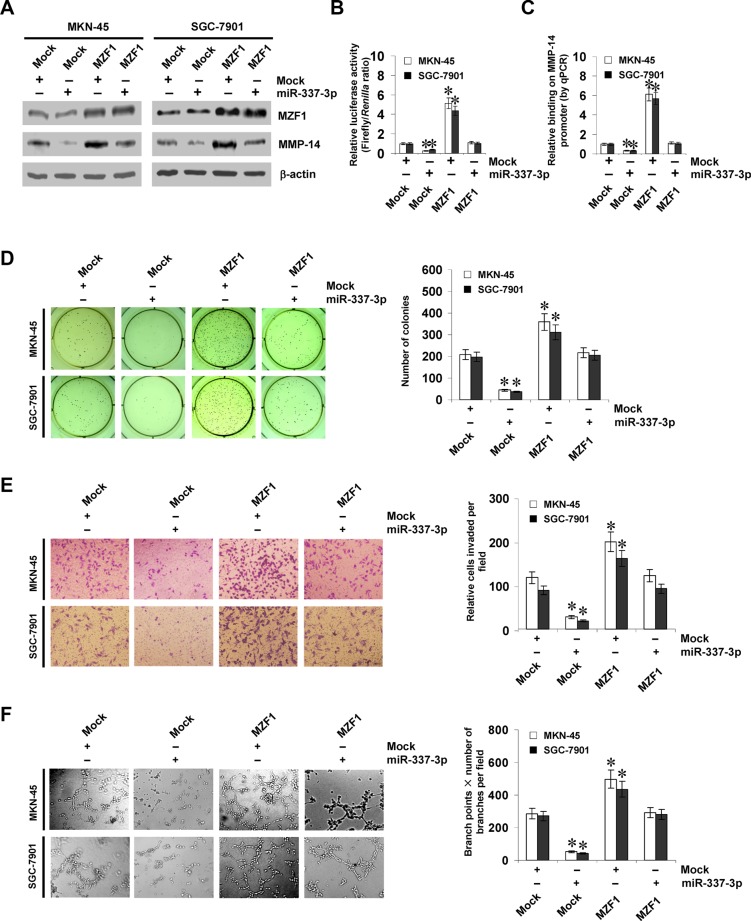
Since above evidence indicated that miR-337-3p attenuated the binding of MZF1 to MMP-14 promoter, we further investigated the effects of miR-337-3p over-expression on MZF1-facilitated MMP-14 expression in gastric cancer cells. Western blot, real-time quantitative RT-PCR, and dual-luciferase assays indicated that ectopic expression of miR-337-3p abolished the enhanced protein and transcript levels of MMP-14 induced by stable transfection of MZF1 (Figure 4A, 4B, and Supplementary Figure S2E). In addition, over-expression of miR-337-3p prevented the enhanced MZF1 enrichment on MMP-14 promoter induced by ectopic expression of MZF1 (Figure 4C). In soft agar assay, MZF1 over-expression promoted the anchorage-independent growth of MKN-45 and SGC-7901 cells, when compared to those stably transfected with empty vector (mock; Figure 4D). In matrigel invasion assay, gastric cancer cells stably transfected with MZF1 presented an increased invasion capacity than mock cells (Figure 4E). The tube formation of endothelial cells was increased by treatment with the medium preconditioned by stable transfection of gastric cancer cells with MZF1 (Figure 4F). Moreover, transfection of miR-337-3p rescued the MKN-45 and SGC-7901 cells from increased growth, invasion, and angiogenesis capability induced by stable over-expression of MZF1 (Figure 4D–4F). On the other hand, we examined the effects of MZF1 knockdown on gastric cancer cells. Stable transfection of sh-MZF1 decreased the transcript and protein levels of MMP-14 (Supplementary Figure S2F, S2G, and S3A), and attenuated the capability of growth (Supplementary Figure S3B and S3C), invasion (Supplementary Figure S3D), and angiogenesis in AGS and SGC-7901 cells (Supplementary Figure S3E). Down-regulation of miR-337-3p via transfection of anti-miR-337-3p inhibitor rescued the AGS and SGC-7901 cells from their changes in these biological features induced by knockdown of MZF1 (Supplementary Figure S3B–S3E). These results indicated that miR-337-3p significantly decreased the growth, invasion, and angiogenesis of gastric cancer cells through repressing MZF1-facilitated MMP-14 expression in vitro.
Figure 4. miR-337-3p suppresses the growth, invasion, and angiogenesis of gastric cancer cells through repressing MZF1-facilitated MMP-14 expression in vitro.
(A and B), western blot and dual-luciferase assays showing the expression of MZF1 and MMP-14 and promoter activity of MMP-14 in MKN-45 and SGC-7901 cells stably transfected with empty vector (mock) or miR-337-3p precursor, and those co-transfected with MZF1. (C), ChIP and qPCR assay indicating the enrichment of MZF1 on MMP-14 promoter in gastric cancer cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1. (D), representation (left) and quantification (right) of soft agar assay showing the anchorage-independent growth of MKN-45 and SGC-7901 cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1. (E), representation (left) and quantification (right) of matrigel invasion assay indicating the invasion capability of gastric cancer cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1. (F), representation (left) and quantification (right) of tube formation assay showing the angiogenic capability of gastric cancer cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1. *P < 0.01 vs. mock.
miR-337-3p attenuates the growth, metastasis, and angiogenesis of gastric cancer cells through repressing MZF1-facilitated MMP-14 expression in vivo
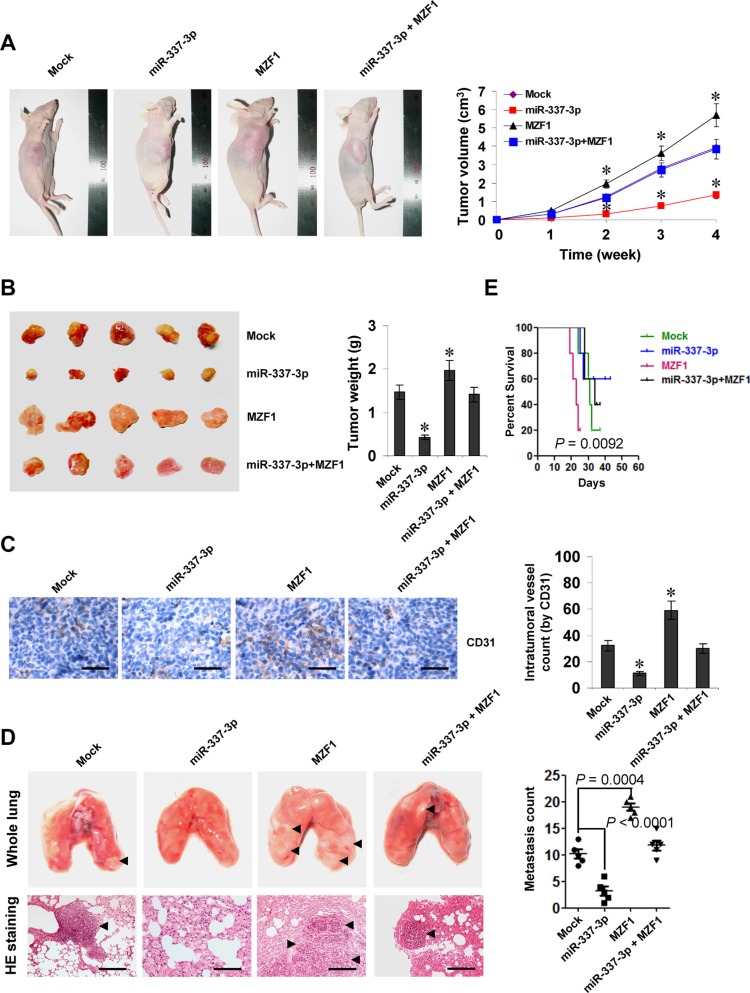
We next investigated the efficacy of miR-337-3p against MZF1-facilitated tumor growth and metastasis in vivo. Stable transfection of MZF1 into SGC-7901 cells resulted in increased growth and tumor weight of subcutaneous xenograft tumors in athymic nude mice, when compared to those stably transfected with empty vector (mock) (Figure 5A, 5B). In addition, stable transfection of MZF1 resulted in increase in CD31-positive microvessels and mean vessel density within tumors (Figure 5C). In the experimental metastasis studies, SGC-7901 cells stably transfected with MZF1 established statistically more lung metastatic colonies than mock group (Figure 5D), and resulted in lower survival probability of nude mice (Figure 5E). Moreover, stable transfection of miR-337-3p precursor into SGC-7901 cells prevented the MZF1-facilitated growth, metastasis and angiogenesis in athymic nude mice (Figure 5A–5E). These results suggested that miR-337-3p could attenuate the MZF1-facilitated growth, metastasis, and angiogenesis of gastric cancer cells in vivo.
Figure 5. miR-337-3p attenuates the growth, metastasis, and angiogenesis of gastric cancer cells through repressing MZF1-facilitated MMP-14 expression in vivo.
(A), tumor growth curve of SGC-7901 cells (1 × 106) stably transfected with empty vector (mock) or miR-337-3p precursor, and those co-transfected with MZF1 in athymic nude mice (n = 5 for each group), after hypodermic injection for 4 weeks. (B), representation (left) and quantification (right) of xenograft tumors formed by hypodermic injection of SGC-7901 cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1. (C), immunohistochemical staining (left) and quantification (right) of CD31 expression within tumors formed by hypodermic injection of SGC-7901 cells stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1. Scale bars: 100 mm. (D), representation (left, arrowhead) and quantification (right) of lung metastasis after injection of SGC-7901 cells (0.4 × 106) stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1 into the tail vein of athymic nude mice (n = 5 for each group). Scale bars: 100 mm. (E), Kaplan–Meier survival plots of nude mice with injection of SGC-7901 cells (0.4 × 106) stably transfected with mock or miR-337-3p precursor, and those co-transfected with MZF1 via the tail vein (n = 5 for each group). *P < 0.001 vs. mock.
MZF1 and miR-337-3p are positively or inversely correlated with MMP-14 levels in gastric cancer tissues
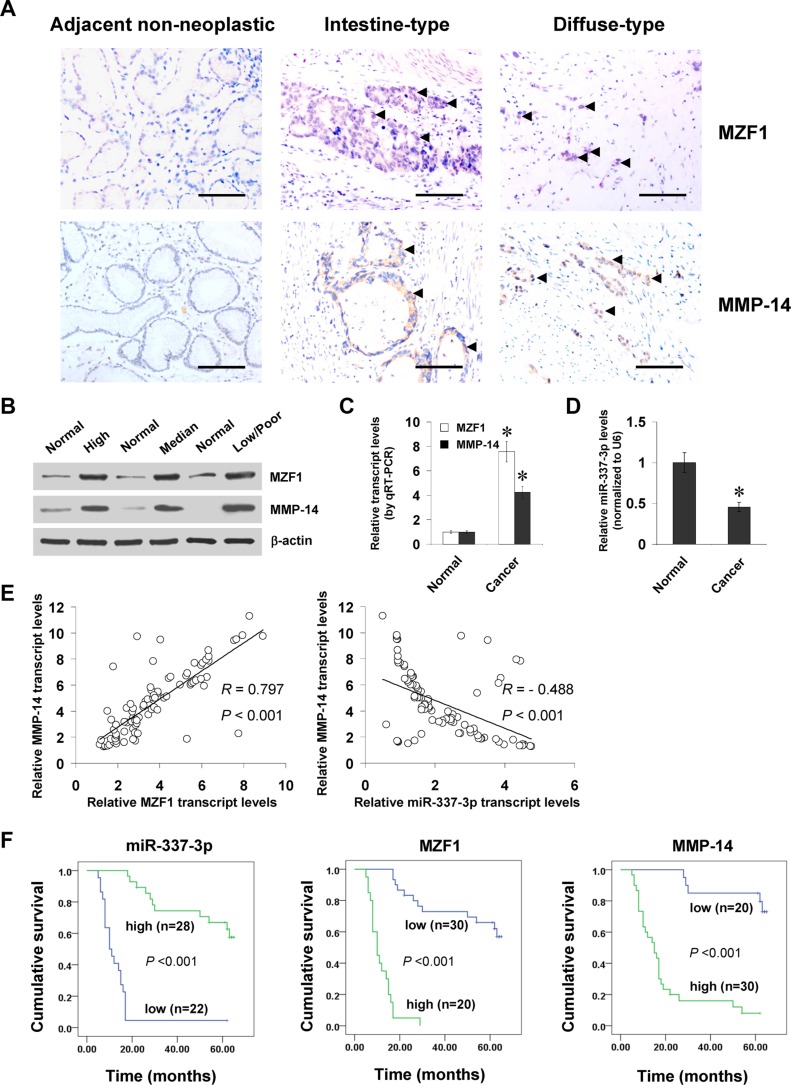
To investigate the expression of MZF1 in gastric cancer tissues, paraffin-embedded sections from 50 well-established primary cases were collected. Immunohistochemical staining revealed that MZF1 was expressed in the nuclei of cancer cells (Figure 6A), and was detected in 30/50 (60.0%) cases, with weak staining in 10, moderate in 8, and intense in 12 (Supplementary Table S1). The MZF1 immunoreactivity was significantly higher in gastric cancer cases with deeper gastric wall invasion (P < 0.001), lymph node metastasis (P < 0.001), distant metastasis (P = 0.029), and advanced tumor-node-metastasis (TNM) stage (P < 0.001) (Supplementary Table S1). Notably, the immunostaining of MZF1 was associated with MMP-14 immunoreactivity in gastric cancer cases (correlation coefficient R = 0.500, P < 0.001, Figure 6A and Supplementary Table S2). Western blot and real-time quantitative RT-PCR were applied to measure the expression levels of MZF1, MMP-14, and miR-337-3p in 90 gastric cancer specimens and normal gastric mucosa. As shown in Figure 6B and 6C, higher protein and transcript levels of MZF1 or MMP-14 were observed in gastric cancer tissues than those in normal gastric mucosa, which was in line with the results from public datasets (Supplementary Figure S4A and S4B). In contrast, miR-337-3p was under-expressed in gastric cancer tissues than in normal gastric mucosa (Figure 6D and Supplementary Figure S4C). Notably, there was a positive correlation between MZF1 and MMP-14 transcript levels in gastric cancer tissues (correlation coefficient R = 0.797, P < 0.001, Figure 6E). Meanwhile, the miR-337-3p expression was inversely correlated with MMP-14 transcript levels in gastric cancer tissues (R = − 0.488, P < 0.001, Figure 6E). Kaplan–Meier survival analysis revealed that patients with high miR-337-3p (P < 0.001) levels or low MZF1 (P < 0.001) and MMP-14 (P < 0.001) expression had greater survival probability than those with low or high expression, respectively (Figure 6F). Significant difference was noted among the survival curves of gastric cancer patients with low or high expression of miR-337-3p, MZF1, or MMP-14 (Supplementary Table S3). Cox regression analysis of these gastric cancer cases indicated that the distant metastasis (hazard ratio HR = 2.052, P = 0.002), TNM stage (HR = 2.722, P = 0.005), MZF1 expression (HR = 3.516, P = 0.018), miR-337-3p levels (HR = 0.393, P = 0.015), and MMP-14 expression (HR = 2.319, P = 0.028), but not patient's age (HR = 1.017, P = 0.765), gender (HR = 1.397, P = 0.675), tumor size (HR = 1.807, P = 0.467), Laurén classification (HR = 1.596, P = 0.545), tumor invasion (HR = 2.218, P = 0.332), or lymph node metastasis (HR = 2.018, P = .431), were independent prognostic factors for gastric cancer patients (Supplementary Table S4). These results indicated the up-regulation of MZF1 and under-expression of miR-337-3p in gastric cancer tissues, which was positively and inversely correlated with the MMP-14 levels, respectively.
Figure 6. MZF1 and miR-337-3p are positively or inversely correlated with MMP-14 levels in gastric cancer tissues.
(A), immunohistochemical staining showing the expression of MZF1 and MMP-14 in the tumor cells of gastric cancer specimens (arrowheads, brown). Scale bars: 100 mm. (B), western blot assay indicating the protein levels of MZF1 and MMP-14 in gastric cancer tissues with different differentiation, and those in normal gastric mucosa. (C and D), real-time quantitative RT-PCR showing the transcript levels of MZF1, MMP-14, and miR-337-3p in normal gastric mucosa (n = 90) and gastric cancer tissues (n = 90). (E), the correlation between MZF1 or miR-337-3p expression and MMP-14 transcript levels in gastric cancer tissues (n = 90). (F), Kaplan–Meier survival plots of 50 well-defined gastric cancer cases with high or low expression of miR-337-3p, MZF1 or MMP-14. *P < 0.01 vs. normal.
DISCUSSION
MZF-1, a transcription factor of Krüppel family proteins, was originally cloned from the cDNA library of chronic myeloid leukemia [22]. Recent evidence shows that MZF1 exerts oncogenic or tumor suppressive functions in different cell contexts. Over-expression of MZF1 aggressively induces the transformation of NIH3T3 cells [23], and increases the migration and invasion of colorectal cancer cells [24], indicating the oncogenic functions of MZF1 in tumor progression. In contrast, MZF1 may also act as a tumor suppressor in the tumorigenesis. MZF1 inhibits the migratory and invasive capability of cervical cancer cells through reducing MMP-2 expression [25]. Knockout of MZF1 increases the proliferative potentials of hemopoietic progenitors and results in lethal neoplasia in mice [26]. In this study, we demonstrated that MZF1 is highly expressed in gastric cancer, and is an independent prognostic factor for unfavorable outcome of patients. We found that knockdown of MZF1 suppressed the growth, invasion, and angiogenesis of gastric cancer cells, suggesting the oncogenic functions of MZF1 during the progression of gastric cancer.
Human MZF1 gene encodes a 485-amino acid protein that activates or suppresses gene transcription depending on the cellular environment [27]. MZF1 directly binds to the p55PIK promoter to activate its expression, and acts as a growth accelerator in colorectal cancer cells [24]. In lung adenocarcinoma cells, liver kinase B1 loss-induced MZF1 expression promotes the transcription of c-Myc, and is responsible for the growth, migration and invasion of cancer cells [28]. On the other hand, MZF1 directly binds to the promoters of CD34 and c-Myb to suppress their transcription in hematopoietic cells [29]. However, the downstream genes of MZF-1 in gastric cancer still remain unknown. In this study, we demonstrated that MMP-14 was a novel target gene of MZF1 in gastric cancer. First, the expression of MZF1 and MMP-14 was positively correlated in gastric cancer tissues and cell lines. Second, the activity of MMP-14 promoter luciferase reporter was responsive to MZF1 over-expression and knockdown. Third, mutation of MZF1 binding site abolished the regulatory effects of MZF1 on the MMP-14 promoter luciferase reporter. Fourthly, ChIP and qPCR assay indicated the binding of MZF1 to MMP-14 promoter. Finally, endogenous MMP-14 expression, both transcript and protein, was increased or decreased by over-expression and knockdown of MZF1 in gastric cancer cells, suggesting that MZF1 may facilitate the MMP-14 expression by activating transcription.
Canonically, microRNAs (miRNAs), a class of small non-coding RNAs with 22 to 25 nucleotides in length, inhibit gene expression at the post-transcriptional levels [30]. Recent evidence shows that endogenous miRNAs are able to recognize complementary genomic sites within human gene promoters, and participate in the heterochromatin formation and regulation of gene transcription [31, 32]. For example, let-7 forms complex with AGO2 to repress the transcription of retinoblastoma 1/E2F downstream genes in senescence [33]. miR-709 is able to form epigenetic silencing complexes with H3K27me3 and AGO1, and represses the transcription of early growth response 2 [34]. In addition, miR-10a binds to the homologous region within homeobox D4 promoter to repress its expression at the transcriptional levels [35]. It has been indicated that miR-337-3p is under-expressed and associated with the lymph node metastasis of gastric cancer [36], and is able to directly bind to the MMP-14 promoter to repress its transcription in neuroblastoma [20]. However, the functions and downstream gene of miR-337-3p in gastric cancer still remain to be elucidated. In this study, we noted the adjacent binding sites of miR-337-3p and MZF1 within MMP-14 promoter. Our findings showed that miR-337-3p was an independent prognostic factor for favorable outcome of gastric cancer, and suppressed the expression of MMP-14 through repressing the enrichment of MZF1 on MMP-14 promoter in gastric cancer cells. The fact that over-expression of miR-337-3p was sufficient to prevent the gastric cancer cells from MZF1-facilitated biological behaviors indicates that miR-337-3p exerts its tumor suppressive functions, at least in part, through repressing the MZF1 activity in gastric cancer.
Since AGO2 is essential for miRNA-directed implementation of silent-state chromatin modification at gene promoters [33], we further observed the functions of AGO2 in miR-337-3p-repressed MMP-14 expression in gastric cancer. We found that AGO2 was enriched surrounding the binding site of miR-337-3p within MMP-14 promoter in gastric cancer cells. In addition, RNase H treatment (specifically degrades the RNA present in RNA-DNA hybrid) [32] attenuated the miR-337-3p-induced enrichment of AGO2, indicating the direct binding of miR-337-3p to MMP-14 promoter. Knockdown of AGO2 abolished the miR-337-3p-induced binding of repressive epigenetic markers, accompanying by increased MZF1 enrichment on MMP-14 promoter. We believe that miR-337-3p/AGO2 complexes may bring in co-repressors such as histone methyltransferases to repress the binding of transcription factor MZF1, which warrants further investigation.
In summary, we have shown that MZF1 is highly expressed and directly binds to the MMP-14 promoter to facilitate its transcription in gastric cancer. Meanwhile, miR-337-3p is under-expressed in human gastric cancer, and suppresses the transcription of MMP-14 via epigenetically suppressing the binding of MZF1 to its promoter, resulting in decreased growth, invasion, metastasis, and angiogenesis of gastric cancer cells in vitro and in vivo. Although MZF1 and miR-337-3p are found to be independent prognostic factors, a larger series of clinical specimens are needed to further explore the cooperative effects of high MZF1 expression and low miR-337-3p levels on the outcome of gastric cancer patients. This study extends our knowledge about the regulation of MMP-14 at transcriptional level, and suggests that MZF1 and miR-337-3p may be of potential values as novel therapeutic targets for human gastric cancer.
MATERIALS AND METHODS
Cell culture
Human gastric cancer cell lines AGS (CRL-1739), SGC-7901, MKN-28 and MKN-45, normal gastric epithelial GES-1 cells, and human endothelial cell line HUVEC (CRL-1730) were obtained from the American Type Culture Collection (Rockville, MD) and Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cell lines were authenticated by the provider, used within 6 months after resuscitation of frozen aliquots, and grown in RPMI1640 medium (Life Technologies, Inc., Gaithersburg, MD) supplemented with 10% fetal bovine serum (Life Technologies, Inc.), penicillin (100 U/ml), and streptomycin (100 mg/ml). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 and applied for transfection.
Gene over-expression and knockdown
Human MZF1 expression vector was kindly provided by Dr. Jiawei Zhou (Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China) [37]. The oligonucleotides encoding shRNAs specific for MZF1 and AGO2 (Supplementary Table S5) were subcloned into the Bam H I and Hind III restrictive sites of GV102 (Genechem Co., Ltd, Shanghai, China). The MZF1 or MZF1 shRNA vectors were transfected into cancer cells with Lipofectamine 2000 (Life Technologies, Inc.), and stable cell lines were screened by administration of puromycin (Invitrogen, Carlsbad, CA). The pcDNA3.1 and sh-Scb were applied as controls (Supplementary Table S5).
Western blot
Cellular protein was extracted with 1× cell lysis buffer (Promega, Madison, WI). Western blot was performed as previously described [15, 20, 38–47], with antibodies specific for MZF1 (Santa Cruz Biotechnology, Santa Cruz, CA), MMP-14 (Abcam Inc, Cambridge, MA), VEGF (Santa Cruz Biotechnology), AGO2 (Cell Signaling Technology, Inc., Danvers, MA), and β-actin (Santa Cruz Biotechnology).
Real-time quantitative RT-PCR
Total RNA was isolated with RNeasy Mini Kit (Qiagen Inc., Valencia, CA). The reverse transcription reactions were conducted with Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN). Real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) and primers indicated in Supplementary Table S6. The transcript levels were analyzed by 2−ΔΔCt method.
miRNA prediction and expression detection
miRNA binding sites within the MMP-14 promoter were analyzed using the algorithm microPIR [48]. The mature miR-337-3p levels were determined using Bulge-LoopTM miRNAs qPCR Primer Set (RiboBio Co. Ltd, Guangzhou, China). After cDNA was synthesized with a miRNA-specific stem-loop primer, the quantitative PCR was performed with the specific primers (Supplementary Table S6). The miRNA levels were normalized as to those of U6 snRNA.
Over-expression and knockdown of miRNA
According to the pre-miR-337-3p (5′-GAACGGCT TCATACAGGAGTT-3′) sequence documented in the miRNA Registry database [49], oligonucleotides encoding the precursor of miR-337-3p (Supplementary Table S5) were subcloned into pcDNA3.1(−) (Genechem Co., Ltd, Shanghai, China). The plasmids pcDNA3.1 and pcDNA3.1-miR-337-3p were transfected into cancer cells, and stable cell lines were screened by administration of neomycin (Invitrogen, Carlsbad, CA). Anti-miR-337-3p or negative control inhibitors (RiboBio Co. Ltd) were transfected into confluent cells with Lipofectamine 2000 (Life Technologies, Inc.).
Luciferase reporter assay
Human MMP-14 promoter luciferase reporter construct was kindly provided by Dr. Jouko Lohi (University of Helsinki, Finland) [11]. Mutation of MZF1 or miR-337-3p binding site was established with GeneTailorTM Site-Directed Mutagenesis System (Invitrogen) and PCR primers (Supplementary Table S5). Dual-luciferase assay was performed as previously described [15, 20, 38–40, 43–47, 50]. For MMP-14 promoter activity, the luciferase signal was normalized by firefly/Renilla ratio.
Nuclear run-on assay
Nuclear run-on assay was performed based on the incorporation of biotin-16-uridine-5′-triphosphate (biotin-16-UTP) into nascent transcripts as previously described [20, 43, 46, 50]. Briefly, nuclei of 5 × 106 cancer cells were isolated and consequently incubated in reaction buffer containing rNTPs and biotin-16-UTP (Roche, Indianapolis, IN) at 30°C for 45 min. The reaction was stopped by adding RNase-free DNase I, and nuclei were lysed and treated with proteinase K. Total RNA was extracted using Trizol (Invitrogen), and biotinylated nascent RNA was purified using agarose-conjugated streptavidin beads (Invitrogen) for real-time quantitative RT-PCR assay.
Chromatin immunoprecipitation
ChIP assay was performed according to the instructions of EZ-ChIP kit (Upstate Biotechnology, Temacula, CA) [15, 21, 40, 44, 45, 47, 50], with antibodies for MZF1, AGO2, H3K9me2, H3K27me3, and H3K4me3 (Upstate Biotechnology, Temacula, CA). Lysates were treated with either RNase H (10 U) or RNase A (20 μg) prior to immunoprecipitation. DNA was sonicated into fragments of an average size of 200 bp. Real-time qPCR was performed with SYBR Green PCR Master Mix and primer sets indicated in Supplementary Table S6. The amount of immunoprecipitated DNA was calculated in reference to a standard curve, and the results were normalized to input DNA.
Cell viability assay
Cancer cells were cultured in 96-well plates at 5×103 cells per well. Cell viability was monitored by the 2-(4,5-dimethyltriazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma, St. Louis, MO) colorimetric assay [50]. All experiments were done with 6–8 wells per experiment and repeated at least three times.
Soft agar assay
Cancer cells at 5 × 103 per well were mixed with 0.05% Nobel agar (Fisher Scientific, Pittsburgh, PA) in growth medium and plated onto 6-well plates containing a solidified bottom layer (0.1% Noble agar in growth medium). After the incubation of cells for 21 days, the number of cell colonies was counted under the microscope, and the cells were fixed with 100% methanol and stained with 0.5% crystal violet dye [38, 40, 43, 47].
Cell invasion assay
Matrigel invasion assay was performed using membranes coated with Matrigel matrix (BD Science, Sparks, MD). Homogeneous single cell suspensions (1 × 105 cells/well) were added to the upper chambers and allowed to invade for 24 hrs at 37°C in a CO2 incubator. Invaded cells were stained with 0.1% crystal violet for 10 min at room temperature. Quantification of invaded cells was performed according to published criteria [15, 20, 21, 38, 39, 41–47, 51].
Tube formation assay
Fifty microliters of growth factor-reduced matrigel were polymerized on 96-well plates. HUVECs were serum starved in RPMI1640 medium for 24 hrs, suspended in RPMI1640 medium preconditioned with cancer cells, added to the matrigel-coated wells at the density of 5×104 cells/well, and incubated at 37°C for 18 hrs. Quantification of anti-angiogenic activity was calculated as previously described [15, 20, 38, 40, 41, 43, 46].
In vivo growth and metastasis assay
All animal experiments were approved by the Animal Care Committee of Tongji Medical College (approval number: Y20080290). For the in vivo tumor growth studies, 2-month-old male nude mice (n = 5 per group) were injected subcutaneously in the upper back with 1×106 cancer cells stably transfected with empty vector, MZF1, or miR-337-3p precursor. One month later, mice were sacrificed and examined for tumor weight and gene expression. The experimental metastasis (0.4×106 tumor cells per mouse, n = 5 per group) studies were performed with 2-month-old male nude mice as previously described [15, 20, 38–40, 43–47].
Patient tissue samples
Approval to conduct this study was obtained from the Institutional Review Board of Tongji Medical College (approval number: 2011-S085). Fresh specimens of 90 well-established primary gastric cancer cases were obtained from the Department of Surgery, Union Hospital, Tongji Medical College. Their pathological diagnosis was proved by at least two pathologists. The demographic and clinicopathological data of subtotal 50 patients were summarized in Supplementary Table S1. Adjacent gastric mucosa specimens that contained no macroscopic tumor were also obtained, and the non-neoplastic areas were subsequently verified by microscopic histology to be free of tumor infiltration. The fresh tumor and adjacent normal gastric specimens were collected and stored at −80°C until use.
Immunohistochemistry
Immunohistochemical staining was performed as previously described [15, 20, 38–40, 43–47, 52], with antibodies specific for MZF1 (Santa Cruz Biotechnology; 1:200 dilution), MMP-14 (Abcam Inc; 1:200 dilution), and CD31 (Santa Cruz Biotechnology; 1:200 dilution). The negative controls included parallel sections treated without primary antibody or with rabbit polyclonal IgG (Abcam Inc.). The immunoreactivity in each tissue section was assessed by at least two pathologists. The degree of positivity was determined according to the percentage of positive tumor cells.
Statistical analysis
Unless otherwise stated, all data were shown as mean ± standard error of the mean (SEM). The χ2 analysis and Fisher exact probability analysis were applied to compare the gene expression in cancer tissues. Pearson's coefficient correlation was applied for analyzing the relationship among gene expression. The log-rank test and Cox regression models were used to assess survival difference and hazard ratios. The Bonferroni method was applied to analyze the difference of survival curves. Difference of tumor cells was determined by t test or analysis of variance (ANOVA).
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
We are grateful for Drs. Jiawei Zhou and Jouko Lohi for providing vectors.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This work was supported by the National Natural Science Foundation of China (81272779, 81372667, 81372401, 81472363, 81402301, 81402408, 81572423), Fundamental Research Funds for the Central Universities (2012QN224, 2013ZHYX003, 01-18-530112, 01-18-530115), and Natural Science Foundation of Hubei Province (2014CFA012).
REFERENCES
- 1.Terry MB, Gaudet MM, Gammon MD. The epidemiology of gastric cancer. Semin Radiat Oncol. 2002;12:111–127. doi: 10.1053/srao.30814. [DOI] [PubMed] [Google Scholar]
- 2.Durães C, Almeida G, Seruca R, Oliveira C, Carneiro F. Biomarkers for gastric cancer: prognostic, predictive or targets of therapy? Virchows Arch. 2014;464:367–378. doi: 10.1007/s00428-013-1533-y. [DOI] [PubMed] [Google Scholar]
- 3.Seiki M. Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 2003;194:1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]
- 4.Maruyama Y, Ono M, Kawahara A, Yokoyama T, Basaki Y, Kage M, Aoyagi S, Kinoshita H, Kuwano M. Tumor growth suppression in pancreatic cancer by a putative metastasis suppressor gene Cap43/NDRG1/Drg-1 through modulation of angiogenesis. Cancer Res. 2006;66:6233–6242. doi: 10.1158/0008-5472.CAN-06-0183. [DOI] [PubMed] [Google Scholar]
- 5.Yang B, Gao J, Rao Z, Shen Q. Clinicopathological and prognostic significance of α5β1- integrin and MMP-14 expressions in colorectal cancer. Neoplasma. 2013;60:254–261. doi: 10.4149/neo_2013_034. [DOI] [PubMed] [Google Scholar]
- 6.Têtu B, Brisson J, Wang CS, Lapointe H, Beaudry G, Blanchette C, Trudel D. The influence of MMP-14, TIMP-2 and MMP-2 expression on breast cancer prognosis. Breast Cancer Res. 2006;8:R28. doi: 10.1186/bcr1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adley BP, Gleason KJ, Yang XJ, Stack MS. Expression of membrane type 1 matrix metalloproteinase (MMP-14) in epithelial ovarian cancer: High level expression in clear cell carcinoma. Gynecol Oncol. 2009;112:319–324. doi: 10.1016/j.ygyno.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández-Pérez M, El-hajahmad M, Massaro J, Mahalingam M. Expression of gelatinases (MMP-2, MMP-9) and gelatinase activator (MMP-14) in actinic keratosis and in in situ and invasive squamous cell carcinoma. Am J Dermatopathol. 2012;34:723–728. doi: 10.1097/DAD.0b013e31824b1ddf. [DOI] [PubMed] [Google Scholar]
- 9.Imanishi Y, Fujii M, Tokumaru Y, Tomita T, Kanke M, Kanzaki J, Kameyama K, Otani Y, Sato H. Clinical significance of expression of membrane type 1 matrix metalloproteinase and matrix metalloproteinase-2 in human head and neck squamous cell carcinoma. Hum Pathol. 2000;31:895–904. doi: 10.1053/hupa.2000.9756. [DOI] [PubMed] [Google Scholar]
- 10.He L, Chu D, Li X, Zheng J, Liu S, Li J, Zhao Q, Ji G. Matrix metalloproteinase-14 is a negative prognostic marker for patients with gastric cancer. Dig Dis Sci. 2013;58:1264–1270. doi: 10.1007/s10620-012-2513-9. [DOI] [PubMed] [Google Scholar]
- 11.Lohi J, Lehti K, Valtanen H, Parks WC, Keski-Oja J. Structural analysis and promoter characterization of the human membrane-type matrix metalloproteinase-1 (MT1-MMP) gene. Gene. 2000;242:75–86. doi: 10.1016/s0378-1119(99)00549-1. [DOI] [PubMed] [Google Scholar]
- 12.Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2 alpha in von Hippel-Lindau renal cell carcinoma. Oncogene. 2004;24:1043–1052. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu H, Hu L, Yu L, Wang X, Urvalek AM, Li T, Shen C, Mukherjee D, Lahiri SK, Wason MS, Zhao J. KLF8 and FAK cooperatively enrich the active MMP14 on the cell surface required for the metastatic progression of breast cancer. Oncogene. 2014;33:2909–2917. doi: 10.1038/onc.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowden Dahl KD, Zeineldin R, Hudson LG. PEA3 is necessary for optimal epidermal growth factor receptor–stimulated matrix metalloproteinase expression and invasion of ovarian tumor cells. Mol Cancer Res. 2007;5:413–421. doi: 10.1158/1541-7786.MCR-07-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang X, Zhao X, Qu H, Li D, Yang D, Pu J, Mei H, Zhao J, Huang K, Zheng L, Tong Q. Hepatocyte nuclear factor 4 alpha promotes the invasion, metastasis and angiogenesis of neuroblastoma cells via targeting matrix metalloproteinase 14. Cancer Lett. 2015;359:187–197. doi: 10.1016/j.canlet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 17.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 18.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 19.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H, Jiao W, Zhao J, Huang K, Zheng L, Tong Q. miRNA-337-3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget. 2015;6:22452–22466. doi: 10.18632/oncotarget.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, Li D, Xiang X, Tong L, Qi M, Pu J, Huang K, Tong Q. Methyl jasmonate abolishes the migration, invasion and angiogenesis of gastric cancer cells through down-regulation of matrix metalloproteinase 14. BMC Cancer. 2013;13:74. doi: 10.1186/1471-2407-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hromas R, Collins SJ, Hickstein D, Raskind W, Deaven LL, O'Hara P, Hagen FS, Kaushansky K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J Biol Chem. 1991;266:14183–14187. [PubMed] [Google Scholar]
- 23.Hromas R, Morris J, Cornetta K, Berebitsky D, Davidson A, Sha M, Sledge G, Rauscher F., 3rd Aberrant expression of the myeloid zinc finger gene, MZF-1, is oncogenic. Cancer Res. 1995;55:3610–3614. [PubMed] [Google Scholar]
- 24.Deng Y, Wang J, Wang G, Jin Y, Luo X, Xia X, Gong J, Hu J. p55PIK transcriptionally activated by MZF1 promotes colorectal cancer cell proliferation. Biomed Res Int. 2013;2013:868131. doi: 10.1155/2013/868131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai SJ, Hwang JM, Hsieh SC, Ying TH, Hsieh YH. Overexpression of myeloid zinc finger 1 suppresses matrix metalloproteinase-2 expression and reduces invasiveness of SiHa human cervical cancer cells. Biochem Biophys Res Commun. 2012;425:462–467. doi: 10.1016/j.bbrc.2012.07.125. [DOI] [PubMed] [Google Scholar]
- 26.Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, Broxmeyer HE, Hromas R, Pandolfi PP. Mzf1 controls cell proliferation and tumorigenesis. Genes Dev. 2001;15:1625–1630. doi: 10.1101/gad.902301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris JF, Hromas R, Rauscher FJ. Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol Cell Biol. 1994;14:1786–1795. doi: 10.1128/mcb.14.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai LH, Wu JY, Cheng YW, Chen CY, Sheu GT, Wu TC, Lee H. The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene. 2015;34:1641–1649. doi: 10.1038/onc.2014.118. [DOI] [PubMed] [Google Scholar]
- 29.Perrotti D, Melotti P, Skorski T, Casella I, Peschle C, Calabretta B. Overexpression of the zinc finger protein MZF1 inhibits hematopoietic development from embryonic stem cells: correlation with negative regulation of CD34 and c-myb promoter activity. Mol Cell Biol. 1995;15:6075–6087. doi: 10.1128/mcb.15.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mei H, Lin ZY, Tong QS. The roles of microRNAs in neuroblastoma. World J Pediatr. 2014;10:10–16. doi: 10.1007/s12519-014-0448-2. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Sætrom P, Snøve O, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39:5682–5691. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adilakshmi T, Sudol I, Tapinos N. Combinatorial action of miRNAs regulates transcriptional and post-transcriptional gene silencing following in vivo PNS injury. PLoS One. 2012;7:e39674. doi: 10.1371/journal.pone.0039674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibition of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Wang J, Yang Y, Hao B, Wang R, Li Y, Wu Q. Loss of has-miR-337-3p expression is associated with lymph node metastasis of human gastric cancer. J Exp Clin Cancer Res. 2013;32:76. doi: 10.1186/1756-9966-32-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X, Zhang X, Shao W, Yin Y, Zhou J. Crucial roles of MZF-1 in the transcriptional regulation of apomorphine-induced modulation of FGF-2 expression in astrocytic cultures. J Neurochem. 2009;108:952–961. doi: 10.1111/j.1471-4159.2008.05845.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, Huang K, Zheng L, Tong Q. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene. 2014;33:387–397. doi: 10.1038/onc.2012.574. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Qi M, Li S, Qi T, Mei H, Huang K, Zheng L, Tong Q. microRNA-9 targets matrix metalloproteinase 14 to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells. Mol Cancer Ther. 2012;11:1454–1466. doi: 10.1158/1535-7163.MCT-12-0001. [DOI] [PubMed] [Google Scholar]
- 40.Li D, Mei H, Qi M, Yang D, Zhao X, Xiang X, Pu J, Huang K, Zheng L, Tong Q. FOXD3 is a novel tumor suppressor that affects growth, invasion, metastasis and angiogenesis of neuroblastoma. Oncotarget. 2013;4:2021–2044. doi: 10.18632/oncotarget.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng L, Pu J, Qi T, Qi M, Li D, Xiang X, Huang K, Tong Q. miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesis of gastric cancer cells. Mol Cancer Res. 2013;11:182–193. doi: 10.1158/1541-7786.MCR-12-0534. [DOI] [PubMed] [Google Scholar]
- 42.Zheng L, Qi T, Yang D, Qi M, Li D, Xiang X, Huang K, Tong Q. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One. 2013;8:e55719. doi: 10.1371/journal.pone.0055719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, Li D, Li S, Mao L, Huang K, Tong Q. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet. 2015;24:2539–2551. doi: 10.1093/hmg/ddv018. [DOI] [PubMed] [Google Scholar]
- 44.Li D, Mei H, Pu J, Xiang X, Zhao X, Qu H, Huang K, Zheng L, Tong Q. Intelectin 1 suppresses the growth, invasion and metastasis of neuroblastoma cells through up-regulation of N-myc downstream regulated gene 2. Mol Cancer. 2015;14:47. doi: 10.1186/s12943-015-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li D, Zhao X, Xiao Y, Mei H, Pu J, Xiang X, Jiao W, Song H, Qu H, Huang K, Zheng L, Tong Q. Intelectin 1 suppresses tumor progression and is associated with improved survival in gastric cancer. Oncotarget. 2015;6:16168–16182. doi: 10.18632/oncotarget.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang X, Mei H, Qu H, Zhao X, Li D, Song H, Jiao W, Pu J, Huang K, Zheng L, Tong Q. miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. Biochim Biophys Acta. 2015;1852:1743–1754. doi: 10.1016/j.bbadis.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 47.Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, Qu H, Huang K, Zheng L, Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2015 doi: 10.1038/onc.2015.422. [DOI] [PubMed] [Google Scholar]
- 48.Piriyapongsa J, Bootchai C, Ngamphiw C, Tongsima S. microPIR: an integrated database of microRNA target sites within human promoter sequences. PLoS One. 2012;7:e33888. doi: 10.1371/journal.pone.0033888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang G, Zheng L, Pu J, Mei H, Zhao J, Huang K, Zeng F, Tong Q. Small RNAs targeting transcription start site induce heparanase silencing through interference with transcription initiation in human cancer cells. PLoS One. 2012;7:e31379. doi: 10.1371/journal.pone.0031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng L, Jiang G, Mei H, Pu J, Dong J, Hou X, Tong Q. Small RNA interference-mediated gene silencing of heparanase abolishes the invasion, metastasis and angiogenesis of gastric cancer cells. BMC Cancer. 2010;10:33. doi: 10.1186/1471-2407-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng LD, Tong QS, Tang ST, Du ZY, Liu Y, Jiang GS, Cai JB. Expression and clinical significance of heparanase in neuroblastoma. World J Pediatr. 2009;5:206–210. doi: 10.1007/s12519-009-0039-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.