Abstract
Overexpression and/or hyperactivation of cyclin-dependent kinases (CDKs) are common features of most cancer types. CDKs have been shown to play important roles in tumor cell proliferation and growth by controlling cell cycle, transcription, and RNA splicing. CDK4/6 inhibitor palbociclib has been recently approved by the FDA for the treatment of breast cancer. CDK11 is a serine/threonine protein kinase in the CDK family and recent studies have shown that CDK11 also plays critical roles in cancer cell growth and proliferation. A variety of genetic and epigenetic events may cause universal overexpression of CDK11 in human cancers. Inhibition of CDK11 has been shown to lead to cancer cell death and apoptosis. Significant evidence has suggested that CDK11 may be a novel and promising therapeutic target for the treatment of cancers. This review will focus on the emerging roles of CDK11 in human cancers, and provide a proof-of-principle for continued efforts toward targeting CDK11 for effective cancer treatment.
Keywords: CDK11, CDKs inhibitor, cell cycle, therapeutic target, cancer therapy
INTRODUCTION
Malignant diseases are characterized by uncontrolled cell proliferation and growth [1]. Currently, surgery, radiotherapy, and chemotherapy are still the main modalities in the treatment of cancer [2–4]. Chemotherapy, as one of the basic treatment strategies, is applied in most cancer therapies. However, cancers usually have innate or acquired ability to develop resistance to many anti-cancer drugs, especially in the late stages [5, 6]. Additionally, most conventional chemotherapy drugs applied in the clinic are non-specific cytotoxic agents and will cause serious adverse effects to patients, thus identifying and developing novel therapeutic strategies is an urgent need for cancer treatment.
Cyclin-dependent kinases (CDKs) are serine/threonine kinases that play a critical role in the regulation of cell cycle progression, as well as cellular transcription. Distinct CDKs are activated upon binding with their corresponding cyclin partners. Each kinase in this family is coordinated in an orchestrated way, and responsible for particular aspects of the cellular events. Aberrant expression or altered activity of distinct CDK complexes results in escape of cells from the cell cycle control and leads to malignant transformation. Therefore, the inhibition of CDKs in malignant cells provides a promising approach in the defense against cancer. Recently, many selective CDK inhibitors targeting specific CDKs were developed, which represent promising anti-cancer drugs due to their strong anti-proliferative efficacy combined with a relative low direct cytotoxicity [7–14]. Notably, palbociclib (IBRANCE®), a dual CDK4/6 inhibitor, recently received accelerated approval by the Food and Drug Administration (FDA) for clinical breast cancer treatment due to its potent and selective inhibitory effect on estrogen receptor (ER) positive/human epidermal growth factor receptor 2 (HER2) negative breast cancer [15–18]. These developments suggest a promising application of CDK inhibitors as a novel therapeutic strategy in the treatment of human cancer.
CDK11 is a serine/threonine protein kinase in the CDK family. CDK11 also plays a crucial role in cancer cell proliferation and growth. Recent studies have found that the overexpression and activation of CDK11 is crucial in the growth and proliferation of cancer cells, including breast cancer, multiple myeloma, osteosarcoma, and other types of cancer, which has suggested that CDK11 may be a novel potential therapeutic target [19–22]. In this review, we discuss the specificity of CDK11 functions, regulation, and interactions in cancers, as well as the potential of targeting CDK11 in cancer treatment.
THE BIOLOGY AND DEREGULATION OF CDKS IN HUMAN CANCERS
CDKs are a family of serine/threonine (Ser/Thr) protein kinases, composed of a catalytic kinase subunit and a regulatory cyclin subunit, which play crucial roles in cell cycle progression and transcriptional regulation in response to extracellular and intracellular signals [23]. CDKs are characterized by requirement of protein cyclin subunits for enzymatic activity, even though some CDKs can also play other roles without the involvement of cyclins [24]. Until now, based on the sequence similarity and nomenclature, the human CDKs family include members from CDK1 to CDK20 with specific or redundant roles in many aspects of cell growth and proliferation [25]. The evolutionary relationships among these CDK subfamilies have been identified [26].
Generally, CDKs are divided into cell cycle-related subfamilies and transcription-related subfamilies in regard to their sequencing and main functional roles [26]. Classical cell cycle CDKs, including CDK1, CDK2, CDK4, and CDK6, mainly regulate the transitions between different phases of the cell cycle [27–29]. The cyclin C-CDK3 complex helps the cells to efficiently exit the G0 state and enter the G1 phase. In addition, CDK5, CDK11, CDK14, CDK15, CDK16, CDK17, and CDK18 also belong to the cell cycle-related CDK subfamilies, which play different functional roles by regulating diverse cell cycle progress [30–38] (Table 1). On the other hand, transcription-related CDKs subfamilies function mainly via influencing transcription by phosphorylating the carboxy-terminal domain (CTD) of RNA polymerase II (RNAP II), which contains 52 tandem repeats of the consensus heptapeptide amino acid sequence (YSPTSPS) [39, 40]. Specifically, the transcription-related CDKs subfamilies are comprised of CDK7, CDK8, CDK9, CDK11, CDK12, CDK13, CDK19, and CDK20, which participate in different transcription regulation and exert diverse cellular functions [27, 39–49] (Table 1).
Table 1. Members of CDK family and their functions in cancers.
| CDK Family | Cyclins and Cyclin-like Partners | Gene Locus | Functions in Cancers | Inhibitors |
|---|---|---|---|---|
| CDK1 | A1, A2, B1, B2, (B3), D, E | 10q21.2 | promotes cancer cell cycle G2/M transition and proliferation | Pirarubicin, Flavopiridol, Dinaciclib, Seliciclib, Roscovitine, Milciclib, Roniciclib, AZ703, UCN-01, P276-00, AT7519, AZD5438, SCH727965, RGB-286638 |
| CDK2 | A1, A2, B1, B3, D, E1, E2, Cables1, SpdYA, SpdYC |
12q13.2 | promotes cancer cell cycle G1/S transition and proliferation | Seliciclib, Flavopiridol, Roscovitine, MLN4924, Dinaciclib, Roniciclib, AZ703, UCN-01, SNS-032, AT7519, SCH727965, RGB-286638, AZD5438 |
| CDK3 | A1, A2, E1, E2, C, Cables1 | 17q25.1 | helps cancer cells to efficiently exit the G0 state and enter the G1 phase, facilitates cell proliferation | TG-02, AT-7519, RGB-286638 |
| CDK4 | D1, D2, D3 | 12q14.1 | promotes cancer cell G1 phase progression and proliferation | Palbociclib, Flavopiridol, Abemaciclib, Dinaciclib, Ribociclib, Milciclib, Roniciclib, P276-00, LY2835219, AT7519, MM-D37K, RGB-286638, AZD5438 |
| CDK5 | p35, p39 (D-, E and G-type cyclins), Cables1 |
7q36.1 | unknown | Roscovitine, Flavopiridol, Milciclib, SCH727965, AZD5438, RGB-286638 |
| CDK6 | D1, D2, D3 | 7q21.2 | promotes cancer cell G1 phase progression and proliferation | Palbociclib, Flavopiridol, Abemaciclib, Ribociclib, LY2835219, AT7519, RGB-286638 |
| CDK7 | H | 5q13.2 | promotes cell cycle progress, RNA transcription, and cancer cell proliferation | Roscovitine, Flavopiridol, Roscovitine, Milciclib, Roniciclib, SNS-032, AT7519, RGB-286638, AZD5438 |
| CDK8 | C, (K) | 13q12.13 | activates RNA transcription and promotes cancer cell proliferation | unknown |
| CDK9 | K, T1, T2 | 9q34.11 | promotes RNA transcription elongation and cancer cell proliferation | Flavopiridol, Roscovitine, Roniciclib, SNS-032, AZD5438, P276-00, AT7519, SCH727965, RGB-286638 |
| CDK10 | M | 16q24.3 | promotes RNA transcription and cancer cell proliferation | unknown |
| CDK11 | D3, L1, L2 | 1p36.33 | transcription, RNAsplicing; cell cycle: G2/M | unknown |
| CDK12 | L1, L2, K | 17q12 | promotes RNA transcription elongation, splicing, and cancer cell proliferation | unknown |
| CDK13 | L1, L2, K | 7p14.1 | promotes RNA transcription elongation, splicing, and cancer cell proliferation | unknown |
| CDK14 | D3, Y | 7q21.13 | links cell cycle regulators and Wnt signaling, promotes cancer cell proliferation, migration, and invasion | unknown |
| CDK15 | unknown | 2q33.1 | promotes cancer cell cycle progression and proliferation | unknown |
| CDK16 | Y, p35, Cables1 | Xp11.3 | promotes cancer cell cycle progression and proliferation | unknown |
| CDK17 | Cables1 | 12q23.1 | unknown | unknown |
| CDK18 | K, A | 1q32.1 | unknown | unknown |
| CDK19 | C | 6q21 | activates RNA transcription and promotes cancer cell proliferation | unknown |
| CDK20 | H | 9q22.1 | unknown | Unknown |
In malignant cells, altered expression of CDKs and their modulators, including overexpression of cyclins and loss of expression of CDK inhibitors, results in deregulated CDK activity, providing a selective growth advantage. In human cancers, owing to various genetic and epigenetic events, CDKs are often overexpressed and/or overactive, bringing about loss of checkpoint integrity and ultimately resulting in uncontrolled cell proliferation [27, 50–53]. For example, the cyclin D/CDK4/CDK6/RB pathway is hyperactive in various malignancies, such as melanoma, glioblastoma, osteosarcoma, lymphomas, breast and cervical cancers, squamous cell cancer, etc [54]. In the transcriptional CDK subfamily CDK9, aberrant activation has also been observed in several primary tumors, including myeloma, prostate cancer, and lung cancer [55].
Because of their critical roles in cell cycle progression and cellular transcription, as well as in the deregulation of human cancer, CDKs comprise an attractive set of targets for novel anticancer drug development [27]. The best known example is the recently FDA-approved CDK4/6 inhibitor palbociclib as the initial endocrine-based therapy for postmenopausal women with ER+/HER2- metastatic breast cancer. Among other members of the CDK family, CDK11 is involved in both cell cycle control and RNA transcription regulation. CDK11 is functionally relevant to many biologic processes, such as RNA transcription and splicing, mitosis, autophagy, and apoptosis [38, 45, 47, 56, 57]. Recently, distinct and unique biologic roles of CDK11 have been discovered in human cancers and in other human diseases [58].
DISTINCT SPECIFICITY OF CDK11
Unlike other CDKs encoded only by a single gene, CDK11, formerly known as PITSLRE, is encoded by two highly homologous genes, CDC2L1 (also known as CDK11B) and CDC2L2 (also known as CDK11A, nonexistent in mouse) in humans. These two genes are localized in a genomic region that spans about 140 kb on human chromosome 1 band p36.3 [59]. In mouse, there is only one gene encoding CDK11 [25]. In human, both of the CDC2L genes contain 20 exons and 19 introns that encode almost identical protein kinases named CDK11A and CDK11B.
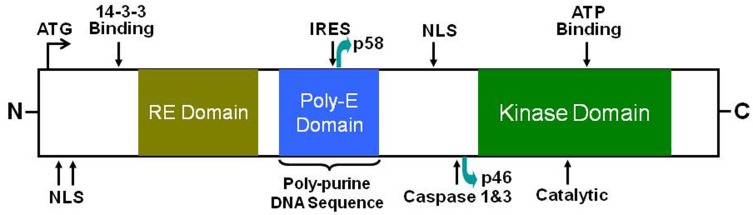
CDK11 is composed of an N-terminal regulatory region, which has multiple nuclear localization signals (NLS) and a 14-3-3 consensus site, and a carboxy-terminal (C-terminal) catalytic domain that is responsible for its kinase activity [40, 60]. There are two separate domains, an arginine/glutamic acid domain (RE domain) and a poly-glutamic acid domain (poly-E domain) located in the center of the CDK11 protein (Figure 1) [40]. The RE domains are linked to association with RNA processing factors and poly-E domains are emerging as potential cytoskeletal interacting domains that support RE domain function and aide in keeping these proteins subnuclear. The most important conserved amino acids in CDK11 are the PSTAIRE-helix and three phosphorylation sites, which are involved in the activation and repression of CDK kinase activity [40].
Figure 1. Schematic diagram of the full length CDK11 protein kinase.
CDK11 is composed of an N-terminal regulatory region, which has multiple nuclear localization signals (NLS) and a 14-3-3 consensus site, and a carboxy-terminal (C-terminal) catalytic domain that is responsible for its kinase activity. There are two separate domains, an RE domain and a poly-E domain located in the center of the CDK11 protein. The full-length CDK11p110 isoform contains an IRES and a caspase-3 site, which leads to the generation of a larger CDK11p58 and a smaller CDK11p46 isoform, respectively (adapted from Trembley et. al., 2004.). NLS, nuclear localization signal; RE, arginine (R) and glutamic (E) acid residues; IRES, internal ribosomal entry site.
CDK11 binds to L-type cyclins and participates in the coordination between transcription and RNA processing, particularly alternative splicing [61]. The functions of CDK11 have been proved to be linked with RNA transcription and processing, regulation of cell cycle, neuronal function, and apoptosis [38, 40, 47, 56, 58]. The potential for CDK11 to regulate these diverse cellular activities is unique in the CDK family and highlights that CDK11 may exert critical regulatory roles in human tumorigenesis and malignant characteristics of cancer cells.
DIFFERENT ISOFORMS OF CDK11
Due to the distinct structure and alterative RNA splicing, the CDC2L gene can produce three different CDK11 isoforms, a larger 110 kDa protein isoform, a mitosis-specific 58 kDa isoform, and a smaller apoptosis-specific 46 kDa isoform (Table 2). The larger CDK11p110 isoform is coded by the full-length CDK11 mRNA and contains an internal ribosome entry site (IRES), which leads to the generation of the CDK11p58 isoform during the G2/M phase of the cell cycle. In response to apoptotic signaling, both CDK11p110 and CDK11p58 isoforms can be cleaved by caspases 1 and 3 and produce the smaller CDK11p46 isoform (Figure 1) [62–64]. These different protein kinase isoforms play diverse cellular functions, including RNA transcription and processing, mitosis, and apoptosis. The larger CDK11p110 kinase is ubiquitously and constantly expressed throughout the cell cycle. Using subcellular fractionation techniques, the CDK11p110 isoform is proven to be a nuclear protein, which localizes to both splicing factor compartments and to the nucleoplasm [65]. On the other hand, the CDK11p58 protein is specifically translated from an internal ribosome entry site and expressed transiently only in the G2/M phase of the cell cycle [66]. Due to the fact that CDK11p58 is produced during a very narrow window of mitosis, it is much more difficult to detect than CDK11p110; its detection depends primarily on the mitotic characteristics of a particular cell type [40, 66]. Although CDK11p58 shares the same sequences, including the kinase domain at the C-terminus of CDK11p110, the two isoforms possess different functions. The CDK11p110 isoform is mainly associated with RNA transcription and splicing [40, 45, 60, 61, 67–69], while CDK11p58 isoform is involved in mitosis [70–77]. In contrast to CDK11p110 and CDK11p58, CDK11p46 localizes to the cytoplasm when ectopically expressed. Like other CDKs, the activation of CDK11 also needs to binding with special cyclins. Cyclins that bind CDK11p110, CDK11p58, and CDK11p46 are L-type cyclins, which are encoded by two cyclin L genes encoding a total of six isoforms [61]. CDK11p58 also binds cyclin D3 [78]. Several non-cyclin partners have also been proposed to interact with CDK11p46, such as eukaryotic initiation factor 3 p47 protein (eIF3 p47) and Ran-binding protein (RanBPM) [79, 80].
Table 2. Biological characteristics of three main CDK11 isoforms.
| CDK11 isoforms | Molecular Weight | Cyclin Partners | Subcellular Localization | Cell Cycle Expression | Expression in Cancer | Cellular Functions |
|---|---|---|---|---|---|---|
| CDK11p110 | 110 kDa | cyclins L1 and L2 | nucleus | throughout cell cycle | highly expressed | transcription RNA splicing hedgehog signaling pathway Wnt/β-catenin signaling pathway |
| CDK11p58 | 58 kDa | cyclins L1, L2 and D3 | nucleus and cytoplasm | transiently in G2/M phase | unknown | mitosis process centrosome maturation bipolar spindle assembly |
| CDK11p46 | 46 kDa | cyclins L1 and L2 | cytoplasm | transiently | unknown | apoptosis autophagy |
FUNCTIONS OF CDK11 IN NORMAL CELLS
The CDK11 null cells exhibit proliferative defects, mitotic arrest, and apoptosis, thus suggesting that CDK11 kinase is critical for embryonic development and cellular viability [73]. CDK11 knockout mice display an earlier lethality phenotype during the blastocyst stage of embryonic development. The larger CDK11p110 isoform, ubiquitously and constantly expressed throughout the cell cycle, is implicated in pre-mRNA splicing and transcription regulation by interacting with numerous proteins involved in the production of RNA transcripts during proliferation, including the largest subunit of RNA polymerase II and casein kinase II [81]. In addition, CDK11p110 has also been identified as a positive regulator of hedgehog signaling pathway [82, 83] as well as a modulator of the Wnt/β-catenin signaling cascade [84], both of which play vital roles in the regulation of embryonic development and adult homeostasis [85–87].
UPSTREAM AND DOWNSTREAM REGULATIONS IN CDK11 EXPRESSION AND FUNCTIONS
The upstream regulation of CDK11 expression has been studied to identify and characterize the CDK11 promoters. As described above, CDK11 is encoded by two highly homologous genes known as CDC2L1 and CDC2L2. The CDC2L1 gene is regulated by a basal promoter region that is between nucleotides −152 and +11 of the 5′ region of the CDC2L1 gene while a region between nucleotides −145 and +10 of the 5′ region of CDC2L2 is confirmed to be critical for basal transcription of the CDC2L2 gene [88, 89]. In the promoter of the CDC2L1 gene, there are transcription factor binding sites for Ets-1 and Skn-1 that are necessary for CDC2L1 gene expression [88]. However, CDC2L2 gene expression is mainly regulated by Ets-1 and CREB [89].
Checkpoint kinase 2 (CHK2) and casein kinase 2 (CK2) are confirmed to be the upstream regulators in the CDK11 signaling pathway, which interact with CDK11 via phosphorylating the serine 737 and serine 227 sites, respectively [67, 81, 90].CDK11 exerts its functional roles by regulating the expression and/or functions of downstream genes. However, the specific downstream signaling pathway mediated by CDK11 has not been fully documented. Interestingly, a kinase siRNA screen identified CDK11 as a crucial regulator of the Hedgehog pathway [83]. CDK11 was confirmed to directly participate in the Hedgehog signaling pathway by functioning downstream of Smo and upstream of the Glioma-associated (Gli) transcription factors. More specifically, CDK11 interacts with the negative regulator Suppressor of Fused (Sufu) protein and relieves its inhibition of Gli, thus activating the Hedgehog signaling pathway, which is associated with developmental abnormalities and cancer [91, 92]. In addition, CDK11 has also been proved to be a positive modulator of the Wnt/β-catenin signaling cascade, whose dysregulation contributes to the development of cancer, using a kinase-targeted high-throughput siRNA screen [84]. Nevertheless, the molecular mechanisms on which CDK11 regulate the Wnt/β-catenin signaling pathway remain unclear.
CDK11 INTERACTING PROTEINS AND THE ROLES OF CDK11 IN TRANSCRIPTION AND RNA PROCESSING
The transcription process of mRNA requires the interaction of a set of general transcription factors and a wide range of gene-specific factors. The mediator complex is one type of large protein complexes, which play critical roles in RNA production and processing. Human mediator complex consists of RNAP II and several positive and negative transcription and RNA processing factors. CDK11 is proven to be a crucial part of the mediator complex. CDK11/cyclin L controls the assembly of the RNAP II mediator complex in fission yeast [69]. To be note, the absence of two conserved domains in the fission yeast CDK11 and cyclin orthologs and the reported essential roles of some CDK11 isoforms in unrelated processes in metazoans propose that CDK11 is essential for human beings gene transcription and processing [71, 74, 76].
In the regulation of RNA production, CDK11 could directly phosphorylate the CTD of RNAP II and regulates transcription initiation and elongation as well as RNA processing. In addition, CDK11 also exert its roles in the regulation of transcription and/or transcript processing via interacting with numerous transcription regulators and RNA splicing factors.
Several CDK11 interacting protein partners have been identified in the transcription and RNA processing by implementing yeast two-hybrid interactive screens, tandem affinity purification, mass spectrometry analyses, immunoblot, and immunoprecipitation (Figure 2, Table 3). The first noteworthy finding is that cyclins L1 and L2 have been well documented as critical cyclin partner proteins of CDK11, which is necessary for the activation of the CDK11p110 protein isoform [61]. In addition, CDK11 also interacts with splicing factors, such as RNA-binding protein (RNPS1) and 9G8 [60, 65], as well as multiple transcriptional initiation and elongation factors, including RNA polymerase elongation factor 2 (ELL2), transcription factor IIF (TFIIF), transcription factor IIS (TFIIS), and facilitates chromatin transcription (FACT) [45]. RNA binding motif protein 15 B (RBM15B/OTT3) has been shown to interact with CDK11 as a competitor and antagonizes the positive effect of CDK11 in splicing [68]. CDK11 exerts the biological functions in cell cycle and apoptosis by interacting with several protein partners, including 14-3-3 protein and heat shock protein 70/90 (Hsp70/90) [93, 94].
Figure 2. Interacting proteins with CDK11 in transcription and RNA processing.
The currently identified CDK11 interacting proteins in the transcription and RNA processing are illustrated in this figure. Among which, the protein kinases CHK2 and CK2 activate CDK11 by phosphorylating the serine 737 and serine 227 sites of the CDK11 kinase, respectively. Cyclin L acts as a crucial protein partner of the CDK11p110 isoform, while cyclin D3 is essential for functioning for the CDK11p58 isoform. RNPSI, 9G8, ELL2, TFIIF, TFIIS, FACT, 14-3-3, and Hsp90 interact with and are subsequently phosphorylated by CDK11. On the other hand, RBM15B/OTT3 interacts with CDK11 as a competitor and antagonizes the positive effect of CDK11 in RNA spicing. Sufu can be negatively regulated by CDK11 and relieve its inhibition on Gli, thus activating the Hedgehog signaling pathway. CHK2, checkpoint kinase 2; CK2, casein kinase 2; RNPS1, RNA-binding protein with serine-rich domain; ELL2, RNA polymerase elongation factor 2; TFIIF, transcription factor IIF; TFIIS, transcription factor IIS; FACT, facilitates chromatin transcription; RBM15B/OTT3, RNA binding motif protein 15 B; Hsp90, heat shock protein 90; Sufu, Suppressor of fused.
Table 3. Characteristics of CDK11 interactiving partners.
| Interacting Partner | Subcellular Localization | Cellular Functions | Cancer Association | References |
|---|---|---|---|---|
| CHK2 | nucleus | responses to genotoxic stress | mutation or low expression in breast, prostate, ovarian, colon, kidney, thyriod, bladder, lung cancer as well as sarcomas and leukemias | 67 |
| CK2 | nucleus | promotes cell proliferation and growth, suppresses cell apoptosis | overexpression or hyperactivation in lung, breast, prostate, gastric, kidney cancer as well as AML, CLL and lymphomas | 81; 90 |
| cyclin L1 | nucleus | regulates transcription and RNA splicing | overexpressed in human head and neck cancer, and is associated with lymph node metastases, also amplify in uterine cervical carcinoma and associates with poor prognosis | 59; 61; 88 |
| cyclin L2 | nucleus | functions as a regulator of the pre-mRNA splicing process, modulates the expression of apoptotic and antiapoptotic proteins | overexpression of cyclin L2 inhibits cancer cell growth, induces apoptosis and cell cycle arrest, and enhances chemosensitivity | 59; 61; 89 |
| cyclin D3 | nucleus and cytoplasm | forms a complex with CDK4 or CDK6, phosphorylates and inhibits RB protein and regulates the cell cycle G1/S transition | overexpressed and associated with a worse prognosis in patients with malignant melanoma, breast cancer, and non-Hodgkin's lymphoma | 78 |
| RNPS1 | nucleus | regulates RNA splicing, promotes mRNA nuclear export and translation, as well as maintenance of postsplicing surveillance | overexpressed in mouse submandibular gland adenocarcinoma | 60; 65 |
| 9G8 | nucleus and cytoplasm | regulates RNA splicing, promotes nucleocytoplasmic export of mRNA and translate | not confirmed | 60 |
| ELL2 | nucleus | promotes transcription initiation and elongation, directs immunoglobulin secretion, inhibits cell growth and survival, induces cell cycle arrest and apoptosis | increased expression in leukemia, mutation associated with multiple myeloma and salivary gland carcinoma | 45 |
| TFIIF | nucleus and cytoplasm | promotes transcription elongation by interacting with RNAPII throughout the elongation phase | unknown | 45 |
| TFIIS | nucleus and cytoplasm | promotes transcription elongation by enhancing the intrinsic endonucleolytic cleavage activity of RNAPII | unknown | 45 |
| FACT | nucleus | modulates nucleosome stability and chromatin remodeling, promotes DNA replication, recombination, and repair, as well as transcript elongation | overexpressed in breast carcinoma, non-small-cell lung cancer, renal cell carcinoma, and prostatic, pancreatic, and colorectal adenocarcinomas | 45 |
| RBM15B/OTT3 | nucleus and cytoplasm | inhibits spliceosomal E complex formation, regulates RNA splicing and mRNA export | unknown | 68 |
| 14-3-3 | nucleus | regulates cell cycle, protein trafficking, and steroidogenesis, promotes cell proliferation, inhibits cell apoptosis | overexpressed in breast, lung, liver, head and neck cancers, as well as glioma and astrocytoma | 94 |
| Hsp90 | cytoplasm and cell membrane | maintains normal tissue homeostasis | overexpression and/or hyperactivation in almost all human cancers | 93 |
| Sufu | nucleus and cytoplasm | promotes embryonic development | deletion, mutation or underexpression in lung, breast, prostate cancer, as well as in medulloblastoma | 91; 92 |
Currently, there is no specific CDK11 inhibitor, which impedes the CDK11-targeted cancer therapy testing in preclinical models. However, the identification and characterization of these CDK11 interactors may facilitate the understanding of the mechanisms underlying CDK11 functions, as well as the development of therapeutic strategies for cancer. Indeed, some inhibitors targeting these CDK11 interactors have been successfully developed and used in preclinical evaluation or clinical trials, which may provide novel candidate compounds for cancer therapy. For example, CX-4945, a potent and selective small molecule inhibitor of CK2 and a confirmed CDK11 interacting protein, has been investigated for the treatment of prostate cancer [95]. Another small molecular compound, AZD7762, a CHK1/CHK2 (another validated CDK11 partner) inhibitor, has been evaluated in the treatment in patients with advanced solid tumors [96].
EXPRESSION AND THERAPEUTIC POTENTIALS OF CDK11 IN DIFFERENT HUMAN CANCERS
Due to their critical roles in the growth and proliferation of many types of human cancer, CDKs comprise an attractive set of targets for novel anticancer drugs development. Many CDKs inhibitors are currently being assessed in preclinical or in clinical trial investigations for cancer therapy; among them, a CDK4/6 selective inhibitor palbociclib has recently been approved for breast cancer treatment due to its selected and promising inhibitory effect on ER+/HER2- breast cancer [15–18]. CDK11 function is a critical regulator of cell cycle progression and RNA transcription, and recent studies have suggested that CDK11 also plays important roles in several types of human cancers (Table 4).
Table 4. Functions of CDK11 in different human cancers.
| Human Cancers | Study Technique | Functions in Tumors |
|---|---|---|
| Breast Cancer | immunohistochemistry; RNAi; immunofluorescence assay; cell viability assay; cell colony formation assay; cell migration assay; western blot; flow cytometry analysis; xenograft RNAi studies | promotes breast cancer cell proliferation, growth, migration and cell cycle progression; inhibits breast cancer cell apoptosis, negatively correlated with breast cancer patient clinical prognosis |
| Osteosarcoma | systematic Kinome shRNA screening; CRISPR, RNAi; cell proliferation assay; apoptosis assay; western blot; immunofluorescence assay; immunohistochemistry; xenograft RNAi studies | promotes osteosarcoma cell proliferation, growth; inhibits osteosarcoma cell apoptosis; negatively correlated with osteosarcoma patient clinical prognosis |
| Liposarcoma | immunohistochemistry; RNAi; cell proliferation assay; western blot; immunofluorescence assay; chemotherapeutic response assay | promotes liposarcoma cell growth, survival; inhibits liposarcoma cell apoptosis; desensitizes liposarcoma cell to chemotherapy |
| Multiple Myeloma | high-throughput siRNA screening;RNA microarray hybridization | promotes myeloma cell proliferation and survival |
| Colon Cancer | high-throughput RNAi screening | acts as a positive modulator of the Wnt/β-catenin pathway in colon cancer |
| Cervical Cancer | RNAi; microtubule regrowth assays | promotes centrosome maturation and bipolar spindle morphogenesis in cervical cancer cells |
| Acute Myeloid Leukemia (AML) | CRISPR-based knockout with RNAi | crucial for TSC2-deficient AML cell growth |
CDK11 IN BREAST CANCER
Recently, the expression and association of CDK11 in human breast cancer has been explored [21]. Immunohistochemical and Western blot assay have revealed that CDK11 is highly expressed in both breast tumor tissues and cell lines. Elevated CDK11 expression in breast cancer tissues significantly correlates with poor differentiation, and is also associated with advanced TNM stage and poor clinical prognosis for breast cancer patients. In vitro knockdown of CDK11 by siRNA significantly inhibits cell growth and migration, and dramatically induces apoptosis in breast cancer cells. Similar results have also been found almost simultaneously by another investigation group, which also showed that CDK11 expression is essential for the maintenance of the aggressive characteristic of triple negative breast cancer (TNBC) [97]. Immunohistochemical analysis of TNBC patient tissues showed that 100% of tumors stained positive for CDK11 with high nuclear intensity compared to normal tissue. The Cancer Genome Atlas analysis by comparing basal to other breast cancer subtypes, and to normal breast tissues revealed statistically significant differences in CDK11 expression. Downregulation of CDK11 in breast cancer cells resulted in significant loss of cell viability and clonal survival, reduced CDK11 relevant mRNA and protein expression, and induced cell death changes [97]. Furthermore, in vivo treatment with tenfibgen-siCDK11 nanocapsules caused MDA-MB-231 xenograft tumor shrinkage, loss of proliferation, and decreased expression of targeted genes. These findings suggest that CDK11 is critical for the survival and proliferation of breast cancer cells, which highlight that CDK11 may be a promising target for therapeutic development in breast cancer.
CDK11 IN MULTIPLE MYELOMA
A kinase-wide RNAi lethality study using high-through kinome siRNA screening identified CDK11 as a vulnerable kinase in human multiple myeloma, which proposes that inhibition of CDK11 represents a uniquely targeted novel therapeutic strategy in human multiple myeloma [22]. To better define potential drug targets in myeloma disease, another investigation conducted a focused RNAi lethality screening and consistently found that CDK11 is a crucial survival gene in multiple myeloma [98]. To further assess CDK11 for myeloma-selective vulnerability, the study evaluated the expression of CDK11 in human multiple myeloma, and showed that CDK11 is significantly upregulated in expression in primary multiple myeloma tissues, as compared with normal human primary tissues. Collectively, CDK11 is overexpressed and functions as a critical survival gene in human multiple myeloma, which suggests that CDK11 may represent a promising druggable target for human multiple myeloma therapy.
CDK11 IN OSTEOSARCOMA
Osteosarcoma is the most common primary malignant tumor of bone. A kinase shRNA screening first identified CDK11 as essential for the survival of osteosarcoma cells [19]. Osteosarcoma cells display high levels of CDK11 expression, and CDK11 knockdown inhibited cell growth and induced apoptosis in osteosarcoma cells [19]. Moreover, immunohistochemical analysis showed that osteosarcoma patients with high CDK11 expression are associated with significantly shorter survival than patients with low CDK11 expression. Systemic in vivo administration of in vivo ready siRNA of CDK11 reduced tumor growth in an osteosarcoma subcutaneous xenograft model. Furthermore, a recent study using a novel robust and highly efficient genome editing tool, the clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9) system, to silence endogenous CDK11 DNA found that CDK11 knockout significantly reduced osteosarcoma cell viability, proliferation, migration, invasion, and induced cell death [99]. All these results show that CDK11 signaling is essential in osteosarcoma cell growth and survival, further elucidating the regulatory mechanisms controlling the expression of CDK11 and ultimately indicate that developing a CDK11 inhibitor may provide therapeutic benefit against osteosarcoma.
CDK11 IN LIPOSARCOMA
Similarly, the functional and therapeutic relevance of CDK11 as a putative target in liposarcoma, another type of mesenchymal tissue-originated malignancy, was also studied. Immunohistochemical analysis of liposarcoma tissue microarray (TMA) showed that CDK11 is highly expressed in liposarcoma tissues as compared with benign lipoma tissues. CDK11 knockdown by synthetic siRNA or lentiviral shRNA decreased cell proliferation and induced apoptosis in liposarcoma cells. Moreover, CDK11 knockdown enhanced the cytotoxic effect of doxorubicin in liposarcoma cells [20]. These results suggest that CDK11 may be a promising therapeutic target for liposarcoma treatment. Future studies on the signaling pathway and molecular mechanisms of CDK11 and cell growth in liposarcoma are required.
CDK11 IN OTHER TYPE OF HUMAN CANCERS
The pioneer studies has also confirmed that CDK11 is ubiquitously expressed many human cancer cell lines, such as Jurkat, Cem C7, HeLa, HEK 293, K562, suggesting the potential functional roles of CDK11 in carcinogenesis [40, 100]. Furthermore, an unbiased high-throughput RNAi screening found that CDK11 is a positive modulator of the Wnt/β-catenin pathway in colon cancer [84]. In cervical cancer, CDK11 knockdown by RNAi in HeLa cells induced abnormal spindle assembly, mitotic arrest by checkpoint activation, and cell death [71]. More recently, by using novel CRISPR-based knockouts with RNAi technology, CDK11 has been identified is the crucial gene in cells lacking either the tuberous sclerosis complex 1 (TSC1) or TSC2, but not that of wild-type cells. Mutation or aberrant inhibition of the TSC complex is common in various human tumor cancers. Knockdown of CDK11 showed growth-inhibiting effects in mammalian TSC2-deficient cell lines, including human tumor-derived (AML) cells. TSC1 and TSC2, function together in an evolutionarily conserved protein complex that is a point of convergence for major cell signaling pathways that regulate mTOR complex 1 (mTORC1) [101].
Taken together, CDK11 is usually overexpressed and/or activated in several types of human malignancies, and the overexpression and/or hyperactivation is highly associated with poor outcomes in cancer patients. Consequently, CDK11 may present as a selective and specific target for cancer therapy. The downregulation of expression and activity of CDK11 may be a novel therapeutic modality in cancer treatment.
CDK11 IN OTHER HUMAN DISEASES
In addition to human cancer, CDKs play important roles in other types of human diseases. Among them, CDK7, CDK9 and CDK13 have been found to be critical for HIV replication inside cells. However, the function of CDK11 remains largely unknown. Recently, CDK11 has been found to regulate the cleavage and polyadenylation (CPA) of all viral transcripts [102, 103]. CDK11 was found to be associated with the TREX/THOC, which recruited this kinase to DNA. Once at the viral genome, CDK11 phosphorylated serines at position 2 in the CTD of RNAP II, which increased levels of CPA factors at the HIV 3′ end. Higher levels of CDK11 increased the length of HIV poly(A) tails and the stability of mature viral transcripts. These data suggests that CDK11 also plays an important role in the transcriptional regulation of HIV mRNA.
CONCLUSIONS
Significant evidences have shown that overexpression and hyperactivation of CDKs are common in cancers, and targeting CDK has emerged as a highly promising therapeutic strategy. Specifically, overexpression of CDK11 has been shown to be associated with different cancers recently. As CDK11 has been found to play a crucial role in cancer cell proliferation and growth, CDK11 represents a promising target for novel anti-cancer drug development.
Acknowledgments
This work was supported by a Joint Research Fund devoted to clinical pharmacy and precision medicine (Z.D and Q.K). Support has also been provided by the National Natural Science Foundation of China (No.: 81402266 and 81372875), the Gattegno and Wechsler funds. Dr. Zhou is supported by a scholarship from the China Scholarship Council.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
REFERENCES
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Du LL, DeFoe M, Ruzinova MB, Olsen JR, Wang-Gillam A. Perioperative Therapy for Surgically Resectable Pancreatic Adenocarcinoma. Hematol Oncol Clin N. 2015;29:717–26. doi: 10.1016/j.hoc.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oseledchyk A, Zivanovic O. Intraoperative Hyperthermic Intraperitoneal Chemotherapy in Patients With Advanced Ovarian Cancer. Oncology (Williston Park) 2015:29. [PubMed] [Google Scholar]
- 4.Stone JB, DeAngelis LM. Cancer-treatment-induced neurotoxicity-focus on newer treatments. Nature reviews Clinical oncology. 2016;13:92–105. doi: 10.1038/nrclinonc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisberg E, Manley PW, Cowan-Jacob SW, Hochhaus A, Griffin JD. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nature reviews Cancer. 2007;7:345–356. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nature reviews Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 7.Rath SL, Senapati S. Molecular basis of differential selectivity of cyclobutyl-substituted imidazole inhibitors against CDKs: insights for rational drug design. PloS one. 2013;8:e73836. doi: 10.1371/journal.pone.0073836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 9.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33:1890–1903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 10.Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Therapeut. 2013;138:255–271. doi: 10.1016/j.pharmthera.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24:1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 12.Malinkova V, Vylicil J, Krystof V. Cyclin-dependent kinase inhibitors for cancer therapy: a patent review (2009 - 2014) Expert opinion on therapeutic patents. 2015;25:953–970. doi: 10.1517/13543776.2015.1045414. [DOI] [PubMed] [Google Scholar]
- 13.Law ME, Corsino PE, Narayan S, Law BK. Cyclin-Dependent Kinase Inhibitors as Anticancer Therapeutics. Molecular pharmacology. 2015;88:846–852. doi: 10.1124/mol.115.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Martinez C, Gelbert LM, Lallena MJ, de Dios A. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorganic & medicinal chemistry letters. 2015;25:3420–3435. doi: 10.1016/j.bmcl.2015.05.100. [DOI] [PubMed] [Google Scholar]
- 15.Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T, Schmidt M, Shparyk Y, Thummala AR, Voytko NL, Fowst C, Huang X, Kim ST, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. The Lancet Oncology. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 16.Mangini NS, Wesolowski R, Ramaswamy B, Lustberg MB, Berger MJ. Palbociclib: A Novel Cyclin-Dependent Kinase Inhibitor for Hormone Receptor-Positive Advanced Breast Cancer. The Annals of pharmacotherapy. 2015;49:1252–60. doi: 10.1177/1060028015602273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J. Palbociclib: a first-in-class CDK4/CDK6 inhibitor for the treatment of hormone-receptor positive advanced breast cancer. Journal of hematology & oncology. 2015;8:98. doi: 10.1186/s13045-015-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner NC, Ro J, Andre F, Loi S, Verma S, Iwata H, Harbeck N, Loibl S, Huang Bartlett C, Zhang K, Giorgetti C, Randolph S, Koehler M, Cristofanilli M. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. The New England journal of medicine. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 19.Duan Z, Zhang J, Choy E, Harmon D, Liu X, Nielsen P, Mankin H, Gray NS, Hornicek FJ. Systematic kinome shRNA screening identifies CDK11 (PITSLRE) kinase expression is critical for osteosarcoma cell growth and proliferation. Clinical cancer research. 2012;18:4580–4588. doi: 10.1158/1078-0432.CCR-12-1157. [DOI] [PubMed] [Google Scholar]
- 20.Jia B, Choy E, Cote G, Harmon D, Ye S, Kan Q, Mankin H, Hornicek F, Duan Z. Cyclin-dependent kinase 11 (CDK11) is crucial in the growth of liposarcoma cells. Cancer letters. 2014;342:104–112. doi: 10.1016/j.canlet.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Han C, Li D, Yu Z, Li F, An Q, Bai H, Zhang X, Duan Z, Kan Q. Cyclin-dependent kinase 11(p110) (CDK11(p110)) is crucial for human breast cancer cell proliferation and growth. Scientific reports. 2015;5:10433. doi: 10.1038/srep10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiedemann RE, Zhu YX, Schmidt J, Yin H, Shi CX, Que Q, Basu G, Azorsa D, Perkins LM, Braggio E, Fonseca R, Bergsagel PL, Mousses S, Stewart AK. Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood. 2010;115:1594–1604. doi: 10.1182/blood-2009-09-243980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 24.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 25.Malumbres M, Harlow E, Hunt T, Hunter T, Lahti JM, Manning G, Morgan DO, Tsai LH, Wolgemuth DJ. Cyclin-dependent kinases: a family portrait. Nature cell biology. 2009;11:1275–1276. doi: 10.1038/ncb1109-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malumbres M. Cyclin-dependent kinases. Genome biology. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. Journal of clinical oncology. 2006;24:1770–1783. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 28.Aleem E, Arceci RJ. Targeting cell cycle regulators in hematologic malignancies. Frontiers in cell and developmental biology. 2015;3:16. doi: 10.3389/fcell.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cadoo KA, Gucalp A, Traina TA. Palbociclib: an evidence-based review of its potential in the treatment of breast cancer. Breast Cancer (Dove Med Press) 2014;6:123–133. doi: 10.2147/BCTT.S46725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SH, Ryan TA. CDK5 serves as a major control point in neurotransmitter release. Neuron. 2010;67:797–809. doi: 10.1016/j.neuron.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song WJ, Son MY, Lee HW, Seo H, Kim JH, Chung SH. Enhancement of BACE1 Activity by p25/Cdk5-Mediated Phosphorylation in Alzheimer's Disease. PloS one. 2015;10:e0136950. doi: 10.1371/journal.pone.0136950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su SC, Tsai LH. Cyclin-dependent kinases in brain development and disease. Annual review of cell and developmental biology. 2011;27:465–491. doi: 10.1146/annurev-cellbio-092910-154023. [DOI] [PubMed] [Google Scholar]
- 33.Duan C, Liu Y, Lu L, Cai R, Xue H, Mao X, Chen C, Qian R, Zhang D, Shen A. CDK14 Contributes to Reactive Gliosis via Interaction with Cyclin Y in Rat Model of Spinal Cord Injury. Journal of molecular neuroscience. 2015 doi: 10.1007/s12031-015-0639-x. [DOI] [PubMed] [Google Scholar]
- 34.Park MH, Kim SY, Kim YJ, Chung YH. ALS2CR7 (CDK15) attenuates TRAIL induced apoptosis by inducing phosphorylation of survivin Thr34. Biochemical and biophysical research communications. 2014;450:129–134. doi: 10.1016/j.bbrc.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 35.Mikolcevic P, Sigl R, Rauch V, Hess MW, Pfaller K, Barisic M, Pelliniemi LJ, Boesl M, Geley S. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Molecular and cellular biology. 2012;32:868–879. doi: 10.1128/MCB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, Johansson A, Rudan I, Aulchenko YS, Kirichenko AV, Janssens AC, Jansen RC, Gnewuch C, Domingues FS, Pattaro C, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS genetics. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuda S, Kominato K, Koide-Yoshida S, Miyamoto K, Isshiki K, Tsuji A, Yuasa K. PCTAIRE kinase 3/cyclin-dependent kinase 18 is activated through association with cyclin A and/or phosphorylation by protein kinase A. The Journal of biological chemistry. 2014;289:18387–18400. doi: 10.1074/jbc.M113.542936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohno S, Shibayama M, Sato M, Tokunaga A, Yoshida N. Polypyrimidine tract-binding protein regulates the cell cycle through IRES-dependent translation of CDK11(p58) in mouse embryonic stem cells. Cell Cycle. 2011;10:3706–3713. doi: 10.4161/cc.10.21.17903. [DOI] [PubMed] [Google Scholar]
- 39.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes & development. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 40.Trembley JH, Loyer P, Hu D, Li T, Grenet J, Lahti JM, Kidd VJ. Cyclin dependent kinase 11 in RNA transcription and splicing. Progress in nucleic acid research and molecular biology. 2004;77:263–288. doi: 10.1016/S0079-6603(04)77007-5. [DOI] [PubMed] [Google Scholar]
- 41.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, Chheda MG, Tamayo P, Finn S, Shrestha Y, Boehm JS, Jain S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Feng D, Wang Q, Abdulla A, Xie XJ, Zhou J, Sun Y, Yang ES, Liu LP, Vaitheesvaran B, Bridges L, Kurland IJ, Strich R, Ni JQ, Wang C, Ericsson J, et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. The Journal of clinical investigation. 2012;122:2417–2427. doi: 10.1172/JCI61462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu DS, Zhao R, Hsu EL, Cayer J, Ye F, Guo Y, Shyr Y, Cortez D. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO reports. 2010;11:876–882. doi: 10.1038/embor.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guen VJ, Gamble C, Flajolet M, Unger S, Thollet A, Ferandin Y, Superti-Furga A, Cohen PA, Meijer L, Colas P. CDK10/cyclin M is a protein kinase that controls ETS2 degradation and is deficient in STAR syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19525–19530. doi: 10.1073/pnas.1306814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trembley JH, Hu D, Hsu LC, Yeung CY, Slaughter C, Lahti JM, Kidd VJ. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. The Journal of biological chemistry. 2002;277:2589–2596. doi: 10.1074/jbc.M109755200. [DOI] [PubMed] [Google Scholar]
- 46.Cheng SW, Kuzyk MA, Moradian A, Ichu TA, Chang VC, Tien JF, Vollett SE, Griffith M, Marra MA, Morin GB. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Molecular and cellular biology. 2012;32:4691–4704. doi: 10.1128/MCB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Cheng C, Shao B, Wu X, Ji Y, Lu X, Shen A. LPS-stimulating astrocyte-conditioned medium causes neuronal apoptosis via increasing CDK11(p58) expression in PC12 cells through downregulating AKT pathway. Cellular and molecular neurobiology. 2013;33:779–787. doi: 10.1007/s10571-013-9945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukhopadhyay A, Kramer JM, Merkx G, Lugtenberg D, Smeets DF, Oortveld MA, Blokland EA, Agrawal J, Schenck A, van Bokhoven H, Huys E, Schoenmakers EF, van Kessel AG, van Nouhuys CE, Cremers FP. CDK19 is disrupted in a female patient with bilateral congenital retinal folds, microcephaly and mild mental retardation. Human genetics. 2010;128:281–291. doi: 10.1007/s00439-010-0848-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng H, Cheng AS, Tsang DP, Li MS, Go MY, Cheung YS, Zhao GJ, Ng SS, Lin MC, Yu J, Lai PB, To KF, Sung JJ. Cell cycle-related kinase is a direct androgen receptor-regulated gene that drives beta-catenin/T cell factor-dependent hepatocarcinogenesis. The Journal of clinical investigation. 2011;121:3159–3175. doi: 10.1172/JCI45967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canavese M, Santo L, Raje N. Cyclin dependent kinases in cancer: potential for therapeutic intervention. Cancer biology & therapy. 2012;13:451–457. doi: 10.4161/cbt.19589. [DOI] [PubMed] [Google Scholar]
- 51.Chen TJ, Lee SW, Lin LC, Lin CY, Chang KY, Li CF. Cyclin-dependent kinase 4 overexpression is mostly independent of gene amplification and constitutes an independent prognosticator for nasopharyngeal carcinoma. Tumour biology. 2014;35:7209–7216. doi: 10.1007/s13277-014-1884-2. [DOI] [PubMed] [Google Scholar]
- 52.Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacology & therapeutics. 2013;138:255–271. doi: 10.1016/j.pharmthera.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 53.Park S, Lee J, Do IG, Jang J, Rho K, Ahn S, Maruja L, Kim SJ, Kim KM, Mao M, Oh E, Kim YJ, Kim J, Choi YL. Aberrant CDK4 amplification in refractory rhabdomyosarcoma as identified by genomic profiling. Scientific reports. 2014;4:3623. doi: 10.1038/srep03623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nature reviews Drug discovery. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- 55.Romano G, Giordano A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle. 2008;7:3664–3668. doi: 10.4161/cc.7.23.7122. [DOI] [PubMed] [Google Scholar]
- 56.Loyer P, Trembley JH, Katona R, Kidd VJ, Lahti JM. Role of CDK/cyclin complexes in transcription and RNA splicing. Cellular signalling. 2005;17:1033–1051. doi: 10.1016/j.cellsig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson S, Croft DR, O'Prey J, Meedendorp A, O'Prey M, Dufes C, Ryan KM. The cyclin-dependent kinase PITSLRE/CDK11 is required for successful autophagy. Autophagy. 2011;7:1295–1301. doi: 10.4161/auto.7.11.16646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bajic VP, Su B, Lee HG, Kudo W, Siedlak SL, Zivkovic L, Spremo-Potparevic B, Djelic N, Milicevic Z, Singh AK, Fahmy LM, Wang X, Smith MA, Zhu X. Mislocalization of CDK11/PITSLRE, a regulator of the G2/M phase of the cell cycle, in Alzheimer disease. Cellular & molecular biology letters. 2011;16:359–372. doi: 10.2478/s11658-011-0011-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gururajan R, Lahti JM, Grenet J, Easton J, Gruber I, Ambros PF, Kidd VJ. Duplication of a genomic region containing the Cdc2L1-2 and MMP21-22 genes on human chromosome 1p36. 3 and their linkage to D1Z2. Genome research. 1998;8:929–939. doi: 10.1101/gr.8.9.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu D, Mayeda A, Trembley JH, Lahti JM, Kidd VJ. CDK11 complexes promote pre-mRNA splicing. The Journal of biological chemistry. 2003;278:8623–8629. doi: 10.1074/jbc.M210057200. [DOI] [PubMed] [Google Scholar]
- 61.Loyer P, Trembley JH, Grenet JA, Busson A, Corlu A, Zhao W, Kocak M, Kidd VJ, Lahti JM. Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. The Journal of biological chemistry. 2008;283:7721–7732. doi: 10.1074/jbc.M708188200. [DOI] [PubMed] [Google Scholar]
- 62.Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ. PITSLRE protein kinase activity is associated with apoptosis. Molecular and cellular biology. 1995;15:1–11. doi: 10.1128/mcb.15.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beyaert R, Kidd VJ, Cornelis S, Van de Craen M, Denecker G, Lahti JM, Gururajan R, Vandenabeele P, Fiers W. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. The Journal of biological chemistry. 1997;272:11694–11697. doi: 10.1074/jbc.272.18.11694. [DOI] [PubMed] [Google Scholar]
- 64.Ariza ME, Broome-Powell M, Lahti JM, Kidd VJ, Nelson MA. Fas-induced apoptosis in human malignant melanoma cell lines is associated with the activation of the p34(cdc2)-related PITSLRE protein kinases. The Journal of biological chemistry. 1999;274:28505–28513. doi: 10.1074/jbc.274.40.28505. [DOI] [PubMed] [Google Scholar]
- 65.Loyer P, Trembley JH, Lahti JM, Kidd VJ. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. Journal of cell science. 1998;111:1495–1506. doi: 10.1242/jcs.111.11.1495. [DOI] [PubMed] [Google Scholar]
- 66.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Molecular cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 67.Choi HH, Choi HK, Jung SY, Hyle J, Kim BJ, Yoon K, Cho EJ, Youn HD, Lahti JM, Qin J, Kim ST. CHK2 kinase promotes pre-mRNA splicing via phosphorylating CDK11(p110) Oncogene. 2014;33:108–115. doi: 10.1038/onc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loyer P, Busson A, Trembley JH, Hyle J, Grenet J, Zhao W, Ribault C, Montier T, Kidd VJ, Lahti JM. The RNA binding motif protein 15B (RBM15B/OTT3) is a functional competitor of serine-arginine (SR) proteins and antagonizes the positive effect of the CDK11p110-cyclin L2alpha complex on splicing. The Journal of biological chemistry. 2011;286:147–159. doi: 10.1074/jbc.M110.192518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drogat J, Migeot V, Mommaerts E, Mullier C, Dieu M, van Bakel H, Hermand D. Cdk11-cyclinL controls the assembly of the RNA polymerase II mediator complex. Cell reports. 2012;2:1068–1076. doi: 10.1016/j.celrep.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 70.Franck N, Montembault E, Rome P, Pascal A, Cremet JY, Giet R. CDK11(p58) is required for centriole duplication and Plk4 recruitment to mitotic centrosomes. PloS one. 2011;6:e14600. doi: 10.1371/journal.pone.0014600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Petretti C, Savoian M, Montembault E, Glover DM, Prigent C, Giet R. The PITSLRE/CDK11p58 protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO reports. 2006;7:418–424. doi: 10.1038/sj.embor.7400639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu D, Valentine M, Kidd VJ, Lahti JM. CDK11(p58) is required for the maintenance of sister chromatid cohesion. Journal of cell science. 2007;120:2424–2434. doi: 10.1242/jcs.007963. [DOI] [PubMed] [Google Scholar]
- 73.Li T, Inoue A, Lahti JM, Kidd VJ. Failure to proliferate and mitotic arrest of CDK11(p110/p58)-null mutant mice at the blastocyst stage of embryonic cell development. Molecular and cellular biology. 2004;24:3188–3197. doi: 10.1128/MCB.24.8.3188-3197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yokoyama H, Gruss OJ, Rybina S, Caudron M, Schelder M, Wilm M, Mattaj IW, Karsenti E. Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. The Journal of cell biology. 2008;180:867–875. doi: 10.1083/jcb.200706189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, Yaffe MB. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 77.Rakkaa T, Escude C, Giet R, Magnaghi-Jaulin L, Jaulin C. CDK11(p58) kinase activity is required to protect sister chromatid cohesion at centromeres in mitosis. Chromosome research. 2014;22:267–276. doi: 10.1007/s10577-013-9400-x. [DOI] [PubMed] [Google Scholar]
- 78.Zhang S, Cai M, Xu S, Chen S, Chen X, Chen C, Gu J. Interaction of p58(PITSLRE), a G2/M-specific protein kinase, with cyclin D3. The Journal of biological chemistry. 2002;277:35314–35322. doi: 10.1074/jbc.M202179200. [DOI] [PubMed] [Google Scholar]
- 79.Shi J, Feng Y, Goulet AC, Vaillancourt RR, Sachs NA, Hershey JW, Nelson MA. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. The Journal of biological chemistry. 2003;278:5062–5071. doi: 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- 80.Mikolajczyk M, Shi J, Vaillancourt RR, Sachs NA, Nelson M. The cyclin-dependent kinase 11(p46) isoform interacts with RanBPM. Biochemical and biophysical research communications. 2003;310:14–18. doi: 10.1016/j.bbrc.2003.08.116. [DOI] [PubMed] [Google Scholar]
- 81.Trembley JH, Hu D, Slaughter CA, Lahti JM, Kidd VJ. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. The Journal of biological chemistry. 2003;278:2265–2270. doi: 10.1074/jbc.M207518200. [DOI] [PubMed] [Google Scholar]
- 82.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nature genetics. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Evangelista M, Lim TY, Lee J, Parker L, Ashique A, Peterson AS, Ye W, Davis DP, de Sauvage FJ. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Science signaling. 2008;1:ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- 84.Naik S, Dothager RS, Marasa J, Lewis CL, Piwnica-Worms D. Vascular Endothelial Growth Factor Receptor-1 Is Synthetic Lethal to Aberrant {beta}-Catenin Activation in Colon Cancer. Clinical cancer research. 2009;15:7529–7537. doi: 10.1158/1078-0432.CCR-09-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Developmental cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development. 2014;141:3445–3457. doi: 10.1242/dev.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kahle A, Feng Y, M AN. Isolation and characterization of the human Cdc2L1 gene promoter. Gene. 2005;344:53–60. doi: 10.1016/j.gene.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 89.Feng Y, Goulet AC, Nelson MA. Identification and characterization of the human Cdc2l2 gene promoter. Gene. 2004;330:75–84. doi: 10.1016/j.gene.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 90.Sachs NA, Vaillancourt RR. Cyclin-dependent kinase 11(p110) activity in the absence of CK2. Biochimica et biophysica acta. 2003;1624:98–108. doi: 10.1016/j.bbagen.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Kugler MC, Joyner AL, Loomis CA, Munger JS. Sonic hedgehog signaling in the lung. From development to disease. American journal of respiratory cell and molecular biology. 2015;52:1–13. doi: 10.1165/rcmb.2014-0132TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 93.Mikolajczyk M, Nelson MA. Regulation of stability of cyclin-dependent kinase CDK11p110 and a caspase-processed form, CDK11p46, by Hsp90. The Biochemical journal. 2004;384:461–467. doi: 10.1042/BJ20040848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feng Y, Qi W, Martinez J, Nelson MA. The cyclin-dependent kinase 11 interacts with 14-3-3 proteins. Biochemical and biophysical research communications. 2005;331:1503–1509. doi: 10.1016/j.bbrc.2005.04.078. [DOI] [PubMed] [Google Scholar]
- 95.Pierre F, Chua PC, O'Brien SE, Siddiqui-Jain A, Bourbon P, Haddach M, Michaux J, Nagasawa J, Schwaebe MK, Stefan E, Vialettes A, Whitten JP, Chen TK, Darjania L, Stansfield R, Bliesath J, et al. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Molecular and cellular biochemistry. 2011;356:37–43. doi: 10.1007/s11010-011-0956-5. [DOI] [PubMed] [Google Scholar]
- 96.Seto T, Esaki T, Hirai F, Arita S, Nosaki K, Makiyama A, Kometani T, Fujimoto C, Hamatake M, Takeoka H, Agbo F, Shi X. Phase I, dose-escalation study of AZD7762 alone and in combination with gemcitabine in Japanese patients with advanced solid tumours. Cancer chemotherapy and pharmacology. 2013;72:619–627. doi: 10.1007/s00280-013-2234-6. [DOI] [PubMed] [Google Scholar]
- 97.Kren BT, Unger GM, Abedin MJ, Vogel RI, Henzler CM, Ahmed K, Trembley JH. Preclinical evaluation of cyclin dependent kinase 11 and casein kinase 2 survival kinases as RNA interference targets for triple negative breast cancer therapy. Breast cancer research. 2015;17:19. doi: 10.1186/s13058-015-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tiedemann RE, Zhu YX, Schmidt J, Shi CX, Sereduk C, Yin H, Mousses S, Stewart AK. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer research. 2012;72:757–768. doi: 10.1158/0008-5472.CAN-11-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feng Y, Sassi S, Shen JK, Yang X, Gao Y, Osaka E, Zhang J, Yang S, Yang C, Mankin HJ, Hornicek FJ, Duan Z. Targeting CDK11 in osteosarcoma cells using the CRISPR-Cas9 system. Journal of orthopaedic research. 2015;33:199–207. doi: 10.1002/jor.22745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiang J, Lahti JM, Grenet J, Easton J, Kidd VJ. Molecular cloning and expression of alternatively spliced PITSLRE protein kinase isoforms. The Journal of biological chemistry. 1994;269:15786–15794. [PubMed] [Google Scholar]
- 101.Housden BE, Valvezan AJ, Kelley C, Sopko R, Hu Y, Roesel C, Lin S, Buckner M, Tao R, Yilmazel B, Mohr SE, Manning BD, Perrimon N. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Science signaling. 2015;8:rs9. doi: 10.1126/scisignal.aab3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pak V, Eifler TT, Jager S, Krogan NJ, Fujinaga K, Peterlin BM. CDK11 in TREX/THOC Regulates HIV mRNA 3′ End Processing. Cell host & microbe. 2015;18:560–570. doi: 10.1016/j.chom.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cary DC, Fujinaga K, Peterlin BM. Molecular mechanisms of HIV latency. The Journal of clinical investigation. 2016:1–7. doi: 10.1172/JCI80565. [DOI] [PMC free article] [PubMed] [Google Scholar]