Abstract
Aldehyde dehydrogenase family 1, member L1 (ALDH1L1) is a recently characterized pan-astrocytic marker that is more homogenously expressed throughout the brain than the classic astrocytic marker, glial fibrillary acidic protein. We generated putative promoter sequence variants of the rat ALDH1L1 gene for use in adeno-associated viral vector-mediated gene transfer, with an aim to achieve selective regulation of transgene expression in astrocytes in the rat brain. Unexpectedly, ALDH1L1 promoter variants mediated transcriptional activity exclusively in neurons in the substantia nigra pars compacta as assessed by luciferase reporter expression at 3 weeks postvector infusion. This selectivity for neurons in the substantia nigra pars compacta also persisted in the context of adeno-associated viral serotype 5, 8 or 9 vector-mediated gene delivery. An in vivo promoter comparison showed the highest performing ALDH1L1 promoter variant mediated higher transgene expression than the neuronal-specific synapsin 1 and tyrosine hydroxylase promoters. The ALDH1L1 promoter was also transcriptionally active in dentate granule neurons following intrahippocampal adeno-associated viral vector infusion, whereas transgene expression was detected in both striatal neurons and astrocytes following vector infusion into the striatum. Our results demonstrate the potential suitability of the ALDH1L1 promoter as a new tool in the development of gene therapy and disease modelling applications.
Introduction
To date, clinically-approved adeno-associated viral (AAV) vectors for central nervous system (CNS) gene therapy almost exclusively target neurons, and astrocytes represent a largely unexplored therapeutic target.1–5 In the human brain, an abundance of astrocytes (~1.4 astrocytes to every neuron) regulate health and function of the CNS, and their dysfunction contributes to disease progression in neurodegenerative diseases.6 Given that disease pathogenesis is dictated by complex neuronal-glia interactions, and chronic neurodegeneration is characterized by substantial neuronal loss and astrogliosis, astrocytes may represent a promising additional cellular target for CNS gene therapy. A greater understanding of the diverse molecular expression profiles of neurons and glia, coupled with a rapidly expanding repertoire of novel viral serotypes, have assisted the development of viral vectors that exhibit diverse tropisms and efficient transduction in the CNS.
Effective astrocyte-targeting has been achieved by coupling astrocyte tropic lentiviral7 and AAV serotypes with the classic astrocyte-specific glial fibrillary acidic protein (GFAP) promoter.8–10 However, it is increasingly evident that astrocytes are a diverse population of cells that exhibit extensive molecular heterogeneity; and GFAP, traditionally considered a pan-astrocytic marker is one such heterogeneously expressed molecule, as reflected by its region-dependent expression.11–14 Genome-wide transcriptional profiling has shown that the aldehyde dehydrogenase family 1, member L1 (ALDH1L1) is more homogenously and selectively expressed in astrocytes throughout the brain in a pattern more consistent with pan-astrocyte expression than GFAP.14 GFAP mRNA is predominantly expressed in white matter, while cellular expression of the protein is detected in the cell body and the main astrocytic processes. In contrast, ALDH1L1 mRNA is more extensively expressed throughout the CNS and its protein expression extends throughout the cell body to the finer astrocytic processes. Indeed, ALDH1L1 labels both GFAP-positive and GFAP-negative astrocytes.14 A bacterial artificial chromosome transgenic mouse that expresses enhanced green fluorescent protein (eGFP) under the control of the ALDH1L1 genomic promoter replicates the astrocyte-specific pattern of expression of endogenous ALDH1L1.13 Furthermore, nucleotide sequences that range from 300 to 1,500 bp in size, from the region immediately upstream of the transcription start site of the ALDH1L1 gene have been shown to exhibit transcriptional activity in A549 lung carcinoma cells in vitro, in a manner dependent on the presence of exon 1 of the gene.15 These studies suggest the potential of ALDH1L1 promoter sequences for applications targeting viral vector-mediated gene delivery to astrocytes.
Based on these findings, we aimed to characterize four putative ALDH1L1 promoter sequence variants for utilization in AAV vector-mediated gene transfer to astrocytes in the rat substantia nigra pars compacta (SNpc) brain region. Unexpectedly, we found that all four promoter variants directed transgene expression exclusively in neurons in the rat substantia nigra, with the neuronal tropism of the ALDH1L1 promoters persisting in the context of AAV serotypes, 5, 8, and 9. Moreover, the ALDH1L1(long)exon1 promoter exhibited significantly greater transcriptional activity in SNpc neurons compared with the commonly used neuronal-specific synapsin 1 (Syn) and tyrosine hydroxylase (TH) promoters. While the neuronal-specific pattern of transgene expression was maintained following AAV9 vector infusion into the rat hippocampus, eGFP transgene expression was found in both neurons and astrocytes in the striatum following intrastriatal vector infusion. Our results suggest that the ALDH1L1 promoters could be a useful addition to the arsenal of tools used in the development of gene therapy and disease modelling applications.
Results
ALDH1L1 promoter variants coupled with AAV9 selectively target neurons in the SNpc
Previous studies characterizing the potential utility of ALDH1L1 regulatory sequences in gene delivery applications prompted us to investigate cell type-specificity and activity of ALDH1L1 promoter sequences with and without exon 1 in the context of AAV vector-mediated gene expression in vivo. Firstly, to determine whether 5ʹ sequences upstream of the ALDH1L1 transcription start site exhibit efficient transcriptional activity in the absence of exon 1, we generated AAV expression plasmids (Figure 1) expressing a luciferase (Luc) reporter gene under control of -931 bp ALDH1L1(short) (ALDH1L1(S)) and -1,974 bp ALDH1L1(long) (ALDH1L1(L)) nucleotide fragments relative to the ALDH1L1 transcriptional start site (0 bp) (Figure 1). To determine the influence of exon 1, that may contain enhancer sequences,15 a +138 bp sequence spanning exon 1 of the ALDH1L1 gene was cloned into the above promoter constructs to generate additional ALDH1L1(short)exon1 (ALDH1L1(S)ex1)- and ALDH1L1(long)exon1 (ALDH1L1(L)ex1)- Luc plasmids (Figure 1). The four ALDH1L1 promoter variants were packaged into AAV serotype 9 (AAV9) vectors. To characterize their tropism in the rat SNpc, titer-matched AAV9 vectors (2 × 109 genomes) were unilaterally injected into the rat SNpc (n = 3 rats per vector), and cellular transduction patterns of transgene expression were analyzed at 3 weeks by immunohistochemical analysis. An additional subgroup of rats injected with an AAV9 vector expressing a yellow fluorescent protein reporter gene (YFP) under the control of a 2.2 kb GFAP promoter (AAV9-GFAP-YFP) that targets astrocytes as well as neurons in the rat hippocampus10 was included to enable comparisons between the cellular transduction patterns mediated by the different promoters.
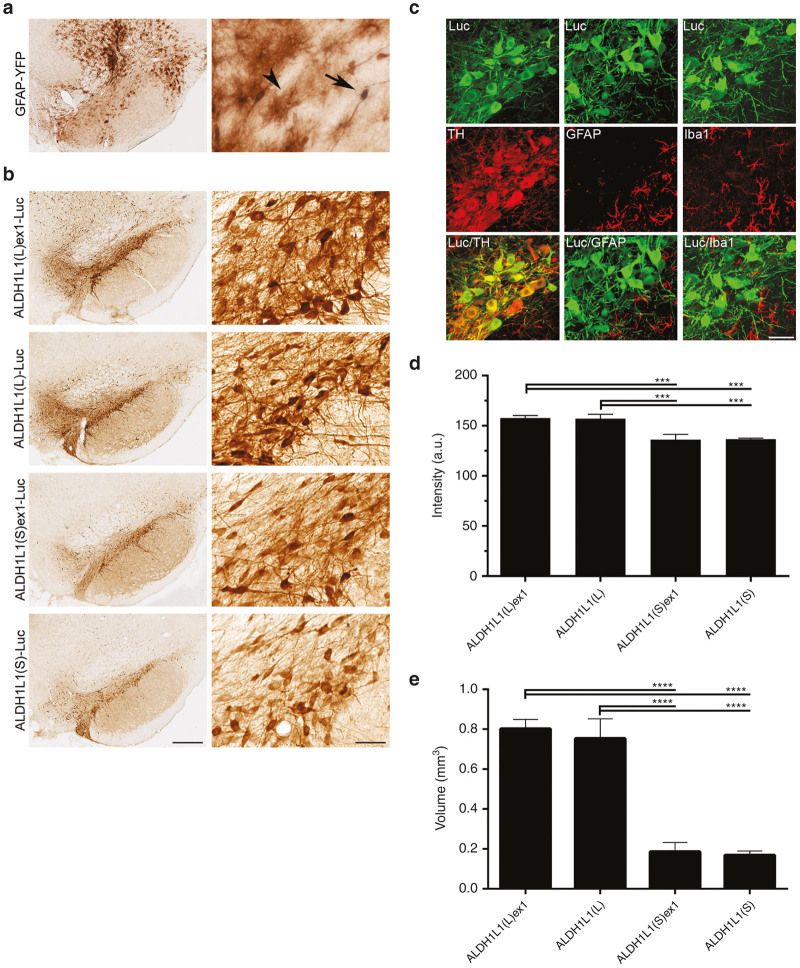
Figure 1.
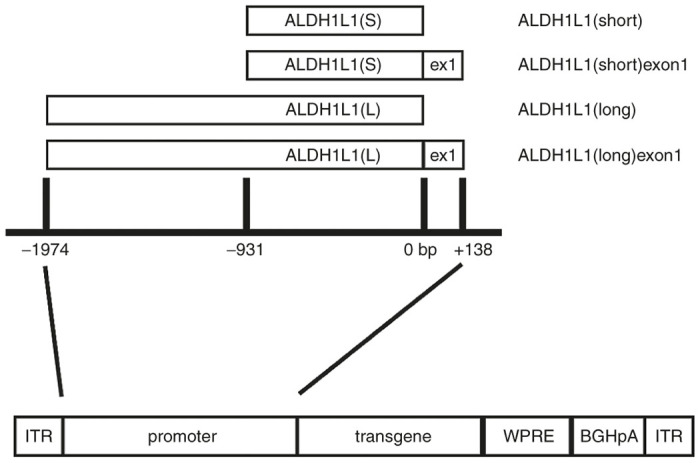
Schematic representation of the ALDH1L1 promoter sequences relative to the transcription start site (0 bp), and the AAV expression cassette configuration (ITR, inverted terminal repeat; WPRE, woodchuck post-transcriptional regulatory element; BGHpA, bovine growth hormone polyadenylation signal). ALDH1L1, aldehyde dehydrogenase family 1, member L1.
Consistent with our previous observations, YFP immunoreactivity was detected in both “star-like” astrocytes with highly ramified processes, as well as neurons in rats injected with the AAV9-GFAP-YFP vector (Figure 2a), indicating that while the GFAP promoter can direct expression in astrocytes, nonspecific transcriptional activity also occurs in a subset of neurons. Unexpectedly, all four ALDH1L1 promoter sequences targeted Luc expression exclusively to nigral neurons as determined by the morphology of transgene-expressing cells (Figure 2b). Luc was expressed throughout neuronal cell bodies and their fibers, while there was no evidence of transgene expression in astrocytes. Selective neuronal transgene expression was confirmed by double-immunofluorescent labelling using antibodies to cell-specific markers. Luc-positive cells exclusively colocalized with the dopamine neuronal marker TH, but not with the astrocytic and microglial markers, GFAP and ionized calcium binding adapter molecule 1 (Iba1), respectively (Figure 2c). Due to weak immunoreactivity achieved with an anti-ALDH1L1 antibody, an anti-GFAP antibody was used for labelling astrocytes (results not shown).
Figure 2.
ALDH1L1 promoter variants selectively target transgene expression to nigral neurons. (a) Immunolabelling with anti-GFP antibody showing dYFP expression in neurons (arrow) and astrocytes (arrowhead) in a representative AAV9-GFAP-YFP injected brain. (b) Immunolabelling with anti-Luc antibody showing promoter length-dependent neuronal-specific Luc immunoreactivity in representative nigral sections injected with ALDH1L1(L)ex1-, ALDH1L1(L)-, ALDH1L1(S)ex1-, or ALDH1L1(S)-coupled AAV9 vector constructs. (c) Double-immunofluorescent labelling of Luc expressing cells with TH, GFAP, and Iba1. Neuronal-specific activity of ALDH1L1 promoters represented by colocalization of ALDH1L1(L)ex1-regulated Luc expression in neurons, but not astrocytes and microglia. (d) Intensity of Luc immunoreactivity in SNpc. (e) Total volume of transgene expression within the SNpc, SNpr and VTA. Bars represent the mean ± SEM for n = 3; One-way analysis of variance (ANOVA) **P < 0.01, ***P < 0.001. Scale bars for a and b 500 µm for low-power image and 100 µm for high-power inset, and c 50 µm. AAV, adeno-associated viral; ALDH1L1, aldehyde dehydrogenase family 1, member L1; GFAP, glial fibrillary acidic protein; GFP, green fluorescent protein; Iba1, ionized calcium binding adapter molecule 1; Luc, luciferase; SEM, standard error of mean; SNpc, substantia nigra pars compacta; SNpr, SN pars reticulata; VTA, ventral tegmental area; YFP, yellow fluorescent protein.
The levels of transgene expression appeared to positively correlate with the sequence length of the promoters, with the two promoters containing the 1,974 bp ALDH1L1(L) sequence mediating considerably higher transgene expression in comparison to the shorter counterparts containing the 931 bp ALDH1L1(S) sequence (Figure 2b,d), as determined by semiquantitative analysis of the density of Luc immunoreactivity in the SNpc. Both ALDH1L1(L)ex1 and ALDH1L1(L) promoters mediated significantly higher transgene expression compared with ALDH1L1(S)ex1, or ALDH1L1(S) sequences (One-way analysis of variance (ANOVA); F3,8 = 10.56; P < 0.01). Inclusion of exon 1 in the ALDH1L1(L) and ALDH1L1(S) promoters did not result in any further increase in transgene expression. In addition to the selective nigral transgene expression achieved with all ALDH1L1 promoter variants, ALDH1L1(L)ex1- and ALDH1L1(L)-regulated Luc expression spread into the neighboring ventral tegmental area (VTA), SN pars reticulata (SNpr), and the midbrain reticular nucleus (Supplementary Figure S1), while transgene expression was more restricted to the SNpc with ALDH1L1(S)ex1 and ALDH1L1(S) (Figure 2b). Quantification of total volume of transduced brain region further confirmed the sequence length-dependent activity of the ALDH1L1 promoters. The highest volume of transgene expression within the SNpc, SNpr, and VTA was achieved with ALDHL1(L)ex1 and ALDH1L1(L) (Figure 2e) compared with both ALDH1L1(S)ex1 and ALDH1L1(S) (One-way ANOVA; F3,8 = 33.73; P < 0.001). Inclusion of exon 1 in the long or short ALDH1L1 promoters also had no effect on volume of transgene expression.
Neurotropism of the ALDH1L1 promoters persists in the context of astrocyte-tropic AAV serotypes 5, 8, and 9
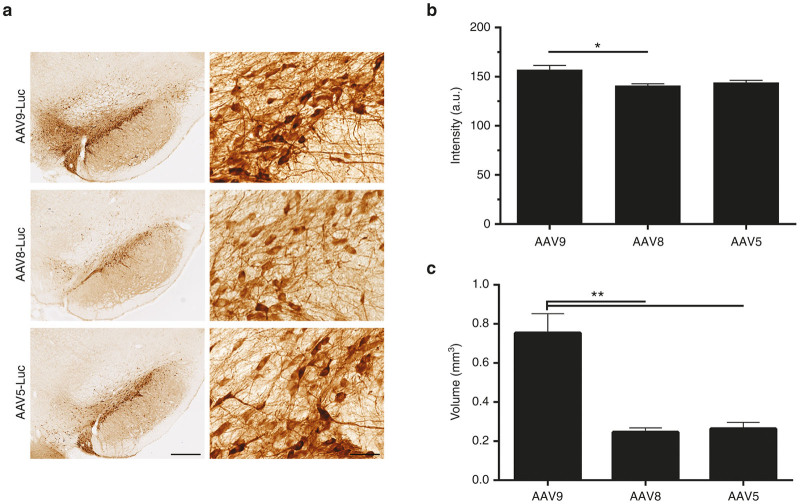
To determine whether we could direct cell-specificity of the ALDH1L1 promoters toward astrocytes in an AAV serotype-dependent manner, their properties in the context of two additional astrocyte-tropic serotypes, AAV5 and AAV8 were subsequently investigated. Similar to AAV9, these serotypes have previously exhibited astrocytic tropism in various regions in the rodent and nonhuman primate CNS.8,9,16 Selective neuronal tropism of the ALDH1L1 promoters persisted in the context of AAV5, and AAV8 at 3 weeks after intranigral infusion of titer-matched vectors (2 × 109 genomes, n = 3 per vector), based on Luc-immunoreactive cell morphology, as represented in the context of ALDH1L1(L)-mediated transgene expression (Figure 3a). In the SNpc, AAV9-mediated Luc expression was higher compared with AAV8 (One-way ANOVA; F2,6 = 7.98; P < 0.05) (Figure 3b), while there was no difference in the intensity of Luc immunoreactivity between AAV9 and AAV5 vector-injected SNpc. Our results suggest that although a larger transduction volume was achieved with AAV9 in comparison with AAV8 and AAV5 (One-way ANOVA; F2,6 = 24.99; P < 0.01), more restricted targeting of SNpc dopaminergic neurons might be better achieved with AAV8 and AAV5 vectors (Figure 3c).
Figure 3.
ALDH1L1 promoter variants coupled with AAV serotypes 9, 8, and 5 direct transgene expression to nigral neurons. (a) Neuronal Luc expression mediated by ALDH11L1 promoters in the context of AAV9, AAV8, and AAV5 (represented by ALDH1L1(L) vector constructs). (b) Intensity of Luc immunoreactivity in the SNpc. (c) Total volume of transduction (mm3) within the SNpc, SNpr, and VTA. Bars represent mean ± SEM, n = 3, One-way analysis of variance (ANOVA), *P < 0.05, **P < 0.01. Scale bars for (a) 500 µm for low-power image and 100 µm for high-power inset. AAV, adeno-associated viral; ALDH1L1, aldehyde dehydrogenase family 1, member L1; Luc, luciferase; SEM, standard error of mean; SNpc, substantia nigra pars compacta; SNpr, SN pars reticulata; VTA, ventral tegmental area.
ALDH1L1(long)exon1 mediates superior nigral transgene expression in comparison to commonly used neuronal promoters, Syn and TH
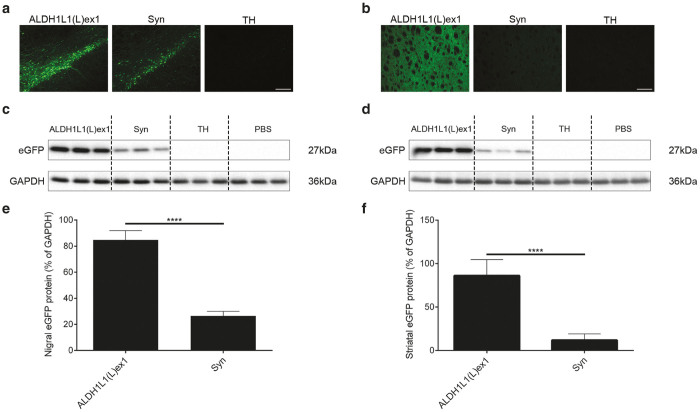
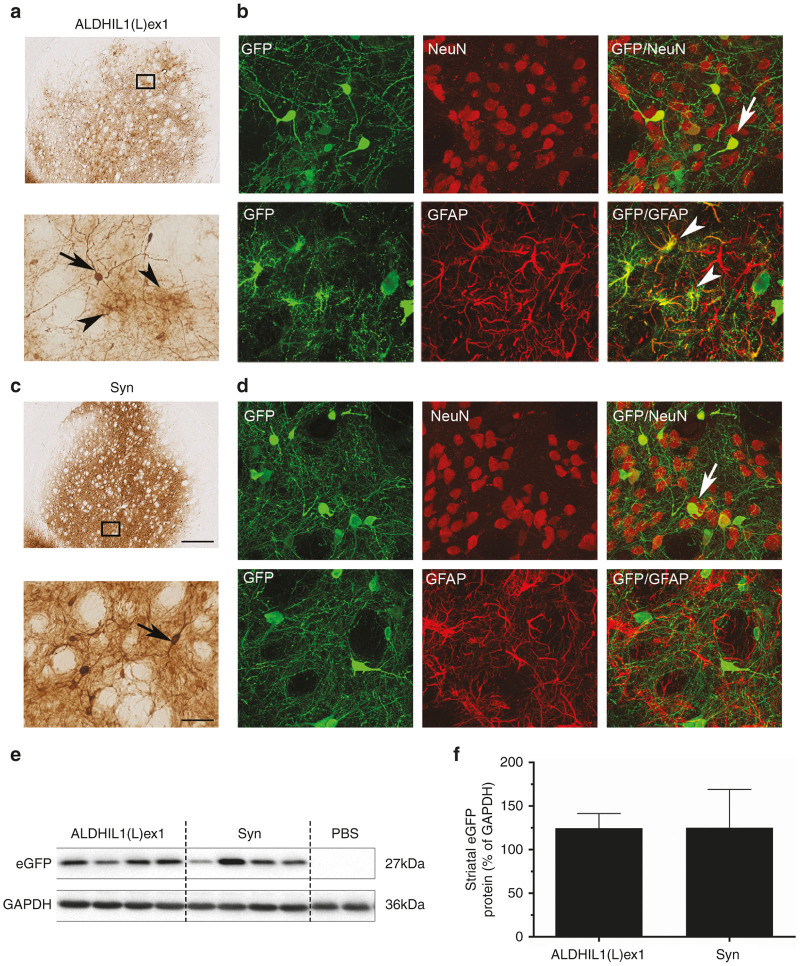
To determine whether the ALDH1L1(L)ex1 promoter variant could potentially mediate superior neuronal transgene expression than commonly used neuronal specific promoters, we conducted a comparison between the ALDH1L1(L)ex1 and a human Syn promoter or rat TH promoter in the context of AAV9, the serotype that mediated the highest levels of transgene expression. We generated additional AAV plasmids expressing eGFP reporter gene under the control of these promoters and packaged AAV9 vectors. Titer-matched AAV9 vectors (2 × 109 genomes) were unilaterally injected into the rat SNpc (n = 10 per vector), and transgene expression levels were analyzed at 3 weeks postvector infusion. eGFP fluorescence in fixed nigral sections (Figure 4a), and semiquantitative Western blot analysis of nigral lysates (Figure 4c,e) indicated that ALDH1L1(L)ex1 mediated significantly higher levels of eGFP expression in the nigra in comparison to Syn (ALDH1L1(L)ex1 84.75 ± 3.18% versus Syn 26.40 ± 1.64%; unpaired t-test, t8 = 16.30; P < 0.0001), while TH-regulated eGFP expression was barely detectable, although prolonged exposure of the blots confirmed low level eGFP expression (Supplementary Figure S2). The levels of eGFP expression in the dopaminergic neuronal terminal fields in the striatum paralleled that in the nigra, with the intensity of signal strongest with the ALDH1L1 promoter (ALDH1L1(L)ex1 86.14 ± 8.22% versus Syn 11.76 ± 3.28%, TH-regulated eGFP expression undetected; unpaired t-test, t8 = 8.41; P < 0.0001) confirming high transcriptional activity of the ALDH1L1(L)ex1 promoter (Figure 4b,d,f).
Figure 4.
ALDH1L1(L)ex1 mediates significantly higher levels of nigral and striatal transgene expression in comparison to Syn and TH promoters, following intranigral AAV vector infusion. Representative images showing (a) nigral and (b) striatal eGFP fluorescence in rats injected with AAV9 vectors expressing eGFP under control of ALDH1L1(L)ex1, Syn or TH promoters. Representative western blot analysis of (c) nigral and (d) striatal homogenates from rats injected with PBS vehicle, ALDH1L1(L)ex1, Syn, and TH AAV vectors probed with anti GFP antibody. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the loading control. Semiquantitative analysis of band intensities from western blots of (e) nigral and (f) striatal lysates. Bars represent the mean ± SEM, n = 5 per vector, unpaired Student’s t-test, ****P < 0.0001. Scale bar for a and b 200 µm. AAV, adeno-associated viral; ALDH1L1, aldehyde dehydrogenase family 1, member L1; eGFP, enhanced green fluorescent protein; PBS, phosphate buffered saline; SEM, standard error of mean; SNpc, substantia nigra pars compacta; Syn, synapsin 1; TH, tyrosine hydroxylase.
ALDH1L1(L)ex1 promoter directs transgene expression in hipppocampal dentate granule neurons but both neurons and astrocytes in the rat striatum
Next, we asked whether the neuronal specificity of ALDH1L1(L)ex1 promoter activity observed is also maintained in other brain regions such as the striatum and hippocampus. AAV9-ALDH1L1(L)ex1-eGFP or AAV9-Syn-eGFP vectors (2 × 109 genomes) were unilaterally injected into the hippocampus (n = 8 per vector) or striatum (n = 8 per vector) in subgroups of rats, and transgene expression levels were analyzed at 3 weeks postvector infusion.
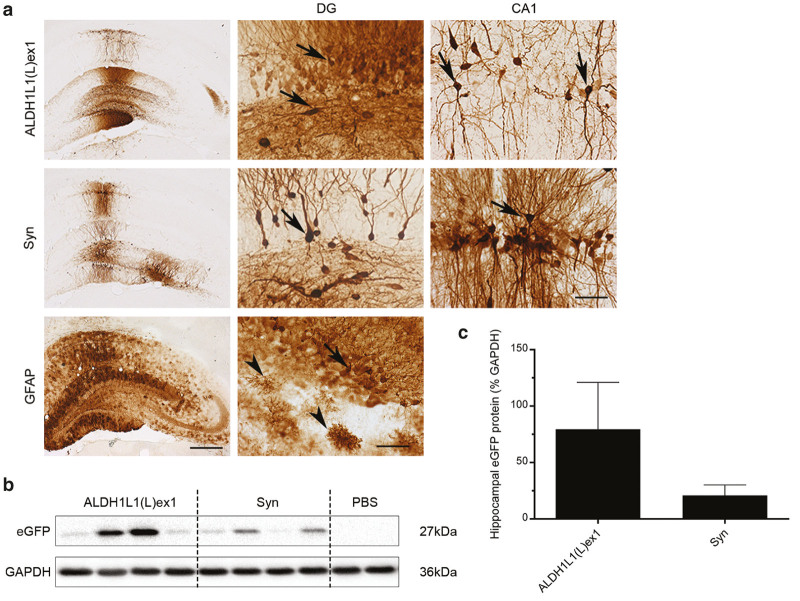
In the hippocampus, both vectors targeted eGFP immunoreactivity predominantly to dentate granule neurons, with a very sparse number of eGFP-expressing CA1 pyramidal neurons also detected (Figure 5a). In comparison to the detection of the transgene in astrocytes in hippocampal sections from a rat that had been injected with an AAV9-GFAP-dYFP10 (Figure 5a), we did not observe any eGFP-expressing cells with an astrocytic morphology in the ALDH1L1-eGFP or Syn-eGFP injected brains (confirmed with double-immunofluorescent labeling; data not shown), consistent with our observations in the substantia nigra. Unexpectedly, there was a wide variability in eGFP expression levels between animals allocated to both immunohistochemistry and Western blot analysis within each treatment group. This contributed to the lack of detection of any differences when comparing Western blot band intensities between treatments (ALDH1L1(L)ex1 79.38 ± 41.62% versus Syn 20.99 ± 9.14%; unpaired t-test, t6 = 1.37) (Figure 5b,c). Although further studies will be required, a general observation across all high expressing samples in both the immunohistochemical and Western blot analysis suggested that the ALDH1L1 promoter produced higher expression levels compared with the Syn promoter (Figure 5b,c).
Figure 5.
ALDH1L1(L)ex1 mediates transgene expression in neurons following intrahippocampal AAV vector infusion. (a) Representative low power images showing eGFP immunoreactivity in the hippocampus of rats injected with AAV9-ALDH1L1(L)ex1 or -Syn vectors, in comparison to staining in a brain injected with AAV9-GFAP-dYFP. High magnification images of eGFP-immunoreactive neurons (arrows) or astrocytes (arrowheads in the GFAP-dYFP injected brain) in the dentate gyrus (DG) or CA1 region from these sections are included. Western blot analysis of (b) hippocampal homogenates from rats injected with PBS vehicle, ALDH1L1(L)ex1, and Syn AAV vectors probed with anti GFP antibody. GAPDH was used as the loading control. (c) Semiquantitative analysis of band intensities from western blots. Bars represent the mean ± SEM, n = 4 per vector. Scale bar for low power images 500 and 200 µm for high power images. AAV, adeno-associated viral; ALDH1L1, aldehyde dehydrogenase family 1, member L1; eGFP, enhanced green fluorescent protein; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; PBS, phosphate buffered saline; SEM, standard error of mean; Syn, synapsin 1; YFP, yellow fluorescent protein.
We found that although eGFP expression levels in the striatum between the two vector treatment groups were comparable by Western blot analysis (ALDH1L1(L)ex1 124.55 ± 16.80% versus Syn 125.06 ± 43.98%; unpaired t-test, t6 = 0.011) (Figure 6), there were significant differences in the transgene expression patterns following intrastriatal infusion of these vectors. As expected, eGFP was exclusively expressed in neurons and surrounding neuropil in the Syn-eGFP vector-injected brains, while eGFP immunoreactivity was more diffuse and found in both neurons as well as astrocytes in the ALDH1L1(L)ex1- vector injected striatum (Figure 6a,b). Neuropil staining was lighter in the ALDH1L1(L)ex1- compared with the Syn-eGFP vector injected striatum. Further studies will be required to determine whether this is reflective of transgene expression in different subpopulations of striatal neurons. Altogether, these results suggest that the ALDH1L1(L)ex promoter is transcriptionally active in neurons and some astrocytes but in a region-dependent manner.
Figure 6.
ALDH1L(L)ex1 targets transgene expression in both neurons and astrocytes following intrastriatal AAV vector infusion. Representative low power images showing eGFP immunoreactivity in striatum from rats injected with (a) AAV9-ALDH1L1(L)ex1 or (c) Syn promoters. High magnification images show eGFP-immunoreactive neurons (arrow) and astrocytes (arrowhead) in a brain injected with a AAV9-ALDH1L1(L)ex1-eGFP, and selectively in neurons (arrow) in a brain injected with c AAV9-Syn-eGFP. (b, d) Double-immunofluorescent colabeling of eGFP expressing cells with NeuN or GFAP showing neuronal (arrow) and astrocytic (arrowhead) transgene expression. Western blot analysis of (e) striatal homogenates from rats injected with PBS vehicle, ALDH1L1(L)ex1, and Syn AAV vectors probed with anti GFP antibody. GAPDH was used as the loading control. (f) Semiquantitative analysis of band intensities from western blots. Bars represent the mean ± SEM, n = 4 per vector. Scale bar for a, c low power images 500 µm and 100 µm for high power images, and b, d 50 µm. AAV, adeno-associated viral; ALDH1L1, aldehyde dehydrogenase family 1, member L1; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; GFAP, glial fibrillary acidic protein; eGFP, enhanced green fluorescent protein; PBS, phosphate buffered saline; SEM, standard error of mean.
Discussion
ALDH1L1 is an astrocyte marker that is more homogenously expressed in astrocytes throughout the brain than the classic astrocyte marker, GFAP14; and indeed, the genomic ALDH1L1 promoter in bacterial artificial chromosome transgenic mice has been shown to target GFP expression selectively to astrocytes in a pattern that parallels the expression of the endogenous ALDH1L1 gene.13 In this study, we generated AAV vectors containing four ALDH1L1 putative promoter sequences and evaluated the ability of these variants to transcriptionally regulate Luc expression in astrocytes. We initially focused on characterization of cellular transduction patterns in the rat SNpc. Unexpectedly, the ALDH1L1 promoter variants targeted transgene expression exclusively to neurons, predominantly in the SNpc and to a lesser extent in neurons in neighboring midbrain regions. The longer ALDH1L1(L)ex1 and ALDH1L1(L) variants mediated higher levels of Luc expression compared with the shorter ALDH1L1(S) promoters, suggesting that the longer promoters contain more DNA elements necessary for increased transcriptional activity. Based on the assumption that the extent of diffusion and spread of the four AAV9-ALDH1L1 vectors in the SN would be similar, more widespread detection of Luc immunoreactive neurons in the two longer ALDH1L1(L) promoter treatment groups at sites distal to the infusion site, where cells would be expected to be transduced with fewer AAV vector genomes, is also indicative of stronger transcriptional activity. This contributed to significantly increased volumes of transduced tissue in the ALDH1L1(L)ex1- and ALDH1L1(L)- vector-injected brains.
Of the native AAV serotypes characterized thus far, AAV9, AAV8, and AAV5 are the most efficient serotypes for astrocyte transduction; we and others have shown that these serotypes exhibit astrocytic tropism in various regions in the rodent and nonhuman primate CNS.8–10,16–18 Consistent with our previous results, we found that AAV9 coupled with the widely used GFAP promoter targeted transgene expression to both nigral neurons and astrocytes confirming that nigral astrocytes are permissive to both AAV9 transduction, and also that an exogenously delivered promoter can be transcriptionally active in these cells. Because astrocytes exhibit a broad molecular and functional heterogeneity under resting and pathological states,11,19–26 such molecular diversity may influence region-specific tropisms and transduction efficacies of viral vectors. A switch to either an AAV8 or AAV5 vector serotype to mediate gene transfer did not alter the neuronal transduction pattern, confirming that a lack of detectable levels of transcriptional activity of the ALDH1L1 promoter in astrocytes contributes to an absence of astrocytic Luc expression. Furthermore, our results suggest that although AAV9 may mediate more efficient neuronal transduction mechanisms in the rat SNpc as assessed by the density of Luc immunoreactivity when compared with the AAV8 vector-injected group, the large difference in transduction volumes between these vector serotypes suggests that more focal targeting of transgene expression to the SNpc would be better achieved with AAV5 or AAV8 vectors. Altogether, we conclude that the ALDH1L1 promoter variants exclusively target neurons in the SNpc.
To broaden the potential utility of these promoters, we determined whether the neuronal transgene expression pattern with the ALDH1L1(L)ex1 promoter was maintained in other brain regions. We found that consistent with that observed following intranigral infusions, transcriptional activity of the ALDH1L1(L)ex1 promoter was restricted to neurons in the hippocampal dentate gyrus using a sensitive anti-GFP antibody and diaminobenzidine (DAB) chromogen to detect eGFP transgene. In contrast, eGFP expression was observed in neurons and astrocytes in the striatum using the same immunodetection method suggesting that striatal astrocytes may possess potential subtype-specific molecular mechanisms that allow more efficient AAV transduction and/ or ALDH1L1(L)ex1 promoter activity. However, the weaker level of transgene expression in astrocytes compared with a moderate level of staining in neurons suggests that targeting of transgene expression in striatal astrocytes may be less efficient than that in neurons. Future work analyzing the neuron to astrocyte transduction ratio, as well as the specific neuronal subtypes that are transduced in the striatum will extend the applicability of the ALDH1L1 promoters to studies characterizing molecular profiles of striatal cell populations.
Additional studies aimed at identifying the additional sequence motifs necessary for cell-specific expression may allow the generation of astrocyte-specific and/or dual neuronal- and astrocyte-transcriptional targeting ALDH1L1 promoters that are tailored for brain regions and transgenes of interest. Deletion analysis of the 2.2 kb GFAP promoter showed that a 45 bp sequence within the 2.2 kb GFAP promoter silences its activity in neurons.12 It is possible that the exclusion of similar sequence motifs that silence neuronal expression and those that activate astrocytic expression from our ALDH1L1 promoters may have resulted in their selective activity in neurons, instead of the astrocyte-specific expression achieved with the full length ALDH1L1 genomic promoter in bacterial artificial chromosome transgenic mice.13
Our results suggest that the ALDH1L1 promoter variants could have utility as additional tools in the development of gene therapy and disease modelling applications. Coupling a neurotropic AAV serotype with a promoter that is highly active and specific to certain neuronal populations is advantageous in achieving optimal levels of transgene expression at the lowest possible vector dose, while restricting therapeutic protein expression to specific CNS nuclei. Constitutively active viral promoters such as the cytomegalovirus or chicken beta-actin/cytomegalovirus hybrid promoters are routinely used to achieve high levels of neuronal transgene expression in the CNS.16,27–32 However, one disadvantage of viral promoters is that they are susceptible to epigenetic modification such as promoter methylation that silences transcriptional activity over time.33 Alternatively, endogenous neuronal promoters such as neuron-specific enolase, platelet-derived growth factor-β chain, Syn, and TH, in the context of AAV serotypes have been used to target neurons in various CNS regions.27,28,34–36 In the SNpc, Syn, and TH promoters have been shown to target transgene expression to dopaminergic neurons.35,36 Here we show in a comparative analysis of various neuronal promoters that the ALDH1L1(L)ex1 promoter variant mediated significantly higher levels of transgene expression compared with the Syn and TH promoters, suggesting its potential utility for driving therapeutic transgene expression in nigral dopaminergic neurons.
Neuronal-specific transgene expression was also observed in the hippocampus. The reasons for variability in hippocampal transgene expression levels within each vector treatment group are unclear given that we observed quite marked consistency in expression levels following intranigral or intrastriatal vector infusion. Nevertheless, there is some indication that the ALDH1L1 promoter may also drive higher levels of transcriptional activity in dentate granule cell neurons compared with the Syn promoter although this will require further confirmation. Future studies will assess whether injection coordinates for hippocampal infusions need optimization.
In conclusion, our results describe new promoters that could have broad utility for gene therapy and disease modelling applications. AAV vectors have been shown to be safe in human gene therapy but the modest packaging capacity of AAV restricts the size of the therapeutic gene cassettes to <4.7 kb. Thus the size of the ALDH1L1(L) promoter variants could place limitations on the size of transgenes that can be delivered using an AAV vector system. We found that inclusion of exon1 did not lead to any appreciable increase in transcriptional activity in the rat brain in contrast to that observed in previous studies,15 suggesting it could be excluded. Because smaller promoters considerably ease the size restriction on therapeutic genes that can be cloned into AAV expression cassettes, future studies should examine whether the two short ALDH1L1 promoters may potentially exhibit greater activity than Syn in various brain regions, and whether transcriptional regulation under the ALDH1L1 promoters is also altered under disease conditions. It may also be useful to investigate the effect of different routes of vector administration18 as well as higher vector titers8 which may also affect cellular transduction patterns.
Materials and Methods
Development of AAV plasmids
Short (S) 931 bp and long (L) 1,974 bp cDNA sequences immediately upstream of (or relative to) the transcription start site of the rat ALDH1L1 gene (GenBank accession number AC_000072.1) were generated using the reverse primer 5ʹ-ATAGAGCTCGGCAGAAGCTCCGGTCTTAT-3ʹ, and forward primers, 5ʹ-ATACTCGAGGGATCCTCCAGGACAGAGCT-3ʹ and 5ʹ-ATACTCGAGTTCACTCTCTACTGCCTGAC-3ʹ, respectively. These were cloned into the poly-linker site of an AAV plasmid containing a truncated woodchuck hepatitis post-transcriptional regulatory element and short bovine growth hormone polyadenylation signal flanked by AAV2 inverted terminal repeats. To insert exon 1 (ex1) (138bp) into the plasmid constructs, a cDNA sequence containing exon 1 were generated using forward primer 5ʹ-ATACTCGAGGGATCCTCCAGGACAGAGCT-3ʹ and reverse primer 5ʹ-ATATCGCGACCAAGGTCAAAGACAGGAAG-3ʹ. cDNA for firefly luciferase (Luc) or eGFP from Aequorea victoria were subsequently cloned into the poly-linker site to yield ALDH1L1(L)-Luc, ALDH1L1(S)-Luc, ALDH1L1(L)ex1-Luc, ALDH1L1(S)ex1-Luc, and ALDH1L1(L)ex1-eGFP. Yellow fluorescent protein (YFP) cDNA was cloned into the poly-linker site of an AAV expression plasmid under the control of the 2.2 kb human GFAP promoter (GenBank accession no. M67446). eGFP was cloned into the poly-linker site of an AAV expression plasmid under the control of a 803 bp rat tyrosine hydroxylase (GenBank accession no. EU240461.1) and a 619 bp human synapsin 1 (GenBank accession no. NG_008437.1) promoters, and containing a woodchuck hepatitis post-transcriptional regulatory element and short bovine growth hormone polyadenylation signal flanked by AAV2 inverted terminal repeats.
AAV vector packaging
The AAV expression plasmids were packaged into AAV serotype 5, 8 and 9 vectors and purified by iodixanol density ultracentrifugation, and genomic titers were determined by real-time Polymerase chain reaction using primers to woodchuck hepatitis post-transcriptional regulatory element as described previously.8 Bands corresponding only to the viral capsid protein VP1, -2 and -3 were visible by Coomassie blue staining on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. The vector stock titers (vector genomes/ml) generated were as follows: ALDH1L1(L)-Luc, 9.1 × 1012; ALDH1L1(S)-Luc, 4.1 × 1012; ALDH1L1(L)ex1-Luc, 2.7 × 1012; ALDH1L1(S)ex1-Luc, 6.9 × 1012; ALDH1L1(L)ex1-eGFP, 2.7 × 1012; Syn-eGFP, 2.34 × 1012; and TH-eGFP, 2.33 × 1012. All vector stocks were diluted to 1.0 × 1012 for the brain infusions.
Vector infusions
All animal experiments were conducted under guidelines and approval by The University of Auckland Animal Ethics Committee. Male Sprague–Dawley rats (250–300 g) were anaesthetized with an intraperitoneal (i.p) dose (70 mg/kg) of pentobarbital, and positioned in a Kopf stereotaxic frame (David Kopf Instruments, Tujunga, CA). Subgroups of rats were randomly assigned to receive 2 μl of AAV vector or 1× phosphate-buffered saline (PBS) vehicle infused unilaterally into the nigra (coordinates: anterior-posterior (AP) −5.3 mm, medial-lateral (ML) + 2.3 mm, dorsal-ventral (DV) −7.6 mm from dural surface, bregma = 0), hippocampus (coordinates: AP −3.6 mm, ML +2.1 mm, DV −4.3 mm from skull surface) or striatum (AP +1.4 mm, ML −2.5 mm, DV −5.5 mm from skull surface)37 at a rate of 70 nl/min for the nigral infusions and 200 nl/min for the hippocampal and striatal infusions regulated by a microinfusion pump (World Precision Instruments, Sarasota, FL).
Immunohistochemistry
Rats were killed 3 weeks postvector infusion by an i.p. overdose of pentobarbital (100 mg/kg), and transcardially perfused with 100 ml of 0.9% (w/v) saline, followed by 4% (w/v) paraformaldehyde in 0.1 mol/l phosphate buffer, pH 7.4 (Sigma-Aldrich, St Louis, MO). The brains were postfixed in 4% paraformaldehyde overnight at 4°C, followed by cryoprotection in 30% (w/v) sucrose in PBS for 72 hours at 4ºC. Forty micron coronal brain sections were cut and used for immunohistochemistry as described previously.8 Endogenous peroxidase activity was blocked by incubating sections in 1% (v/v) H2O2 in 50% methanol for 20 minutes, followed by an overnight incubation with rabbit anti-Luc (1:1,000; 70C-CR2029RAP; Fitzgerald, North Acton, MA), or rabbit anti-GFP antibody (1:50,000; ab290; Abcam, Cambridge, UK) diluted in PBS containing 0.2% (v/v) Triton X-100 and 10% (v/v) horse serum. Sections were then incubated with biotinylated secondary antibodies, goat anti-rabbit (1:250; Sigma-Aldrich) for 3 hours, followed by 2 hours incubation with ExtrAvidin Peroxidase (1:250; SigmaAldrich). Immunoreactivity was visualized by incubation with diaminobenzidine(DAB) in 0.1 mol/l phosphate buffer containing 0.01% H2O2 (v/v), and images were captured on an Olympus AX70 microscope (Olympus, Centre Valley, PA) using a CX9000 digital camera and StereoInvestigator software (MBF Biosciences, Williston, VT).
For eGFP immunodetection in the promoter comparison study, the biotin/avidin amplification step was omitted to reduce the intensity of eGFP immunoreactivity. Following the overnight incubation with rabbit anti-GFP antibody (1:50,000; ab290; Abcam), these sections were incubated with horseradish peroxidase-conjugated goat anti-rabbit (1:500; A0545; Sigma Aldrich) for 2 hours. Immunoreactivity was visualized by incubation with diaminobenzidine (DAB) in 0.1 mol/l phosphate buffer containing 0.01% H2O2 (v/v).
Immunofluorescence
For immunofluorescence labeling, 0.01% H2O2 treatment was omitted, and sections were incubated with rabbit anti-Luc (1:500; 70C-CR2029; Fitzgerald) or rabbit anti-GFP (1:50,000; ab290; Abcam) in combination with either mouse anti-GFAP (1:50 000; G3893; Sigma Aldrich), mouse anti-TH (1:500; MAB318; Millipore, Billerica, MA), mouse anti-NeuN (1:1000; MAB377; Millipore), or goat anti-Iba1 (1:2,000; ab5076; Abcam, Cambridge, UK) for 48 hours at 4°C, and subsequently incubated with donkey anti-rabbit Alexa-488 secondary antibody (1:1,000; Invitrogen) in combination with either donkey anti-mouse Alexa-594 (1:1,000; Invitrogen) or biotinylated donkey anti-goat secondary antibody (1:1,000; Jackson ImmunoResearch, Westgrove, PA) for 24 hours at 4°C. For Iba1 detection, sections were further incubated with streptavidin Alexa-594 (1:1,000; Invitrogen) for 24 hours at 4°C. Images were acquired on an Olympus FV1000 confocal laser scanning microscope (Tokyo, Japan) with an oil-immersion x60 objective, and Olympus Fluoview version 3.0 Software.
Volume measurement
The volume of midbrain tissue expressing transgene in the SNpc, SNpr, and VTA by AAV vectors was estimated using the Cavalieri estimator probe on Stereo Investigator 7 (MBF Bioscience). On every sixth 40 μm section (each 240 μm apart), markers were placed at a grid size of 50 μm to outline areas containing Luc-immunoreactive cell bodies and fibers within the SNpc, SNpr, and VTA. The volume of transgene expression was estimated by the software based on the average area of transgene expression and rostro-caudal distance.
Density measurements of transgene staining
Intensity of transgene staining within the SNpc was measured using ImageJ software (ImageJ version 1.48, National Institute of Health). A standardized threshold intensity was set for grayscale images of every sixth 40 μm section (each 240 μm apart) to select Luc immunoreactive neuronal cell bodies and fibers within the outlined SNpc, and the reciprocal chromogen intensity was calculated by subtracting the measured intensity from a maximum intensity value of 255.
Western blotting
Three weeks postinfusion of AAV9-ALDH1L1(L)ex1-eGFP, AAV9-Syn-eGFP, and AAV9-TH-eGFP (n = 4–5 per vector), rats were killed and then nigral, striatal or hippocampal regions were dissected. Each tissue sample was homogenized by sonication in ice-cold lysis buffer (50 mmol/l Tris-HCl, 2 mmol/l ethylenediaminetetraacetic acid (EDTA), 0.05% Triton X-100; pH 7.4) containing protease inhibitors (Complete Protease inhibitor Cocktail; Roche Diagnostics, Mannheim, Germany). Lysates were centrifuged at 8,000g in a microfuge for 10 minutes at 4°C, and supernatants collected for analysis. For western blot analysis of eGFP expression, 20 µg of tissue lysate was resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to Hybond-ECL membranes (GE Healthcare, Uppsala, Sweden). The membranes were blocked with 5% (w/v) skim milk powder in Tris-buffered saline containing 0.05% (v/v) Tween 20 (20 mmol/l Tris, 0.5 mol/l NaCl, 0.05% Tween 20; pH 7.4), and incubated with rabbit anti-GFP antibody (1:50,000; ab290; Abcam) or mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:50,000; ab8245, Abcam) overnight at 4°C. The following day, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit (1:10,000; A0545; SigmaAldrich), or anti-mouse (1:5,000; sc-2005, Santa Cruz Biotechnology, Dallas, TX) antibodies diluted in Tris-buffered saline at room temperature for 2 hours, prior to detection of immunoreactive bands with enhanced chemiluminescence detection reagent (GE Healthcare Life Sciences). Chemiluminescent bands were visualized and gel images acquired using a ChemiDoc MP imaging system (BioRad). Density of eGFP protein band relative to the reference protein, GAPDH was estimated using the Image Lab software (version 4.1; BioRad).
Statistical analysis
Data are expressed as mean ± standard error of mean (SEM) . A one-way ANOVA with Tukey’s post hoc test or unpaired Student’s t-test were performed using SPSS (IBM SPSS, version 21, Chicago, IL) to compare means, with significance levels set at P < 0.05.
Acknowledgments
This work was supported by funding from the Royal Society of New Zealand Marsden Fund to D.Y.. We thank M.. Brenner for providing the 2.2 kb GFAP promoter and the Penn Vector Core for AAV packaging plasmids.
The authors declared no conflict of interest.
References
- Sánchez-Pernaute, R, Harvey-White, J, Cunningham, J and Bankiewicz, KS (2001). Functional effect of adeno-associated virus mediated gene transfer of aromatic L-amino acid decarboxylase into the striatum of 6-OHDA-lesioned rats. Mol Ther 4: 324–330. [DOI] [PubMed] [Google Scholar]
- Muramatsu, S, Fujimoto, K, Ikeguchi, K, Shizuma, N, Kawasaki, K, Ono, F et al. (2002). Behavioral recovery in a primate model of Parkinson’s disease by triple transduction of striatal cells with adeno-associated viral vectors expressing dopamine-synthesizing enzymes. Hum Gene Ther 13: 345–354. [DOI] [PubMed] [Google Scholar]
- Luo, J, Kaplitt, MG, Fitzsimons, HL, Zuzga, DS, Liu, Y, Oshinsky, ML et al. (2002). Subthalamic GAD gene therapy in a Parkinson’s disease rat model. Science 298: 425–429. [DOI] [PubMed] [Google Scholar]
- Kaplitt, MG, Feigin, A, Tang, C, Fitzsimons, HL, Mattis, P, Lawlor, PA et al. (2007). Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: an open label, phase I trial. Lancet 369: 2097–2105. [DOI] [PubMed] [Google Scholar]
- Mittermeyer, G, Christine, CW, Rosenbluth, KH, Baker, SL, Starr, P, Larson, P et al. (2012). Long-term evaluation of a phase 1 study of AADC gene therapy for Parkinson’s disease. Hum Gene Ther 23: 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV and Vinters, HV (2010). Astrocytes: biology and pathology. Acta Neuropathologica 119: 7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson, J, Ericson, C, Jansson, M, Björk, E and Lundberg, C (2003). Targeted transgene expression in rat brain using lentiviral vectors. J Neurosci Res 73: 876–885. [DOI] [PubMed] [Google Scholar]
- Lawlor, PA, Bland, RJ, Mouravlev, A, Young, D and During, MJ (2009). Efficient gene delivery and selective transduction of glial cells in the mammalian brain by AAV serotypes isolated from nonhuman primates. Mol Ther 17: 1692–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkut, A, Tereshchenko, Y, Schulz, JB, Bähr, M and Kügler, S (2012). Efficient gene therapy for Parkinson’s disease using astrocytes as hosts for localized neurotrophic factor delivery. Mol Ther 20: 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, D, Fong, DM, Lawlor, PA, Wu, A, Mouravlev, A, McRae, M et al. (2014). Adenosine kinase, glutamine synthetase and EAAT2 as gene therapy targets for temporal lobe epilepsy. Gene Ther 21: 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz, W and Lang, MK (1998). Immunocytochemical evidence for a distinct GFAP-negative subpopulation of astrocytes in the adult rat hippocampus. Neurosci Lett 257: 127–130. [DOI] [PubMed] [Google Scholar]
- Lee, Y, Messing, A, Su, M and Brenner, M (2008). GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia 56: 481–493. [DOI] [PubMed] [Google Scholar]
- Yang, Y, Vidensky, S, Jin, L, Jie, C, Lorenzini, I, Frankl, M et al. (2011). Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia 59: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy, JD, Emery, B, Kaushal, A, Foo, LC, Zamanian, JL, Christopherson, KS et al. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinik, NV, Krupenko, NI and Krupenko, SA (2011). Epigenetic silencing of ALDH1L1, a metabolic regulator of cellular proliferation, in cancers. Genes Cancer 2: 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis, EA, Vives, KP, Bober, J, Leichtle, S, Leranth, C, Beecham, J et al. (2010). Comparative transduction efficiency of AAV vector serotypes 1-6 in the substantia nigra and striatum of the primate brain. Mol Ther 18: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski, PI, Dong, J, Mungenast, A, Yue, C, Takano, H, Watson, DJ et al. (2010). Selective induction of astrocytic gliosis generates deficits in neuronal inhibition. Nat Neurosci 13: 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust, KD, Nurre, E, Montgomery, CL, Hernandez, A, Chan, CM and Kaspar, BK (2009). Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 27: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab, LT and Pow, DV (2007). Expression of the exon 9-skipping form of EAAT2 in astrocytes of rats. Neuroscience 150: 705–711. [DOI] [PubMed] [Google Scholar]
- Yeh, TH, Lee, DY, Gianino, SM and Gutmann, DH (2009). Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia 57: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X, Taniguchi, K and Kofuji, P (2009). Heterogeneity of Kir4.1 channel expression in glia revealed by mouse transgenesis. Glia 57: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan, MR, Huang, YH, Kim, YS, Dykes-Hoberg, MI, Jin, L, Watkins, AM et al. (2007). Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J Neurosci 27: 6607–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Z, Schools, GP and Kimelberg, HK (2000). Metabotropic glutamate receptors in acutely isolated hippocampal astrocytes: developmental changes of mGluR5 mRNA and functional expression. Glia 29: 70–80. [DOI] [PubMed] [Google Scholar]
- Reuss, B, Leung, DS, Ohlemeyer, C, Kettenmann, H and Unsicker, K (2000). Regionally distinct regulation of astroglial neurotransmitter receptors by fibroblast growth factor-2. Mol Cell Neurosci 16: 42–58. [DOI] [PubMed] [Google Scholar]
- Karavanova, I, Vasudevan, K, Cheng, J and Buonanno, A (2007). Novel regional and developmental NMDA receptor expression patterns uncovered in NR2C subunit-beta-galactosidase knock-in mice. Mol Cell Neurosci 34: 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitting, S, Zou, S, Chen, W, Vo, P, Hauser, KF and Knapp, PE (2010). Regional heterogeneity and diversity in cytokine and chemokine production by astroglia: differential responses to HIV-1 Tat, gp120, and morphine revealed by multiplex analysis. J Proteome Res 9: 1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, RL, Meyer, EM, Peel, AL, Zolotukhin, S, Meyers, C, Muzyczka, N et al. (1998). Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp Neurol 150: 183–194. [DOI] [PubMed] [Google Scholar]
- Paterna, JC, Moccetti, T, Mura, A, Feldon, J and Büeler, H (2000). Influence of promoter and WHV post-transcriptional regulatory element on AAV-mediated transgene expression in the rat brain. Gene Ther 7: 1304–1311. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, L, Chtarto, A, Lehtonen, E, Velu, T, Brotchi, J and Levivier, M (2004). Recombinant AAV-mediated gene delivery to the central nervous system. J Gene Med 6 Suppl 1: S212–S222. [DOI] [PubMed] [Google Scholar]
- Van der Perren, A, Toelen, J, Carlon, M, Van den Haute, C, Coun, F, Heeman, B et al. (2011). Efficient and stable transduction of dopaminergic neurons in rat substantia nigra by rAAV 2/1, 2/2, 2/5, 2/6.2, 2/7, 2/8 and 2/9. Gene Ther 18: 517–527. [DOI] [PubMed] [Google Scholar]
- Watakabe, A, Ohtsuka, M, Kinoshita, M, Takaji, M, Isa, K, Mizukami, H et al. (2015). Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci Res 93: 144–157. [DOI] [PubMed] [Google Scholar]
- Burger, C, Gorbatyuk, OS, Velardo, MJ, Peden, CS, Williams, P, Zolotukhin, S et al. (2004). Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther 10: 302–317. [DOI] [PubMed] [Google Scholar]
- Brooks, AR, Harkins, RN, Wang, P, Qian, HS, Liu, P and Rubanyi, GM (2004). Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med 6: 395–404. [DOI] [PubMed] [Google Scholar]
- Nathanson, JL, Yanagawa, Y, Obata, K and Callaway, EM (2009). Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neurosci 161: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, MS, Hong, SJ, Huh, Y and Kim, KS (2009). Expression of transgenes in midbrain dopamine neurons using the tyrosine hydroxylase promoter. Gene Ther 16: 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decressac, M, Kadkhodaei, B, Mattsson, B, Laguna, A, Perlmann, T and Björklund, A (2012). α-Synuclein-induced down-regulation of Nurr1 disrupts GDNF signaling in nigral dopamine neurons. Sci Transl Med 4: 163ra156. [DOI] [PubMed] [Google Scholar]
- Paxinos, G, and Watson, C (1986). The Rat Brain in Stereotaxic Coordinates, 2nd edn, Academic Press New York: New York, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.