SUMMARY
Nuclear mRNA export is highly regulated to ensure accurate cellular gene expression. Viral inhibition of cellular mRNA export can enhance viral access to the cellular translation machinery and prevent anti-viral protein production but is generally thought to be nonselective. We report that ORF10 of Kaposi's sarcoma associated herpesvirus (KSHV), a nuclear DNA virus, inhibits mRNA export in a transcript-selective manner to control cellular gene expression. Nuclear export inhibition by ORF10 requires an interaction with an RNA export factor, Rae1. Genome-wide analysis reveals a subset of cellular mRNAs whose nuclear export is blocked by ORF10 with the 3' untranslated regions (3' UTRs) of ORF10-targeted transcripts conferring sensitivity to export inhibition. The Rae1-ORF10 interaction is important for the virus to express viral genes and produce infectious virions. These results suggest that a nuclear DNA virus can selectively interfere with RNA export to restrict host gene expression for optimal replication.
Keywords: Herpesvirus, KSHV, ORF10, Rae1, Nup98, mRNA nuclear export
Introduction
For eukaryotes, nuclear export of mRNAs is an important step in the gene expression pathway to ensure translation in the cytoplasm. mRNA export has three main stages: first, export factors are recruited to messenger ribonucleoparticles (mRNPs); second, through export factors, mRNPs are docked to the nuclear pore complex (NPC) and transported through the NPC central channel; and third, mRNPs are released into the cytoplasm for translation (Carmody and Wente, 2009; Muller-McNicoll and Neugebauer, 2013). The major export factor for mRNAs is a heterodimer comprised of NXF1 (nuclear export factor 1, also known as TAP) and NXT1 (NTF2-related export protein NXF1, also known as p15). The NXF1-NXT1 (or TAP-p15) heterodimer docks mRNP to the nuclear pore complex via direct binding to the phenylalanine-glycine (FG) repeats of nucleoporins (Nups). The NPC is composed of ~30 Nups; around one third of Nups contain FG repeats (Strambio-De-Castillia et al., 2010). The FG repeats of Nups located within the central channel of the pore form a permeability barrier for selective nuclear transport. Interactions of the NXF1-NXT1 export factor with FG-Nups overcome the permeability barrier at the pore and allow the translocation of export factor/mRNP complexes to the cytoplasm (Grunwald et al., 2011; Powers and Forbes, 2012).
Viral inhibition of cellular mRNA export can grant virus greater access to the translation machinery as well as prevent cells from producing proteins that antagonize viral replication. Indeed, a number of viral proteins suppress host mRNA export (Yarbrough et al., 2014). These viral inhibitors, identified in both DNA and RNA viruses, employ distinct mechanisms to disrupt the RNA transport process. The best studied viral inhibitor is matrix protein (M) encoded by an RNA virus, vesicular stomatitis virus (VSV). The inhibitory effect of M protein is through its interaction with Rae1, and consequently with the Rae1-Nup98 complex (Faria et al., 2005; von Kobbe et al., 2000). Rae1 is an RNA export factor that contributes to the mRNA transport process by interacting with a 98-kD FG-Nup, Nup98 (Pritchard et al., 1999). The ability of Rae1 to form a complex with the major RNA export factor, NXF1, and with Nup98 has implicated Rae1 in facilitating the targeting of mRNPs to NPC (Blevins et al., 2003).
Herpesviruses are large DNA viruses that have two distinct life cycle phases: lytic replication and latency. During lytic replication, they operate a highly regulated viral gene expression program, resulting in the production of infectious virions. Herpesviruses are known for manipulating cellular mRNA export machinery. This is because most of their transcripts do not have introns and splicing is a crucial event for recruiting the NXF1-NXT1 RNA export factor to mRNPs. The viral RNA binding protein, ICP27 of herpes simplex virus-1 (HSV-1) and its homologues in other herpesviruses, interact with the cellular proteins that recruit the NXF1-NXT1 heterodimers, thereby linking the viral transcripts to the cellular RNA export pathway (Chen et al., 2002; Koffa et al., 2001; Malik et al., 2004). Nevertheless, this manipulation of the pathway has not been shown to block cellular mRNA export.
Nuclear export has been generally thought to be constitutive and universal for all mRNAs. However, increasing evidence indicates that mRNA export can be selectively regulated by specific export factors to control distinct biological processes (Wickramasinghe and Laskey, 2015). Inhibiting selective mRNA export may provide a strategy for a nuclear replicating DNA virus, such as a herpesvirus, to control certain cellular processes without disrupting export of their own transcripts. Here we report that ORF10 of Kaposi's sarcoma associated-herpesvirus (KSHV), a gamma-herpesvirus, selectively blocks nuclear export of a subset of cellular mRNAs by interacting with the Rae1-Nup98 pathway. Through a reporter assay and RNA sequencing, we find that export inhibition of ORF10 is transcript-selective. Furthermore, the 3' untranslated regions (3'UTRs) of the ORF10-targeted genes are able to sensitize the reporter transcript to export inhibition by ORF10. We then show that both knocking down Rae1 expression and disrupting the interaction of ORF10 with Rae1 attenuate KSHV gene expression during viral lytic replication. Our study reveals a strategy employed by KSHV to regulate host gene expression.
Results
ORF10 induces nuclear accumulation of poly(A)+ RNA
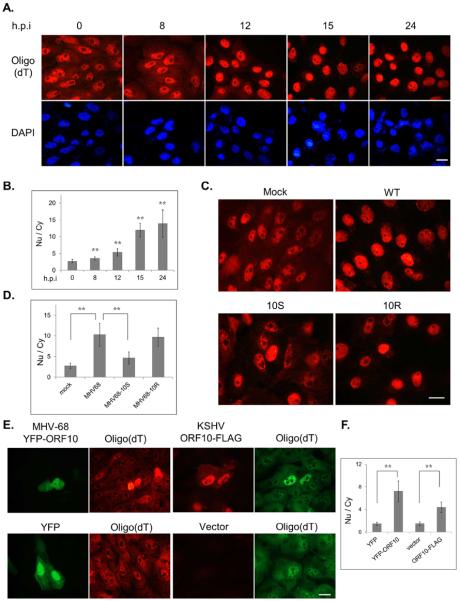
Several viruses retain host mRNA in the nucleus as a mechanism to control cellular gene expression. To determine whether infection of gamma-herpesviruses affects mRNA distribution, we employed a rodent gamma-herpesvirus, murine gammaherpesvirus-68 (MHV-68), which readily infects a variety of cell lines and has been widely regarded as a model to study fundamental aspects of gammaherpesvirus biology. Compared to mock-infected cells, MHV-68 infection resulted in a loss of cytoplasmic poly(A)+ RNA signal with a concomitant increase in nuclear labeling (Figs. 1A & 1B). This change in poly(A)+ RNA distribution could be observed as early as 12 hours after infection. The nuclear poly(A)+ RNA signal continued to increase at later times as demonstrated by the increasing nuclear to cytoplasmic (Nu/Cy) signal ratio. We and another group previously identified several MHV-68 and KSHV proteins that down-regulate the type I interferon response (Bisson et al., 2009; Leang et al., 2011). One of these ORFs, ORF10, was found to interact with the Rae1-Nup98 complex through co-IP coupled with mass spectrometry. The interaction between MHV-68 ORF10 and Rae1 during infection was further confirmed by western blotting (Fig. S1A). The Rae1-Nup98 pathway is exploited by the VSV M protein to block mRNA export (Faria et al., 2005; von Kobbe et al., 2000). To test whether ORF10 is responsible for the altered poly(A)+ RNA distribution during MHV-68 infection, we constructed a mutant viral genome with the insertion of triple stop codons at ORF10 (10S) to eliminate ORF10 expression via homologous recombination in E. coli (described in Material and Methods). As a control, we used the same recombination technique to reintroduce the wild-type sequence to the 10S viral genome and made a revertant virus of 10S (10R). Compared to the cells infected with WT or 10R, the 10S-infected cells showed detectable cytoplasmic poly(A)+ RNA with a less intense nuclear signal (Fig. 1C). Accordingly, the Nu/Cy ratio of poly(A)+ RNA signal in the 10S-infected cells was largely reduced to a level close to mock-infected cells (Fig. 1D). This result indicates that ORF10 is required for the effect of MHV-68 on poly(A)+ RNA distribution. We also tested whether expression of ORF10 alone is able to induce mRNA nuclear accumulation. For this experiment, we included ORF10 from a human gammaherpesvirus, KSHV, as well. Both ORF10 homologues were capable of inducing nuclear accumulation of poly(A)+ RNA signals (Figs. 1E and 1F), indicated by bright nuclear signals that appeared only in ORF10-expressing cells. Therefore, the transfection studies support the hypothesis that ORF10 of both KSHV and MHV-68 inhibits mRNA trafficking out of the nucleus. For the following mechanistic studies, we focused on KSHV ORF10.
Fig. 1.
ORF10 induces poly(A)+ RNA accumulation in the nucleus. Poly(A)+ RNA was detected by in situ oligo(dT) hybridization. The poly(A)+ RNA signals of all cells (n>24) across four fields were quantified with ImageJ, and the Nu/Cy signal ratio was calculated and plotted as mean ± SD. (A) SLK cells were infected with MHV-68 for indicated hours, and the Nu/Cy signal ratio of poly(A)+ RNA was shown in (B). (C) SLK cells were mock infected (M) or infected with the wild-type MHV-68 (WT), the ORF10-null mutant (10S) or the revertant (10R) virus at an MOI of 3 for 15 hours and the Nu/Cy signal ratio of poly(A)+ RNA was shown in (D). (E) SLK cells were transfected with YFP-ORF10, ORF10-FLAG, or empty vectors, and the Nu/Cy signal ratio of poly(A)+ RNA was shown in (E). Scale bar: 20 μm.
ORF10 interacts with Rae1, and further forms complex with Nup98
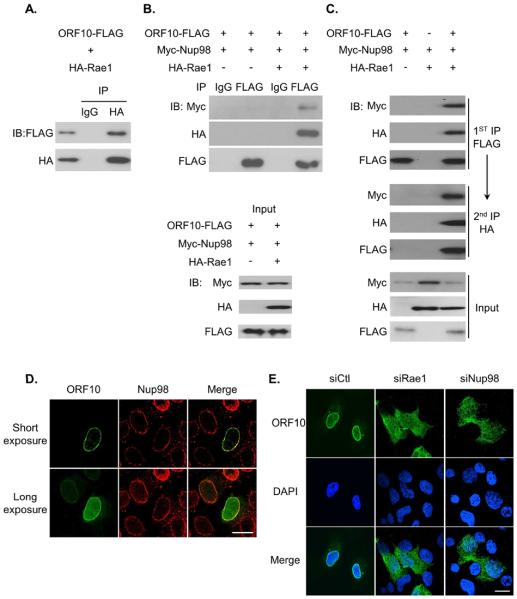
A recent global analysis of virus-host interactions identified Rae1 and Nup98 among potential interacting partners of KSHV ORF10 (Davis et al., 2015). Our data on MHV-68 ORF10 (Fig. S1A) agree with this result. To examine the interaction of KSHV ORF10 and the Rae1-Nup98 pathway, we performed co-immunoprecipitation (co-IP) with FLAG-tagged KSHV ORF10 and HA-tagged Rae1. The ORF10-Rae1 interaction was readily detectable (Fig. 2A). In addition, the localization of HA-Rae1 largely overlapped with ORF10-FLAG (Fig. S1B). On the other hand, co-IP of ORF10-FLAG and Myc-Nup98 yielded very little interaction between these two proteins without HA-Rae1 (Fig. 2B). However, when all three tagged proteins, ORF10, Rae1, and Nup98, were co-expressed in cells, a significant amount of Myc-Nup98 was found in association with ORF10 (Fig. 2B). Because Rae1 forms a complex with Nup98, these co-IP results indicate that ORF10 interacts with Nup98 through Rae1. Using a sequential IP assay we demonstrated that ORF10, Rae1 and Nup98 form a protein complex together (Fig. 2C). Our data also suggest that ORF10 does not prevent Rae1 from interacting with Nup98.
Fig. 2.
ORF10 interacts with Rae1, and further forms a complex with Nup98. (A) ORF10 interacts with Rae1 in co-IP. (B) Rae1 mediates the interaction between ORF10 and Nup98. (C) ORF10, Rae1 and Nup98 form a complex in sequential IP. (D) Localization of ORF10 during MHV-68 infection. SLK cells were infected with the FLAG-tagged ORF10 MHV-68 virus (MHV68-F10) and after 16 hours, cells were fixed and subjected to IFA with αFLAG and αNup98 antibody. (E) Nuclear envelope enrichment of ORF10 requires Rae1 and Nup98. SLK cells were transfected with control siRNA (siCtl), siRNA against Rae1 or Nup98 for 2 days, followed by infection with the MHV68-F10 virus, 16 hours later, cells were fixed and subjected to IFA with αFLAG antibody. Scale bar: 20 μm. See also Fig. S1.
Rae1, though shuttles between the nucleus and cytoplasm, predominantly resides in the nucleus. Rae1 appears to have preferential nuclear envelope localization, due to its interaction with Nup98 at the NPC (Pritchard et al., 1999). To examine the localization of ORF10 in the context of infection, we turned to MHV-68, using a recombinant virus that has the FLAG tag fused to the N-terminus of ORF10 in the genome. FLAG-ORF10 had obvious nuclear envelope localization coinciding with endogenous Nup98 in the context of MHV-68 infection (Fig. 2D). We further examined the role of Rae1 and Nup98 in ORF10 localization by siRNA knockdown. We found that the nuclear envelope localization of ORF10 is dependent on the presence of Rae1 and Nup98 (Figs. 2E, S1C and S1D). In addition, much more prominent cytoplasmic staining of ORF10 was detected in cells knocked down with Rae1 or Nup98. These localization studies performed in the context of infection support that ORF10 interacts with the Rae1-Nup98 complex at the nuclear envelope and the association with the complex leads to a dominant nuclear localization of ORF10.
ORF10 induces poly(A)+ RNA nuclear accumulation as a consequence of interacting with Rae1
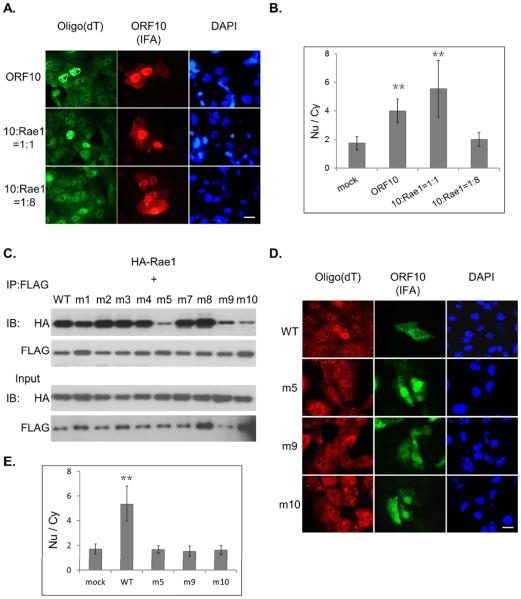
The Rae1-Nup98 interaction has been implicated previously in mRNA export (Pritchard, 1999). Our hypothesis is that, through binding to the Rae1-Nup98 complex, ORF10 interferes with the function of the complex, thereby inhibiting mRNA export and the resulting nuclear accumulation of mRNA. To test this hypothesis, we first examined whether an excess of Rae1 was able to thwart the effect of ORF10 on mRNA distribution. As shown in Figs. 3A & 3B, ORF10-induced nuclear accumulation of poly(A)+ RNA was counteracted by transfection of a large excess of the Rae1-expression plasmid. Next, we sought to determine the functional consequences of the ORF10 mutants that can no longer interact with Rae1, with the prediction that these ORF10 mutants would lose the ability to alter mRNA distribution. To generate site-specific ORF10 mutants, we exploited the fact that both KSHV and MHV-68 ORF10 interact with Rae1. We reasoned that the amino acid residues responsible for interacting with Rae1 are conserved between these two homologues, which share 18% amino acid sequence identity. Therefore, we introduced alanine substitutions at some of those identical residues and generated nine KSHV ORF10 mutants in total (Table S1). Three of these nine ORF10 mutants, m5, m9 and m10, while expressed at the level comparable to the WT protein, lost the bulk of their ability to interact with Rae1 (Fig. 3C). Localization of these three mutants were also examined by co-confocal microscopy and they were still able to localize in the nucleus, but all of them lost nuclear envelop enrichment and produced more cytoplasmic staining compared to wild type ORF10 (Fig. S2). As shown by in situ hybridization, these three mutants failed to retain poly(A)+ RNA in the nucleus (Figs. 3D & 3E), indicating that the interaction with Rae1 is required for ORF10 to induce nuclear mRNA accumulation.
Fig. 3.
The interaction with Rae1 is required for ORF10 to induce nuclear accumulation of poly(A)+ RNA. (A) Increasing the expression of Rae1 reverts nuclear poly(A)+ RNA accumulation induced by ORF10. SLK cells were co-transfected with a fixed amount (100ng) of ORF10-FLAG and increasing amount of HA-Rae1, and then subjected to in situ oligo(dT) hybridization and IFA with αFLAG antibody. (B) The Nu/Cy signal ratio of poly(A)+ RNA in (A). ORF10-negative cells were quantified as mock. Signal in all cells across four fields was quantified with ImageJ, and data are represented as mean ± SD. (C) Site-specific mutagenesis of ORF10 identified the mutations at three sites that disrupt the interaction with Rae1. (D) Distribution of poly(A)+ RNA in cells expressing WT or loss-of-interaction mutants of ORF10 was examined by in situ oligo(dT) hybridization and ORF10 expression examined by IFA with αFLAG antibody. (E) The Nu/Cy signal ratio of poly(A)+ RNA in (D) determined as in (B). Scale bar: 20 μm. See also Fig. S2.
ORF10 selectively blocks mRNA export
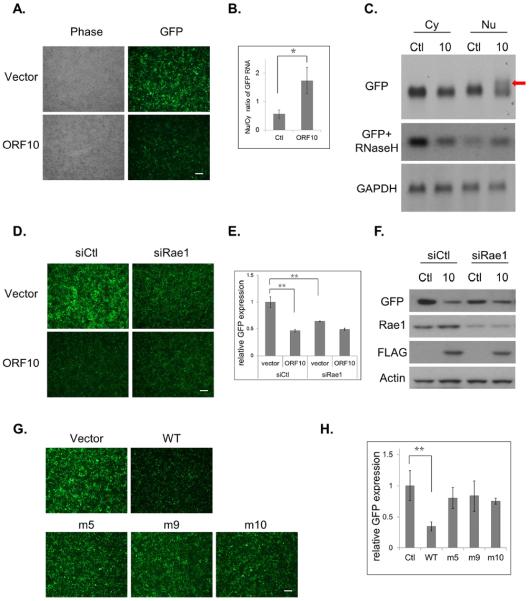
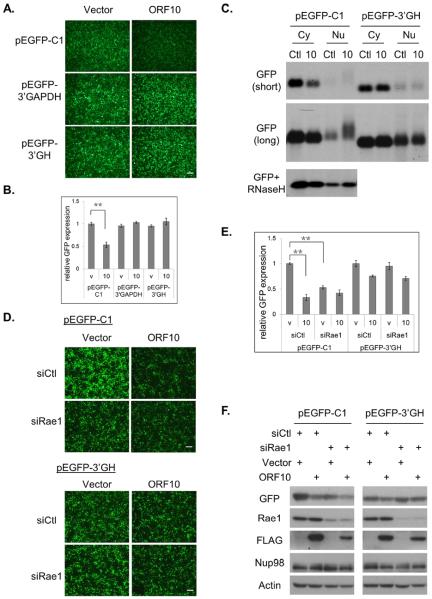
To further study the role of ORF10 and Rae1 in mRNA export, we employed a green fluorescent protein (GFP) expression plasmid, pEGFP-C1, as a reporter. Transcription of GFP from pEGFP-C1 is driven by the cytomegalovirus promoter and is terminated by the simian virus 40 (SV40) early polyadenylation signal. KSHV ORF10 significantly down-regulated GFP expression (Fig. 4A). Consistent with ORF10 inhibition on nuclear export of the GFP transcript, the Nu/Cy ratio of GFP transcripts was increased (Fig. 4B), When there is a defect or block on mRNA export, hyperadenylation is often observed (Lee and Glaunsinger, 2009). Therefore, we also carried out northern blot analysis to examine whether hyperadenylation occurred to the GFP transcript. Larger species of GFP transcripts were observed only in the nuclear fraction of ORF10-transfected cells (Fig. 4C), and were largely diminished when pre-treated with oligo(dT) plus RNaseH. The increased Nu/Cy ratio of GFP RNA and hyperadenylation of the GFP transcript in ORF10-transfected cells provide evidence for the inhibitory effect of ORF10 on nuclear export of GFP RNA, leading to the reduction in GFP protein expression.
Fig.4.
ORF10 inhibits gene expression through blocking mRNA export. (A) ORF10 inhibits GFP reporter expression. 293T cells were transfected with pEGFP-C1 and ORF10-FLAG or vector control, and two days later, GFP protein expression was visualized under fluorescence microscope. (B) ORF10 inhibits nuclear export of GFP mRNA. The Cy and Nu RNAs were purified from transfected cells in (A), and the GFP transcript was quantified by RT-qPCR. The Nu/Cy ratio was plotted as mean ± SD (n=3). (C) ORF10 induces hyperadenylation of nuclear GFP RNA. The Cy and Nu RNAs from transfected cells in (A) were subjected to northern blotting with the labeled GFP or GAPDH probe. (D) Rae1 knockdown reduces GFP reporter expression. (E) Quantification of the fluorescence intensity in (D). (F) Expression of indicated proteins was analyzed by western blotting. (G) The loss-of-interaction ORF10 mutants failed to reduce GFP expression. (H) Quantification of the fluorescence intensity in (G). All fluorescence data with error bars are plotted as mean ± SD (n=5). Scale bar: 100 μm.
Next we used the reporter to examine the role of Rae1 in ORF10-mediated inhibition. Knocking down Rae1 expression by siRNA reduced GFP reporter expression in vector-transfected cells (Figs. 4D, 4E & 4F), indicating that the reporter expression is dependent on Rae1. Additional ORF10 expression did not further lower GFP expression, consistent with our hypothesis that ORF10 inhibition on mRNA export requires the presence of Rae1. The three ORF10 mutants that cannot interact with Rae1 also failed to down-regulate GFP reporter expression (Figs. 4G & 4H).
The intense nuclear poly(A)+ RNA signals in ORF10-expresising cells (Fig. 1E) can be due to a general block on all mRNA export or hyperadenylation of a subset of mRNAs whose export is inhibited. In the above northern blot analysis (Fig. 4C), we noticed that the nuclear fraction of the GAPDH transcript was not hyperadenylated in the presence of ORF10, in contrast to the GFP transcript. This observation led us to postulate that the inhibitory effect of ORF10 on mRNA export is not global, but is mRNA selective. We first analyzed possible sequence elements that define the GFP transcript as a target for ORF10 inhibition. The replacement of the EGFP coding sequence with the dsRED coding sequence still rendered the reporter expression inhibited by ORF10 (data not shown). When the 3'UTR region, mostly derived from the SV40 early poly(A) signal region, was replaced with other 3'UTRs, such as that of human GAPDH (pEGFP-GAPDH) or human growth hormone gene (pEGFP-GH), reporter expression was no longer affected by ORF10 (Figs. 5A & 5B). We further analyzed the GFP transcript by northern blots. The transcript expressed from the pEGFP-C1 reporter that has the SV40 poly(A) signal was reduced in the cytoplasmic fraction of ORF10-transfected cells (Fig. 5C, short exposure), and was also hyperadenylated in the nuclear fraction (Fig. 5C, long exposure and RNase H). Such effects of ORF10 on the GFP transcript were not observed when the pEGFP-GH reporter was used. These results indicate that the SV40 poly(A) signal region is critical for ORF10-mediated export inhibition.
Fig 5.
ORF10 inhibits gene expression via the 3'UTR region. (A) ORF10 inhibit GFP expression from pEGFP-C1, but not from pEGFP-3'GAPDH or pEGFP-3'GH. (B) Quantification of the fluorescence intensity in (A). (C) The Cy and Nu RNAs were purified from transfected cells, and analyzed by northern blotting with the labeled GFP probe. Images from both short and long exposure are shown. (D) Impact of Rae1 knockdown on GFP reporter expression. (E) Quantification of the fluorescence intensity in (D). (F) Expression of indicated proteins was analyzed by western blotting. Scale bar: 100 μm. All data with error bars are plotted as mean ± SD (n=5).
We next determined whether the dependence of reporter protein expression on Rae1 had similar selectivity for 3'UTR. Rae1 knockdown markedly reduced GFP expression from pEGFP-C1 but not from the pEGFP-GH reporter (Figs. 5D &5E). We also confirmed the fluorescence data by western blot analysis (Fig. 5F). Taken together, these results demonstrate that the inhibitory effects of both ORF10 and Rae1 knockdown on reporter protein expression were selective for the SV40 poly(A) signal region. While the exact mechanism of the selectivity for the SV40 poly(A) signal region remains to be determined, the results provide evidence for the hypothesis that export inhibition of ORF10 is transcript-selective.
Global identification of cellular genes targeted by ORF10
To investigate whether ORF10 selectively affects the export of a subset of mRNAs, we performed RNA sequencing to globally analyze the impact of ORF10 on mRNA export (Figs. S3A and S3B). Raw reads were aligned to human genome assembly, and the RNA levels of individual genes were expressed as reads per million total reads (RPMs) (Table S2). Of 12,181 genes examined, 2,971 (~24%) were significantly down-regulated by more than 50% in the cytoplasmic RNA samples of ORF10-transfected cells (p<0.05 and with the difference in RPM > 2). In contrast, only 59 genes were up-regulated by more than 50% (p<0.05 and with the difference in RPM > 2). We further calculated the relative Nu/Cy ratios by dividing the cytoplasmic RPMs with the nuclear RPMs of individual genes. As shown in Fig. S3C, ORF10 expression increased the relative Nu/Cy ratio of the positive control, GFP, but not the ratios of several house-keeping genes, including GAPDH. Among the 2,971 genes whose cytoplasmic RNAs were reduced by ORF10, 25% of them (749 genes) also had increased Nu/Cy ratios in ORF10-transfected cells (p<0.05). These results support the hypothesis that ORF10 inhibits mRNA export in a transcript-selective manner. We focused on the 686 genes (Table S3) that have more than a 50% increase in their Nu/Cy ratios and performed gene ontology enrichment analysis using DAVID bioinformatics. We found that this group of genes is significantly enriched for the following biological processes: mitosis, gene silencing, DNA metabolic process, chromosome organization, cell cycle, and transcription regulation (Fig. S3D and Table S4).
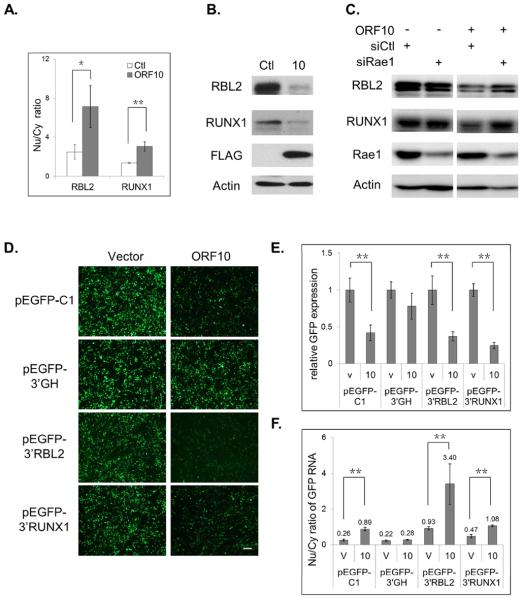
Next, we validated the RNA-seq data for two ORF10 candidate targets, retinoblastoma-like protein 2 (RBL2) and runt-related transcription factor 1 (RUNX1), using real-time PCR (Fig. 6A) and western blotting (Fig. 6B). The Nu/Cy RNA ratios of both genes were increased by ORF10, confirming the RNA-seq data and supporting the interpretation that their mRNA export was inhibited by ORF10. We also examined the role of Rae1 in this selective inhibition of ORF10. Rae1 knockdown by siRNA significantly impaired the ability of ORF10 to down-regulate the protein expression of RBL2 and RUNX1 (Fig. 6C). In addition, we also noted that knocking down Rae1 expression alone did not affect RBL2 or RUNX1 expression, indicating that their RNA export does not depend on Rae1. We also determined whether RBL2 and RUNX1 expression is affected in the context of MHV-68 lytic replication. Infection with the WT or 10R virus led to a pronounced reduction in both RBL2 and RUNX1 proteins; moreover, their expression was partially restored with infection of the ORF10-null (10S) mutant virus (Fig. S3E). This result supports the model in which during MHV-68 lytic replication, ORF10 inhibits mRNA export to down-regulate expression of host proteins such as RBL2 and RUNX1.
Fig. 6.
ORF10 inhibits mRNA export of cellular genes. (A) The Cy and Nu RNAs were purified from transfected cells and quantified for the transcript level of RUNX1 and RBL2 by RT-qPCR. The Nu/Cy ratio was plotted as mean ± SD (n=3). (B) Expression of RUNX1 and RBL2 proteins in transfected cells was analyzed by western blotting. (C) An inducible cell line of ORF10 was transfected with Rae1 siRNA or control siRNA and then treated with tetracycline to induce ORF10 expression. Two days later, expression of RBL2 and RUNX1 were analyzed by western blotting. (D) The GFP constructs fused with the 3'UTR of RBL2 (pEGFP-3'RBL2) or RUNX1 (pEGFP-3'RUNX1) are sensitive to ORF10 inhibition. (E) Quantification of the fluorescence intensity in (D). (F) The Nu/Cy ratio of GFP RNA in cells of (D) is plotted as mean ± SD (n=3). Scale bar: 100 μm. See also Figs. S3, S4 and S5, Tables S2, S3 and S4.
Furthermore, we used the GFP reporter assay to test whether the 3'UTRs of RBL2 and RUNX1 were capable of sensitizing the GFP transcript to ORF10 inhibition. The 3'UTR region downstream of the RBL2 translation stop codon was able to subject the GFP expression to ORF10 down-regulation (Figs. 6D and 6E); additionally, there was a 266% increase in the Nu/Cy ratio of GFP RNA (Fig. 6F). The 3'UTRs of RUNX1 are complex, ranging from 150 to 4300 nucleotides, and there are three major poly(A) signals at the 3' end of the gene (Levanon et al., 2001). The 3'UTR region that contains the two distal poly(A) signals were able to support comparable GFP expression to other GFP reporters, and also sensitized GFP protein expression as well as GFP RNA export to ORF10 inhibition, just as the RBL2 3'UTR did. The results indicate that the 3'UTRs of RUNX1 and RBL2 contain elements that select them as targets for ORF10.
Function of ORF10 in the context of KSHV lytic replication
After determining the function of ORF10 in blocking cellular mRNA nuclear export, we examined the role of ORF10 during KSHV lytic replication. We generated an ORF10 null KSHV mutant (10S) by introducing a translation stop codon immediately after its 41st amino acid, and also constructed the corresponding revertant mutant (10R) (Figs. S6A & S6B). The KSHV BAC plasmids of 10S and 10R were introduced into iSLK-puro cells to produce the latent KSHV cell lines. Viral lytic replication was examined after induction of KSHV reactivation in the latent cells lines. There were about ten times less virions produced from lytic replication of 10S than from WT or 10R (Fig. S6C); however, viral DNA replication after induction of KSHV reactivation was comparable among three viruses (Fig. S6D). Next we analyzed the viral protein expression of 10S, 10R, and WT. Herpesviral genes are expressed in a cascade manner with an order of immediate early (IE), early (E), and late (L) genes. The late genes are classified by their dependence for expression on viral DNA replication. All three viruses had similar expression of RTA (an IE gene) and K8 (an E gene), but the 10S virus expressed significantly less late gene products, e.g., K8.1, ORF26, and ORF45 (Fig. S6E). Without a normal expression level of late genes, which mainly encode structural components of virions, the production of infectious virions will be reduced, as observed for the 10S virus. The study of KSHV lytic replication indicate that ORF10, while not required for the expression of immediate early genes, for expression of early genes, or for viral DNA replication, plays a critical role in the expression of late genes and is, therefore, essential for efficient virion production.
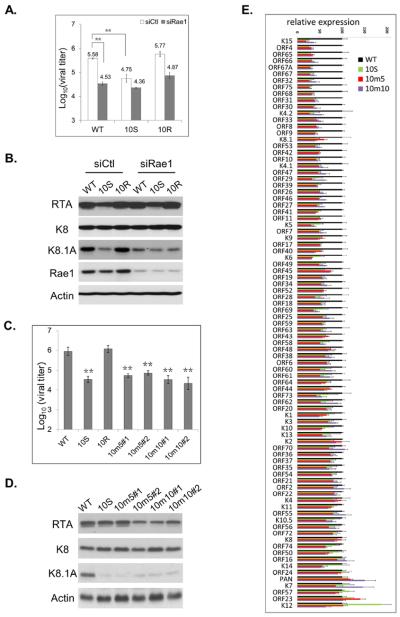
Our model proposes that ORF10 inhibition of mRNA export requires interaction with Rae1. Thus, we sought to examine the function of Rae1 during KSHV lytic replication. Knocking down Rae1 expression with siRNA significantly reduced virion production of WT and 10R to a level similar to the 10S virus (Fig. 7A). In addition, Rae1 knockdown impaired the protein expression of the late viral K8.1 gene, but not expression of the immediate early RTA or the early K8 gene during lytic replication of the WT or 10R virus (Fig. 7B). The effects of Rae1 knockdown on KSHV lytic replication resemble those of eliminating ORF10 expression from the virus. This result is consistent with our model that ORF10 functions through Rae1. To further demonstrate that the interaction with Rae1 is important for the function of ORF10, we introduced two individual site-specific mutations (m5 and m10) that abolished the interaction with Rae1 into the ORF10 coding region of the viral genome. Similar to the 10S virus and to the effects of Rae1 knockdown, both of the loss-of-interaction mutants produced ten times less infectious virions and expressed much less K8.1A, but expressed comparable levels of RTA and K8 proteins to WT (Fig. 7C & 7D).
Fig. 7.
Efficient KSHV late gene expression requires ORF10 and its interaction with Rae1. (A) KSHV-latently infected iSLK cells, iSLK-WT, iSLK-10S and iSLK-10R, were transfected with Rae1 siRNA or control siRNA and then treated with doxycycline and sodium butyrate to induce viral lytic replication. The supernatants were collected to quantify infectious virions. Data are plotted as mean ± SD (n=3). (B) The lysates were harvested from cells in (A) for analyzing viral and cellular protein expression. (C) Infectious virion production from KSHV WT or ORF10 mutants, including 10S, two clones of 10m5, and two clones of 10m10. (D) Viral and cellular protein expression. (E) Genome-wide analysis of viral lytic gene expression for WT and ORF10 mutants. Total RNAs were purified from iSLK-WT, −10S, −10m5#1 and 10m10#1 cells, and subjected to RNA sequencing. The relative viral gene expression to the WT virus was plotted as means ± SD (n=3). See also Fig S6, Tables S5 and S6.
The western blotting results indicate that ORF10 plays a critical role in regulating late gene expression. To globally examine the viral gene expression during lytic replication of 10S and the two loss-of-interaction mutants (10m5 and 10m10), we performed RNA-seq (Table S5). In general, the viral gene expression was comparable among three ORF10 mutants (10S, 10m5 and 10m10) and most of the viral genes were expressed at levels less than WT (Fig. 7E). After ranking viral genes based on their levels of reduction in the ORF10 mutants, we found that the most down-regulated genes are those classified as late genes (top part of Fig. 7E and Table S6), while those less-affected genes belong to the IE, E, and latent gene categories (bottom part of Fig. 7E). We also found good correlation in reduction of viral gene expression between the loss-of-expression mutant (10S) and the loss-of interaction mutants (10m5 and 10m10) (Fig. S6F). Collectively, we showed that ORF10, in combination with Rae1, is critical for KSHV lytic replication and plays a specific role in regulating late gene expression.
Discussion
A variety of strategies are employed by various viruses to suppress host gene expression. Here, we present the identification of ORF10 encoded by KSHV as a selective inhibitor of mRNA nuclear export. ORF10 expression increases the Nu/Cy ratio of a subset of cellular mRNAs. We show that the 3'UTRs of the ORF10-targeted transcripts contain elements that select the transcripts as targets for ORF10. RNA export inhibition by ORF10 is mediated by the interaction with a cellular mRNA export factor, Rae1. Rae1, interacting with Nup98, was previously implicated in mRNA export (Pritchard et al., 1999). ORF10 interacts with Rae1, which in turns interacts with Nup98. Enriched nuclear rim localization of ORF10 also requires the presence of Rae1 and Nup98. Furthermore, ORF10 inhibition on mRNA nuclear transport is disrupted both by mutations that diminish the interaction with Rae1 and by Rae1 knockdown. While ORF10 blocks mRNA export of a subset of cellular genes, potentially hundreds of them, our RNA-seq experiment did not detect any cellular transcript whose export is significantly inhibited by Rae1 knockdown (data not shown). Further RT-PCR and western blot results confirmed that neither of the two confirmed ORF10-targeted genes, RBL2 and RUNX1, depends on Rae1 to export their mRNAs for translation (Figs. 6C S4A, S4B and S4C). Based on these results, we propose that ORF10 interacts with the Rae1-Nup98 complex at the nuclear rim, most likely at NPCs, to block export of mRNAs that are associated with the complex; binding to Rae1 allows ORF10 to gain access to a specific subset of mRNAs and inhibit their exit from the nucleus. Thus, our working model of ORF10 also implicates a role of Rae1 in regulating mRNA export of selective transcripts.
In addition, we demonstrate that ORF10 and Rae1 are both important for KSHV to efficiently express its late genes during lytic replication. The consequences of Rae1 knockdown on KSHV lytic replication resemble those of removing ORF10 expression from the virus. The loss-of-interaction ORF10 mutant viruses also displayed similar lytic replication phenotypes to the ORF10-null mutant. Our findings suggest that ORF10, through interacting with Rae1, selectively blocks RNA export of a subset set of mRNAs to facilitate viral late gene expression.
Rae1 and mRNA export
Rae1 is highly conserved among all eukaryotes from yeast to mammals. The first implication for a function of Rae1 in mRNA export came from studies in yeast (Brown et al., 1995; Murphy et al., 1996). Yeasts with disruption of Rae1 gene accumulate poly(A)+ RNA in the nucleus. Subsequent studies in metazoa further supported the idea that Rae1 functions as an mRNA export factor (Kraemer and Blobel, 1997; Pritchard et al., 1999). Still, the precise role of Rae1 in mRNA export remains unclear. Rae1-null mice are embryonic lethal, but poly(A)+ RNA distribution appears normal in cultured Rae1−/− blastocysts (Babu et al., 2003), in contrast to the observations in Rae1-deficient yeast. It is further shown that during mitosis, Rae1 in complex with Nup98, plays an essential role in mitotic spindle assembly and mitotic checkpoint regulation (Babu et al., 2003). Through binding to Rae1, VSV M protein blocks mRNA export during interphase (Faria et al., 2005; von Kobbe et al., 2000), and interferes with mitotic progression during mitosis (Chakraborty et al., 2009). Our study on ORF10 highlights the importance of Rae1 and its interaction with Nup98 in mRNA export; two viruses from very different families both target Rae1 to inhibit mRNA export. It seems clear that the Rae1-Nup98 interaction plays an important, but as yet not well characterized, role in regulating mRNA trafficking. One possibility is that Rae1, while not absolutely required, facilitates transport of a subset of mRNAs. Viruses that target Rae1 may specifically regulate this subset of mRNAs to achieve optimal replication. The implication is that the transcripts regulated by Rae1 may encode proteins that have functions in the viral life cycle. For example, the studies using mouse embryonic fibroblasts with reduced expression of Rae1 or Nup98 have implicated that the Rae1-Nup98 pathway is important for expression of several immune-related genes, which are known to play a role in host antiviral responses (Satterly et al., 2007).
Transcript-selective regulation
Transcript-selective export pathways have been previously demonstrated. Often these selectively exported transcripts enrich for certain functional categories, such as DNA repair, transcription, and cell proliferation (Wickramasinghe and Laskey, 2015). It has been proposed that expression of functionally related genes can be coordinately regulated at a post-transcriptional level to facilitate efficient responses to cellular needs (Keene, 2007). Therefore, export of mRNAs that function in the same biological process may be co-regulated. RNA-binding proteins of mRNP complexes are postulated to coordinate post-transcriptional regulation by specifically recognizing and interacting with an RNA sequence and/or a structural element within a subset of mRNAs, analogous to transcriptional factors controlling multiple promoters in concert. In such a model, Rae1, which was shown previously to have RNA-binding ability (Kraemer and Blobel, 1997) or an RNA-binding protein that interacts with Rae1, can facilitate the transport of a subset of mRNAs by interacting with Nup98. The selective interaction of Rae1 with transcripts is demonstrated by our RNA-IP study (Fig. S5). Both ORF10 and Rae1 were associated with the ORF10-targeted GFP reporter transcript, but not with cellular GAPDH or actin transcripts (Figs. S5A and S5C). Knocking down Rae1 diminished the interaction between ORF10 and its target transcript (Fig. S5B), indicating a pivotal role of Rae1 in bridging ORF10 and mRNAs.
Our data also showed that ORF10 forms a complex with Rae1 and Nup98, so ORF10 does not appear to dissociate Rae1 from Nup98. A previous structure study of VSV M-Rae1-Nup98 has suggested that M protein mimics phosphate backbone of nucleic acid to compete for nucleic acid binding to Rae1 (Quan et al., 2014). However, our RNA-IP result supports a different mechanism of action for ORF10. ORF10 interacts with its target transcript through Rae1 (Fig. S5B). Unlike VSV M, ORF10 does not displace the transcript from Rae1; instead, ORF10 hijacks Rae1, gaining access to the transcripts whose nuclear export is regulated by Rae1. In this model, knockdown of Rae1 expression alone would not have effects on ORF10-targeted transcripts, but would alleviate the export inhibition imposed by ORF10. This is exactly what we observed in our siRNA knockdown studies (Fig. 6C): while the expression of two ORF10-targeted genes, RUNX1 and RBL2, does not require Rae1, ORF10 depends on Rae1 to inhibit their expression.
With the use of ORF10, we systematically analyzed the transcript selectivity of export inhibition. Nuclear RNA export of 686 of the total 12,181 genes examined in our RNA-seq is potentially inhibited by ORF10. These ORF10-regulated transcripts enrich for certain functional categories, including cell cycle, chromosome organization, mitosis, and transcription regulation. Furthermore, we presented evidence to support specific recognition of RNA elements within 3'UTR of mRNA for ORF10-mediated export inhibition. Using a GFP reporter construct, we showed that the 3'UTRs of two ORF10-targeted genes, RBL2 and RUNX1, render nuclear export of GFP transcripts sensitive to ORF10 inhibition (Fig. 6F). Furthermore, nuclear export of these GFP transcripts fused with 3'UTR of RUNX1 and RBL2 is also reduced by Rae1 knockdown (Figs. S4D and S4E), in contrast to RBL2 and RUNX1 transcripts. We speculate that for most of cellular mRNAs, the function of Rae1 in export is redundant and complemented by other proteins associated with mRNAs, but not for the GFP transcript whose processing and maturation is less complicated than cellular mRNAs. For example, the GFP transcript does not undergo splicing and yet splicing plays a major role in recruiting the heterodimeric NXF1-NXT1 export factor. Nevertheless, the GFP reporter provides a powerful tool to further delineate the 3'UTR and to investigate the role of Rae1 in mRNA export. As the poly(A) signal and the mechanism of 3'-end processing are well conserved among transcripts, we consider that specific RNA elements for selective inhibition exist within the 3'UTRs of ORF10-sensitive transcripts. Sequence analysis of the ORF10-targeted transcripts did not reveal any consensus linear sequence that can distinguish targeted from non-targeted ones. Although RNA secondary structures often serve as recognition motifs, their prediction is challenging. In the future, we will utilize the GFP reporter to identify more ORF10-sensitive 3'UTRs and perform detailed mutagenesis studies to dissect the RNA elements that mediate transcript-selective regulation.
Inhibition of RNA export and KSHV lytic replication
Viral inhibition of cellular gene expression is thought to suppress host anti-viral activities. KSHV lytic replication results in a widespread shutoff of host gene expression. One contributing viral gene to host shutoff is SOX, which induces global cytoplasmic RNA degradation (Glaunsinger and Ganem, 2004). Reduced mRNA in the cytoplasm leads to re-localization of cytoplasmic poly(A)-binding protein 1 (PABP1 or PABPC1) to the nucleus, causing nuclear retention of cellular mRNAs (Kumar and Glaunsinger, 2010). Because both eliminating ORF10 expression and disrupting ORF10 interaction with Rae1 impact KSHV lytic replication, mRNA export inhibition of ORF10 is not redundant with host shutoff activity of SOX. Still, it is unclear how the ORF10-Rae1 interaction facilitate viral late gene expression, Previous studies by us and by others have implicated several viral genes, including ORF18, 24, 30, 31, 34, 66, as late gene transcription activators (LTAs) (Aubry et al., 2014; Gong et al., 2014; Wu et al., 2009). Without normal expression of LTAs, transcription of late genes will be affected. Our genome-wide RNA-seq analysis of viral transcript levels (Fig. 7E) indicates that three LTAs (ORF66, ORF30 and ORF31) are among the viral genes whose transcription is greatly reduced in the absence of ORF10. We also confirmed that the KSHV ORF10-null mutant produces much less ORF31 protein than wild-type virus (Fig. S6G). Since ORF10 inhibits mRNA export of many cellular genes, further investigation will be required to identify the genes that regulate transcription of LTAs, and whether they are part of host anti-viral responses.
In summary, we propose that ORF10 of KSHV inhibits RNA export by interacting with the Rae1-Nup98 complex to facilitate viral late gene expression. While Rae1 is not absolutely essential for RNA export, we hypothesize that through interaction with Rae1, ORF10 gains access to a specific subset of mRNAs whose export is regulated by Rae1 and then blocks their export through interfering with the function of Rae1-Nup98 complex. Accordingly, using ORF10 as a tool, we identified potential cellular transcripts whose export may be selectively regulated by Rae1. Our results not only support the idea that Rae1 plays a previously unrecognized role in viral replication, but also offer a system for unraveling the function of Rae1.
Experimental Procedures
Cells
HEK 293T (293T), SLK (clear-cell renal-cell carcinoma cell line), Vero, iSLK-puro and iSLK-KSHV-BAC16 cells (Brulois et al., 2012) were cultured in DMEM medium supplemented with 10% fetal bovine serum and 1% Pen-Strep. iSLK-puro cells were also supplied with 1 μg/ml puromycin and 250 μg/ml G418. iSLK-KSHV-BAC16 and its derivatives were also maintained in the presence of 1 μg/ml puromycin, 250 μg/ml G418 and 1,200 μg/ml hygromycin B.
Plasmids
All primers used for cloning are listed in Table S1. Detailed cloning procedures are described in Supplemental Experimental Procedures.
Immunofluorescence and confocal microscope
Transfected or infected SLK cells were fixed in 2% paraformaldehyde, permeabilized with 0.1% Triton-X100, and then blocked with 3% BSA and 10% FBS. FLAG-tagged ORF10 protein was detected with mouse anti-FLAG antibody (Sigma); Nup98 was detected with anti-Nup98 antibody (Abcam, ab50610); HA-tagged Rae1 protein was detected with rabbit anti-HA antibody (Sigma); DAPI was used for staining of DNA. Samples were examined with confocal microscopy using a Nikon Eclipse Ti imaging system.
Poly(A) mRNA in situ hybridization
Infected or transfected cells were fixed with paraformaldehyde, permeabilized with methanol, and then poly(A)+ RNA was detected with fluorophore labeled oligo(dT) probe using a method as previously described (Lee and Glaunsinger, 2009) with some modifications. See details in Supplemental Experimental Procedures.
Cytoplasmic and nuclear RNA fractionation
Cytoplasmic and nuclear RNA fractionation was performed via combined use of centrifugation and extraction buffers according to a previous described method with some modifications (Wang et al., 2006).
Northern blotting
Cytoplasmic and nuclear RNAs were denatured, separated on a 1% agarose gel containing 2% formaldehyde, and then transferred onto Hybond-N+ nylon membrane (GE Amersham). Northern blotting was carried out using DIG-High prime DNA labeling and detection starter kit II (Roche) with probes against the GFP or GAPDH coding region. RNase H digestion with oligo(dT) primer was performed using a previously described method (Lee and Glaunsinger, 2009).
Reverse transcription and real-time PCR
RNA was first treated with DNase I, and then subjected to reverse transcription using SuperScript III reverse transcriptase (Life technologies) and random hexamers. Real-time PCR was then performed with the primers listed in Table S1. To determine KSHV genome copy numbers, total DNA was subjected to real-time PCR using primers for ORF59.
Co-IP, sequential IP and western blot
Immunoprecipitation and western blot analysis were performed as described (tools.thermofisher.com) with some modifications. See details in Supplemental Experimental Procedures.
MHV-68 and KSHV mutagenesis
MHV-68 and KSHV mutations were generated via BAC homologous recombination in E. coli (Gong et al., 2014; Wu et al., 2011). See details in Supplemental Experimental Procedures.
Directional RNA-seq library preparation and sequencing
RNA-seq libraries were prepared using a modified method based on ScriptSeq mRNA-Seq library preparation kit (Epicentre) (Gong et al., 2014). Multiplex sequencing was performed by 50 bp single-end read with Illumina HiSeq 2000 machine. See details in Supplemental Experimental Procedures. Raw read data is available from NCBI Gene Expression Omnibus under accession number GSE82241.
Statistical analysis
All numerical data were calculated and plotted with mean+/− SD. Results were analyzed by unpaired Student's t test. Differences were considered statistically significant when p<0.05 (*) or p<0.01 (**).
Supplementary Material
Acknowledgement
We thank Professors, Harvey Herschman, and Guillaume Chanfreau, for their critical reviews and comments, members of the Ren Sun and Ting-Ting Wu labs for discussions. This work was supported by NIH grants DE023591 and CA177322.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions All experiments were performed by D.G. with the help of Y.H.K., Y.X., Y.X., K.K.L and D.Z.. Y.D., J.F., N.F., S.S., X.D., S.K.C., T.M.R., N.J.K, R.S. and T.T.W provided reagents and advice. D.G. and T.T.W. designed the experiment, analyzed the data, and wrote the manuscript.
Supplemental information Supplemental information for this article includes 6 figures, 6 tables, and supplemental experimental procedures
References
- Aubry V, Mure F, Mariame B, Deschamps T, Wyrwicz LS, Manet E, Gruffat H. Epstein-Barr Virus Late Gene Transcription Depends on the Assembly of a Virus-Specific Preinitiation Complex. Journal of virology. 2014;88:12825–12838. doi: 10.1128/JVI.02139-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu XS, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson SA, Page AL, Ganem D. A Kaposi's sarcoma-associated herpesvirus protein that forms inhibitory complexes with type I interferon receptor subunits, Jak and STAT proteins, and blocks interferon-mediated signal transduction. Journal of virology. 2009;83:5056–5066. doi: 10.1128/JVI.02516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. Journal of Biological Chemistry. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. The Journal of biological chemistry. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, et al. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. Journal of virology. 2012;86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122:1933–1937. doi: 10.1242/jcs.041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Seemann J, Mishra RK, Wei JH, Weil L, Nussenzveig DR, Heiber J, Barber GN, Dasso M, Fontoura BM. Vesicular stomatitis virus inhibits mitotic progression and triggers cell death. EMBO reports. 2009;10:1154–1160. doi: 10.1038/embor.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IHB, Sciabica KS, Sandri-Goldin RM. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. Journal of virology. 2002;76:12877–12889. doi: 10.1128/JVI.76.24.12877-12889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, et al. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Molecular cell. 2015;57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Molecular cell. 2005;17:93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Molecular cell. 2004;13:713–723. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- Gong D, Wu NC, Xie Y, Feng J, Tong L, Brulois KF, Luan H, Du Y, Jung JU, Wang CY, et al. Kaposi's Sarcoma-Associated Herpesvirus ORF18 and ORF30 Are Essential for Late Gene Expression during Lytic Replication. Journal of virology. 2014;88:11369–11382. doi: 10.1128/JVI.00793-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Koffa MD, Clements JB, Izaurralde E, Wadd S, Wilson SA, Mattaj IW, Kuersten S. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. Embo Journal. 2001;20:5769–5778. doi: 10.1093/emboj/20.20.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Glaunsinger BA. Nuclear Import of Cytoplasmic Poly(A) Binding Protein Restricts Gene Expression via Hyperadenylation and Nuclear Retention of mRNA. Mol Cell Biol. 2010;30:4996–5008. doi: 10.1128/MCB.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang RS, Wu TT, Hwang S, Liang LT, Tong L, Truong JT, Sun R. The anti-interferon activity of conserved viral dUTPase ORF54 is essential for an effective MHV-68 infection. PLoS pathogens. 2011;7:e1002292. doi: 10.1371/journal.ppat.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Glaunsinger BA. Aberrant Herpesvirus-Induced Polyadenylation Correlates With Cellular Messenger RNA Destruction. PLoS biology. 2009;7 doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Glusman G, Bangsow T, Ben-Asher E, Male DA, Avidan N, Bangsow C, Hattori M, Taylor TD, Taudien S, et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33. doi: 10.1016/s0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. Journal of Biological Chemistry. 2004;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Molecular biology of the cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ. Nuclear Transport: Beginning to Gel? Curr Biol. 2012;22:R1006–R1009. doi: 10.1016/j.cub.2012.10.037. [DOI] [PubMed] [Google Scholar]
- Pritchard CEJ, Fornerod M, Kasper LH, van Deursen JMA. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–253. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan B, Seo HS, Blobel G, Ren Y. Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 * Nup98) Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9127–9132. doi: 10.1073/pnas.1409076111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang YM, Faria PA, Levay A, Levy DE, Fontoura BMA. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Bio. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, van Deursen JM, Rodrigues JP, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo-Fonseca M, Izaurralde E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Molecular cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39:356–362. doi: 10.1016/j.ymeth.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Bio. 2015;16:431–442. doi: 10.1038/nrm4010. [DOI] [PubMed] [Google Scholar]
- Wu TT, Liao HI, Tong L, Leang RS, Smith G, Sun R. Construction and characterization of an infectious murine gammaherpesivrus-68 bacterial artificial chromosome. Journal of biomedicine & biotechnology. 2011;2011:926258. doi: 10.1155/2011/926258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Park T, Kim H, Tran T, Tong L, Martinez-Guzman D, Reyes N, Deng H, Sun R. ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. Journal of virology. 2009;83:2265–2273. doi: 10.1128/JVI.01785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough ML, Mata MA, Sakthivel R, Fontoura BMA. Viral Subversion of Nucleocytoplasmic Trafficking. Traffic. 2014;15:127–140. doi: 10.1111/tra.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.